Summary
Targeted cellular auxin distribution is required for morphogenesis and adaptive responses of plant organs. In Arabidopsis thaliana (Arabidopsis), this involves the prototypical auxin influx facilitator AUX1 and its LIKE‐AUX1 (LAX) homologs, which act partially redundantly in various developmental processes. Interestingly, AUX1 and its homologs are not strictly essential for the Arabidopsis life cycle. Indeed, aux1 lax1 lax2 lax3 quadruple knock‐outs are mostly viable and fertile, and strong phenotypes are only observed at low penetrance.
Here we investigated the Brachypodium distachyon (Brachypodium) AUX1 homolog BdAUX1 by genetic, cell biological and physiological analyses.
We report that BdAUX1 is essential for Brachypodium development. Bdaux1 loss‐of‐function mutants are dwarfs with aberrant flower development, and consequently infertile. Moreover, they display a counter‐intuitive root phenotype. Although Bdaux1 roots are agravitropic as expected, in contrast to Arabidopsis aux1 mutants they are dramatically longer than wild type roots because of exaggerated cell elongation. Interestingly, this correlates with higher free auxin content in Bdaux1 roots. Consistently, their cell wall characteristics and transcriptome signature largely phenocopy other Brachypodium mutants with increased root auxin content.
Our results imply fundamentally different wiring of auxin transport in Brachypodium roots and reveal an essential role of BdAUX1 in a broad spectrum of developmental processes, suggesting a central role for AUX1 in pooideae.
Keywords: AUX1, auxin, Brachypodium, monocotyledon, seminal root
Introduction
Modulation of auxin activity through differential auxin distribution plays a central role in developmental and adaptive growth processes (Benjamins & Scheres, 2008; Zazimalova et al., 2010). It is largely achieved through plasma membrane‐integral auxin efflux carriers, the PIN‐FORMED (PIN) proteins, whose polar cellular localization can lead to asymmetric auxin secretion. Coordination of PIN polarity across cell files thus can promote targeted, so‐called polar auxin transport at the tissue and organ level (Benjamins & Scheres, 2008; Zazimalova et al., 2010). In contrast to the carrier requirement for auxin efflux, cellular auxin influx can occur through diffusion, because in the acidic environment of the apoplast auxin is mostly protonated and thus lipophilic enough to cross the plasma membrane (Zazimalova et al., 2010). Nevertheless, dedicated auxin influx facilitators, AUX1 and the LIKE AUX1 (LAX) proteins that accelerate auxin uptake have been identified (Maher & Martindale, 1980; Bennett et al., 1996; Marchant et al., 2002; Yang et al., 2006; Peret et al., 2012). Their differential expression, as well as often polar localization, can modulate polar auxin transport to reinforce or attenuate local auxin accumulations. Arabidopsis thaliana (Arabidopsis) mutants in the prototypical auxin influx facilitator AUX1 have been identified because of their root agravitropism (Maher & Martindale, 1980), which can be rescued by addition of the lipophilic auxin analog 1‐naphthylacetic acid (1‐NAA) (Swarup et al., 2001). Mutants in the three AUX1 homologs, LAX1‐3, display either no, or less conspicuous phenotypes (Ugartechea‐Chirino et al., 2010; Vandenbussche et al., 2010; Peret et al., 2012). However, corresponding multiple mutants reveal (partially) redundant roles of AUX1 and LAX1‐3, for instance in phyllotaxis (Bainbridge et al., 2008) and embryogenesis (Robert et al., 2015), although mutant phenotypes are not always fully penetrant. Moreover, AUX1 and LAX1‐3 proteins are not fully interchangeable in every cellular context (Peret et al., 2012).
Compared to the well characterized roles of AUX1/LAX1‐3 in Arabidopsis, little is known about the developmental role of auxin influx facilitators in monocotyledons (Balzan et al., 2014). Yet, AUX1 homologs can be readily identified, since they are highly conserved. For example, in rice (Oryza sativa) and the more distantly related panicoid grasses maize (Zea Mays L.) and Setaria viridis (Setaria), five AUX1 homologs have been identified (Zhao et al., 2012; Huang et al., 2017). In maize, the closest AtAUX1 homolog has 73% sequence identity (Hochholdinger et al., 2000). Functional studies of mutants in AUX1 homologs in maize and Setaria demonstrated involvement of those genes in inflorescence development and root gravitropism (Huang et al., 2017). Also, the OsAUX1 gene has subsequently been implicated in lateral root formation and shoot elongation (Zhao et al., 2015), as well as seminal root elongation and root hair elongation (Yu et al., 2015). Although rice, maize and Setaria can be considered model systems for the grasses, it remains unclear whether findings from these species can be directly transferred to other groups within the poaceae. One such group is the pooideae, which comprise the major cereal crops wheat, rye and barley. The monocotyledon Brachypodium distachyon (Brachypodium) is a model species for these temperate cereals (Brkljacic et al., 2011; Girin et al., 2014). AUX1 homologs can be readily identified in the Brachypodium genome. However, unlike rice, maize or Setaria with five homologs, Brachypodium only possesses three AUX1 homologs, which display almost sequence identity with their Arabidopsis counterparts (Supporting Information Fig. S1). Nevertheless, slightly divergent N‐ and C‐termini and the gene sequences allow the assignment of clear one‐to‐one homologies in sequence similarity analyses (Fig. 1a). Here we investigated the developmental role of the closest AUX1 homolog of Brachypodium, the Brachypodium distachyon AUX1 (BdAUX1) gene. We report that BdAUX1 loss‐of‐function results in counter‐intuitive root phenotypes and reveals its essential role in a broad spectrum of developmental processes, suggesting a more central and diversified role for AUX1 in pooideae.
Figure 1.
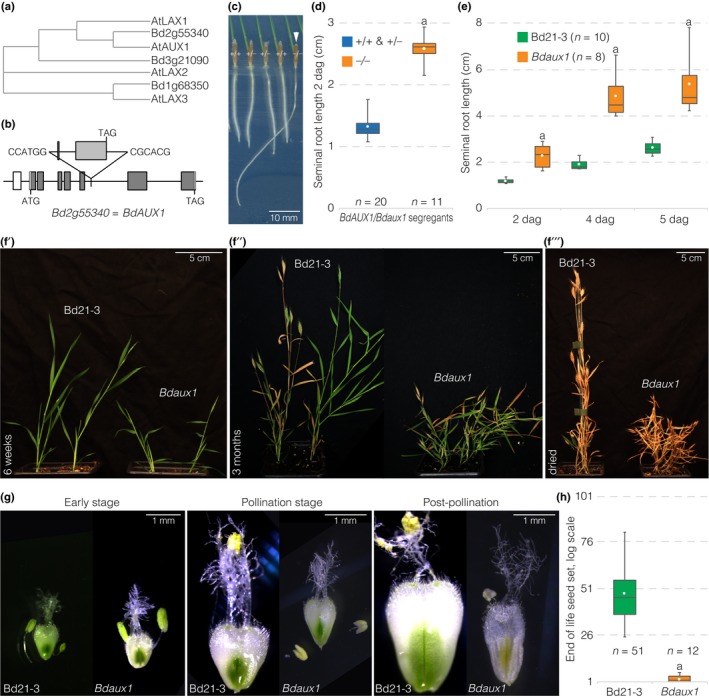
Root and shoot phenotypes of the Bdaux1 mutant. (a) Sequence similarity (Clustal alignment, neighbor joining, with distance correction) of Arabidopsis and Brachypodium AUX1 homologs. (b) Schematic presentation of the T‐DNA insertion line for BdAUX1. (c) Representative seedlings (2‐d‐old) segregating in the progeny of a heterozygous BdAUX1/Bdaux1 (±) mother plant, genotypes are indicated. (d, e) Seminal root length of indicated genotypes (dag, days after germination). (f) Shoot development of Bdaux1 plants in comparison to its Bd21‐3 wild type background at different stages of the life cycle. (g) Different stages of flower development in Bdaux1 plants as compared to Bd21‐3. (h) End of life seed set in indicated genotypes. Box plots display second and third quartiles, maximum, minimum and mean (white dot). Statistically significant differences are indicated (Student's t‐test; a, P < 0.001).
Materials and Methods
Plant materials, genotyping and growth conditions
The Bdtar2l hypo mutant has been described before (Pacheco‐Villalobos et al., 2013). The Bdaux1 mutant line JJ5658 was obtained from a Brachypodium T‐DNA insertion library (Bragg et al., 2012). RT‐PCR was performed to verify that the T‐DNA insertion indeed leads to a truncated BdAUX1 mRNA. To this end, the following oligonucleotides were used: F1 5′‐ATG GTG CCG CGC GAG CAT G‐3′, located at the start‐codon; R1 5′‐GCA TGA TCT CCA CTG TGA CG‐3′, at the border of the T‐DNA insertion; R2 5′‐GGT GAA GCT GAC GAG TAG CG‐3′, located 285 bp before the STOP‐codon; and R3 5′‐ GAT CCG GTA GTT GTG GAA GG‐3′, located 160 bp before the T‐DNA insertion (see Fig. S2A). Bdaux1 CRISPR mutants were obtained directly as homozygotes from transformations (see below, ‘Transformation’) and could not be amplified due to their sterility. Bdtar2l hypo Bdaux1 double mutants were obtained by crossing. For tissue culture, seeds were sterilized as described (Bragg et al., 2012) and stratified for 3 d at 4°C before transfer to plates with half‐strength Murashige‐Skoog (MS) media (2.45 g l−1 MS salts with vitamins, 0.3% sucrose, 1% agar, pH 5.7) placed vertically at a slight angle to prevent roots from growing into the media or the air. Unless indicated otherwise, analyses were performed on 2‐d‐old seedlings raised as previously described (continuous light of 100–120 μE intensity, 22°C, PhilipsF17T8/TL741 fluorescent light bulbs) (Pacheco‐Villalobos et al., 2013). Roots that had grown into the media or the air were excluded from analysis. For gravitropism assays, seeds were grown for 1 d on vertically oriented plates, which were then rotated 90° and seedlings were left to grow for another 2 d. Root length was measured using Fiji software (https://imagej.net/Fiji?Downloads). For auxin analysis, cell wall analysis and RNAseq, 1 cm seminal root segments harvested 2–3 mm above the root tip were used (Pacheco‐Villalobos et al., 2016). Genotyping of Bdtar2l hypo was performed as described (Pacheco‐Villalobos et al., 2013). For Bdaux1 genotyping, the wild type allele was monitored with primers 5′‐GTG AAC TTT CCA CAC TGA GC‐3′ and 5′‐TCA CAA GAG CTG GGC AAT GG‐3′, and the T‐DNA insertion with 5′‐GTG AAC TTT CCA CAC TGA GC‐3′ and 5′‐CAG GAA TTC ATG CCG ACA GC‐3′. Double mutants were genotyped with the same methodology for both T‐DNA insertions.
Plasmid construction
To create a vector with kanamycin resistance, the nptII sequence was amplified with primers 5′‐CCA CTC GAG GAT CTC CAC TCT AGT CGA G‐3′ and 5′‐TGT CTC GAG TTG AAC GAT CGG GGA TCC‐3′. The fragment was digested with XhoI and cloned into XhoI‐digested pCAMBIA1305.1 to replace the hygromycin‐resistance gene to give pCAMBIA1305.1‐nptII. Next, BdAUX1::BdAUX1 was amplified in three pieces from genomic DNA with primers 5′‐CAT GAT TAC GAA TTC GAG CTC GTC ACT TAA TCT CGT C‐3′ and 5′‐CGA ATT TCC TCT CTG TCT CC‐3′ for piece 1, 5′‐GGA GAC AGA GAG GAA ATT CG‐3′ and 5′‐CAA TGC ACC TCA TCG TTC CA‐3′ for piece 2, and 5′‐CAA TGC ACC TCA TCG TTC CA‐3′ and 5′‐GGA AAT TCG AGC TGG TCA CCT AGC AAG CAT TAC TGG GTT‐3′ for piece 3. The fragments were combined into SacI–SalI‐digested pCAMBIA1305.1‐nptII using Gibson ligation. BdAUX1::NLS3xVENUS was created by insertion of amplified NLS3xVENUS into HindIII–Pml I‐digested pCAMBIA1305.1‐nptII. The BdAUX1 promoter was then amplified with primers 5′‐CTA GAG CTC TGG ACG TGG TTT TGT CCT AG‐3′ and 5′‐ACG CGT CGA CAT CTC TTC AAC GCG CTG TC‐3′, and inserted in front of NLS3xVENUS using SacI and SalI digestion. For BdAUX1 localization, a GFP fusion tag was added to the protein. To this end, BdAUX1 promoter was amplified with primers 5′‐GCG ACT GTG CCA ACA CCC‐3′ and 5′‐GCC CTT GCT CAC CAT CTC TTC AAC GCG CTG TCC TC‐3′, the transcript region was amplified with primers 5′‐GTC GAC TCT AGA GGA TCC ATG GTG CCG CGC GAG CAT‐3′ and 5′‐TTT TTC CTC GGG TTA GTT AAT TAA TTC‐3′, and GFP was amplified from pVec8GFP with primers 5′‐ATG GTG AGC AAG GGC GAG G‐3′ and 5′‐ATC CTC TAG AGT CGA CCT TGT ACA GCT CGT CCA TGC‐3′. The three fragments were then combined into XmaI–PacI‐digested pCAMBIA1305.1‐nptII in a Gibson reaction. The BdAUX1 CRISPR/Cas9 cassette was created by amplifying the Zea mays UBIQUITIN (UBQ) promoter (Bragg et al., 2012) using primers 5′‐GAG CTC CAG CTT GCA TGC CTG CAG TG‐3′ and 5′‐GAG CTC TCT AGA GTC GAC CTG CAG AA‐3′ and ligation of the fragment into SacI‐digested pCAMBIA1305.1. A Brachypodium‐optimized Cas9 with FLAG‐tag and nuclear localization signal (Methods S1), followed by a multiple cloning site, was synthesized and cloned behind the UBQ promoter after KpnI and BsteII digestion, to create vector p5Cas. Next, a 770 bp cassette containing a Brachypodium U6 promoter, BsaI restriction sites, tracrRNA, a rice U6 promoter, BtgZI restriction sites and tracrRNA was synthesized (see Methods S1) and cloned into BamHI–EcoRI‐digested pDONR221. This allowed two sgRNA sequences to be added, using BsaI and BtgZI restriction sites, respectively. The Bdaux1 knock‐out cassette was then assembled by annealing, phosphorylating and ligating the following primer pairs into the BsaI–BtgZI‐digested pDONR vector: 5′‐TCT CGT CAC CAG CTT CCT CTG GCA‐3′ and 5′‐AAA CTG CCA GAG GAA GCT GGT GAC‐3′ for sgRNA1, and 5′‐GTG TGA TCC GGT AGT TGT GGA AGG‐3′ and 5′‐AAA CCC TTC CAC AAC TAC CGG ATC‐3′ for sgRNA2. The sgRNA cassette was then isolated and ligated into p5Cas via BamHI–HindIII restriction digest. Target specificity of the sgRNA was checked bioinformatically (http://bioinfogp.cnb.csic.es/tools/breakingcas/?gset=8x2_GENOMES_EnsemblGenomes_39).
Transformation
For Brachypodium transformations (Pacheco‐Villalobos et al., 2013) the Agrobacterium tumefaciens strain GV3101 pMP90 was used. BdAUX1::NLS‐3XVENUS, BdAUX1::BdAUX1 and BdAUX1::GFP‐BdAUX1 transformants in Bd21‐3 and Bdaux1 were selected on media with 400 μg ml−1 paramomycin and 600 μg ml−1 CuSO4. Regeneration media contained 50 μg ml−1 paramomycin and 600 μg ml−1 CuSO4. Transformants for the CRISPR/Cas9 BdAUX1 knock out construct were selected on hygromycin as described (Pacheco‐Villalobos et al., 2013), with the addition of 600 μg ml−1 copper sulfate (CuSO4) to the regeneration media.
Metabolic analyses, qPCR and RNAseq
For auxin measurements, three independent batches of two replicates each, containing 20 pooled 1‐cm root segments per genotype were analyzed as described (Pacheco‐Villalobos et al., 2013, 2016). For cell wall polysaccharide quantifications, three independent pools of 100 to 120 segments per genotype were collected and freeze‐dried overnight. The monosaccharide composition and glycosidic linkages of the wall material was analyzed as described (Pacheco‐Villalobos et al., 2016). qPCR on Brachypodium AUX1 homologs was performed as described normalizing against UBIQUITIN CONJUGATING ENZYME 18 (BdUBC18) (Pacheco‐Villalobos et al., 2013). The following specific primers were used: 5′‐CCA TGT CAT CCA GTG GTT CG‐3′ and 5′‐GAT GAG CTG GAT GAC GGA GC‐3′ for Bradi1g68350; 5′‐CGT CAT CCA GTG GTT TGA GG‐3′ and 5′‐CAG CCG ATG AGC TGG ATC AC‐3′ for Bradi3g21090. For RNAseq, two independent pools of segments were collected from 12 roots per genotype. RNAseq was performed as described (Pacheco‐Villalobos et al., 2016). The raw data have been deposited in the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra/) under accession SRP137652.
Microscopy
For microscopic imaging, seminal roots of 2‐d‐old seedlings were fixed 1 wk in 4% (w/v) paraformaldehyde in 1× phosphate‐buffered saline (PBS) solution (pH 6.9). Roots were then washed two times in 1× PBS before transfer into ClearSee solution for at least one month, which was necessary to quench the challenging autofluorescence of Brachypodium roots. ClearSee solution was changed weekly. Then, 2–3 d before imaging, roots were stained with 0.2% Calcofluor White (in ClearSee) solution for 1–2 h with gentle shaking, next washed in ClearSee solution until imaging. Root hairs were imaged in differential interference contrast using a Leica DM5000 microscope. For meristem analyses, stained roots were mounted in ClearSee solution and imaged with Zeiss 880 or LSM710 inverted confocal microscopes using ×40 oil objectives. For Calcofluor imaging, roots were excited with a 405 nm laser and emission signal was captured over 410–509 nm. GFP was imaged with sequential scans using the 518 nm Argon laser and a 493–523 nm emission spectrum to reduce background. NLS‐3×VENUS was imaged as a sequential scan and excited with a 488 nm laser, emission was recorded at 519–572 nm to reduce background. Cell length measurements were performed with Fiji software.
Microtome sectioning and analysis
Seminal roots of 2‐d‐old seedlings were fixed overnight at 4°C in 1% glutaraldehyde, 4% formaldehyde and 50 mM sodium phosphate buffer (pH 7.2). Roots were dehydrated for at least 1 h each in 15%, 30%, 50%, 70%, 85% and 100% ethanol (EtOH). Samples were pre‐incubated and embedded in TechnoVit 7100 solution as described (Pacheco‐Villalobos et al., 2013). 0.3‐μm sections were obtained on a Leica RM2255 microtome. Sections were stained with 0.1% toluidine blue before visualization with a Leica DM5000 microscope. Cell numbers were counted in one representative image per root using the cell counter plugin of ImageJ software. (https://imagej.nih.gov/ij/plugins/cell-counter.html)
Results and Discussion
To investigate the role of auxin influx facilitators in Brachypodium, we obtained a T‐DNA insertion line in Bradi2g55340 (BdAUX1 hereafter), the closest homolog of Arabidopsis AUX1 (AtAUX1) in Brachypodium. In this Bdaux1 mutant allele, BdAUX1 is disrupted by an insertion in the 6th intron, which leads to a truncated mRNA (Figs 1b, S2). Plants that were homozygous for this insertion displayed agravitropic roots (Fig. 1c), similar to Ataux1 loss‐of‐function mutants (Maher & Martindale, 1980; Bennett et al., 1996). Thus, the T‐DNA insertion apparently results in BdAUX1 loss of function. However, unlike Ataux1 mutants, Bdaux1 mutant roots were considerably longer than those of their wild type siblings or the corresponding Bd21‐3 wild type background line (Figs 1c–e, S3A). Quantitative RT‐PCR (qPCR) suggested that this phenotype was not due to possible (over)compensatory up‐regulation of the two other AUX1 homologs in Brachypodium (Fig. S2B). Bdaux1 plants also displayed a dwarf shoot phenotype with aberrant flower development (Fig. 1f,g). Bdaux1 mutants were thus sterile (Fig. 1h) and could not be maintained as homozygotes in practice. Both the root and shoot phenotypes could be complemented by introduction of transgenes that expressed either BdAUX1 or GFP‐BdAUX1 fusion protein under control of the native BdAUX1 promoter (BdAUX1::BdAUX1 and BdAUX1::GFP‐BdAUX1) into the Bdaux1 background (Fig. S3B,C). Moreover, the mutant phenotypes were also observed in Bdaux1 homozygous knock out plants that were generated by the CRISPR/Cas9 technique (Bdaux1 CRISPR). This included the severe shoot phenotype and infertility (Fig. S3C,D), which also precluded recovery of the lines. Therefore, Bdaux1 loss‐of‐function was causative for the observed mutant phenotype.
A more detailed characterization of the mutants revealed that their increased root elongation could be attributed to increased mature cell length (Fig. 2a). Moreover, Bdaux1 roots were markedly thinner than wild type roots (Fig. 2b). Although the number of cell files was significantly reduced in every tissue except xylem and phloem (Fig. 2c), this alone could not entirely account for the overall reduction in root thickness. Rather, cells generally appeared slightly smaller in radial sections (Fig. 2b), and at the same time, root hairs were markedly shorter, reduced in number and appeared later than in wild type (Fig. 2d). Therefore, the Bdaux1 root elongation phenotype was apparently caused by overall higher cellular anisotropy. Interestingly, it thus resembles the roots of hypomorphic mutants in the Brachypodium TAR2‐LIKE (TAR2L) gene (Pacheco‐Villalobos et al., 2013). Bdtar2l hypo mutants are partially impaired in a rate‐limiting step of auxin biosynthesis, which results in higher cellular auxin levels in the root because of the particular regulatory wiring in Brachypodium (Pacheco‐Villalobos et al., 2013, 2016). To further explore the similarity between Bdaux1 and Bdtar2l hypo mutant roots, we also determined cellular auxin levels in Bdaux1 root tips. Indeed, we again observed increased auxin levels (Fig. 3a). This result was surprising, given the Arabidopsis precedent that AUX1 is needed for efficient shoot to root mobilization of auxin, and Ataux1 mutants therefore have reduced, rather than increased, auxin levels in the root (Marchant et al., 2002). In Bdtar2l hypo plants, the root phenotype was also associated with slight alterations in cell wall composition, notably a reduction in 1,3‐galactosyl and 1,2‐galactosyl residues, suggesting an altered arabinogalactan structure, and an increase in 1,4‐glucosyl residues (Pacheco‐Villalobos et al., 2016). Similar changes were observed in Bdaux1 root tips (Figs 3b, S3E), again confirming similarity with Bdtar2l hypo plants. Finally, a survey of the Bdaux1 transcriptome in elongating root tip segments revealed a number of differentially expressed genes, mostly in cell wall modifiers (Table S1), which were c. 10‐fold over‐represented (P = 2.33E‐5). Again, this observation matches what has been described for Bdtar2l hyporoot segments (Pacheco‐Villalobos et al., 2016), although the scope of transcriptional changes was less dramatic in Bdaux1. A notable commonality was the strong upregulation of expansins, which are thought to be primary targets of auxin‐induced cell elongation (Cosgrove, 2005). Confirming the qPCR analysis, no differential expression of the two other AUX1 homologs was observed in the Bdaux1 transcriptome (Table S2). In summary, in many ways Bdaux1 roots phenocopy Bdtar2l hypo roots.
Figure 2.
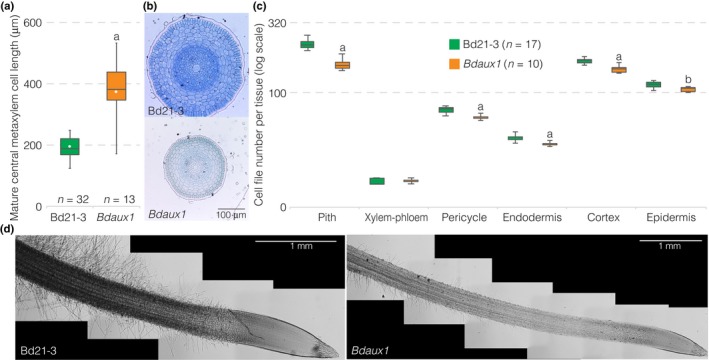
Cellular root phenotypes of the Brachypodium Bdaux1 mutant. (a) Mature central metaxylem cell length in indicated genotypes. (b) Histological cross‐sections (toluidine blue‐stained) through 2‐d‐old roots of indicated genotypes, taken from the mature part of the root, above the elongation zone. (c) Quantification of cell files in different mature tissue layers of indicated genotypes (2‐d‐old roots). (d) Illustration of root hair development in indicated genotypes (light microscopy, differential interference contrast; composite images). Box plots display second and third quartiles, maximum, minimum and mean (white dot). Statistically significant differences are indicated (Student's t‐test: a, P < 0.001; b, P < 0.03).
Figure 3.
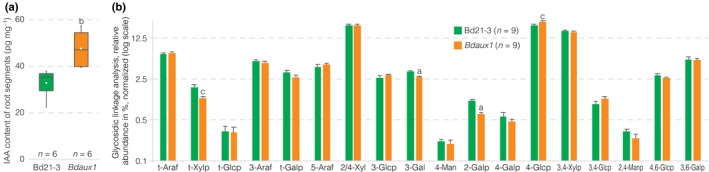
Metabolic Brachypodium Bdaux1 phenotypes. (a) Free auxin (indole‐3‐acetic acid, IAA) content of 1‐cm root elongation zone segments (2‐d‐old roots) of indicated genotypes. (b) Glycosidic linkage analysis of wall material of 1‐cm root elongation zone segments (2‐d‐old roots) of indicated genotypes (error bars, + standard error (SE)). Box plots display second and third quartiles, maximum, minimum and mean (white dot). Statistically significant differences are indicated (Student's t‐test: a, P < 0.001; c, P < 0.05).
Similarities with Bdtar2l mutants could also be observed in the shoot. In mutants of the hypomorphic Bdtar2l hypo allele, the root phenotype is accompanied by a slight reduction in leaf blade length and width (Pacheco‐Villalobos et al., 2013). However, in mutants of the null allele Bdtar2l qnull, the root phenotype is weaker and transient, while the shoot displays a dwarf phenotype that is accompanied by severely reduced fertility (Pacheco‐Villalobos et al., 2013). Thus, the shoot phenotype of Bdtar2l qnull plants is similar to Bdaux1 plants. The strongly reduced fertility of Bdaux1 appeared to be due to delayed development of anthers as compared to gynoecia as well as poor pollen viability (Fig. 1g). Nevertheless, because plants heterozygous for Bdaux1 were similar to wild type, we could create double mutants with the Bdtar2l hypo allele. Overall, the phenotype of these Bdaux1 Bdtar2l hypo double mutants appeared to be additive as compared to their segregating single mutants and wild type siblings (with the caveat that background loci might modulate the phenotypes to some degree because the two single mutants had different wild type parents). The dwarfism of Bdaux1 plants was more exaggerated in Bdaux1 Bdtar2l hypo double mutants (Fig. S3B,F), and the double mutant roots were thinner than in either single mutant and longer than in Bdtar2l hypo alone (Fig. S3G). This could be attributed to an even higher mature cell length, and an additional reduction in cell files (Fig. S2H). However, unlike the single mutants, the double mutants displayed a reduced root meristem size that was accompanied by slight changes in root meristem organization, such as an apparently smaller quiescent center (Fig. S3I,J). Overall, the data suggest parallel impacts of BdAUX1 and BdTAR2L mutation that reinforce each other. This is also consistent with the absence of significant expression changes in rate‐limiting auxin biosynthesis genes in Bdaux1 (Table S2).
The BdAUX1::GFP‐BdAUX1 plants, as well as BdAUX1::NLS‐3XVENUS plants, allowed us to assess the expression pattern of BdAUX1 in the root. AtAUX1 is expressed specifically in the Arabidopsis root protophloem, epidermis and root cap‐columella (Marchant et al., 2002). BdAUX1 transcriptional and translational reporters displayed similar expression patterns, with the exception of expression in the root cap. Moreover, unlike AtAUX1, BdAUX1 was also expressed throughout the stele and in the outer cortex layers (Fig. 4a–c). Thus, the expression pattern of BdAUX1 encompasses the combined domains of AtAUX1, AtLAX2 and AtLAX3 (Peret et al., 2012) with the exception of the root cap, and therefore, possibly, their combined functions in these tissues. Consistent with its homology to AtAUX1, GFP‐BdAUX1 protein was localized at the plasma membrane, in a typically polar fashion (Fig. 4d,e). In the stele, the orientation was generally shootward (Fig. 4e), while in the outer cell layers, BdAUX1 polar localization appeared mostly rootward (Fig. 4f). However, in the later epidermis, BdAUX1 was detected on both the apical and basal sides of the cell, as well as facing inside (Fig. 4g). In summary, the localization is consistent with a role of BdAUX1 in promoting auxin transport from the shoot to the root tip, and in evacuating auxin from the tip via the epidermis. Notably, despite the increased auxin level in Bdaux1 root tips (Fig. 3a), the Bdaux1 root agravitropism could be somewhat rescued by application of 1‐NAA (Fig. 5a), similar to Ataux1 (Swarup et al., 2001). However, 1‐NAA levels that rescued agravitropism did not restore normal root elongation (Fig. 5b), which was always higher in Bdaux1 than in Bd21‐3, indicating that the roles of BdAUX1 in cell elongation and gravitropism are physiologically separable.
Figure 4.
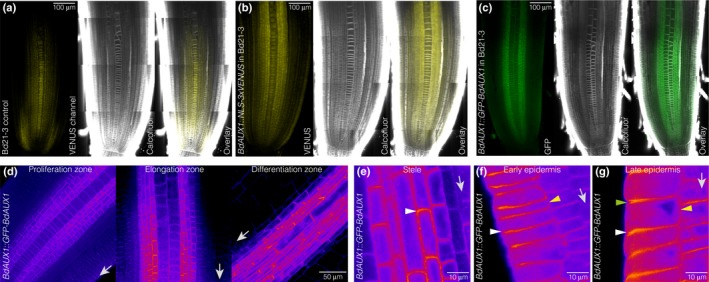
Brachypodium BdAUX1 expression. (a) Confocal microscopy of a 2‐d‐old Bd21‐3 root meristem after ClearSee and calcofluor staining (white), illustrating background fluorescence (yellow) in the VENUS channel. Please note that autofluorescence of Brachypodium roots cannot be fully eliminated (see the Materials and Methods section). (b) Expression pattern of a BdAUX1 transcriptional reporter (nuclear‐localized VENUS fluorescence, yellow). (c) Expression pattern of a GFP‐BdAUX1 translational reporter fusion protein (plasma membrane‐localized green fluorescence). (d) Expression level and cellular localization of GFP‐BdAUX1 fusion protein (magenta fluorescence) in different parts of a 2‐d‐old root meristem (arrows point towards root tip). (e) Cellular localization of GFP‐BdAUX1 fusion protein (magenta fluorescence) in the stele, showing shootward polar accumulation of BdAUX1 (arrowhead) (arrow points towards root tip). (f) Cellular localization of GFP‐BdAUX1 fusion protein (magenta fluorescence) in the early epidermis, showing rootward polar accumulation of BdAUX1 (white arrowhead) and absence from inward facing side (yellow arrowhead) (arrow points towards root tip). (g) Cellular localization of GFP‐BdAUX1 fusion protein (magenta fluorescence) in the late epidermis, showing both rootward (white arrowhead) and shootward polar accumulation (green arrowhead), as well as inward facing localization (yellow arrowhead) of BdAUX1 (arrow points towards root tip). (a–c) are composite images.
Figure 5.
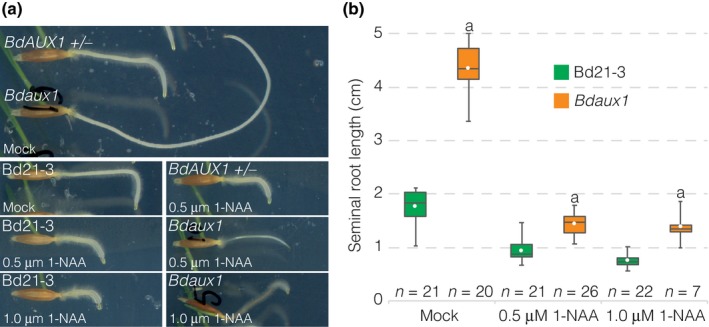
Rescue of Brachypodium Bdaux1 agravitropism. (a) Response of indicated genotypes to a 90° change in the gravity vector (3‐d‐old roots, plates were turned when they were 1‐d‐old), in the absence or presence of 1‐NAA. (b) Root length of indicated genotypes in the absence or presence of 1‐NAA. Box plots display second and third quartiles, maximum, minimum and mean (white dot). Statistically significant differences are indicated (Student's t‐test: a, P < 0.001; b, P < 0.01; c, P < 0.05).
In summary, our detailed analyses of Bdaux1 mutants revealed phenotypes that are counterintuitive with respect to the expectations set by the precedent of corresponding Arabidopsis mutants. However, interestingly, an exaggerated root elongation phenotype has also been described for Osaux1 mutants (Yu et al., 2015), although it has not been noticed by others working with the same lines (Zhao et al., 2015). Moreover, Osaux1 mutants also display slightly reduced shoot organ elongation (Zhao et al., 2015). Yet, compared to the Bdaux1 mutants, these phenotypes appear relatively mild, and no flower development or reproductive phenotypes were reported. Likewise, AUX1 mutants in maize and Setaria also display apparently milder inflorescence and root phenotypes than BdAUX1 (Huang et al., 2017). Possibly, this reflects partial genetic redundancy in rice, maize and Setaria, which contain two more AUX1 homologs than Brachypodium, including close OsAUX1, ZmAUX1 and SvAUX1 homologs (Zhao et al., 2012, 2015; Huang et al., 2017). Thus, the auxin uptake facilitator network in Brachypodium might be less complex than in other grasses, confirming once more that the regulatory wiring of auxin biosynthesis or transport can vary between species, and thus can trigger distinct physiological and morphological consequences if tampered with (Pacheco‐Villalobos et al., 2013; O'Connor et al., 2014). In summary, our data suggest that in Brachypodium, BdAUX1 primarily assures correct local auxin accumulation and has a broad role in root and shoot development. This role is apparently broader than the role of AtAUX1 in Arabidopsis, and could potentially encompass activities of AtLAX homologs (Marchant et al., 2002). However, a detailed analysis of the other Brachypodium AUX1 homologs will be required to conclusively resolve whether this is indeed the case.
Author contributions
A.v.d.S. and C.S.H. designed the study and wrote the paper. A.v.d.S., C.V., K.L., M.P. and C.S.H. designed experiments. A.v.d.S. and C.V. performed experiments. J.B. and J.V. provided crucial reagents.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Clustal protein sequence alignment of Arabidopsis and Brachypodium AUX1 homologs.
Fig. S2 Expression analysis of Brachypodium BdAUX1 and other AUX1 homologs.
Fig. S3 Various genetic and physiological analyses of Brachypodium BdAUX1.
Table S1 List of differentially expressed genes in Bdaux1 root segments (P < 0.01)
Table S2 Comparison of RNAseq analyses of Bdaux1 and Bd21‐3 root segments
Methods S1 DNA sequences of oligonucleotides and the CRISPR/Cas9 cassette used in this study.
Acknowledgements
The authors would like to thank the Lausanne Genomic Technologies Facility for RNAseq services, and A. Amiguet‐Vercher and R. Granbom for excellent technical support. This work was funded by Swiss National Science Foundation grant CR32I3_156724 awarded to C.S.H. and SystemsX funding. Experiments performed by C.V. and M.P. were supported by CEPLAS (Cluster of Excellence on Plant Sciences – Deutsche Forschungsgemeinschaft EXC1028). K.L. was supported by the Swedish Governmental Agency for Innovation Systems (VINNOVA) and the Swedish Research Council (VR). The work conducted by the US DOE Joint Genome Institute is supported by the Office of Science of the US Department of Energy under Contract no. DE‐AC02‐05CH11231.
References
- Bainbridge K, Guyomarc'h S, Bayer E, Swarup R, Bennett M, Mandel T, Kuhlemeier C. 2008. Auxin influx carriers stabilize phyllotactic patterning. Genes & Development 22: 810–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzan S, Johal GS, Carraro N. 2014. The role of auxin transporters in monocots development. Frontiers in Plant Science 5: 393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamins R, Scheres B. 2008. Auxin: the looping star in plant development. Annual Review of Plant Biology 59: 443–465. [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA. 1996. Arabidopsis AUX1 gene: a permease‐like regulator of root gravitropism. Science 273: 948–950. [DOI] [PubMed] [Google Scholar]
- Bragg JN, Wu J, Gordon SP, Guttman ME, Thilmony R, Lazo GR, Gu YQ, Vogel JP. 2012. Generation and characterization of the Western Regional Research Center Brachypodium T‐DNA insertional mutant collection. PLoS ONE 7: e41916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brkljacic J, Grotewold E, Scholl R, Mockler T, Garvin DF, Vain P, Brutnell T, Sibout R, Bevan M, Budak H et al 2011. Brachypodium as a model for the grasses: today and the future. Plant Physiology 157: 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. 2005. Growth of the plant cell wall. Nature Reviews Molecular Cell Biology 6: 850–861. [DOI] [PubMed] [Google Scholar]
- Girin T, David LC, Chardin C, Sibout R, Krapp A, Ferrario‐Mery S, Daniel‐Vedele F. 2014. Brachypodium: a promising hub between model species and cereals. Journal of Experimental Botany 65: 5683–5696. [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Wulff D, Reuter K, Park WJ, Feix G. 2000. Tissue‐specific expression of AUX1 in maize roots. Journal of Plant Physiology 157: 315–319. [Google Scholar]
- Huang P, Jiang H, Zhu C, Barry K, Jenkins J, Sandor L, Schmutz J, Box MS, Kellogg EA, Brutnell TP. 2017. Sparse panicle1 is required for inflorescence development in Setaria viridis and maize. Nature Plants 3: 17054. [DOI] [PubMed] [Google Scholar]
- Maher EP, Martindale SJ. 1980. Mutants of Arabidopsis thaliana with altered responses to auxins and gravity. Biochemical Genetics 18: 1041–1053. [DOI] [PubMed] [Google Scholar]
- Marchant A, Bhalerao R, Casimiro I, Eklof J, Casero PJ, Bennett M, Sandberg G. 2002. AUX1 promotes lateral root formation by facilitating indole‐3‐acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14: 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor DL, Runions A, Sluis A, Bragg J, Vogel JP, Prusinkiewicz P, Hake S. 2014. A division in PIN‐mediated auxin patterning during organ initiation in grasses. PLoS Computational Biology 10: e1003447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco‐Villalobos D, Diaz‐Moreno SM, van der Schuren A, Tamaki T, Kang YH, Gujas B, Novak O, Jaspert N, Li Z, Wolf S et al 2016. The effects of high steady state auxin levels on root cell elongation in Brachypodium. Plant Cell 28: 1009–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco‐Villalobos D, Sankar M, Ljung K, Hardtke CS. 2013. Disturbed local auxin homeostasis enhances cellular anisotropy and reveals alternative wiring of auxin‐ethylene crosstalk in Brachypodium distachyon seminal roots. PLoS Genetics 9: e1003564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peret B, Swarup K, Ferguson A, Seth M, Yang Y, Dhondt S, James N, Casimiro I, Perry P, Syed A et al 2012. AUX/LAX genes encode a family of auxin influx transporters that perform distinct functions during Arabidopsis development. Plant Cell 24: 2874–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert HS, Grunewald W, Sauer M, Cannoot B, Soriano M, Swarup R, Weijers D, Bennett M, Boutilier K, Friml J. 2015. Plant embryogenesis requires AUX/LAX‐mediated auxin influx. Development 142: 702–711. [DOI] [PubMed] [Google Scholar]
- Swarup R, Friml J, Marchant A, Ljung K, Sandberg G, Palme K, Bennett M. 2001. Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes & Development 15: 2648–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugartechea‐Chirino Y, Swarup R, Swarup K, Peret B, Whitworth M, Bennett M, Bougourd S. 2010. The AUX1 LAX family of auxin influx carriers is required for the establishment of embryonic root cell organization in Arabidopsis thaliana . Annals of Botany 105: 277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F, Petrasek J, Zadnikova P, Hoyerova K, Pesek B, Raz V, Swarup R, Bennett M, Zazimalova E, Benkova E et al 2010. The auxin influx carriers AUX1 and LAX3 are involved in auxin–ethylene interactions during apical hook development in Arabidopsis thaliana seedlings. Development 137: 597–606. [DOI] [PubMed] [Google Scholar]
- Yang Y, Hammes UZ, Taylor CG, Schachtman DP, Nielsen E. 2006. High‐affinity auxin transport by the AUX1 influx carrier protein. Current Biology 16: 1123–1127. [DOI] [PubMed] [Google Scholar]
- Yu C, Sun C, Shen C, Wang S, Liu F, Liu Y, Chen Y, Li C, Qian Q, Aryal B et al 2015. The auxin transporter, OsAUX1, is involved in primary root and root hair elongation and in Cd stress responses in rice (Oryza sativa L.). Plant Journal 83: 818–830. [DOI] [PubMed] [Google Scholar]
- Zazimalova E, Murphy AS, Yang H, Hoyerova K, Hosek P. 2010. Auxin transporters–why so many? Cold Spring Harbor Perspectives in Biology 2: a001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Ma T, Wang X, Deng Y, Ma H, Zhang R, Zhao J. 2015. OsAUX1 controls lateral root initiation in rice (Oryza sativa L.). Plant, Cell & Environment 38: 2208–2222. [DOI] [PubMed] [Google Scholar]
- Zhao H, Ma H, Yu L, Wang X, Zhao J. 2012. Genome‐wide survey and expression analysis of amino acid transporter gene family in rice (Oryza sativa L.). PLoS ONE 7: e49210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Clustal protein sequence alignment of Arabidopsis and Brachypodium AUX1 homologs.
Fig. S2 Expression analysis of Brachypodium BdAUX1 and other AUX1 homologs.
Fig. S3 Various genetic and physiological analyses of Brachypodium BdAUX1.
Table S1 List of differentially expressed genes in Bdaux1 root segments (P < 0.01)
Table S2 Comparison of RNAseq analyses of Bdaux1 and Bd21‐3 root segments
Methods S1 DNA sequences of oligonucleotides and the CRISPR/Cas9 cassette used in this study.


