Significance
Skin pigmentation provides first-line protection against UV radiation (UVR) that increases the risk of skin cancers. However, mechanisms underlying this process remain poorly understood. Here, we identified the autophagic tumor suppressor UVRAG as a bona fide player in melanosome biogenesis by targeting biogenesis of lysosome-related organelles complex 1 (BLOC-1) independently of autophagy. UVRAG maintains the localization and stability of BLOC-1 to facilitate the sorting/delivery of melanogenic cargoes. Reduced levels of UVRAG rendered cells unresponsive to UVR–α-MSH–MITF signaling and defective melanocyte development in vivo. Moreover, UVRAG-mediated melanogenesis and tanning response were impaired in oncogene-driven melanoma. This study represents a description of a noncanonical role of autophagy factor in melanogenic remodeling and also provides mechanistic insights into UVRAG in pigmentation disorder and UV-associated cancer.
Keywords: UVRAG, MITF, BLOC-1, melanosome, BRAF
Abstract
UV-induced cell pigmentation represents an important mechanism against skin cancers. Sun-exposed skin secretes α-MSH, which induces the lineage-specific transcriptional factor MITF and activates melanogenesis in melanocytes. Here, we show that the autophagic tumor suppressor UVRAG plays an integral role in melanogenesis by interaction with the biogenesis of lysosome-related organelles complex 1 (BLOC-1). This interaction is required for BLOC-1 stability and for BLOC-1–mediated cargo sorting and delivery to melanosomes. Absence of UVRAG dispersed BLOC-1 distribution and activity, resulting in impaired melanogenesis in vitro and defective melanocyte development in zebrafish in vivo. Furthermore, our results establish UVRAG as an important effector for melanocytes’ response to α-MSH signaling as a direct target of MITF and reveal the molecular basis underlying the association between oncogenic BRAF and compromised UV protection in melanoma.
UV radiation (UVR) from sunlight has been epidemiologically identified as a leading risk factor for melanoma and other skin cancers (1). Skin pigmentation serves as the main UV-protective mechanism afforded by pigment-producing cells called melanocytes (2). UV-exposed keratinocytes produce and secrete α-melanocyte-stimulating hormone (α-MSH), which binds to the melanocortin 1 receptor (MC1R) on the surface of melanocytes to activate cAMP cascades and induces lineage-restricted microphthalmia-associated transcription factor (MITF) expression (2). Consequently, UV-blocking melanin production is ramped up in lysosome-related organelle known as melanosomes. The melanosomes are transported to neighboring keratinocytes, where they form a cap over the sun-exposed side of the nuclei for protection against UVR. Impaired melanosome function and melanin production allow rampant UVR penetration, leading to accrual of DNA mutagenesis and cancer predisposition (2). Understanding how cell pigmentation is regulated in melanocytes and its environmental adaptation to UV-induced cutaneous α-MSH (the suntan response) is critical not only for melanocyte biology but also for its role in skin cancer, including life-threatening melanoma.
The genetic program of pigmentation and suntan response is centrally controlled by MITF, which belongs to a family of basic helix–loop–helix zipper (bHLH-Zip) transcription factors and regulates the expression of many genes involved in melanocyte differentiation and function (3). This includes but is not limited to the core enzymes for melanin biosynthesis such as tyrosinase (TYR), tyrosinase-related protein 1 (TYRP1), and dopachrome tautomerase (DCT), as well as melanosome structural proteins such as amyloid protein PMEL (4). Melanin is synthesized exclusively within the melanosomes. Melanosome biogenesis/maturation is regulated by specific transport machinery and accompanied by selective cargo delivery (5). Notably, the early step of cargo delivery to melanosomes has been shown to require the biogenesis of lysosome-related organelles complex 1 (BLOC-1), which consists of eight subunits named BLOS1, BLOS2, BLOS3, Pallidin, Snapin, Muted, Cappuccino, and Dysbindin (6), and is found to be associated with tubular endosomes (7, 8). Although the specific function of each individual subunit remains to be established, dysfunction of BLOC-1 has been shown to confer the Hermansky–Pudlak syndrome (HPS) phenotype in humans and pigmentation abnormalities in mice (9). However, how BLOC-1 is recruited to endosomes and eventually mediates cargo delivery during melanosome biogenesis remains largely unknown.
The tumor suppressor protein UV-radiation resistance associated gene (UVRAG) was initially identified as a promoter of Beclin1–PI3KC3-mediated autophagy (10) and later found to be a multivalent trafficking adaptor that plays a prominent role in maintaining energy homeostasis (11–13). UVRAG also patrols genetic integrity in response to different types of DNA damage (14, 15). A dominant negative mutation in UVRAG is a natural cause of increased growth rate and tumor metastasis in some human cancers (16). Importantly, all of the different activities of UVRAG are functionally independent, suggesting a biological connection and coordinated regulation of UVRAG in different processes. Interestingly, a previous report (17) had linked UVRAG polymorphisms to susceptibility to vitiligo, a skin pigmentary disorder that is characterized by aberrant melanogenesis. However, little is known about UVRAG in cell pigmentation and melanocyte biology. In our continuing effort to characterize the function of UVRAG, we report here that UVRAG plays a crucial role in melanocyte function by targeting BLOC-1. Loss of UVRAG destabilizes BLOC-1 and disrupts BLOC-1 function in cargo sorting and delivery for melanin production, leading to defective melanogenesis and impaired pigmentation. Furthermore, expression of UVRAG is specifically activated by MITF in melanocytes in response to α-MSH signaling, which is suppressed by oncogenic BRAF. Thus, our study reveals a mechanism for UVRAG in melanogenesis and its significance in UV-induced adaptive skin pigmentation.
Results
UVRAG Is Required for Cell Pigmentation.
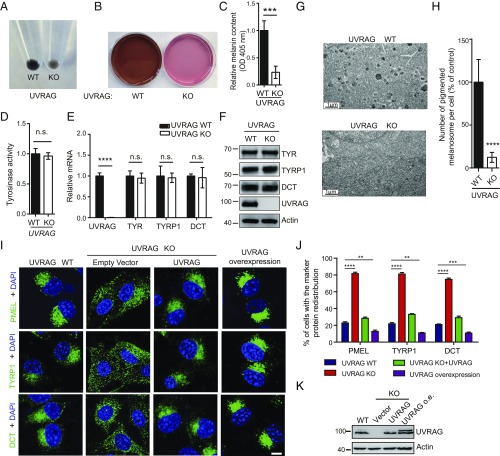
To examine the function of UVRAG in melanocytes, we used CRISPR-Cas9 genome editing (18) to specifically knock out (KO) the UVRAG gene in B16 melanoma cells (SI Appendix, Fig. S1A). Compared with control cells expressing wild-type (WT) UVRAG, we observed significant whitening of UVRAG KO cells (Fig. 1A). Both the pellet and the culture medium of UVRAG KO cells were hypopigmented with reduced melanin contents (OD at 405 nm) in the lysate (Fig. 1 A–C). To validate that the observed cell whitening was due to UVRAG deletion introduced by CRISPR-Cas9, we used lentiviruses encoding shRNA to knock down (KD) UVRAG in B16 cells and observed a similar whitening phenotype (SI Appendix, Fig. S1B). Conversely, cells with stable overexpression of Flag-UVRAG were visibly hyperpigmented (SI Appendix, Fig. S1 E and F). Of note, the darkness of melanocytes results from the synthesis of melanin by TYR (3). Interestingly, gain- and loss of function of UVRAG did not readily affect tyrosinase activity in these cells (Fig. 1D and SI Appendix, Fig. S1G). Nor did they affect mRNA (Fig. 1E and SI Appendix, Fig. S1 C and H) and protein levels (Fig. 1F and SI Appendix, Fig. S1 D and I) of melanogenic genes, including TYR, TYRP1, and DCT. To understand how UVRAG regulates cell pigmentation, we investigated the consequence of UVRAG suppression to the melanosomes, the cellular sites for melanin synthesis (5), by electron microscopy. As shown in Fig. 1G, UVRAG KO melanocytes had much fewer melanosomes in the cytoplasm compared with the control (Fig. 1 G and H). Additionally, UVRAG deficiency resulted in a redistribution of the melanosome precursor marker protein PMEL and the enzymatic machinery TYRP1 and DCT from a juxtanuclear punctate staining to a more diffuse pattern in the melanocyte cytoplasm (Fig. 1I). Such mislocalization of melanosome markers was reversed by reexpression of WT UVRAG in UVRAG KO cells (Fig. 1 I–K). Consistently, ectopic expression of UVRAG markedly increased the perinuclear puncta staining of melanosomes (Fig. 1 I–K). These results indicate that UVRAG is required for the dynamic integrity and pigmentation of melanosomes.
Fig. 1.
UVRAG is required for melanogenesis. (A) Effect of UVRAG on the pigmentation of B16 melanoma cells. Equal number of UVRAG wild-type (WT) and knockout (KO) cells were pelleted by centrifugation after a 6-d culture. (B) Photographs of UVRAG WT and KO B16 cell culture media. (C) Relative melanin contents of the cell lysates of UVRAG WT and KO B16 cells (6-d culture), measured as optical density (OD) at 405 nm. Data are mean ± SD from three independent experiments. (D) Tyrosinase activity in extracts from cells in A, measured by in vitro l-DOPA assay. Data are mean ± SD from three independent experiments. (E) Quantitative RT-PCR showing the relative levels of mRNA expression for TYR, TYRP1, and DCT in UVRAG WT and KO cells in A. Data are mean ± SD from three independent experiments. (F) Western blot analysis of the relative expression of the indicated proteins in cells in A. Actin serves as a loading control. (G and H) Melanosome morphology upon UVRAG deletion. UVRAG WT and KO B16 melanoma cells were subjected to electron microscopy analysis (G). Compared with UVRAG WT cells, the darkly pigmented melanosomes were barely detected in cells lacking UVRAG. The number of pigmented melanosomes per cell was quantified in UVRAG KO melanocytes and normalized to that of WT control (H). Data represent mean ± SD (n = 30–40 cells obtained by gathering data from three independent experiments). (I–K) UVRAG deficiency leads to redistribution of melanosome markers. UVRAG WT (first column), UVRAG KO B16 cells complemented with empty vector (second column) or UVRAG (third column), and B16 cells stably expressing Flag-UVRAG (fourth column) were stained for PMEL, TYRP1, and DCT (I). Nuclei were stained with DAPI (blue). The percentage of cells with redistribution of the indicated proteins was quantified in J. Data represent mean ± SD (n = 200 cells obtained from three independent experiments). The expression of UVRAG in these cells is shown in K. o.e., overexpression. (Scale bars, 20 μm.) n.s., not significant; **P < 0.01; ***P < 0.001; ****P < 0.0001.
UVRAG Interacts with BLOC-1 to Regulate Melanosomal Cargo Sorting.
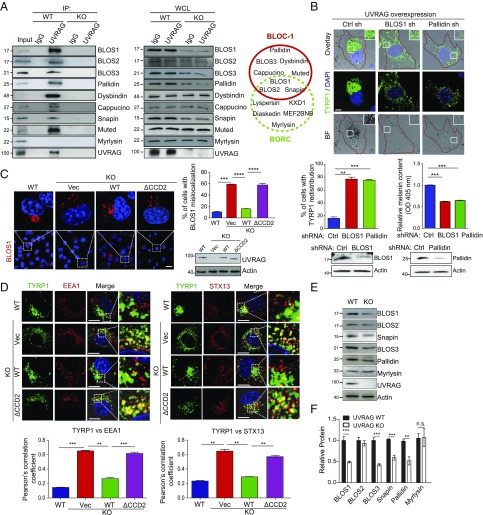
To understand the mechanism by which UVRAG regulates melanogenesis, we conducted a yeast two-hybrid screen to identify melanosome-related UVRAG-interacting proteins by using the full-length UVRAG as bait. The results of the screen are summarized in SI Appendix, Table S1 and can be seen to include a number of known UVRAG-interacting proteins such as BECN1 and CEP63 (10, 15). In addition, five independent isolates of a clone encoding the BLOS1 protein were identified. BLOS1 was previously identified as a key subunit of the BLOC-1 complex, which also includes BLOS2, Snapin, BLOS3, Pallidin, Dysbindin, Cappucino, and Muted (6). Indeed, Snapin was another candidate in our screening (SI Appendix, Table S1). Given previous studies showing the activity of BLOC-1 in melanosomal cargo sorting (8, 19), we chose to investigate the role of BLOS1 and by extension the BLOC-1 complex in UVRAG-mediated melanogenesis. An interaction between recombinant UVRAG and BLOS1 was detected in vitro, supporting their direct interaction (SI Appendix, Fig. S2A). Endogenous UVRAG readily coimmunoprecipitated with BLOS1 and other components of BLOC-1, while no interaction was detected in UVRAG KO cells (Fig. 2A). Consistently, we observed that a portion of endogenous UVRAG colocalized with BLOC-1 proteins in the perinuclear region and the cytoplasmic puncta that were costained with the recycling endosomal marker STX13 (SI Appendix, Fig. S2B). This was in agreement with the previous observations demonstrating the function of BLOC-1 on tubular endosomes in melanogenesis (7, 8). Interestingly, significant costaining was also detected for UVRAG and BLOC-1 with the premelanosome marker PMEL (SI Appendix, Fig. S2C). Of note, the BLOC-1 subunits BLOS1, BLOS2, and Snapin are also part of BLOC-one related complex (BORC), which consists of eight small proteins but plays a distinct role in lysosome positioning and cell migration (Fig. 2A) (20). However, little, if any, UVRAG interacted with the BORC-specific subunit Myrlysin (Fig. 2A). Moreover, depletion of BLOS1 or Pallidin abolished the ability of UVRAG in promoting the punctate accumulation of TYRP1 and cell pigmentation (Fig. 2B). By contrast, abrogation of autophagy by depletion of the autophagy-essential genes Atg16, Atg5, or Beclin1 or by treatment of cells with the autophagy inhibitors bafilomycin A1 (Baf A1) or 3-methyladenenine (3-MA) did not readily alter the effect of UVRAG on the redistribution of melanosome marker protein and melanization (SI Appendix, Fig. S2 D–F). A previous study showed that mTORC1 signaling is involved in regulation of melanogenesis (21) and that UVRAG is phosphorylated by mTORC1 at Ser498, which inhibits the autophagy function of UVRAG (22). In agreement, treatment of cells with the mTOR inhibitor Torin1 suppressed the phosphorylation of UVRAG at S497, the mouse equivalent of human UVRAG S498 (22). However, it did not affect its interaction with BLOC-1 (SI Appendix, Fig. S2G). Similar results were also obtained when cells were treated with 3-MA (SI Appendix, Fig. S2H), which inhibits Vps34-mediated PtdIns3P production during autophagy (23). Together, these results indicate that BLOC-1 is required for UVRAG function in melanogenesis and pigmentation, independent from the autophagy machinery and its upstream signaling.
Fig. 2.
UVRAG interacts with BLOC-1 and regulates BLOC-1 complex stability, distribution, and cargo-sorting activity in melanosome biogenesis. (A) Interaction between UVRAG and the BLOC-1 complex proteins. Whole cell lysates (WCLs) of UVRAG WT and KO B16 melanocytes were used for immunoprecipitation (IP) with control IgG or anti-UVRAG antibody, followed by immunoblotting (IB) with the indicated antibodies. Right shows endogenous protein expression. As input, 5% of the WCLs was used. Note that UVRAG does not interact with the BORC-specific protein Myrlysin. The protein composition of BLOC-1 and BORC is also shown at Right. (B) BLOC-1 is required for UVRAG-mediated cell pigmentation. B16 melanoma cell stably expressing UVRAG was transfected with control shRNA or shRNA against BLOS1 or Pallidin, and then immunostained with anti-TYRP1 for bright-field (BF) and confocal microscopy (Top). Nuclei were stained with DAPI (blue). The redistribution of TYRP1 (Middle Left) and the relative melanin contents (Middle Right) in the cell lysates in shRNA-transfected cells was quantified. Endogenous protein expression is also shown (Bottom). Means were calculated from the data collected from three independent experiments (n = 200). (Scale bar, 10 μm.) (C) Cytoplasmic dispersion of BLOS1 in UVRAG KO and mutant cells. Representative images of BLOS1 staining in UVRAG WT, KO, and UVRAG KO B16 cells complemented with UVRAG WT or ΔCCD2 are shown (Left). The percentage of cells with BLOS1 dispersion was quantified (Right). Means were calculated from the data collected from three independent experiments (n = 200). Endogenous protein expression is also shown. (Scale bar, 10 μm.) (D) Deletion of UVRAG caused aberrant accumulation of TYRP1 in EEA1- and STX13-positive endosomes. UVRAG WT, KO, and KO B16 cells complemented with UVRAG WT or ΔCCD2 were stained for TYRP1 along with EEA1 or STX13. Nuclei were stained with DAPI, followed by confocal microscopy. The degree of colocalization of TYRP1 with the indicated markers was quantified (Bottom). Means were calculated from the data collected from three independent experiments (n = 50). (Scale bars, 20 μm.) (E and F) Western blot analysis (E) and densitometric quantification (F) of the relative expression of BLOC-1 in UVRAG WT and KO cells. Note decreased levels of BLOC-1 proteins in UVRAG KO melanocytes. Means were calculated from the data collected from three independent experiments. n.s., not significant; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Using several UVRAG deletion mutants, we identified residues 217–234 in the coiled-coil domain (CCD) of UVRAG as required for BLOS1 interaction (SI Appendix, Fig. S3 A–C). Given that UVRAG CCD is also engaged in binding Beclin1 (10), we examined whether the Beclin1-binding activity of UVRAG is affected by the Δ217–234 (referred to as ΔCCD2) mutation. Both UVRAG WT and ΔCCD2, but not ΔCCD, coimmunoprecipitated with Beclin1 (SI Appendix, Fig. S3C). Furthermore, UVRAG ΔCCD2 enhanced Torin1-induced autophagy to a similar extent as WT, as judged by increased levels of the autophagosome-associated lipidated LC3 (LC3-II) and the levels of p62 (an autophagic substrate) (24) degradation (SI Appendix, Fig. S3D). These results indicate that the ΔCCD2 mutant, albeit defective in BLOS1 interaction, remains competent for autophagy and that UVRAG–BLOC-1 mediated melanogenesis is distinguished from its previously defined autophagy activity.
To examine how UVRAG regulates BLOC-1 in melanosome biogenesis, we found that deletion of UVRAG resulted in relocation of the BLOC-1 proteins from juxtanuclear to a more diffuse cytoplasmic punta pattern, whereas the BORC-specific protein, Myrlysin, remained unaffected (Fig. 2C and SI Appendix, Fig. S4A). Reintroducing UVRAG, but not the ΔCCD2 mutant defective in BLOS1 binding, clearly suppressed the aberrant distribution of BLOC-1 proteins in UVRAG KO cells, indicating the specific effect of UVRAG in regulating subcellular localization of BLOC-1 (Fig. 2C and SI Appendix, Fig. S4A). As seen with BLOC-1 deficiency (7, 8), UVRAG knockout caused missorting and aberrant accumulation of melanogenic cargoes TYRP1 and DCT to the early/recycling endosomes, which was subsequently rescued by reexpression of UVRAG but not ΔCCD2 (Fig. 2D and SI Appendix, Fig. S4B). This is consistent with the notion that BLOC-1 is required for melanosome-directed cargo sorting from early tubular carriers to melanosomes (7, 8). Furthermore, UVRAG deficiency caused a marked decrease in overall protein levels of BLOC-1 subunits, but not BORC-related Myrlysin (Fig. 2 E and F). Of note, BLOC-1 protein mRNA levels were not altered by UVRAG deletion (SI Appendix, Fig. S4C). Indeed, cycloheximide (CHX) treatment studies revealed that UVRAG KO accelerated BLOC-1 protein turnover in melanocytes, while the BORC-related Myrlysin remains unaffected (SI Appendix, Fig. S4 D–G). BLOC-1 protein stability is regulated by its complex formation (25, 26). We asked whether UVRAG coordinates the complex assembly of BLOC-1. We found that UVRAG expression resulted in increased coimmunoprecipitation of BLOC-1 proteins both in vivo (SI Appendix, Fig. S4H) and in vitro using recombinant proteins (SI Appendix, Fig. S2A). Together, these results indicate that UVRAG interaction is required for the early stage of melanosome formation by regulating BLOC-1 complex stability, subcellular distribution, and cargo-sorting function.
UVRAG Is Essential for Melanocyte Development in Vivo.
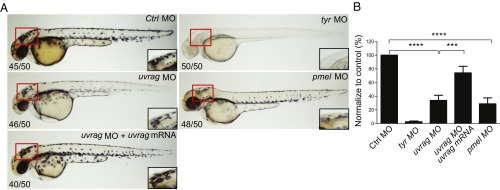
To determine whether the observed melanogenic activity of UVRAG is conserved in vivo, we knocked down uvrag by using specific antisense morpholino (MO) oligonucleotide targeting the uvrag transcript in zebrafish, an ideal model for studying melanocyte differentiation (27) (SI Appendix, Fig. S5). Relative to fish embryos injected with a nontargeting MO, embryos injected with the uvrag-specific MO showed a significant decrease in the formation of pigmented melanocytes (Fig. 3). A similar reduction was also observed in pmel- and tyr-knockdown fish, although the latter showed a more severe phenotype (Fig. 3). The specificity of the MOs was demonstrated by the inhibition of a GFP reporter plasmid containing the target sequence (SI Appendix, Fig. S5B), and by the rescue of pigmentation phenotype by coinjection of human uvrag mRNA lacking the morpholino target sequences into the uvrag-knockdown fish (Fig. 3). Notably, the effect of uvrag and pmel silencing on melanocyte pigmentation was very similar to the pmel-mutant zebrafish with fading vision and defective melanosome biogenesis (28). These results indicate that UVRAG plays an important role in melanocyte development in zebrafish in vivo.
Fig. 3.
UVRAG is required for melanocyte development in zebrafish in vivo. (A) Representative images of 48 h postfertilization (hpf) zebrafish embryos injected with control morpholino (MO) or MO targeting uvrag, pmel, or tyr. Rescue of uvrag MO-mediated depigmentation is shown by coinjection of human uvrag mRNA. Box highlights area of enlargement visualized in Insets. Numbers on each panel indicate the number of embryos showing the representative phenotype per total number of embryos examined. (B) Quantitative measurement shows significantly decreased pigmentation in uvrag MO and pmel MO group compared with the control group. Means were calculated from the data collected from three independent experiments (n = 50). ***P < 0.001; ****P < 0.0001.
UVRAG Is a Direct Transcriptional Target of MITF.
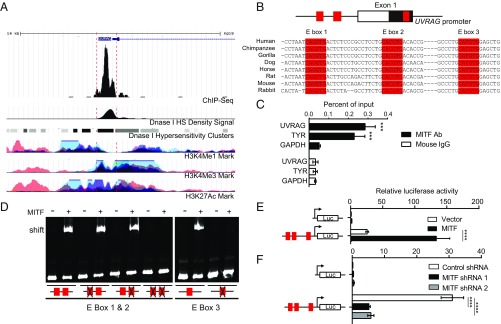
Melanocyte pigmentation is primarily controlled by MITF (29). Overexpression of MITF induced the expression of UVRAG at both mRNA and protein levels (SI Appendix, Fig. S6 A and B), whereas MITF knockdown by two distinct shRNA targeting two different sites of the MITF transcript reduced UVRAG expression in human melanocytes (SI Appendix, Fig. S6 C and D). To test whether UVRAG is a MITF-responsive gene, we analyzed the chromatin immunoprecipitation-sequencing (ChIP-seq) dataset of MITF in melanocytes (30) and found that the proximal 1-kb promoter region of the UVRAG gene contains consensus and transcriptionally active MITF-binding elements in human melanocytes (Fig. 4A). MITF regulates gene transcription by binding to the E-box (CANNTG) element in MITF-responsive genes (31). We identified three distinct E boxes on the UVRAG promoter, located at −585, −563, and +94 from the transcriptional start site, respectively (Fig. 4B and SI Appendix, Fig. S6E). ChIP analysis using an anti-MITF antibody revealed high MITF occupancy at the UVRAG promoter in melanocytes, as seen with its association with the TYR promoter, a known target of MITF (Fig. 4C). Furthermore, electrophoretic mobility shift assay (EMSA) demonstrated a supershift of the UVRAG promoter probes by purified recombinant MITF proteins (SI Appendix, Fig. S6F), which had sequence specificity for each individual E-box consensus probe (Fig. 4D). Mutations in both E-box 1 and adjacent E-box 2 probes or E-box 3 probe prevented MITF binding (Fig. 4D). These results confirmed direct MITF binding to all three of the E boxes of the UVRAG promoter.
Fig. 4.
UVRAG is a transcriptional target of MITF. (A) University of California Santa Cruz genome browser view shows the 10-kb region around the UVRAG promoter. The ChIP-seq signals for MITF (30), and peaks of H3K4Me1, H3K4Me3, H3K27Ac, and DNase I hypersensitivity (HS) density in human melanocytes are extracted from the ENCODE dataset and annotated. (B) Structure of UVRAG promoter showing the location of exon 1 and the three E boxes of the MITF-binding sites (E-box 1, E-box 2, and E-box 3; in red) highly conserved through different species as indicated. (C) ChIP analysis shows MITF occupancy on the UVRAG promoter in human melanocytes. The TYR and GAPDH promoters were used as the positive and negative control, respectively. ***P < 0.001. (D) Electrophoretic mobility shift assay (EMSA) shows the direct association of purified recombinant MITF protein with three E-box sequences in the UVRAG promoter, but not with the E-box mutated sequences. EMSA probes containing either the WT or the mutant E-box sequence are indicated. (E) Dual luciferase reporter assay showing the activity of the UVRAG promoter in response to transfection of MITF in 293T cells. Red boxes indicate the location of three E boxes. Data are mean ± SD from three independent experiments. (F) Knockdown of MITF strongly suppresses UVRAG promoter activity in melanocytes. The UVRAG promoter–reporter was transfected into human melanocytes expressing MITF-specific shRNA and luciferase activity was measured 48 h posttransfection. Results are expressed as mean ± SD from three independent experiments. ****P < 0.0001.
To determine whether MITF binding regulates the activity of the UVRAG promoter, we performed luciferase reporter assays using the UVRAG promoter carrying all three E boxes. As shown in Fig. 4E, ectopic expression of MITF drastically induced activity of the UVRAG promoter by more than fourfold. Conversely, depletion of endogenous MITF in melanocytes by two distinct shRNAs abolished the basal transcriptional activity of the UVRAG promoter (Fig. 4F). To examine which of the consensus elements was mainly responsible for transcription activation of UVRAG by MITF, a series of luciferase reporters were tested for MITF responsiveness with different combinations of mutations at each individual E box (SI Appendix, Fig. S6G). We found that mutation of any of the three E boxes led to a 30% reduction of MITF responsiveness of the UVRAG promoter, while mutations of two or all three E boxes could repress transactivation to as little as 5–15% of the normal level (SI Appendix, Fig. S6G). These results indicate that all three E-box sites act cumulatively to allow UVRAG expression at the highest levels and that UVRAG is a bona fide target gene of MITF.
MITF Induction of UVRAG Is Required for UV-Induced Tanning.
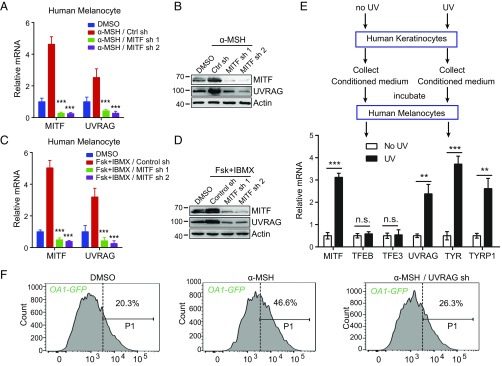
MITF-mediated melanogenesis is induced in UV-tanning response (32). UV exposure triggers keratinocytes to secrete α-MSH, which engages the MC1R on melanocytes and activates cAMP signaling and MITF expression (2). To investigate the role of UVRAG in this pathway, we examined the response of UVRAG to α-MSH signaling. Human melanocytes were treated with α-MSH or cAMP agonists forskolin and IBMX. As shown in Fig. 5 A–D, α-MSH and cAMP signaling dramatically induced UVRAG expression in melanocytes within 2 h of treatment, concomitant with the up-regulation of MITF as previously reported (33). Depletion of MITF almost completely abrogated the induction of UVRAG in α-MSH–stimulated and forskolin + IBMX-stimulated cells (Fig. 5 A–D). Similar results were also achieved in B16 melanoma cells, whereby α-MSH drastically increased both mRNA and protein levels of UVRAG in a MITF-dependent manner (SI Appendix, Fig. S7 A and B). Unlike MITF, knockdown of TFEB and TFE3 had minimal effect on α-MSH–induced UVRAG expression (SI Appendix, Fig. S7 C–F). Indeed, expression of TFEB and TFE3 was not altered by α-MSH signaling either (SI Appendix, Fig. S7 C–F). These data indicate that MITF is selectively required for the induction of UVRAG in response to α-MSH/cAMP signaling.
Fig. 5.
MITF induces UVRAG expression in response to α-MSH. (A and B) UVRAG mRNA expression in human melanocytes expressing MITF-specific shRNA and treated with α-MSH for 2 h (A). The relative expression of MITF and UVRAG in cells in A after α-MSH treatment is shown in B. Data are mean ± SD from three independent experiments. (C and D) UVRAG mRNA expression in human melanocytes expressing MITF-specific shRNA and treated with forskolin (FSK) + IBMX for 2 h (C). The relative expression of MITF and UVRAG in cells in C after FSK + IBMX treatment is shown in D. Data are mean ± SD from three independent experiments. (E) Melanogenic gene expression in human melanocytes induced by conditioned media collected from UV-irradiated human keratinocytes. Keratinocytes were UVB (10 mJ/cm2) irradiated for 16 h and medium was collected. Naïve human melanocytes were then incubated for 5 h with the collected medium from mock- and UV-treated cells and then evaluated for gene expression as indicated. (F) B16 cells stably expressing melanosome marker OA1-GFP were transfected with control shRNA or UVRAG-specific shRNA and treated with α-MSH. The resulted cells were subjected to flow cytometry analysis. The portion of cells with the fluorescence intensity greater than the indicated threshold (P1 gate) are indicated. n.s., not significant; **P < 0.01; ***P < 0.001.
To directly test the effect of UV-induced factors secreted from keratinocytes, human keratinocytes were subjected to UVB (10 mJ/cm2), and the culture media from these cells was collected and incubated with human melanocytes (Fig. 5E). As seen with treatment of α-MSH, the conditioned media induced the expression of MITF threefold and UVRAG more than twofold (Fig. 5E). Again, no change was detected for the expression of MITF-related family member TFEB and TFE3 (Fig. 5E). Notably, depletion of UVRAG abolished the effect of α-MSH and cAMP agonists (forskolin + IBMX) in promoting melanosome biogenesis (Fig. 5F and SI Appendix, Fig. S7G), as illustrated by the fluorescent melanosomal marker ocular albinism 1 (OA1)-GFP (34). Likewise, the melanogenic effect of MITF was strongly abrogated in UVRAG-deficient melanocytes, as exemplified by the mislocalization of TYRP1 and cell hypopigmentation (SI Appendix, Fig. S7 H–J). Hence, UVRAG represents an integral molecule in UV-induced melanogenesis.
Oncogenic BRAF Inhibits UVRAG Expression and Melanogenesis in Melanoma.
UV-induced melanogenesis is strongly associated with the risk of melanoma (35, 36). Given that the oncogenic BRAF(V600E) is most prevalent in melanoma and is considered a driving force for melanoma development (37, 38), we attempted to further investigate the potential role of BRAF(V600E) in UVRAG-mediated melanogenesis and protective tanning response. As shown in Fig. 6A, ectopic expression of BRAF(V600E) in human melanocytes resulted in more than 50% reduction of UVRAG mRNA and other melanogenic genes TYR and DCT. Correspondingly, α-MSH–stimulated accrual of melanosomes and melanin production as observed in normal melanocytes was drastically suppressed by BRAF(V600E) expression (Fig. 6 B–D). These results suggest that oncogenic BRAF negatively regulates UV-induced melanogenesis. Indeed, α-MSH–induced melanosome accumulation was compromised in BRAF(V600E)-positive melanoma cell lines compared with that in the MeWo cell line that is BRAF WT (SI Appendix, Fig. S8A). Moreover, analysis of the TCGA provisional melanoma patient datasets (39–42) also revealed that BRAF(V600E) melanomas had significantly reduced expression of melanogenic genes including UVRAG (t test, P = 0.0006), PMEL (t test, P = 0.0053), and DCT (t test, P = 0.0011) (SI Appendix, Fig. S8 B–D). To examine whether impaired melanogenesis is BRAF mediated, A375 melanoma cells were treated with PLX4720, a selective BRAF(V600E) inhibitor (43). As expected, PLX4720 reverted the effect of BRAF(V600E) and induced both mRNA and protein expression of melanogenic genes, including UVRAG (Fig. 6 E and F). Analogous results were also obtained in other BRAF(V600E) melanoma cell lines, but not in MeWo (SI Appendix, Fig. S8E). Removing MITF abolished the ability of PLX4720 to up-regulate UVRAG and OA1-GFP–labeled melanosomes, suggesting that the up-regulation is MITF dependent (Fig. 6G and SI Appendix, Fig. S8F). Removing UVRAG resulted in similar effect in PLX4720-treated cells (SI Appendix, Fig. S8G). In fact, PLX4720 treatment enhanced MITF expression (Fig. 6 E and F), an observation consistent with the previous report (37). To further validate that oncogenic BRAF regulates melanogenesis in a MITF-dependent manner, we analyzed the MITF ChIP-seq dataset in human melanocytes stably expressing BRAF(V600E) and in COLO829 melanoma cells positive for BRAF(V600E) (30). As shown in Fig. 6H, expression of BRAF(V600E) in melanocytes drastically reduced the binding of MITF at the UVRAG sites, whereas BRAF inhibition by PLX4032, another BRAF-specific inhibitor (44), led to a marked enrichment of MITF at these sites. Similar observations were detected for other melanogenic machinery such as PMEL, TYR, and DCT (SI Appendix, Fig. S8 H–J). These findings suggest that individuals harboring BRAF(V600E) mutations might be less competent in UV-induced melanogenesis and therefore more susceptible to UV-induced skin damage than individuals with WT BRAF, which may contribute to its oncogenic potential in skin cancer.
Fig. 6.
Oncogenic BRAF(V600E) inhibits UVRAG expression and attenuates the tanning ability of melanocytes. (A) Quantitative RT-PCR showing the relative level of mRNA expression for UVRAG, TYR, and DCT in human melanocytes expressing empty vector or BRAF(V600E). Data are mean ± SD from three independent experiments. (B–D) BRAF(V600E) inhibits α-MSH–induced melanogenesis. Confocal microscopy of the distribution pattern of PMEL in human melanocytes expressing vector or BRAF(V600E) upon α-MSH treatment (B). The relative intensity of PMEL was quantified (C). The melanin contents of the lysates of cells in B were measured OD at 405 nm (D). Data are mean ± SD from three independent experiments. (E and F) Quantitative RT-PCR showing the relative mRNA expression for MITF, UVRAG, TYR, and DCT in A375 cells treated with PLX4720 (1 μM) for 48 h (E). Western blot analysis of the indicated proteins in cells in E is shown in F. (G) A375 melanoma cells expressing control shRNA or MITF-specific shRNA were treated with PLX4720 (1 μM) and subjected to flow cytometry analysis using OA1-mCherry as a marker of melanosomes. (H) ChIP-seq analysis of MITF occupancy on the UVRAG promoter in melanocytes transduced with either empty vector (first panel) or BRAF(V600E) (second panel); and in BRAF(V600E)-positive COLO829 melanoma cells treated with either DMSO (third panel) or PLX4032 (fourth panel). The ChIP-seq original data are from a previous report (30). **P < 0.01; ***P < 0.001; ****P < 0.0001.
Discussion
UV-induced melanogenesis and pigmentation is the first line of defense against environmental UVR that increases the risk of developing skin cancers (2). We have identified the pivotal role of autophagic tumor suppressor, UVRAG, in the tanning response to UV light as a downstream effector of the α-MSH/cAMP signaling pathway and as a major transcriptional target of MITF. UVRAG mediates melanosome biogenesis by interaction with BLOC-1. Removal of UVRAG resulted in destabilization and mislocalization of BLOC-1, leading to the missorting of melanosomal cargo proteins. To the best of our knowledge, these findings represent the description of an autophagy-related factor directly functioning in melanogenic membrane remodeling and cell pigmentation, which reveals a broader functional scope for UVRAG and also provides mechanistic insights into the role of UVRAG in pigmentation disorder and skin cancer.
The multitasking UVRAG has been found to be associated with different subcellular compartments to regulate cellular pathways in a coordinated fashion. We have previously discovered that UVRAG associates with Bif-1 and Beclin1 to promote autophagy (10, 13), while endosomal UVRAG targets the tethering factor to facilitate endosome maturation (12). Upon DNA damage, UVRAG can be recruited to the damaged sites and participate in the assembly of DNA repair machineries (14, 15). Apart from these findings, the present study provides an advancement in understanding the role of UVRAG in pigmentation biology. Deficiency of UVRAG mislocalized melanogenic machinery proteins and induced hypopigmentation even in the presence of MITF and/or α-MSH/cAMP activation. Notably, autophagy has been implicated in melanocyte function (45). However, we observed that autophagy loss could not forestall cell pigmentation induced by UVRAG expression. In line with this, a previous study also showed that the melanocyte-specific deficiency in autophagy did not cause major defects in melanosome biogenesis, nor did it produce visual changes in skin pigmentation (46). While our observations do not exclude a role for autophagy in pigmentation, such events would appear to be downstream of a critical UVRAG-dependent step in regulating melanogenesis. Further support for UVRAG as a melanogenic molecule was demonstrated in the zebrafish study. The development of pigmented melanocytes in zebrafish was impaired upon UVRAG deficiency. Recovery of UVRAG in UVRAG-deficient melanocytes and fish embryos rescued melanogenesis.
The engagement of UVRAG in melanosome pigmentation was found to be associated with BLOC-1 by increasing the complex stability and functionality in melanosome-related cargo sorting and delivery. Although BLOC-1 shares the BLOS1, BLOS2, and Snapin subunits with the BORC complex (20), UVRAG is not associated with BORC, nor does it cause any change in BORC distribution and stability. Our results thus support the distinct role of BORC and BLOC-1 in melanosome activity as previously reported (20). Although the molecular mechanisms involved in BLOC-1–mediated biogenesis of lysosome-related organelles (LROs) remain unclear, it is proposed that BLOC-1 is required for the sorting of a subset of melanosomal cargoes such as TYRP1 into tubular carriers (8). Consistent with this, inactivation of UVRAG results in mislocalization of the melanogenic enzymes to the early/recycling endosomes, leading to reduced melanosome pigmentation. Intriguingly, in addition to the defective cargo sorting to melanosomes as seen with BLOC-1 inactivation, the distribution pattern of the early melanosome protein PMEL was also affected in the absence of UVRAG. Given that the underlying mechanism of PMEL traffic is not yet understood but is thought to be BLOC-1 independent (8, 47), we speculate that UVRAG-mediated PMEL distribution involves a new mechanism that awaits further investigation. Nevertheless, our results indicate that the UVRAG–BLOC-1 interaction is critically required for complete melanocyte function.
Consistent with its function in regulating melanogenesis, our study demonstrated that UVRAG is transcriptionally controlled by MITF through the three E-box elements in the UVRAG promoter. Notably, in addition to the canonical E-box motif (E-box 2, CACGTG; E-box 3, CATGTG) (48), we were able to detect binding by MITF to the atypical E-box motif (E-box 1, CAGGTG) present in the UVRAG promoter both in vitro and in the reporter assay in vivo. Of course, this does not exclude a role for E-box 1 in recruiting other bHLH E-box binding transcriptional factors; instead, it implies that regulation of gene expression by MITF seems to be more complex and likely executed in a gene context-dependent manner. Unlike MITF that is predominantly expressed in neural crest-derived melanocytes, TFEB and TFE3 are more ubiquitously expressed (49). Both TFEB and TFE3 are implicated in transcriptional activation of autophagy and lysosome biogenesis via binding to a 10 bp motif (GTCACGTGAC), so called the coordinated lysosomal expression and regulation (CLEAR) element, in the target gene promoters (50, 51). This motif contains the E-box binding sequence (CANNTG) for MITF. This then raises a question as to whether the regulation of UVRAG by MITF is equally shared by TFEB and TFE3 in α-MSH–stimulated melanocytes. However, inactivation of TFEB or TFE3 had minimal effect on UVRAG expression after α-MSH stimulation. Indeed, unlike MITF that was drastically up-regulated by α-MSH, the levels of TFEB and TFE3 remained marginally affected. Hence, we propose that UVRAG expression is most substantially controlled by MITF in melanocytes in response to UV tanning, thereby broadening the involvement of MITF as a regulator of pigmentation and potentially other UVRAG-dependent functions in melanocyte and skin cancer development.
Although impaired UV-induced skin pigmentation and oncogenic BRAF signaling are both involved in melanoma, the interplay between these two pathways remains largely unknown. By comparing melanomas with or without BRAF V600E mutation, we found that oncogenic BRAF activation is associated with reduced melanogenic gene expression, including UVRAG, resulting in suppressed melanogenesis that cannot be stimulated by α-MSH. This dampened tanning ability triggered by BRAF(V600E) is largely due to suppressed MITF expression, which can be reversed by BRAF inhibitors. Our findings provide a mechanism to explain previous observations that oncogenic BRAF was correlated with the skin phenotypes that have poor UV protection (38, 52). Given the association of tanning ability with melanoma and other skin cancer risk (35, 53), understanding the complex and synergistic interaction between UVR, BRAF signaling, and melanogenic mechanism thus has significance in skin cancer prevention.
Experimental Procedures
All experiments were independently repeated at least three times. Data are represented as mean ± SD. Statistical significance was calculated using Student’s t test, one-way ANOVA, and two-way ANOVA, using GraphPad Prism 6.0 (GraphPad Software, Inc.). A P value of ≤0.05 was considered statistically significant. Methods for generation of UVRAG knockout cells by CRISPR-Cas9 nickase, melanin determination assay, gene knockdown, ChIP analysis, MO design and injection and EGFP reporter assay, and RNA rescue experiment are provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank D. Hauser and E. Barron for performing electron microscopy, Dr. J. U. Jung for providing imaging support and related reagents, and Dr. Oancea for providing the OA1 plasmid. The results shown here are in part based upon data generated by the The Cancer Genome Atlas Research Network: https://cancergenome.nih.gov/. This work was supported by National Institutes of Health (NIH) Grant R01 CA140964 and Melanoma Research Alliance Grant 509218 (to C.L.), NIH Grant AI073099 (to J. U. Jung), and Shenzhen Science and Technology Program Grant JCYJ20150630145302246 (to H.Z.).
Footnotes
Conflict of interest: The authors declare no conflict of interest.
This article is a PNAS Direct Submission. K.-L.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803303115/-/DCSupplemental.
References
- 1.Aubert B, et al. BABAR Collaboration Measurement of branching fractions and charge asymmetries in B decays to an eta meson and a K* meson. Phys Rev Lett. 2006;97:201802. doi: 10.1103/PhysRevLett.97.201802. [DOI] [PubMed] [Google Scholar]
- 2.Chen H, Weng QY, Fisher DE. UV signaling pathways within the skin. J Invest Dermatol. 2014;134:2080–2085. doi: 10.1038/jid.2014.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin JY, Fisher DE. Melanocyte biology and skin pigmentation. Nature. 2007;445:843–850. doi: 10.1038/nature05660. [DOI] [PubMed] [Google Scholar]
- 4.Theos AC, Truschel ST, Raposo G, Marks MS. The silver locus product Pmel17/gp100/Silv/ME20: Controversial in name and in function. Pigment Cell Res. 2005;18:322–336. doi: 10.1111/j.1600-0749.2005.00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raposo G, Marks MS. Melanosomes–Dark organelles enlighten endosomal membrane transport. Nat Rev Mol Cell Biol. 2007;8:786–797. doi: 10.1038/nrm2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Starcevic M, Dell’Angelica EC. Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1) J Biol Chem. 2004;279:28393–28401. doi: 10.1074/jbc.M402513200. [DOI] [PubMed] [Google Scholar]
- 7.Di Pietro SM, et al. BLOC-1 interacts with BLOC-2 and the AP-3 complex to facilitate protein trafficking on endosomes. Mol Biol Cell. 2006;17:4027–4038. doi: 10.1091/mbc.E06-05-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Setty SR, et al. BLOC-1 is required for cargo-specific sorting from vacuolar early endosomes toward lysosome-related organelles. Mol Biol Cell. 2007;18:768–780. doi: 10.1091/mbc.E06-12-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dell’Angelica EC. The building BLOC(k)s of lysosomes and related organelles. Curr Opin Cell Biol. 2004;16:458–464. doi: 10.1016/j.ceb.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Liang C, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 11.He S, et al. PtdIns(3)P-bound UVRAG coordinates Golgi-ER retrograde and Atg9 transport by differential interactions with the ER tether and the beclin 1 complex. Nat Cell Biol. 2013;15:1206–1219. doi: 10.1038/ncb2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang C, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10:776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi Y, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y, et al. Autophagic UVRAG promotes UV-induced photolesion repair by activation of the CRL4(DDB2) E3 ligase. Mol Cell. 2016;62:507–519. doi: 10.1016/j.molcel.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Z, et al. A dual role for UVRAG in maintaining chromosomal stability independent of autophagy. Dev Cell. 2012;22:1001–1016. doi: 10.1016/j.devcel.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berson A, et al. Toxicity of alpidem, a peripheral benzodiazepine receptor ligand, but not zolpidem, in rat hepatocytes: Role of mitochondrial permeability transition and metabolic activation. J Pharmacol Exp Ther. 2001;299:793–800. [PubMed] [Google Scholar]
- 17.Jeong TJ, et al. Association of UVRAG polymorphisms with susceptibility to non-segmental vitiligo in a Korean sample. Exp Dermatol. 2010;19:e323–e325. doi: 10.1111/j.1600-0625.2009.01039.x. [DOI] [PubMed] [Google Scholar]
- 18.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Setty SR, et al. Cell-specific ATP7A transport sustains copper-dependent tyrosinase activity in melanosomes. Nature. 2008;454:1142–1146. doi: 10.1038/nature07163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pu J, et al. BORC, a multisubunit complex that regulates lysosome positioning. Dev Cell. 2015;33:176–188. doi: 10.1016/j.devcel.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho H, Kapadia R, Al-Tahan S, Ahmad S, Ganesan AK. WIPI1 coordinates melanogenic gene transcription and melanosome formation via TORC1 inhibition. J Biol Chem. 2011;286:12509–12523. doi: 10.1074/jbc.M110.200543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim YM, et al. mTORC1 phosphorylates UVRAG to negatively regulate autophagosome and endosome maturation. Mol Cell. 2015;57:207–218. doi: 10.1016/j.molcel.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelárová H, Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem. 1997;243:240–246. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- 24.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee HH, et al. Assembly and architecture of biogenesis of lysosome-related organelles complex-1 (BLOC-1) J Biol Chem. 2012;287:5882–5890. doi: 10.1074/jbc.M111.325746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X, et al. The BLOC-1 subunit pallidin facilitates activity-dependent synaptic vesicle recycling. eNeuro. 2017;4:ENEURO.0335-16.2017. doi: 10.1523/ENEURO.0335-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Webster GF, et al. Efficacy and tolerability of once-daily tazarotene 0.1% gel versus once-daily tretinoin 0.025% gel in the treatment of facial acne vulgaris: A randomized trial. Cutis. 2001;67(6 Suppl):4–9. [PubMed] [Google Scholar]
- 28.Schonthaler HB, et al. A mutation in the silver gene leads to defects in melanosome biogenesis and alterations in the visual system in the zebrafish mutant fading vision. Dev Biol. 2005;284:421–436. doi: 10.1016/j.ydbio.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 29.Levy C, Khaled M, Fisher DE. MITF: Master regulator of melanocyte development and melanoma oncogene. Trends Mol Med. 2006;12:406–414. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 30.Webster DE, et al. Enhancer-targeted genome editing selectively blocks innate resistance to oncokinase inhibition. Genome Res. 2014;24:751–760. doi: 10.1101/gr.166231.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie X, et al. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.D’Orazio JA, et al. Topical drug rescue strategy and skin protection based on the role of Mc1r in UV-induced tanning. Nature. 2006;443:340–344. doi: 10.1038/nature05098. [DOI] [PubMed] [Google Scholar]
- 33.Shoag J, et al. PGC-1 coactivators regulate MITF and the tanning response. Mol Cell. 2013;49:145–157. doi: 10.1016/j.molcel.2012.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bruder JM, et al. Melanosomal dynamics assessed with a live-cell fluorescent melanosomal marker. PLoS One. 2012;7:e43465. doi: 10.1371/journal.pone.0043465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baccarelli A, Landi MT. [Risk factors of malignant skin melanoma in Italian population: Review of results of a case-control study] Epidemiol Prev. 2002;26:293–299. [PubMed] [Google Scholar]
- 36.McIlwaine WJ, Donnelly MD, Chivers AT, Evans AE, Elwood JH. Certification of death from ischaemic heart disease in Belfast. Int J Epidemiol. 1985;14:560–565. doi: 10.1093/ije/14.4.560. [DOI] [PubMed] [Google Scholar]
- 37.Haq R, et al. Oncogenic BRAF regulates oxidative metabolism via PGC1α and MITF. Cancer Cell. 2013;23:302–315. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edwards RH, et al. Absence of BRAF mutations in UV-protected mucosal melanomas. J Med Genet. 2004;41:270–272. doi: 10.1136/jmg.2003.016667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cerami E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hodis E, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berger MF, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–506. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gillies P, Elwood JM, Hawtin P, Ledwith F. Anxieties in adolescents about unemployment and war. Br Med J. 1985;291:383–384. doi: 10.1136/bmj.291.6492.383-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smalley KS. PLX-4032, a small-molecule B-Raf inhibitor for the potential treatment of malignant melanoma. Curr Opin Investig Drugs. 2010;11:699–706. [PubMed] [Google Scholar]
- 45.Ganesan AK, et al. Genome-wide siRNA-based functional genomics of pigmentation identifies novel genes and pathways that impact melanogenesis in human cells. PLoS Genet. 2008;4:e1000298. doi: 10.1371/journal.pgen.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang CF, et al. Suppression of autophagy dysregulates the antioxidant response and causes premature senescence of melanocytes. J Invest Dermatol. 2015;135:1348–1357. doi: 10.1038/jid.2014.439. [DOI] [PubMed] [Google Scholar]
- 47.Jani RA, Purushothaman LK, Rani S, Bergam P, Setty SR. STX13 regulates cargo delivery from recycling endosomes during melanosome biogenesis. J Cell Sci. 2015;128:3263–3276. doi: 10.1242/jcs.171165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pogenberg V, et al. Restricted leucine zipper dimerization and specificity of DNA recognition of the melanocyte master regulator MITF. Genes Dev. 2012;26:2647–2658. doi: 10.1101/gad.198192.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martina JA, Diab HI, Li H, Puertollano R. Novel roles for the MiTF/TFE family of transcription factors in organelle biogenesis, nutrient sensing, and energy homeostasis. Cell Mol Life Sci. 2014;71:2483–2497. doi: 10.1007/s00018-014-1565-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palmieri M, et al. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet. 2011;20:3852–3866. doi: 10.1093/hmg/ddr306. [DOI] [PubMed] [Google Scholar]
- 51.Sardiello M, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 52.Maldonado JL, et al. Determinants of BRAF mutations in primary melanomas. J Natl Cancer Inst. 2003;95:1878–1890. doi: 10.1093/jnci/djg123. [DOI] [PubMed] [Google Scholar]
- 53.Elwood JM, Gallagher RP, Davison J, Hill GB. Sunburn, suntan and the risk of cutaneous malignant melanoma–The Western Canada Melanoma Study. Br J Cancer. 1985;51:543–549. doi: 10.1038/bjc.1985.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.