Significance
The oxygen-evolving photosystem II (PSII) complex is essential for driving the light reactions of photosynthesis and for producing the oxygen we breathe. Accessory proteins, most of unknown function, are vital for assembling and maintaining PSII activity. Here we provide structural information on Ycf48/HCF136, which is involved in the early steps of PSII assembly and the repair of PSII following damage by light. We show that Ycf48 is a seven-bladed beta-propeller and that a highly conserved arginine patch is important for function. We propose a role for Ycf48 in coordinating the binding of chlorophyll to protein during insertion of chlorophyll-binding proteins into the membrane. Our work provides insights into the assembly of the photosynthetic apparatus used to harvest sunlight.
Keywords: photosystem II, photosynthesis, chlorophyll-binding proteins
Abstract
Robust photosynthesis in chloroplasts and cyanobacteria requires the participation of accessory proteins to facilitate the assembly and maintenance of the photosynthetic apparatus located within the thylakoid membranes. The highly conserved Ycf48 protein acts early in the biogenesis of the oxygen-evolving photosystem II (PSII) complex by binding to newly synthesized precursor D1 subunit and by promoting efficient association with the D2 protein to form a PSII reaction center (PSII RC) assembly intermediate. Ycf48 is also required for efficient replacement of damaged D1 during the repair of PSII. However, the structural features underpinning Ycf48 function remain unclear. Here we show that Ycf48 proteins encoded by the thermophilic cyanobacterium Thermosynechococcus elongatus and the red alga Cyanidioschyzon merolae form seven-bladed beta-propellers with the 19-aa insertion characteristic of eukaryotic Ycf48 located at the junction of blades 3 and 4. Knowledge of these structures has allowed us to identify a conserved “Arg patch” on the surface of Ycf48 that is important for binding of Ycf48 to PSII RCs but also to larger complexes, including trimeric photosystem I (PSI). Reduced accumulation of chlorophyll in the absence of Ycf48 and the association of Ycf48 with PSI provide evidence of a more wide-ranging role for Ycf48 in the biogenesis of the photosynthetic apparatus than previously thought. Copurification of Ycf48 with the cyanobacterial YidC protein insertase supports the involvement of Ycf48 during the cotranslational insertion of chlorophyll-binding apopolypeptides into the membrane.
The photosystem II (PSII) complex found in the thylakoid membranes of cyanobacteria and chloroplasts is a multisubunit pigment–protein complex that uses light energy to drive the oxidation of water to oxygen and the reduction of plastoquinone to plastoquinol in oxygenic photosynthesis (1, 2). PSII is prone to light-induced damage in vivo and is repaired via the specific replacement of damaged protein subunits within the complex, especially the D1 reaction center subunit (3). Recent advances in determining the structures of PSII from both cyanobacteria (4) and chloroplasts (5, 6) make PSII an excellent model system to study both the assembly and repair of a thylakoid membrane protein complex at the molecular level.
PSII in cyanobacteria is assembled in the membrane in a stepwise fashion from smaller pigment–protein modules or subcomplexes composed of a major chlorophyll-binding subunit plus associated low-molecular mass PSII subunits (3, 7). The PSII reaction center (PSII RC) assembly complex is formed from D1/PsbI and D2/Cyt b559 subcomplexes and is converted to the RC47 complex (8) by addition of the CP47 module (9); the monomeric PSII core complex is then formed by attachment of the CP43 module (3, 9). Formation of the holoenzyme involves light-driven assembly of the water-oxidizing Mn4CaO5 cluster, attachment of lumenal extrinsic proteins, and dimerization (reviewed in ref. 10).
Conserved accessory factors optimize assembly and/or repair of PSII following photodamage (3, 11, 12). Of these, Ycf48 (also known as HCF136 in plants) acts early in assembly and is important for formation of the PSII RC assembly complex in both plants (13, 14) and cyanobacteria (15). Ycf48 has been biochemically isolated as a component of cyanobacterial PSII RC assembly complexes (16) and stabilizes unassembled precursor D1 destined for either formation of the PSII RC during assembly or replacement of damaged D1 during PSII repair (15). Ycf48 is able to recognize the C-terminal extension of precursor D1, although this interaction is not crucial for assembly of the PSII RC (15). In the case of chloroplasts, many Ycf48/HCF136 contain a characteristic insertion of 19-aa residues of unknown function (13). Despite its physiological importance, the structural features of Ycf48 that are important for function remain unknown.
We show here that Ycf48, encoded by the thermophilic cyanobacterium Thermosynechococcus elongatus and the thermophilic red alga Cyanidioschyzon merolae, is a seven-bladed beta-propeller, and we identify a highly conserved positively charged region (the so-called “Arg patch”) that is important for attachment of Ycf48 to PSII. Surprisingly, we find that Ycf48 also binds to photosystem I (PSI), consistent with a direct role for Ycf48 in the accumulation of PSI not just PSII. Based on the copurification of Ycf48 with the YidC insertase involved in the insertion of proteins into the thylakoid membrane, and the requirement for Ycf48 for normal chlorophyll biosynthesis, we propose that Ycf48 binds to chlorophyll-binding apopolypeptides as they are released into the membrane, possibly to help coordinate protein insertion and folding with loading of chlorophyll molecules into their binding sites.
Results
Ycf48 Binds to Large PSI and PSII Complexes.
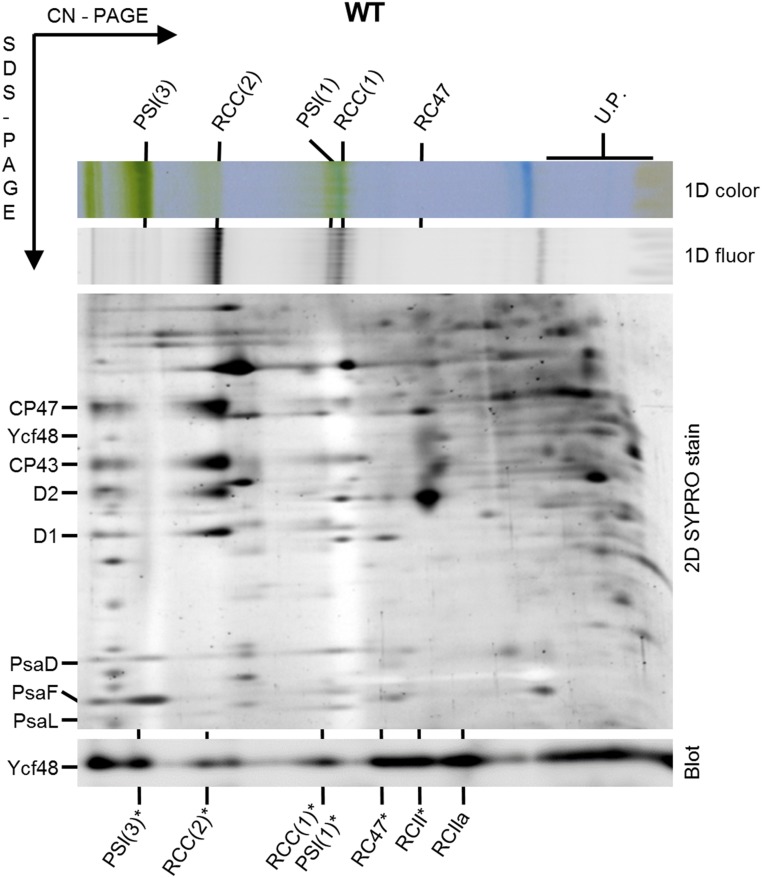
In previous work we used 2D electrophoresis combined with immunoblotting to demonstrate that the Ycf48 protein in the cyanobacterium Synechocystis sp. PCC 6803 (hereafter Synechocystis) is a component of two types of PSII RC assembly complex, designated PSII RCII* and PSII RCa (15–17). PSII RCII* is larger than the PSII RCIIa complex as it also contains the Ycf39/Hlip complex involved in chlorophyll delivery (16). The antibody we originally used to detect Ycf48 was raised against Ycf48 from Arabidopsis (15). To determine whether Ycf48 was also present at low abundance in other complexes, we repeated the analysis using a more sensitive antibody raised against Synechocystis Ycf48 and by using clear native-polyacrylamide gel electrophoresis (CN-PAGE) rather than blue native (BN)-PAGE in the first dimension. Ycf48 was now detected in several larger complexes (Fig. 1). Based on the analysis of mutants unable to assemble PSI (a ΔPSI mutant lacking both the PsaA and PsaB RC subunits) or blocked at various stages of PSII assembly (in strains lacking either CP43 or CP47) (SI Appendix, Fig. S1), we tentatively assigned these larger complexes as monomeric and dimeric core complexes of PSII (18) and monomeric and trimeric complexes of PSI (Fig. 1).
Fig. 1.
Analysis of YCF48-containing complexes in membranes of WT Synechocystis. Membranes isolated from WT cells cultivated at 40 µE m−2⋅s−1 were analyzed by 2D clear native (CN)-SDS/PAGE electrophoresis and the gel was stained by SYPRO Orange, photographed, blotted to a PVDF membrane, and probed with a specific antibody raised to Ycf48 from Synechocystis (segment of the 2D blot with the specific antibody signal is shown). Pigmented complexes were detected in the first dimension by their color (1D color) and fluorescence (1D fluor). Designation of complexes in the SYPRO-stained gel: PSI(1), monomeric PSI complex; PSI(3), trimeric PSI complex; RCC(2) and RCC(1), dimeric and monomeric PSII core complexes, respectively; RC47, PSII core complex lacking CP43; U.P., unassembled proteins. In the case of the immunoblot, the migration of PSI and PSII complexes containing Ycf48 are designated by an asterisk. RCII* and RCIIa are reaction center assembly complexes previously shown to contain Ycf48 (15, 16). Thylakoid membranes containing 4 µg chlorophyll were analyzed.
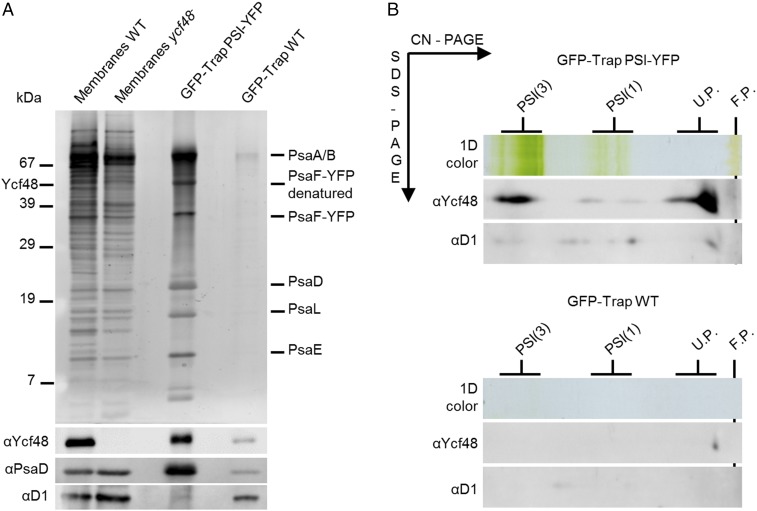
The apparent binding of Ycf48 to PSI was intriguing, as Ycf48 has so far been considered a PSII-specific accessory factor. To provide additional evidence for an interaction with PSI in vivo, PSI complexes were isolated from a Synechocystis strain expressing YFP-tagged PsaF by immunoaffinity chromatography (19). Ycf48 was detected in the YFP-tagged PSI complexes by immunoblotting, especially the trimer, but was barely detectable in the wild-type (WT) control (Fig. 2). Likewise, pulldown experiments, using various types of His-tagged PSII complex (8, 9), confirmed the association of Ycf48 with His-tagged CP47 and with larger PSII complexes containing His-tagged CP47 (SI Appendix, Fig. S2 A and B).
Fig. 2.
Identification of the Ycf48 protein in YFP-tagged PSI complexes isolated by a single step GFP-Trap affinity pulldown. (A) GFP-Trap preparations pulled down from the strain expressing PsaF-YFP (PSI-YFP) and from the WT strain were eluted by hot SDS and analyzed by 1D SDS/PAGE. The gels were stained by SYPRO Orange; blotted to a PVDF membrane; and Ycf48, PsaD, and D1 were consecutively detected using specific antibodies (segment of the blot with the specific antibody signal is shown). A total of 2 μg of chlorophyll was loaded for WT and ycf48− membranes; 15 μL of SDS-eluted pulldowns (50% of the sample) was loaded for each GFP-Trap sample. (B) The 2D CN-SDS/PAGE analysis of the GFP-Trap pulldowns from the PSI-YFP and WT strains eluted by low pH (glycine buffer pH = 2.5) and immediately neutralized by Tris buffer. A total of 15 μL of low pH-eluted pulldowns (50% of the sample) was analyzed by CN-PAGE and lanes were cut and reelectrophoresed in the second dimension. The 2D gels were stained by SYPRO Orange, blotted to a PVDF membrane, and Ycf48 and D1 were consecutively detected using specific antibodies (segment of the blot containing individual antibody signals is shown).
We also obtained experimental support for the lumenal location of cyanobacterial Ycf48 following trypsin digestion of intact right-side-out thylakoids (SI Appendix, Fig. S2C), in agreement with an earlier characterization of the spinach Ycf48 homolog (13). In summary, these results showed that Ycf48 was able to bind to the lumenal side of larger PSI and PSII complexes, not just PSII RC assembly complexes.
Structure of Ycf48 from T. elongatus and C. merolae.
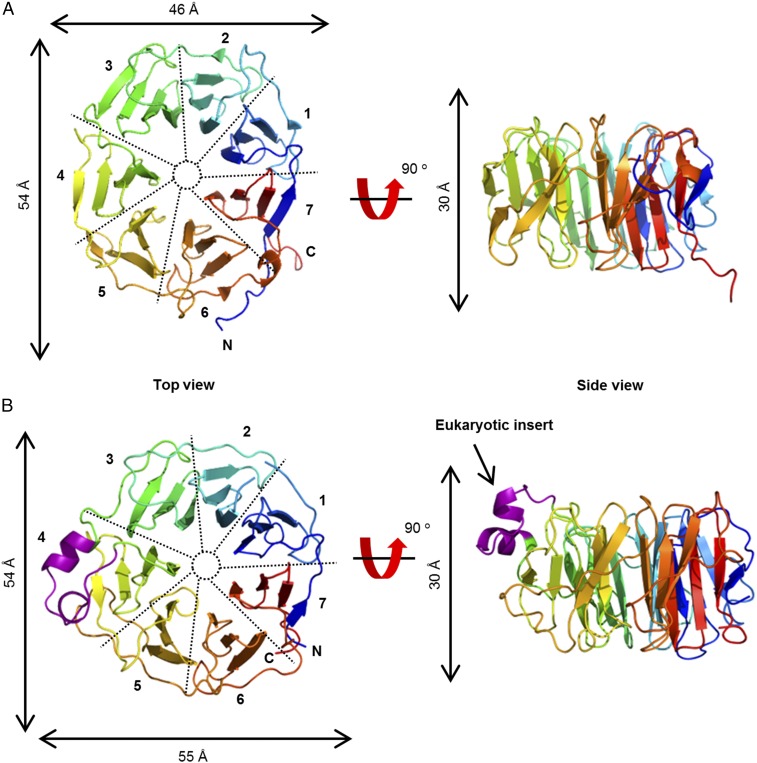
To gain insights into the mechanism of Ycf48 function, we solved the crystal structure of recombinant Ycf48 from the thermophilic cyanobacterium T. elongatus, which is widely used for structural studies (20, 21), to a resolution of 1.5 Å (Fig. 3A). Ycf48 is a 7-bladed beta-propeller, in an approximate torus shape with a width of ∼50 Å and a depth of ∼25 Å. Seven “blades” of four-stranded beta-sheets are arranged around the axis, with a central pore. Ycf48 has the beta-propeller “Velcro,” where the final beta-strand of the C-terminal blade comes from the N-terminal end of the protein. The beta-propeller fold is common, with examples known with 4–10 blades. Some beta-propellers are enzymes, but many have scaffolding roles in larger protein complexes (22). The most similar structure in the Protein Data Bank (PDB) to T. elongatus Ycf48 is an antibiotic-degrading enzyme, virginiamycin B lyase (Vgb) (23), with a root-mean-square deviation (rmsd) of 3.7 Å over 272 residues, but the active site residues of Vgb are not present in Ycf48. We do not predict any enzyme activity for Ycf48.
Fig. 3.
Structure of Ycf48 from cyanobacteria and red algae. Ribbon diagram representation of the structure of Ycf48 from T. elongatus (PDB ID code 2XBG) (A) and C. merolae (PDB ID code 5OJ3) (B), color gradient blue at the N terminus to orange at the C terminus. The 19-residue insert exclusive to eukaryotic Ycf48 is shown in purple. The blades are numbered along the direction of translation, with the “top” of the protein defined as the view that gives an anticlockwise arrangement of the blades.
Eukaryotic Ycf48 sequences have a 19-aa insertion compared with cyanobacterial Ycf48 (13). We also determined the crystal structure of Ycf48 from the thermophilic red alga C. merolae. C. merolae Ycf48 showed the same seven-bladed structure as T. elongatus Ycf48 with the 19-aa insert, consisting of two short alpha-helices and two turns, located between blades 3 and 4 (Fig. 3B). The three other small insertions seen in C. merolae Ycf48 (E161–E164, N322–A324, and N345–S348) compared with T. elongatus Ycf48 are also found in loop regions exposed to the surface (SI Appendix, Fig. S3). Some Ycf48 sequences have inserts in other regions of the protein (SI Appendix, Fig. S3B). Based on the structures reported here, most of these inserts are again predicted to lie in between conserved blade sheet regions (SI Appendix, Fig. S3B).
To test the physiological effect of the 19-residue insert in Synechocystis, which is widely used for mutagenesis studies, we created a mutant, Ycf48-CM, in which the 19-residue insert from C. merolae was inserted into the endogenous Ycf48 protein of Synechocystis. No effect was observed as the Ycf48-CM mutant grew as well as WT at high irradiances (SI Appendix, Fig. S4A), had WT levels of chlorophyll (SI Appendix, Fig. S4B), and could still bind to the photosynthetic complexes as assessed by 2D gel electrophoresis and immunoblotting (SI Appendix, Fig. S4C).
A Conserved Arg Patch Is Important for Binding of Ycf48.
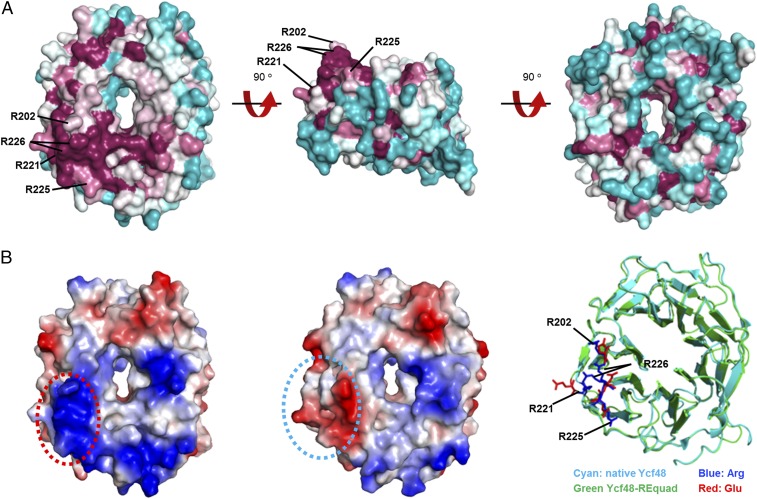
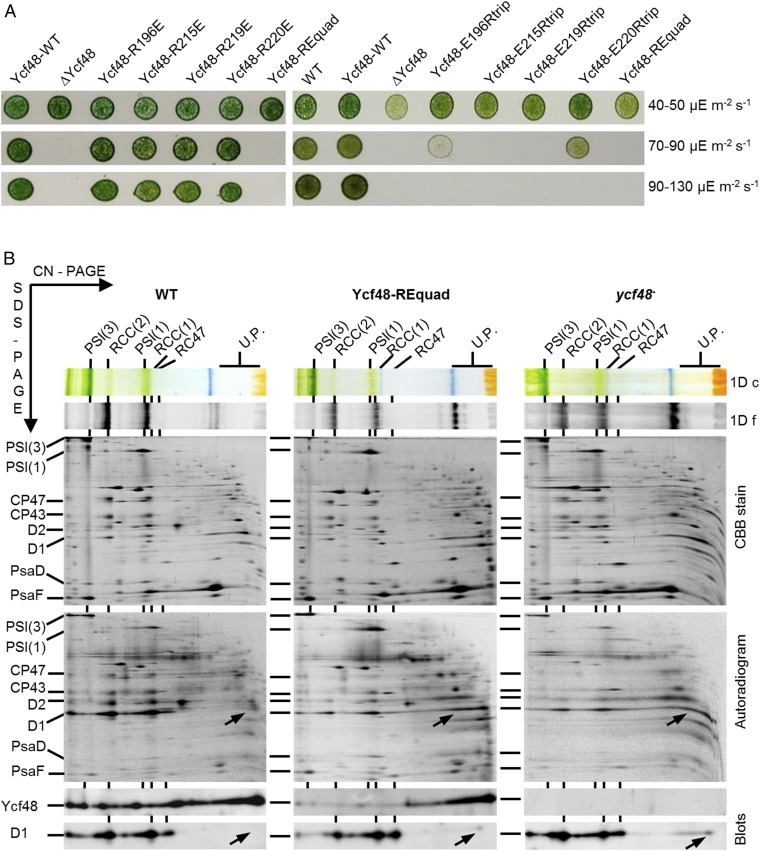
The conservation of the Ycf48 structures was analyzed using ConSurf (24, 25), which maps residue conservation scores to the structure. The most conserved region on the surface of T. elongatus Ycf48 was a patch of four Arg residues (202, 221, 225, and 226; T. elongatus numbering) on blades 4 and 5 on what we designate as the top surface of the beta-propeller (Fig. 4A). To investigate the importance of this region, we made a quadruple mutant of Synechocystis in which all four Arg-patch residues were mutated to Glu (mutant Ycf48-REquad). The homologous Arg residues in Synechocystis are R196, R215, R219, and R220. The Ycf48-REquad mutant was chlorophyll deficient like the ycf48 deletion strain (SI Appendix, Fig. S4B) and was unable to grow photoautotrophically at high irradiance, suggesting loss of function (Fig. 5A and SI Appendix, Fig. S4A). The 2D gel electrophoresis revealed that Ycf48 accumulated in the Ycf48-REquad mutant, but showed impaired binding to the photosynthetic complexes (Fig. 5B). Pulse-labeling cells with [35S]-Met/Cys and subsequent analysis of newly synthesized membrane proteins confirmed that the assembly of PSII complexes was impaired at an early stage of biogenesis in the Ycf48-REquad mutant (Fig. 5B) similar to the ycf48 null mutant (15). Radiolabeled D1 was detected mainly in an unassembled state, whereas in WT more D1 was incorporated into larger PSII complexes. Mutating the four Arg residues to Ala (in mutant Ycf48-RAquad) also increased susceptibility of growth to light stress and reduced levels of chlorophyll per cell, but to a lesser extent than the Ycf48-REquad mutant (SI Appendix, Fig. S4 A and B), despite WT levels of Ycf48 (SI Appendix, Fig. S7D).
Fig. 4.
Structural conservation and charge distribution of Ycf48 across cyanobacteria. (A) Different views of Ycf48 (PDB ID code 2XBG) displaying degree of residue conservation as deduced by a ConSurf (20) analysis of 122 cyanobacteria species; the sequence conservation score ranges from purple (high) to cyan (low). The position of the four conserved Arg residues are indicated with black lines. Two conformations of the side chain of R220 are modeled into the crystal structure. (B) Structural comparison of Ycf48-REquad (PDB ID code 5OJP) with native Ycf48. The charge distribution on the surface of WT containing the four arginine residues (Left) and Ycf48-REquad (Middle) containing the four glutamate substitutions are displayed; note the switch from positive (blue) to negatively charged (red) surface in the Arg-patch region (circled). Right shows the alignment of the alpha-carbons in Ycf48-REquad and native Ycf48 and the side-chain positions of the four Arg (blue) and four Glu substitutions (red).
Fig. 5.
Analysis of Ycf48-RE mutants. (A) Cell cultures were spotted on BG-11 plates and grown at low irradiance (40–50 µE m−2⋅s−1, Top row), semihigh irradiance (70–90 µE m−2⋅s−1, Middle row), and high irradiance (90–130 µE m−2⋅s−1, Bottom row). Strains analyzed included WT, WT control carrying the downstream gentamycin-resistance marker, but expressing native Ycf48 (Ycf48-WT), Ycf48 knockout strain (∆Ycf48), Ycf48 with single Arg replaced with Glu (Ycf48-RxxxE), Ycf48 with all four Arg replaced with Glu (Ycf48-REquad), and Ycf48 with three of the four Arg replaced with Glu (Ycf48-ExxxRtrip, the number representing the Arg not mutated to Glu). Results of two separate growth experiments are shown. (B) Membranes (4 µg chlorophyll each) isolated from WT, Ycf48-REquad, and Ycf48 knockout mutant (ycf48−) subjected to radioactive pulse labeling were analyzed by 2D CN-SDS/PAGE electrophoresis. The gels were either stained by Coomassie Brilliant Blue (CBB), photographed, dried, and exposed to phosphoimager plate, or blotted to a PVDF membrane, and Ycf48 and D1 proteins were sequentially detected using specific antibodies (segments of the blot containing individual antibody signals are shown). Pigmented complexes were detected in the first dimension by their color (1D c) and fluorescence (1D f). Designations of complexes are as in Fig. 1. Arrows designate unassembled mature D1.
To assess possible indirect effects of the mutations on the folding of Ycf48, the structure of recombinant T. elongatus Ycf48-REquad was determined by X-ray crystallography. The structure was similar to the WT (rmsd 0.75 Å), except for local changes around the cluster of four arginines, implying no dramatic changes in the tertiary structure of Ycf48 in the quadruple mutant in vivo (Fig. 4B).
In contrast to the Ycf48-REquad mutant, the single arginine-to-glutamate mutants behaved like the WT control strain and could grow at high irradiance, suggesting that binding of Ycf48 was dependent on two or more Arg residues in the patch (Fig. 5A). The functional importance of each Arg residue was further assessed by mutating each of the four Glu residues in the Ycf48-REquad mutant back to Arg to generate four triple mutants. Growth tests at increasing irradiances revealed that restoring R196 or R220 alone conferred better resistance to light stress than that of R215 and R219, although none of the triple mutants was able to grow when the irradiance reached 90–130 µE m−2⋅s−1, indicating reduced fitness compared with the WT control strain (Fig. 5A). In all triple mutants, Ycf48 was expressed at WT levels (SI Appendix, Fig. S7D).
Analysis of the T. elongatus Ycf48 structure revealed that the equivalent residues to R196 and R220 in Synechocystis sit on the top surface of Ycf48, with side chains pointing upward, while the equivalent residues to R215 and R219 lie slightly to the side (Fig. 4A and SI Appendix, Fig. S5). R220 was observed in two conformations in the electron density, showing flexibility. Overall, our data identify an important role for the Arg patch in binding to both PSI and PSII complexes and provide evidence for differences in the functional importance of the four Arg residues.
Ycf48 Copurifies with YidC and the Phenotype of the Null Mutant Can Be Suppressed by Increased Chlorophyll Availability.
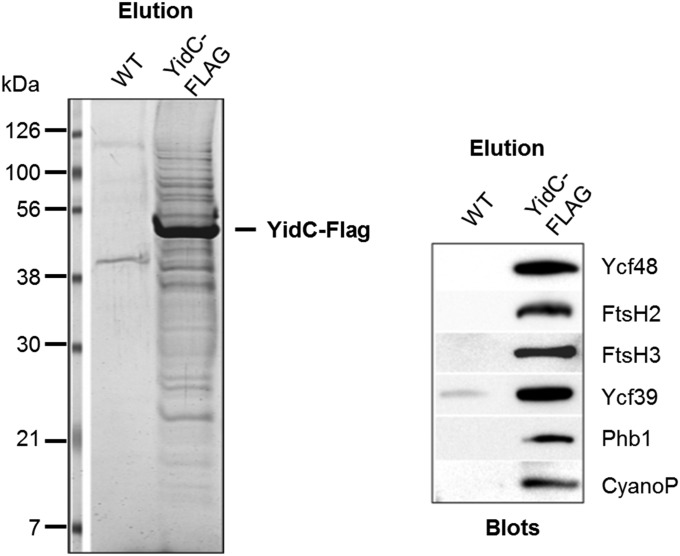
A role for Ycf48 at an early stage of biogenesis of PSII raised the possibility that the blades of Ycf48 might engage with lumenally exposed regions of newly synthesized proteins as they are cotranslationally inserted into membrane. The D1 subunit of PSII enters into the membrane via the ALB3 insertase in chloroplasts (26) and its homolog, YidC, in cyanobacteria (27). To test for the association of Ycf48 and possibly other assembly factors with YidC in Synechocystis, pulldown experiments using an anti-FLAG antibody were performed with detergent-solubilized extracts isolated from a C-terminal FLAG-tagged derivative of YidC (YidC-FLAG) (28). YidC-FLAG was immunopurified and, importantly, both Ycf48 and CyanoP, the latter shown recently to bind to D2 at an early stage of assembly of PSII (29), were coeluted with YidC-FLAG (Fig. 6). FtsH protease subunits (FtsH2 and FtsH3) and the band 7 protein, Phb1, also copurified with YidC-FLAG, which is consistent with earlier experiments showing copurification of FtsH and the band 7 proteins HflK/C with Escherichia coli YidC in cross-linking experiments (30). In contrast, CyanoQ (21), PAM68/Sll0933 (31), Psb27 (32), Psb28 (33), and Psb28-2 (34), which are all involved in PSII assembly/repair, were not detected in the YidC-FLAG eluate by either immunoblotting or mass spectrometry.
Fig. 6.
Ycf48 and CyanoP protein are copurified with FLAG-tagged YidC insertase. The FLAG-specific eluate obtained after purification of membrane proteins from a strain containing FLAG-tagged YidC (YidC-FLAG) or from WT as a negative control was analyzed by SDS/PAGE and the gel stained by Coomassie Brilliant Blue (Left). Equivalent elutions were separated by SDS/PAGE and sequentially immunoblotted with a range of specific antibodies (Right, segment of the blot with signals of each antibody is shown). The most intensively stained band was identified by protein mass spectrometry as the bait (YidC-FLAG).
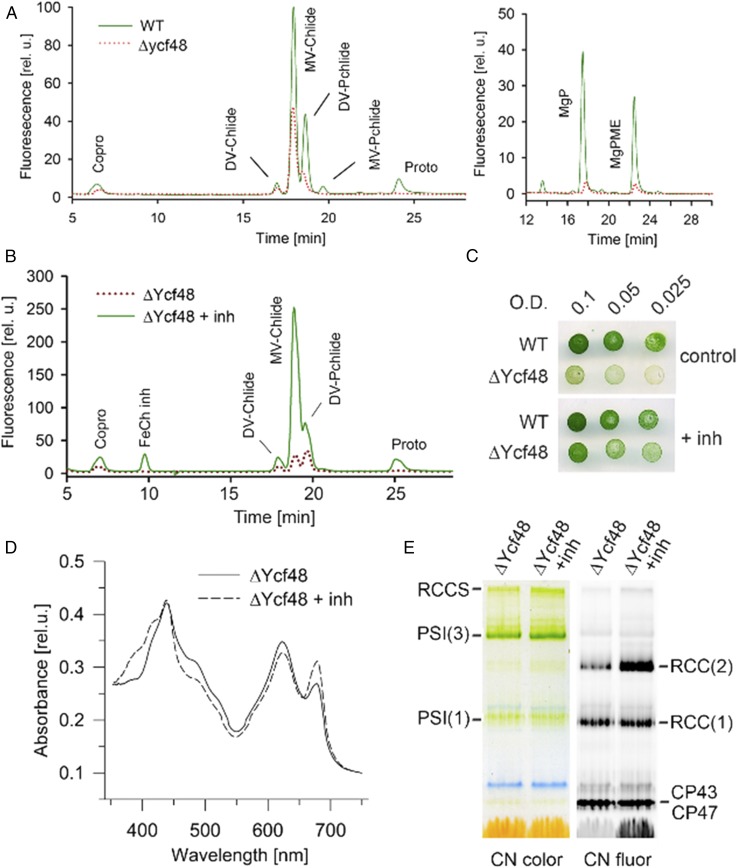
It has been shown recently that the Ycf48 protein is particularly important when chlorophyll availability in the cell is low: a point mutation in the Mg-chelatase enzyme, which produces Mg-protoporphyrin IX for chlorophyll biosynthesis, abolished photoautotrophic growth of a Ycf48 deletion mutant, whereas there was no significant effect on growth in the WT background (35). As these results indicated a tight link between the function of Ycf48 and chlorophyll biosynthesis, we analyzed intermediates of the chlorophyll synthesis pathway in the ycf48 null mutant using HPLC. Our analysis showed that chlorophyll precursors were substantially reduced compared with WT (Fig. 7A).
Fig. 7.
Enhanced chlorophyll biosynthesis restores accumulation of chlorophyll–protein complexes in the Ycf48 mutant. (A) Levels of chlorophyll precursors are dramatically reduced in ΔYcf48. Chlorophyll precursors were extracted from 2 mL of cells with OD750 = 0.4 and quantified by HPLC equipped with two fluorescence detectors set on different excitation and emission wavelengths (chromatograms on the Left and Right, detailed methods described in ref. 43). Coproporhyrinogen III (Copro) and protoporphyrin IX (Proto) are the last common precursors for both chlorophyll and heme synthesis. The first committed intermediate of chlorophyll pathway is Mg-protoporphyrin IX (MgP), which is consequently methylated (MgPME). The MgPME is converted into divinyl protochlorophyllide (DV-Pchlide) and then into divinyl and monovinyl chlorophyllide (DV/MV-Chlide); chlorophyll is finally made by attachment of phytol to the MV-Chlide. (B) Elevated levels of Pchlide and Chlide in ΔYcf48 grown for 24 h in the presence of 0.2 mM N-methyl-mesoporphyrin IX [ferrochelatase inhibitor (inh)]. (C) The growth of ΔYcf48 cells at low irradiance (40–50 µE m−2⋅s−1) in the presence or absence of 0.3 mM N-methyl-mesoporphyrin IX, a ferrochelatase inhibitor. (D) Absorbance spectra of mutant cells before and after 24 h in the presence of 0.2 mM N-methyl-mesoporphyrin IX. (E) CN-PAGE of membrane complexes isolated from the identical cultures shown in D and loaded on an equal cell number basis (∼2 mL of cells, OD750 = 0.5).
To assess the effect of a better chlorophyll supply on the phenotype of the ycf48 null mutant, liquid and solid cultures were supplemented with N-methyl mesoporphyrin IX, a potent and specific inhibitor of the ferrochelatase enzyme, whose partial inhibition has been shown to increase chlorophyll biosynthesis (36). After a 1-d treatment with inhibitor, the ycf48 null mutant accumulated high levels of chlorophyll precursors (Fig. 7B) and showed improved growth (Fig. 7C) and enhanced levels of chlorophyll per cell (Fig. 7D). Analysis by CN-PAGE indicated increased levels of both trimeric PSI and dimeric PSII complexes (Fig. 7E).
Discussion
A number of accessory factors involved in the assembly and maintenance of the thylakoid membrane have now been identified, mainly through genetic studies (reviewed in ref. 12). However, there is little information on the structural features of these proteins that are important for function. We show here that Ycf48 from both cyanobacteria and chloroplasts forms a seven-bladed beta-propeller. The beta-propeller structural motif is found widely in nature both in enzymes and in structural proteins involved in the assembly of larger protein complexes, with individual blades helping to mediate protein–protein interactions (reviewed in ref. 37). This latter function is consistent with the known role for Ycf48 in orchestrating the assembly of PSII in cyanobacteria. Besides binding to unassembled D1 (15), Ycf48 is thought to interact with CyanoP, prebound to D2, to promote assembly of the PSII RC assembly complex (29) and with PAM68/Sll0933 to form larger PSII complexes (31, 38).
Using the Ycf48 crystal structures, we identified a conserved patch of arginine residues as a region to investigate further. A role in binding to the lumenal surfaces of thylakoid membrane complexes was revealed through analysis of a quadruple mutant in which all four Arg residues were replaced by Glu. The lack of a significant growth defect at high irradiances in the single Arg mutants suggests that the functionality of the Arg patch is a collective property of two or more residues. Further analysis on triple Arg-to-Glu mutants in Synechocystis revealed that R196 and R220 are functionally more important than R215 and R219, which suggests that the top surface of the Arg patch has greater importance for binding (Fig. 4A and SI Appendix, Fig. S5).
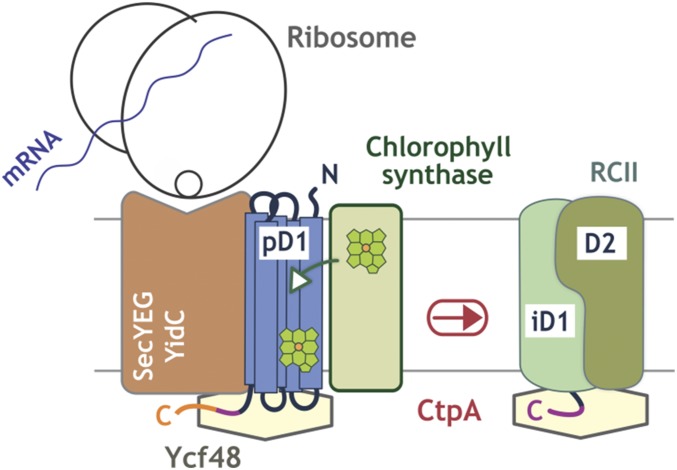
An interaction between Ycf48 and the 16-aa-long C-terminal extension of the precursor of D1, which is proteolytically cleaved after formation of the PSII RC to form an intermediate form of D1 (iD1) with an 8-aa extension (39), has previously been suggested from yeast two-hybrid experiments (15). We have been able to cocrystallize T. elongatus Ycf48 with either the mD1 peptide (NAHNFPLDLA), corresponding to residues 335–344 of the mature D1 protein (SI Appendix, Fig. S6A) or the iD1 peptide (NAHNFPLDLASAESAPVA), corresponding to residues 335–352 of the precursor D1 protein (39) (SI Appendix, Fig. S6 B and C). Three potential binding sites were identified with the binding site for the extended iD1 peptide starting close to the Arg patch (SI Appendix, Fig. S6). However, the physiological relevance of these sites is unknown, as we have so far been unable to find conditions to detect peptide binding to isolated Ycf48 by NMR or isothermal calorimetry to allow the candidate peptide-binding sites to be probed by mutagenesis.
Binding of chlorophyll synthase, the final enzyme involved in chlorophyll biosynthesis, to YidC is essential for cotranslational insertion of chlorophyll into the nascent polypeptide chains of newly synthesized chlorophyll-binding proteins (28). The copurification of Ycf48 with a FLAG-tagged version of Synechocystis YidC (Fig. 6) and partial suppression of the absence of Ycf48 by increased chlorophyll supply (Fig. 7) therefore suggest that Ycf48 is prebound to YidC and might assist YidC during the synthesis of chlorophyll-binding proteins. Extending previous ideas on the role of Ycf48/HCF136 in chloroplasts (14), we suggest that the blades of Ycf48 might interact with emerging lumenal parts of the newly synthesized membrane proteins, possibly via coulombic interactions with the Arg patch, to allow efficient and precise incorporation of pigments and other cofactors into the transmembrane regions (Fig. 8). The 19-residue insert exclusive to eukaryotes does not impair function of Ycf48 in cyanobacteria (SI Appendix, Fig. S4) and so might play a specific role in eukaryotes, such as interacting with Alb3, which has clear sequence differences to YidC in prokaryotes (40).
Fig. 8.
Model for the role of Ycf48 in the synthesis of D1 and assembly of the PSII RCII complex. The potential mechanism of action of Ycf48 in PSII assembly is illustrated schematically. Ycf48 interacts with the C-terminal extension of precursor D1 (pD1) and intermediate D1 (iD1), helping to synchronize D1 insertion via the YidC insertase, chlorophyll loading from chlorophyll synthase, and C-terminal processing by the CtpA protease.
Our detection of Ycf48 in larger PSII core complexes might reflect retention of Ycf48 on the lumenal surface of PSII before light-driven assembly of the oxygen-evolving complex. Control immunoblots showed that neither YidC nor PAM68/Sll0933 was present in large PSII complexes resolved by native gel electrophoresis, suggesting that Ycf48 is not attached to PSII via either of these components. Given that Ycf48 associates with unassembled D1, a plausible site of interaction is between the Arg patch of Ycf48 and the negatively charged residues on the lumenal surface of D1, including those that ultimately ligate the oxygen-evolving Mn4CaO5 cluster. In this scenario, Ycf48 maintains D1 in a competent state for assembly of the Mn cluster and attachment of the lumenal proteins. Alternatively, binding of Ycf48 to larger PSII complexes might reflect an early stage of repair in which Ycf48 associates with damaged PSII.
It is not clear why the Ycf48 knockout mutant exhibits strongly down-regulated chlorophyll biosynthesis, as this seems to be essentially the opposite response that is required if the cell is to accumulate more chlorophyll–protein complexes in the absence of Ycf48. Restricted chlorophyll biosynthesis has, however, been reported for other PSII mutants (36), and so it appears that there is a feedback mechanism that coordinates de novo chlorophyll synthesis with PSII assembly/repair. In this way the levels of free chlorophyll and chlorophyll precursors, which can lead to the light-induced production of reactive oxygen species, can be controlled. Although detection of Ycf48 in PSI complexes implies a direct role for Ycf48 in PSI assembly, the reduced level of PSI (Fig. 7E) in the Ycf48 mutant might also be partly caused by a limitation in chlorophyll supply. Previous work has shown that PSI complexes are the main sink for newly synthesized chlorophyll in Synechocystis (41) and that PSI accumulation is sensitive to the availability of chlorophyll (42).
Materials and Methods
Details of the construction of Synechocystis strains, cultivation conditions, photometric measurements of cells and chlorophyll, HPLC analysis of chlorophyll-derived pigments and precursors, preparation of cellular membranes, isolation of YidC-FLAG and PSI-YFP through affinity chromatography, protein electrophoresis, immunoblotting, and radiolabeling are described in SI Appendix, SI Materials and Methods. Work related to the structural studies of Ycf48, including construction of expression vectors, protein expression, purification, X-ray crystallography, and data processing is also described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Martin Walsh for assistance at the European Synchrotron Radiation Facility Beamline 14 and Diamond Light Source for access to beamlines I03 and I04 (mx7299 and mx12579) that contributed to the results presented here. J.Y., F.M., W.B., P.J.N., J.W.M. and E.C. are grateful to the Biotechnology and Biological Sciences Research Council (BBSRC) for funding this work (Grants BB/I00937X/1 and BB/L003260/1). J.W.M. was funded for part of this work by a BBSRC David Phillips Fellowship (BB/F023308/1). J. Knoppová, M.K.S., A.S., G.P.A., R.S. and J. Komenda were supported by Projects 17-08755S and P501/12/G055 of the Czech Science Foundation and by the Czech Ministry of Education (Projects CZ 1.05/2.1.00/19.0392, LO1416, and LM2015055).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [Ycf48 from T. elongatus (PDB ID code 2XBG), Ycf48 from C. merolae (PDB ID code 5OJ3), Ycf48-REquad (PDB ID code 5OJP), Ycf48-D1 cocrystals (PDB ID code 5OF5), and (PDB ID code 5OJR)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800609115/-/DCSupplemental.
References
- 1.Diner BA, Rappaport F. Structure, dynamics, and energetics of the primary photochemistry of photosystem II of oxygenic photosynthesis. Annu Rev Plant Biol. 2002;53:551–580. doi: 10.1146/annurev.arplant.53.100301.135238. [DOI] [PubMed] [Google Scholar]
- 2.Barber J. Photosystem II: The water splitting enzyme of photosynthesis and the origin of oxygen in our atmosphere. Q Rev Biophys. 2016;49:e14. doi: 10.1017/S0033583516000093. [DOI] [PubMed] [Google Scholar]
- 3.Komenda J, Sobotka R, Nixon PJ. Assembling and maintaining the photosystem II complex in chloroplasts and cyanobacteria. Curr Opin Plant Biol. 2012;15:245–251. doi: 10.1016/j.pbi.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 4.Shen J-R. The structure of photosystem II and the mechanism of water oxidation in photosynthesis. Annu Rev Plant Biol. 2015;66:23–48. doi: 10.1146/annurev-arplant-050312-120129. [DOI] [PubMed] [Google Scholar]
- 5.Wei X, et al. Structure of spinach photosystem II-LHCII supercomplex at 3.2 Å resolution. Nature. 2016;534:69–74. doi: 10.1038/nature18020. [DOI] [PubMed] [Google Scholar]
- 6.van Bezouwen LS, et al. Subunit and chlorophyll organization of the plant photosystem II supercomplex. Nat Plants. 2017;3:17080. doi: 10.1038/nplants.2017.80. [DOI] [PubMed] [Google Scholar]
- 7.Heinz S, Liauw P, Nickelsen J, Nowaczyk M. Analysis of photosystem II biogenesis in cyanobacteria. Biochim Biophys Acta. 2016;1857:274–287. doi: 10.1016/j.bbabio.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Boehm M, et al. Subunit composition of CP43-less photosystem II complexes of Synechocystis sp. PCC 6803: Implications for the assembly and repair of photosystem II. Philos Trans R Soc Lond B Biol Sci. 2012;367:3444–3454. doi: 10.1098/rstb.2012.0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boehm M, et al. Investigating the early stages of photosystem II assembly in Synechocystis sp. PCC 6803: Isolation of CP47 and CP43 complexes. J Biol Chem. 2011;286:14812–14819. doi: 10.1074/jbc.M110.207944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bao H, Burnap RL. Photoactivation: The light-driven assembly of the water oxidation complex of photosystem II. Front Plant Sci. 2016;7:578. doi: 10.3389/fpls.2016.00578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nixon PJ, Michoux F, Yu J, Boehm M, Komenda J. Recent advances in understanding the assembly and repair of photosystem II. Ann Bot. 2010;106:1–16. doi: 10.1093/aob/mcq059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nickelsen J, Rengstl B. Photosystem II assembly: From cyanobacteria to plants. Annu Rev Plant Biol. 2013;64:609–635. doi: 10.1146/annurev-arplant-050312-120124. [DOI] [PubMed] [Google Scholar]
- 13.Meurer J, Plücken H, Kowallik KV, Westhoff P. A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J. 1998;17:5286–5297. doi: 10.1093/emboj/17.18.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plücken H, Müller B, Grohmann D, Westhoff P, Eichacker LA. The HCF136 protein is essential for assembly of the photosystem II reaction center in Arabidopsis thaliana. FEBS Lett. 2002;532:85–90. doi: 10.1016/s0014-5793(02)03634-7. [DOI] [PubMed] [Google Scholar]
- 15.Komenda J, et al. The cyanobacterial homologue of HCF136/YCF48 is a component of an early photosystem II assembly complex and is important for both the efficient assembly and repair of photosystem II in Synechocystis sp. PCC 6803. J Biol Chem. 2008;283:22390–22399. doi: 10.1074/jbc.M801917200. [DOI] [PubMed] [Google Scholar]
- 16.Knoppová J, et al. Discovery of a chlorophyll binding protein complex involved in the early steps of photosystem II assembly in Synechocystis. Plant Cell. 2014;26:1200–1212. doi: 10.1105/tpc.114.123919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobáková M, Tichy M, Komenda J. Role of the PsbI protein in photosystem II assembly and repair in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 2007;145:1681–1691. doi: 10.1104/pp.107.107805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komenda J, et al. Accumulation of the D2 protein is a key regulatory step for assembly of the photosystem II reaction center complex in Synechocystis PCC 6803. J Biol Chem. 2004;279:48620–48629. doi: 10.1074/jbc.M405725200. [DOI] [PubMed] [Google Scholar]
- 19.Strašková A, Knoppová J, Komenda J. Isolation of the cyanobacterial YFP-tagged photosystem I using GFP-Trap. Photosynthetica. 2018;56:300–305. [Google Scholar]
- 20.Michoux F, Takasaka K, Boehm M, Nixon PJ, Murray JW. Structure of CyanoP at 2.8 A: Implications for the evolution and function of the PsbP subunit of photosystem II. Biochemistry. 2010;49:7411–7413. doi: 10.1021/bi1011145. [DOI] [PubMed] [Google Scholar]
- 21.Michoux F, et al. Crystal structure of CyanoQ from the thermophilic cyanobacterium Thermosynechococcus elongatus and detection in isolated photosystem II complexes. Photosynth Res. 2014;122:57–67. doi: 10.1007/s11120-014-0010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaudhuri I, Söding J, Lupas AN. Evolution of the β-propeller fold. Proteins. 2008;71:795–803. doi: 10.1002/prot.21764. [DOI] [PubMed] [Google Scholar]
- 23.Korczynska M, Mukhtar TA, Wright GD, Berghuis AM. Structural basis for streptogramin B resistance in Staphylococcus aureus by virginiamycin B lyase. Proc Natl Acad Sci USA. 2007;104:10388–10393. doi: 10.1073/pnas.0701809104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glaser F, et al. ConSurf: Identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 25.Ashkenazy H, et al. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–W350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ossenbühl F, et al. Efficient assembly of photosystem II in Chlamydomonas reinhardtii requires Alb3.1p, a homolog of Arabidopsis ALBINO3. Plant Cell. 2004;16:1790–1800. doi: 10.1105/tpc.023226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ossenbühl F, Inaba-Sulpice M, Meurer J, Soll J, Eichacker LA. The Synechocystis sp PCC 6803 Oxa1 homolog is essential for membrane integration of reaction center precursor protein pD1. Plant Cell. 2006;18:2236–2246. doi: 10.1105/tpc.106.043646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chidgey JW, et al. A cyanobacterial chlorophyll synthase-HliD complex associates with the Ycf39 protein and the YidC/Alb3 insertase. Plant Cell. 2014;26:1267–1279. doi: 10.1105/tpc.114.124495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knoppová J, Yu J, Konik P, Nixon PJ, Komenda J. CyanoP is involved in the early steps of photosystem II assembly in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2016;57:1921–1931. doi: 10.1093/pcp/pcw115. [DOI] [PubMed] [Google Scholar]
- 30.van Bloois E, et al. Detection of cross-links between FtsH, YidC, HflK/C suggests a linked role for these proteins in quality control upon insertion of bacterial inner membrane proteins. FEBS Lett. 2008;582:1419–1424. doi: 10.1016/j.febslet.2008.02.082. [DOI] [PubMed] [Google Scholar]
- 31.Armbruster U, et al. The Arabidopsis thylakoid protein PAM68 is required for efficient D1 biogenesis and photosystem II assembly. Plant Cell. 2010;22:3439–3460. doi: 10.1105/tpc.110.077453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nowaczyk MM, et al. Psb27, a cyanobacterial lipoprotein, is involved in the repair cycle of photosystem II. Plant Cell. 2006;18:3121–3131. doi: 10.1105/tpc.106.042671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dobáková M, Sobotka R, Tichý M, Komenda J. Psb28 protein is involved in the biogenesis of the photosystem II inner antenna CP47 (PsbB) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 2009;149:1076–1086. doi: 10.1104/pp.108.130039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bečková M, et al. Association of Psb28 and Psb27 proteins with PSII-PSI supercomplexes upon exposure of Synechocystis sp. PCC 6803 to high light. Mol Plant. 2017;10:62–72. doi: 10.1016/j.molp.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 35.Crawford TS, Eaton-Rye JJ, Summerfield TC. Mutation of Gly195 of the ChlH subunit of Mg-chelatase reduces chlorophyll and further disrupts PS II assembly in a Ycf48-deficient strain of Synechocystis sp. PCC 6803. Front Plant Sci. 2016;7:1060. doi: 10.3389/fpls.2016.01060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sobotka R, Komenda J, Bumba L, Tichý M. Photosystem II assembly in CP47 mutant of Synechocystis sp. PCC 6803 is dependent on the level of chlorophyll precursors regulated by ferrochelatase. J Biol Chem. 2005;280:31595–31602. doi: 10.1074/jbc.M505976200. [DOI] [PubMed] [Google Scholar]
- 37.Chen CKM, Chan NL, Wang AHJ. The many blades of the β-propeller proteins: Conserved but versatile. Trends Biochem Sci. 2011;36:553–561. doi: 10.1016/j.tibs.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 38.Rengstl B, Knoppová J, Komenda J, Nickelsen J. Characterization of a Synechocystis double mutant lacking the photosystem II assembly factors YCF48 and Sll0933. Planta. 2013;237:471–480. doi: 10.1007/s00425-012-1720-0. [DOI] [PubMed] [Google Scholar]
- 39.Komenda J, et al. Cleavage after residue Ala352 in the C-terminal extension is an early step in the maturation of the D1 subunit of photosystem II in Synechocystis PCC 6803. Biochim Biophys Acta. 2007;1767:829–837. doi: 10.1016/j.bbabio.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 40.Yen MR, Harley KT, Tseng YH, Saier MH., Jr Phylogenetic and structural analyses of the oxa1 family of protein translocases. FEMS Microbiol Lett. 2001;204:223–231. doi: 10.1111/j.1574-6968.2001.tb10889.x. [DOI] [PubMed] [Google Scholar]
- 41.Kopecná J, Komenda J, Bucinská L, Sobotka R. Long-term acclimation of the cyanobacterium Synechocystis sp. PCC 6803 to high light is accompanied by an enhanced production of chlorophyll that is preferentially channeled to trimeric photosystem I. Plant Physiol. 2012;160:2239–2250. doi: 10.1104/pp.112.207274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kopecná J, Sobotka R, Komenda J. Inhibition of chlorophyll biosynthesis at the protochlorophyllide reduction step results in the parallel depletion of photosystem I and photosystem II in the cyanobacterium Synechocystis PCC 6803. Planta. 2013;237:497–508. doi: 10.1007/s00425-012-1761-4. [DOI] [PubMed] [Google Scholar]
- 43.Pilný J, Kopečná J, Noda J, Sobotka R. Detection and quantification of heme and chlorophyll precursors using a high performance liquid chromatography (HPLC) system equipped with two fluorescence detectors. Bio Protoc. 2015;5:e1390. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.