Abstract
The terminal steps in the biosynthesis of the monoterpenoid indole alkaloids vindoline and minovincinine are catalyzed by separate acetyl coenzyme A-dependent O-acetyltransferases in Madagascar periwinkle (Catharanthus roseus G. Don). Two genes were isolated that had 63% nucleic acid identity and whose deduced amino acid sequences were 78% identical. Active enzymes that were expressed as recombinant His-tagged proteins in Escherichia coli were named minovincinine-19-O-acetyltransferase (MAT) and deacetylvindoline-4-O-acetyltransferase (DAT) because they catalyzed the 19-O-acetylation of indole alkaloids such as minovincinine and hörhammericine and the 4-O-acetylation of deacetylvindoline, respectively. Kinetic studies showed that the catalytic efficiency of recombinant MAT (rMAT) was very poor compared with that of recombinant DAT (rDAT), whose turnover rates for Acetyl-coenzyme A and deacetylvindoline were approximately 240- and 10,000-fold greater than those of rMAT. Northern-blot analyses showed that MAT is expressed in cortical cells of the root tip, whereas DAT is only expressed in specialized idioblast and laticifer cells within light exposed tissues like leaves and stems. The coincident expression of trytophan decarboxylase, strictosidine synthase, and MAT within root cortical cells suggests that the entire pathway for the biosynthesis of tabersonine and its substituted analogs occurs within these cells. The ability of MAT to catalyze the 4-O-acetylation of deacetylvindoline with low efficiency suggests that this enzyme, rather than DAT, is involved in vindoline biosynthesis within transformed cell and root cultures, which accumulate low levels of this alkaloid under certain circumstances.
The Apocynaceae plant family, which contains the important medicinal plant Madagascar periwinkle (Catharanthus roseus G. Don), is characterized by the large variety of monoterpenoid indole alkaloids that it produces. The alkaloid chemistry of many members of this family has been well characterized and several thousand structures have been elucidated. Among these many structures, vinblastine and vincristine from Madagascar periwinkle are of particular importance because of their wide use in cancer chemotherapy. These alkaloids are produced in vivo by the condensation of vindoline and catharanthine. The pharmaceutical value of these dimeric alkaloids, their low abundance, and their cost of production have prompted extensive efforts to generate cost efficient high-yielding cell and organ cultures of Madagascar periwinkle. These efforts successfully produced cell cultures, which accumulated high levels of all the major types of Madagascar periwinkle alkaloids, like serpentine (corynanthe), catharanthine (iboga), and tabersonine (aspidosperma). However, the inability of cell cultures to consistently make vindoline resulted in the ultimate failure to produce dimeric indole alkaloids.
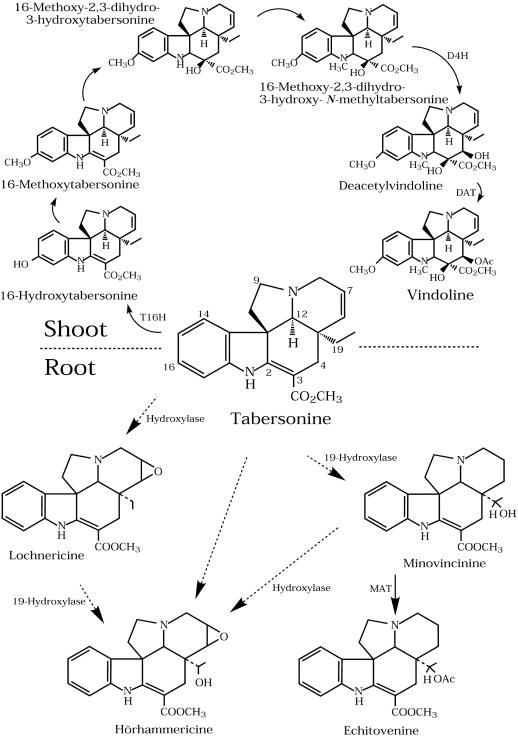
Enzyme and metabolic studies with plants suggested that vindoline biosynthesis is restricted to the aboveground organs and that the pathway beyond tabersonine (Fig. 1) is not expressed in tissue cultures. These results raised the possibility that cell cultures lacked the cell types required to accommodate the late stages of vindoline biosynthesis. Recent experiments (St-Pierre et al., 1999) have shown that the formation of vindoline in intact plants involves at least two separate cell types requiring the translocation of a pathway intermediate. In situ hybridization and immunolocalization studies confirmed that Trp decarboxylase and strictosidine synthase, which are involved in the formation of strictosidine, were only expressed in the epidermis of aerial tissues and in cortical cells of the root apical meristem. In contrast, the expression of desacetoxyvindoline-4-hydroxylase (D4H) and deacetylvindoline-4-O-acetyl-transferase (DAT), which catalyze the last two steps in vindoline biosynthesis (Fig. 1), occurred exclusively in laticifers and idioblasts of aerial tissues.
Figure 1.
Biosynthesis of tabersonine-derived indole alkaloids in Madagascar periwinkle organs. Tabersonine is converted into vindoline via six enzymatic steps, with the terminal hydroxylation (D4H) and O-acetylation (DAT) occurring in specialized cells known as idioblasts and laticifers of leaves and stems [1]. Tabersonine is converted into lochnericine, hörhammericine, and minovincinine via uncharacterized hydroxylations and 19-hydroxy-indole alkaloids are converted into their respective products by O-acetylation (MAT). These tabersonine analogs are known to accumulate under certain conditions within cell cultures (Kutney et al., 1980) and roots (Shanks et al., 1998) of Madagascar periwinkle.
Madagascar periwinkle hairy root cultures, which appear to be more stable than cell cultures have recently been investigated for their ability to produce indole alkaloids. Hairy roots accumulate tabersonine, lochnericine, and hörhammericine (Fig. 1) in addition to serpentine and ajmalicine (Rijhwani et al., 1998; Shanks et al., 1998). Roots isolated from the plant also accumulate the same types of corynanthe (El-Deeb et al., 1957), iboga (Svoboda et al., 1963), and aspidosperma (Nair and Pillay, 1959) alkaloids, but not vindoline. However, low levels of vindoline were recently reported to accumulate in hairy root cultures transformed with Agrobacterium rhizogenes (O'Keefe et al., 1997). In addition, suspension cultures established after leaf disc transformation with either Agrobacterium tumefaciens or A. rhizogenes accumulated catharanthine, as well as low levels of vindoline and also showed a deacetylvindoline-O-acetyltransferase activity, which catalyzes the last step in vindoline biosynthesis (O'Keefe et al., 1997).
The surprising ability of transformed tissues to accumulate vindoline raises the possibility that the differential cell type-specific expression required for this to occur in leaves and stems of Madagascar periwinkle plants may not be absolutely needed under all circumstances. The present report describes the cloning and biochemical characterization of minovincinine-O-acetyltransferase (MAT). This gene, which is only expressed in roots, is a homolog of DAT, which is only expressed in idioblasts and laticifers. Evidence is presented that MAT, whose function is to acetylate minovincinine and/or hörhammericine (Fig. 1), may also be involved in vindoline biosynthesis in the special circumstances created during plant transformation.
RESULTS
Comparison of DAT and MAT Genes and Sequences
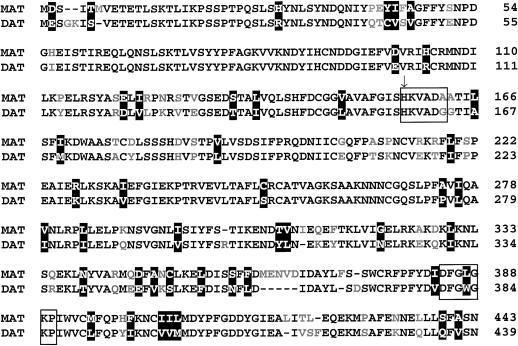
Sequence analysis of gDAT-6 and gDAT-15 revealed open reading frames (ORFs) for putative 439- and 443-amino acid DAT and MAT proteins, respectively. Sequence comparison of gDAT-6 and gDAT-15 showed a 63% nucleic acid identity between these two genes (results not shown) and a 78% amino acid identity between the putative ORFs (Fig. 2). Previous studies with DAT (St-Pierre et al., 1998) showed it to belong to a large family of acyltransferases with a putative active site related to chloramphenicol O-acetyltransferase and dihydrolipoyl acetyltransferase. Members of this gene family that are particularly abundant in plants displayed similar Mrs, as well as the conserved amino acid sequences HXXXDG and DFGWGKP. The MAT gene also belongs to this family since it has a similar Mr to DAT and contains both consensus sequences (Fig. 2). The conserved HIS in the MAT HXXXDA sequence is thought to be involved in binding the Ac coenzyme A (coA) cosubstrate as was shown to be the case with DAT (St-Pierre et al., 1998). In contrast, MAT appears to contain an extra five amino acids (MENVD) near the carboxy terminal end that are not found in DAT and that might be responsible for some of the differences in the kinetic properties of these two enzymes.
Figure 2.
Amino acid alignment of rMAT and rDAT gene products. Identical amino acids are shown in black, conserved substitutions are in white on a black background, and differing amino acids are gray. The boxed residues highlight the conserved HXXXDG active site and DFGWGKP motif and the arrow identifies the active site His residue.
Substrate Specificity and Kinetic Parameters
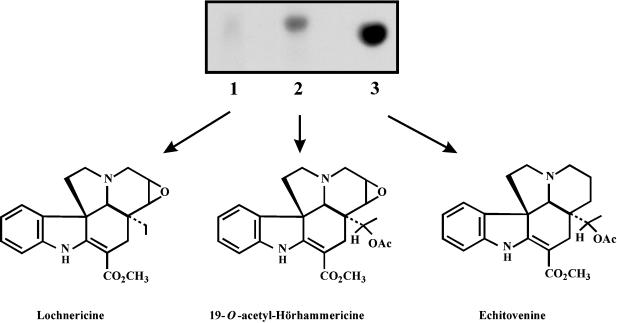
Recombinant MAT (rMAT) and DAT (rDAT) were purified (see “Materials and Methods”) and used in enzyme assays to compare their substrate specificities. The reaction products obtained from radioactive rMAT assays were submitted to Si-Kieselgel thin-layer chromatography (TLC) and autoradiography (Fig. 3). No reaction product was produced with lochnericine as substrate (Fig. 3, lane 1), whereas hörhammericine was converted to radioactive 19-O-acetyl-hörhammericine (Rf = 0.66). Crude Madagascar periwinkle extracts of root or hairy root alkaloids also contained a substrate capable of being acetylated by rMAT. Incubation of these extracts with rMAT protein produced an intense radioactive spot (Rf = 0.62) on the autoradiogram that comigrated with an echitovenine standard (Fig. 3, lane 3). These results suggest that Madagascar periwinkle roots may contain sufficient minovincinine to produce radioactive echitovenine. Attempts to purify this substrate from crude root extracts proved unsuccessful (data not shown), which suggests it is present at very low levels.
Figure 3.
Autoradiogram of reaction products obtained from MAT-catalyzed reactions. Lochnericine (lane 1), hörhammericine (lane 2), or a minovincinine-containing root extract (lane 3) were used as substrates. The reaction products were extracted and chromatographed as described in “Material and Methods.” The structures of the respective acetylated products are shown. Although no acetylated product was observed when the supplied substrate was lochnericine, its structure is included to show the requirement for the hydroxyl group at position 19 for enzyme activity.
The alkaloid substrate specificities of rDAT and rMAT were strikingly different (Table I). Minovincinine-containing hairy root extract and hörhammericine were substrates for rMAT activity, whereas they were not accepted as substrates by rDAT (Table I). In contrast deacetylvindoline (DAV), which is the true substrate of rDAT, was also acetylated by rMAT. The Km of rMAT for DAV was over 8-fold higher than that of rDAT (Table II) and DAT purified from Madagascar periwinkle leaves (Power et al., 1990). The apparent specific activity of the partially purified rDAT (38.1 pkat μg−1) (Table II) was similar to that found previously for homogeneous rDAT (30 pkat μg−1; St-Pierre et al., 1998) and DAT purified from Madagascar periwinkle leaves (36 pkat μg−1; Power et al., 1990). The turnover rate of rDAT for acetyl CoA and DAV was approximately 240- and 10,000-fold greater, respectively, compared with that of rMAT (Vmax/Km; Table II). This low turnover rate is maintained for rMAT with hörhammericine as substrate (Table II) and confirms the low efficiency of this enzyme compared with DAT.
Table I.
Substrate specificities of recombinant forms of DAT and MAT, as determined from TLC scrapings of the major radioactive bands
| Substrateb | Activitya
|
|
|---|---|---|
| rDAT | rMAT | |
| % | ||
| Deacetylvindoline | 100 | 48 |
| Crude hairy root alkaloid | NAc | 100 |
| Tabersonine | NA | NA |
| 16-MeOH-tabersonine | NA | NA |
| 3-OH-tabersonine | NA | NA |
| n-CH3-3-OH-tabersonine | NA | NA |
| 2,3-Dihydro-tabersonine | NA | NA |
| 6,7-Dihydro-3-OH- tabersonine | NA | NA |
| Hörhammericine | NA | 40 |
| Lochnericine | NA | NA |
100% activity refers to a total radioactivity count of 36,367 dpm for rDAT and 2,233 dpm for reactivated rMAT reaction products, respectively.
Assays were carried out as described in experimental procedures, containing either 0.17 μg of rDAT or 6.0 μg of rMAT, respectively, along with 20 μM deacetylvindoline, 20 μL of crude hairy root alkaloid extract, approximately 10 μg of each respective alkaloid, or 20 μg of hörhammericine or lochnericine, respectively, as substrate in a final assay volume of 100 μL containing 1% (v/v) DMSO (final concentration) as the substrate solvent.
NA, Not accepted as a substrate.
Table II.
Kinetic parameters of rMAT and rDAT with respect to various substrates
Tissue-Specific Expression of the MAT Gene in Madagascar Periwinkle
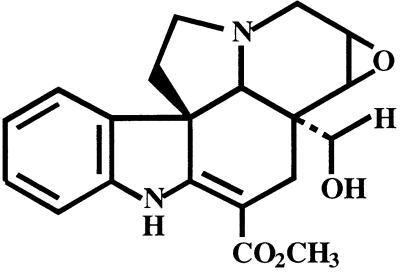
RNA gel-blot analysis revealed that MAT is expressed differently than DAT in various Madagascar periwinkle tissues. The same RNA blot was first probed with the DAT ORF fragment and after stripping was reprobed with the 423-bp HindIII MAT::pBluescript fragment under high stringency conditions (Fig. 4). MAT transcripts were detected only in 5-d-old etiolated seedlings and 14-d-old hairy roots, whereas DAT transcripts that were detected predominantly in leaf tissue and flower petals were very faintly detected in stem. This tissue-specific expression of each transcript corroborates previous studies that located DAT gene expression to laticifers and idioblast cells (St-Pierre et al., 1999). In contrast, MAT gene expression appears to be restricted to roots.
Figure 4.
Northern blots of total RNA isolated from Madagascar periwinkle hairy roots, roots, stems, leaves, flower petals, or etiolated seedlings, respectively. Blots were probed with the DAT ORF fragment and a 423-bp HindIII fragment of MAT::pBluescript under high stringency conditions. RNA was quantified by ethidium bromide staining and 20 μg of total RNA per sample was electrophoresed on a 7% (v/v) formaldehyde agarose gel. RNA was transferred to nitrocellulose for hybridization.
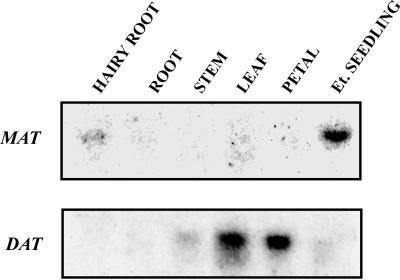
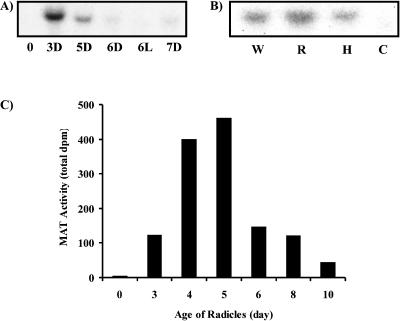
Further studies showed that the MAT gene was already active in 3-d-old etiolated seedlings (Figs. 5A and 3D) and that unlike DAT (St-Pierre et al., 1998), light was not required to activate its expression (Figs. 5A and 6L). The expression of MAT, which was restricted to mostly roots, was also present at low levels in hypocotyls, but not in cotyledons (Fig. 5B). The level of the MAT transcript that was most abundant in 3-d-old etiolated seedlings decreased significantly after 5 d of growth and was virtually non-detectable in 6-d-old seedlings (Fig. 5A). These results were consistent with the appearance of MAT enzyme activity during etiolated seedling development where maximum enzyme activity was found in young 4- to 5-d-old radicles, respectively (Fig. 5C). To further locate the site of MAT gene expression within Madagascar periwinkle roots, 14-d-old lateral hairy roots were divided into sections and were analyzed for the abundance of MAT transcripts (Fig. 6A). These were most abundant within the first full centimeter from the root tip and decreased rapidly in developmentally older hairy root sections. These results were in agreement with RNA in situ hybridization studies, which located MAT gene expression within the cortex and epidermis of tissues near the root tip (Fig. 6B).
Figure 5.
Northern blots of total RNA were isolated from the following: A, 0- to 7-d-old etiolated (D) seedlings, or 5-d-old etiolated seedlings treated with light (L) for of 24 h (6L); and B, 5-d-old whole etiolated seedlings (W), roots (R), hypocotyls (H), and cotyledons (C). Hybridization was carried out under high stringency conditions using a 423-bp HindIII fragment of MAT::pBluescript. RNA was quantified by ethidium bromide staining and 20 μg of total RNA was electrophoresed on a 7% (v/v) formaldehyde agarose gel and transferred to nitrocellulose for hybridization. The blots in A and B were exposed for 3 d, respectively. C, Distribution of MAT activity in 0- to 10-d-old radicles isolated from dark-grown seedlings. MAT activity was determined by liquid scintillation counting of radioactive echitovenine after isolation by TLC (see “Materials and Methods”). Data presented are the average of two trials.
Figure 6.
Tissue- and cell-specific localization of mat. A, Northern blot of total RNA isolated from 0.5-cm sections of lateral hairy root tissue (as shown in the schematic). Hybridization with a 423-bp HindIII fragment of MAT::pBluescipt was carried out under high stringency conditions. B, Localization of mat mRNA by in situ RNA hybridization in hairy roots. The longitudinal section of a 14-d-old lateral hairy root apex was hybridized with antisense RNA for mat as described in “Materials and Methods.” Magnification = 250×. Bar = 100 μm.
Gene Copy Number of MAT
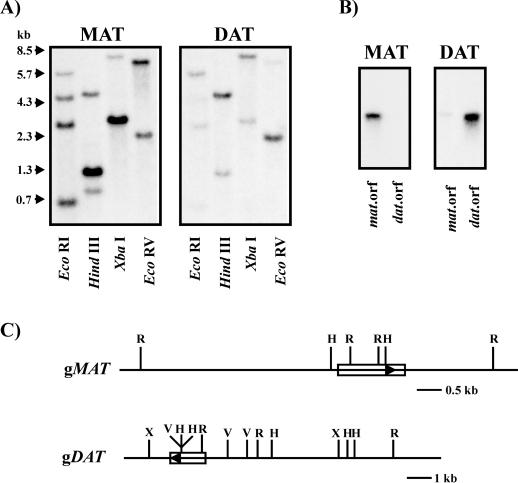
Blots containing EcoRI-, HindIII-, XbaI-, or EcoRV-digested Madagascar periwinkle genomic DNA were probed with either a 423-bp HindIII MAT::pBluescript fragment (Fig. 7A, MAT) or the DAT ORF fragment (Fig. 7A, DAT) at high stringency. The specificity of each probe for their respective, homologous sequences was verified on separate blots containing 100 pg of the MAT ORF and the DAT ORF fragments (Fig. 7B) using conditions similar to those used for the genomic DNA-blot hybridizations. The results indicate that MAT is a single-copy gene and is consistent with the observation that DAT also occurs in a single copy (St-Pierre et al., 1998). The strongly hybridizing EcoRI bands of approximately 0.7, 2.7, and 4.5 kb in the MAT-probed blot correspond to the MAT fragments illustrated in the schematic (Fig. 7C), whereas the 5.7-kb band corresponds to a DAT fragment as observed on the DAT-probed blot (Fig. 7A) and as illustrated on the schematic (Fig. 7C, gDAT). The strongly hybridizing 1.3-kb HindIII band on the MAT-probed blot clearly represents a MAT fragment (Fig. 7, A and C) as it is less intense on the DAT-probed blot (Fig. 7A), whereas the weaker hybridizing band of approximately 4.5 kb represents the corresponding strongly hybridizing DAT fragment (Fig. 7, A, DAT blot and C, gDAT). The presence of a weak approximately 1.0-kb signal in the HindIII-MAT-probed blot suggests it is a cross-hybridizing DAT fragment. However the absence of this signal on the HindIII-DAT-probed blot does not support this conclusion, and therefore, cannot be explained with this data. Although gMAT was not mapped with XbaI (Fig. 7C), a strongly hybridizing 3.0-kb band was observed on the MAT-probed blot (Fig. 7A) along with a weaker hybridizing 8.3-kb band assigned to gDAT, since it is a strong signal on the DAT-probed blot (Fig. 7A). Because an XbaI site had been sequenced, it was possible to map the 8.3-kb band of gDAT (Fig. 7C). In addition, restriction sites for XbaI appear to also occur on gMAT, possibly lying on either side of the ORF accounting for a 3.0-kb fragment. The same may hold true for the presence of a approximately 7.5-kb EcoRV MAT fragment, which gives a strongly hybridizing band with the MAT probe and very weak with the DAT probe (Fig. 7A). Restriction digestion of gMAT with EcoRV was not performed. On the other hand, the approximately 2.3-kb EcoRV fragment (Fig. 7A, MAT blot) clearly corresponds to a DAT EcoRV fragment (Fig. 7, A, DAT blot and C, gDAT). Despite efforts to select a region of low homology between both genes for the design of specific probes, and the use of high stringency conditions, slight cross-hybridization could still be observed. The two genes appear to have different restriction sites as demonstrated in the DNA-blot hybridizations and these structural differences are illustrated in Figure 7C.
Figure 7.
Southern-blot analysis of the MAT and DAT genes in Madagascar periwinkle. A, Genomic DNA isolated from Madagascar periwinkle leaves was restriction digested with EcoRI (R), HindIII (H), XbaI (X), and EcoRV (V). Approximately 12 μg of genomic DNA was electrophoresed per lane, in duplicate, to allow for hybridization under high stringency conditions with the DAT ORF fragment and a 423-bp HindIII fragment of MAT::pBluescript, respectively. B, Southern blots of approximately 100 pg of MAT ORF and DAT ORF fragments illustrating the specificity of the probes used for hybridization. MAT (423-bp HindIII fragment of MAT::pBluescript)and DAT (the DAT ORF fragment). C, Restriction map of the respective genomic clones, gDAT (gDAT-6) and gMAT (gDAT-15), as deduced from the restriction digest patterns of the genomic Southerns. The ORF segments are illustrated as boxes, with arrows indicating the orientation of the ORF in the genomic clones.
DISCUSSION
The cloning and characterization of the DAT gene (St-Pierre et al., 1998) resulted in the isolation of a separate genomic clone (gDAT-15), which is described in the present report. This clone, which harbors the sequence to the partial cDNA clone, A-3 (St-Pierre et al., 1998), is a single-copy (Fig. 7), root-specific gene (Fig. 4–6) sharing 63% nucleic acid identity with the DAT gene and having an ORF encoding a 443-amino acid protein with an estimated molecular mass of approximately 50 kD and 78% amino acid identity with DAT (Fig. 2). The ORF was PCR-amplified, subcloned, and expressed in Escherichia coli as a protein containing a six-HIS extension at its amino terminal. The resulting recombinant protein was named MAT, since it catalyzed the O-acetylation of the 19-hydroxyl group of alkaloids like minovincinine to yield echitovenine and hörhammericine to yield 19-O-acetyl-hörhammericine (Fig. 3; Table I). Echitovenine occurs naturally and was initially isolated from the roots of Catharanthus tricophyllus (Cordell and Farnsworth, 1976).
Kinetic studies demonstrated that the two recombinant proteins (rMAT and rDAT) were quite different with respect to their substrate specificities and catalytic efficiencies. Although rMAT catalyzed the O- acetylation of minovincinine, hörhammericine, and DAV, rDAT only accepted DAV as a substrate (Table I). In addition, the catalytic efficiency of rMAT was very poor compared with rDAT, whose turnover rate for Acetyl CoA and DAV was approximately 240- and 10,000-fold greater than that of rMAT (Table II). In addition, rMAT also exhibited a low turnover rate with respect to hörhammericine that also suggests it to be a poor acetyltransferase for these alkaloid substrates. These findings clearly indicate that two distinct tissue-specific acetyl CoA-dependent O-acetyltransferases are present within Madagascar periwinkle that are responsible for the biosynthesis of 19-O-acetyl-substituted derivatives of tabersonine and vindoline, respectively.
It is evident from northern-blot analyses of different Madagascar periwinkle tissues that MAT transcripts accumulate in young roots, hairy roots tips, and in radicles of etiolated seedlings (Fig. 4–6), as compared with DAT transcripts, which are only found in light exposed tissues (St-Pierre et al., 1998) like leaves and petals (Fig. 4). Furthermore, MAT activity, which peaked in 4- to 5-d-old radicles (Fig. 5C) approximately 24 to 48 h after the optimal levels of transcript accumulation (Fig. 5A) decreased with age. These results, together with the root tip-localized expression of MAT transcripts (Fig. 6) suggest that MAT gene expression is also highly regulated during root development. The presence of different alkaloids and expressed genes within aboveground and underground tissues confirms that distinct tissue-specific pathways are expressed (Fig. 1).
The idioblast and laticifer-specific expression of D4H and DAT, which catalyze the last two steps in vindoline biosynthesis, have been used to speculate about the source of biosynthetic precursors, which permit the formation of final products (St-Pierre et al., 1999). Advanced precursors to vindoline biosynthesis, like tabersonine, might be synthesized in roots to be transported to laticifers and idioblasts in leaves and stems, for elaboration into vindoline. The localization and expression patterns of TDC (St-Pierre et al., 1999), STR1 (St-Pierre et al., 1999), and MAT within cortical root tip cells strongly suggest that the whole pathway leading to the biosynthesis of tabersonine and its substituted analogs occurs within the same root cortical cells. Enzymes like a putative tabersonine 19-hydroxylase and MAT may therefore control the amount of tabersonine available for transport out of the roots for further elaboration (Fig. 1).
The likely presence of the tabersonine pathway, as well as MAT in roots may explain how alkaloids like 19-hydroxytabersonine (Kutney et al., 1980), lochnericine, hörhammericine (Shanks et al., 1998), and echitovenine (Cordell and Farnsworth, 1976) are made. However, previous studies have shown that transformed cell cultures (O'Keefe et al., 1997) and roots (Bhadra et al., 1993) can produce low levels of vindoline under certain circumstances where the vindoline pathway can be induced. Activation of this pathway was suggested by the isolation of an enzyme capable of catalyzing the 4-O-acetylation of DAV to yield vindoline (Bhadra et al., 1993) and the recent cloning and characterization of tabersonine 16-hydroxylase from Madagascar periwinkle cell cultures (Schröder et al., 1999). Hairy root cultures that do not make vindoline were obtained from (Vázquez-Flota et al. (1997) and were shown to have MAT enzyme activity (data not shown). These results, as well as the root specific expression of MAT (Fig. 4–6) and the ability of rMAT to catalyze the 4-O-acetylation of DAV suggest that the enzyme described in (Bhadra et al., 1993) and (O'Keefe et al., 1997) is likely to be MAT rather than DAT.
Recent studies have shown that although roots do contain laticifers, they do not express D4H and DAT, which are required for vindoline biosynthesis (St-Pierre et al., 1999). Studies with young etiolated seedlings show that despite the presence of laticifer cells in mature embryos and in etiolated cotyledons (Vázquez-Flota et al., 2000), light is essential to activate vindoline biosynthesis (Aerts and De Luca, 1992; Vázquez-Flota and De Luca, 1998; Vázquez-Flota et al., 2000). Light treatment of etiolated seedlings did not change the distribution of laticifer and idioblast cells within cotyledons, nor did it change the level or cellular distribution of D4H protein within laticifers and idioblasts (Vázquez-Flota et al., 2000). The presence of inactive D4H protein in idioblasts and laticifers of etiolated seedlings indicates that D4H is expressed properly at early stages of seedling development. The results suggest that light activates the expression of d4h and dat rather than inducing the production of particular cells such as idioblasts and laticifers (Vázquez-Flota et al., 2000). Although this has in part been explained in this report, it remains to be established how root cultures may recreate the complex biological conditions required for vindoline biosynthesis.
MATERIALS AND METHODS
Plant Material
Madagascar periwinkle (Catharanthus roseus G. Don cv Little Delicata) seeds (W.H. Perron, Laval, Quebec, Canada) were sterilized in 70% (v/v) ethanol for 30 s and then washed thoroughly in sterile water. After imbibition for 12 h in sterile water, the seeds were germinated at 25°C in the dark on water-moistened, sterile filter paper (1 layer of Whatman no. 1 filter paper + 3 layers of commercial paper towels) in 9-mm Petri dishes. Seedlings were sectioned into radicles, hypocotyls, and cotyledons prior to harvesting at different times of development. Harvested material was frozen in liquid nitrogen and kept at −80°C until required for analysis. Madagascar periwinkle plants were grown under standard greenhouse conditions and were harvested immediately prior to analysis.
Madagascar periwinkle hairy root cultures (Vázquez-Flota et al., 1994) were grown in the dark in one-half-strength Gamborg B5 medium containing 3% (w/v) Suc on an orbital shaker set at 130 rpm at 25°C. Hairy roots were subcultured every 21 d and analyses were performed with material grown for 14 d.
Alkaloid Extraction
Alkaloids were extracted from Madagascar periwinkle leaves, hairy roots, and roots, respectively, according to the method described in Monforte-González et al. (1992).
Cloning of the MAT Gene
A genomic library prepared from Madagascar periwinkle (Vázquez-Flota et al., 1997) was screened as described previously (St-Pierre et al., 1998). The isolated genomic clone, gDAT-6, corresponded to DAT, which catalyzes the terminal step in vindoline biosynthesis. The genomic clone, gDAT-15, corresponded to a homologous gene whose characterization and biochemical function is the focus of this report. The gDAT-15 clone was digested with EcoRI and three fragments (0.6, 2.4, and 4.0 kb) were subcloned into pBSIISK, and sequenced on both strands using the dideoxynucleotide chain-termination method (Sanger et al., 1977) with a recombinant T7 DNA polymerase (Pharmacia Biotech, Baie d'Urfé, Quebec).
Isolation of DNA and Analysis
Leaf genomic DNA was isolated (Murray and Thompson, 1980), digested with various restriction endonucleases, electrophoresed on 1% (w/v) agarose gels, and transferred to nylon membranes (Hybond-N+, Amersham, Arlington Heights, IL; Sambrook et al., 1989). Membrane hybridization was carried out with either a random primer 32P-labeled 423-bp HindIII MAT::pBluescript fragment or BamHI-PstI digested DAT ORF, respectively, under high stringency conditions (65°C in 250 mm sodium phosphate buffer, pH 8.0, 7% [w/v] SDS, 1% [w/v] bovine serum albumin, and 1 mm EDTA) for 2 d. Blots were washed at 65°C twice with 2× SSC, 0.1% (w/v) SDS and twice with 0.5× SSC, 0.1% (w/v) SDS (Sambrook et al., 1989; 1× SSC = 150 mm NaCl and 15 mm sodium citrate, pH 7.0). Autoradiography of the membranes on x-ray film was performed at −80°C in the presence of intensifying screens.
Total RNA was isolated according to Jones et al. (1985). RNA concentrations were determined spectrophotometrically at 260 nm and adjusted following analysis by agarose gel electrophoresis and staining with ethidium bromide. Electrophoresis of 20 μg of total RNA was carried out on 7.7% (v/v) formaldehyde and 1% (w/v) agarose and transferred to nitrocellulose (BA-85, Schleicher & Schuell, Keene, NH) as described in Sambrook et al. (1989). The membranes were hybridized under high stringency conditions with 32P-labeled probe for 24 h at 65°C, washed, and exposed to film as described for genomic DNA.
In Situ RNA Hybridization
RNA in situ hybridization for mat transcripts within Madagascar periwinkle lateral hairy root tissue was performed as described in St-Pierre et al. (1999).
Expression of MAT in Escherichia coli
The 443-amino acid ORF of the gDAT-15 (MAT) clone was amplified by PCR with Pwo DNA polymerase (Boehringer Mannheim, Laval, Quebec, Canada) and primers MAT5 (5′-GCGGATCCATGGACTCAATAACAATGGTTG-3′) and MAT6 (5′-GCTGCAGAGAGACAATCATGCTGAAACTC-3′) according to the manufacturer's instructions. The primers were designed to incorporate a 5′-BamHI (MAT5) and a 3′-PstI (MAT6) restriction site into the amplified PCR product. Following restriction digestion with the respective endonucleases, the amplified fragment was inserted into the corresponding restriction sites of the pQE30 expression vector (Qiagen, Chatsworth, CA), as well as the pBluescript II SK phagemid (Stratagene, La Jolla, CA). Clone pQE-MAT expressed a MAT polypeptide containing a six-His residue N-terminal extension (HIS(6)-MAT). BB4 E. coli cells (Stratagene) harboring pQE-MAT or pQE30 were grown at 37°C in 25 mL of Luria-Bertani medium to OD600 = 0.6 to 0.7. Expression of the recombinant protein was induced by the addition of isopropyl-1-thio-β-d-galactopyranoside to a final concentration of 2 mm. Cells were collected 3 h post-induction, centrifuged to remove the medium, and frozen at −80°C until used for analysis. The cellular pellet was thawed, washed once, and then resuspended with 2 mL of extraction buffer {100 mm HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid], pH 7.6, 5 mm EDTA, 2 mm dithiothreitol, and 0.5 mm phenylmethylsulfonyl fluoride}, followed by freeze-thawing in liquid N2. The bacterial suspension was sonicated, on ice, using a Branson Sonifier model 250 (Branson Ultrasonic Corporation, Danbury, CT) set at 40% duty cycle with an output control of 4, for three pulses of 30 s with 30-s pauses in between. The cellular debris was removed by centrifugation and the supernatant was desalted on a PD-10 column (Pharmacia Biotech) pre-equilibrated with 100 mm HEPES, pH 7.6, containing 2 mm ascorbate, followed by rechromatography on a PD-10 column pre-equilibrated with 50 mm Na-PO4−, pH 8.0 + 300 mm NaCl + 10% (v/v) glycerol (buffer A). The desalted protein was submitted to nickel-nitrilotriacetic acid metal-affinity chromatography (Qiagen; column dimensions: 10.0 mm i.d. × 3.0 cm) using an fast-protein liquid chromatography system (Pharmacia Biotech) in the presence of buffer A containing 5 mm β-mercaptoethanol (β-ME; buffer B). The column was washed with buffer B containing 5 mm imidazole until the O.D. 280 readout was less than 0.01. Elution of the HIS-tagged protein was carried out using a 30-mL, 5 to 180 mm imidazole linear gradient in buffer A containing 5 mm β-ME. Enzyme assays were carried out as described in the following section and protein purity was estimated by SDS-PAGE on 10% (w/v) gels stained with Coomassie Blue R-250 and silver (Wray et al., 1987).
Enzyme Assays
DAT activity assays were carried out as previously described (St-Pierre et al., 1998). MAT activity assays were performed in a total reaction volume of 100 μL containing crude alkaloids (10 μL) extracted from either root, hairy root, or leaf tissue, respectively; 0.05 μCi of [14C]acetyl coenzyme A (specific activity [sp. act.] 51 mCi mmol−1; Amersham); and 100 mm HEPES, pH 7.6, containing 2 mm ascorbate. The reaction, which was initiated by the addition of protein, was incubated at 37°C for 20 min. The reaction was terminated with the addition of 25 μL 200 mm NaOH, shaken for 1 min, and alkaloid reaction products were extracted in 500 μL ethyl acetate. The radioactivity of reaction products was quantified by liquid scintillation counting of 100 μL of the ethyl acetate fraction and the remaining organic phase was evaporated to dryness. Reaction products were dissolved in 10 μL of MeOH and submitted to TLC on Si-Kieselgel F-254 plates (0.2 mm, E. Merck, Darmstadt). The plates were developed in methanol:ethyl acetate (1:9, v/v), viewed under UV light (λ = 254 and 365 nm, respectively), and exposed to x-ray film for autoradiography at −80°C.
Enzyme Kinetics
Assays were carried out at 37°C in a total volume of 100 μL containing either 0.20 μg of rDAT or 6.0 μg of rMAT for a linear reaction time of 5 or 20 min, respectively. The assays were terminated as previously described. For the generation of saturation curves for rDAT, the following micromolar concentrations of [1-14C]acetylcoenzyme A (sp. act. 51 mCi mmol−1) were used: 3.6, 4.6, 5.6, 6.5, 7.1, 8.1, 10.0, 10.8, 16.3, 16.7, 25.0, 32.5, and 50.0 in the presence of 200 μM DAV. Similarly, the following micromolar concentrations of DAV were used: 16.7, 21.4, 30.0, 37.5, 50.0, 75.0, 150.0, and 300.0 in the presence of 19.6 μM [1-14C]acetylcoenzyme A. Assays were carried out in duplicate. Saturation curves for rMAT were generated using the following micromolar concentrations of [1-14C]acetylcoenzyme A (sp. act. 51 mCi mmol−1): 2.0, 2.8, 3.6, 5.0, 6.3, 8.3, 12.5, 18.0, and 25.0 in the presence of 100 μM DAV. Similarly, the following micromolar concentrations of DAV were used: 136.4, 166.7, 214.3, 300.0, 375.0, 500.0, 750.0, 1,000.0, and 1,500.0 in the presence of 12.7 μM [1-14C]acetylcoA. Assays were carried out in duplicate. The following micromolar concentrations of hörhammericine were used: 166.7, 214.3, 300.0, 500.0, 1,000.0, and 1,500.0 for generating saturation curves for rMAT in the presence of 12.7 μM [1-14C]acetylcoA (sp. act. 59 mCi mmol−1). Assays were carried out in duplicate. Hörhammericine was quantified by HPLC with the photodiode array detector set to monitor the eluate at 254, 300, and 329 nm, respectively, based on the HPLC response factor of tabersonine (Rijhwani and Shanks, 1998).
HPLC analysis was performed using a 600 Multisolvent Delivery System (Waters, Milford, MA) fitted with a 991 Photodiode Array Detector (Waters), a 712 plus Autosampler (Waters) and a C18 (reverse phase) Nucleosil 100 (C18) 3U column (4.6 × 150 mm, Alltech, Mandel Scientific, Guelph, Ontario, Canada) fitted with a guard column (Nucleosil C18; 7.5 × 4.6 mm, Alltech). The solvent system used was as follows: A, 0.2% (w/v) NH4OAc in H2O, pH approximately 7.6; and B, MeCN. The samples were injected onto the pre-equilibrated column (55%, A) at 1.0 mL min−1 followed by a 30-min linear gradient to 30% (A) at 1.0 mL min−1, held at 30% (A) for 10 min, and then increased to 100% (B) in 1 min. Injections of a 5-mm tabersonine/MeOH solution (20 μL) were carried out in triplicate and the corresponding peak areas were determined with the use of the Waters Photo Array Detector software including Stand-alone and Run & Report. Volumes of 10, 20, and 40 μL, respectively, of a approximately 2 mg mL−1 hörhammericine/MeOH solution were injected into the HPLC and their corresponding peak areas determined as described above for the tabersonine sample.
Re-activation of rMAT was necessary for generating the saturation curves for rMAT with hörhammericine, as the activity was quite low. Re-activation was carried out by incubating an aliquot of protein with 1 mm dithiothreitol, on ice, for 60 min, prior to assaying. Saturation curves, Lineweaver-Burk reciprocal plots, and Km and Vmax values were calculated using the ENZFIT software (version 1.05, 1987, Elsevier, Cambridge, UK).
ACKNOWLEDGMENTS
The authors wish to thank Drs. Jaqueline V. Shanks and John Morgan of Rice University (Houston) for their generous gifts of lochnericine, hörhammericine, and echitovenine samples. We gratefully appreciate the help of Zeina Saïkali (Centre de Cancérologie Charles Bruneau, Hôpital Ste-Justine, Montreal) for the use of computers and graphics software. Sylvain Lebeurier is gratefully acknowledged for maintenance of plants in the greenhouse.
Footnotes
This work was supported by a grant from the National Sciences and Engineering Research Council of Canada (to V.D.L.). P.L. was a recipient of a graduate studies scholarship (Bourses Spéciales de la Faculté des Études Supérieurs) from l'Université de Montréal.
LITERATURE CITED
- Aerts RJ, De Luca V. Phytochrome is involved in the light-regulation of vindoline biosynthesis in Catharanthus. Plant Physiol. 1992;100:1029–1032. doi: 10.1104/pp.100.2.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadra R, Vani S, Shanks JV. Production of indole alkaloids by selected hairy root lines of Catharanthus roseus. Biotechnol Bioeng. 1993;41:581–592. doi: 10.1002/bit.260410511. [DOI] [PubMed] [Google Scholar]
- Cordell GA, Farnsworth NR. Catharanthus alkaloids: XXXII. Isolation of alkaloids from Catharanthus trichophyllus roots and structure elucidation of cathaphylline. J Pharm Sci. 1976;65:366–369. doi: 10.1002/jps.2600650312. [DOI] [PubMed] [Google Scholar]
- El-Deeb S, Karawia MS, Abu-Shady H. Vinca rosea Linn: II. Isolation of ajmalicine (d-yohimbine) from the root. Proc Pharm Soc Egypt Sci Ed. 1957;39:201–204. [Google Scholar]
- Jones JDG, Dunsmuir P, Bedbrook J. High level expression of introduced chimeric genes in regenerated transformed plants. EMBO J. 1985;4:2411–2418. doi: 10.1002/j.1460-2075.1985.tb03949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutney JP, Choi LSL, Kolodziejczyk P, Sleigh SK, Stuart KL, Worth BR, Kurz WGW, Chatson KB, Constabel F. Alkaloid production in Catharanthus roseus cell cultures: isolation and characterization of alkaloids from one cell line. Phytochemistry. 1980;19:2585–2595. [Google Scholar]
- Monforte-González M, Ayora-Talavera T, Maldonado-Mendoza IE, Loyola-Vargas VM. Quantitative analysis of serpentine and ajmalicine in plant tissues of Catharanthus roseus and hyoscyamine and scopolamine in root tissues of Datura stramonium by densitometry in thin layer chromatography. Phytochem Anal. 1992;3:117–121. [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair CPN, Pillay PP. Lochnericine: a new alkaloid from Lochnera rosea. Tetrahedron. 1959;6:89–91. [Google Scholar]
- O'Keefe BR, Mahady GB, Gills JJ, Beecher CWW. Stable vindoline production in transformed cell cultures of Catharanthus roseus. J Nat Prod. 1997;60:261–264. [Google Scholar]
- Power R, Kurz WGW, De Luca V. Purification and characterization of acetylcoenzyme A: deacetylvindoline 4-O-acetyltransferase from Catharanthus roseus. Arch Biochem Biophys. 1990;279:370–376. doi: 10.1016/0003-9861(90)90504-r. [DOI] [PubMed] [Google Scholar]
- Rijhwani SK, Shanks JV. Effect of elicitor dosage and exposure time on biosynthesis of indole alkaloids by Catharanthus roseus hairy root cultures. Biotechnol Prog. 1998;14:442–449. doi: 10.1021/bp980029v. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;83:8073–8076. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder G, Unterbusch E, Kaltenbach M, Scmidt J, Strack D, De Luca V, Schröder J. Light-induced cytochrome P450-dependent enzyme in indole alkaloid biosynthesis: tabersonine 16-hydroxylase. FEBS Lett. 1999;458:97–102. doi: 10.1016/s0014-5793(99)01138-2. [DOI] [PubMed] [Google Scholar]
- Shanks JV, Bhadra R, Morgan J, Rijhwani S, Vani S. Quantification of metabolites in the indole alkaloid pathways of Catharanthus roseus: implications for metabolic engineering. Biotechnol Bioeng. 1998;58:333–338. doi: 10.1002/(sici)1097-0290(19980420)58:2/3<333::aid-bit35>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- St-Pierre B, Laflamme P, Alarco AM, De Luca V. The terminal O-acetyltransferase involved in vindoline biosynthesis defines a new class of proteins responsible for coenzyme A-dependent acyl transfer. Plant J. 1998;14:703–713. doi: 10.1046/j.1365-313x.1998.00174.x. [DOI] [PubMed] [Google Scholar]
- St-Pierre B, Vázquez-Flota FA, De Luca V. Multicellular compartmentation of Catharanthus roseus alkaloid biosynthesis predicts intercellular translocation of a pathway intermediate. Plant Cell. 1999;11:887–900. doi: 10.1105/tpc.11.5.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda GH, Oliver AT, Bedwell DR. Alkaloids of Vinca rosea (Catharanthus roseus): XIX. Extraction and characterization of root alkaloids. Lloydia. 1963;26:141–153. [Google Scholar]
- Vázquez-Flota F, De Carolis E, Alarco AM, De Luca V. Molecular cloning and characterization of desacetoxyvindoline 4-hydroxylase, a 2-oxoglutarate dependent dioxygenase involved in the biosynthesis of vindoline in Catharanthus roseus (L.) G. Don Plant Mol Biol. 1997;34:935–948. doi: 10.1023/a:1005894001516. [DOI] [PubMed] [Google Scholar]
- Vázquez-Flota F, Moreno-Valenzuela O, Miranda-Ham ML, Coello-Coello J, Loyola-Vargas VM. Catharanthine and ajmalicine synthesis in Catharanthus roseus hairy root cultures. Plant Cell Tissue Org Cult. 1994;38:273–279. [Google Scholar]
- Vázquez-Flota FA, De Luca V. Developmental and light regulation of desacetoxyvindoline 4-hydroxylase in Catharanthus roseus (L.) G. Don Plant Physiol. 1998;117:1351–1361. doi: 10.1104/pp.117.4.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Flota FA, St-Pierre B, De Luca V (2000) Light activation of vindoline biosynthesis does not require cytomorphogenesis in Catharanthus roseus seedlings. Phytochemistry (in press) [DOI] [PubMed]
- Wray W, Bonlikas T, Wray VP, Hancock R. Silver staining of proteins in polyacrylamide gels. Anal Biochem. 1987;118:197–203. doi: 10.1016/0003-2697(81)90179-2. [DOI] [PubMed] [Google Scholar]