Abstract
Objectives:
Detection and comparison of metallo-β-lactamase (MBL) production in clinical isolates by phenotypic and genotypic measures. The objective of this study is to evaluate clinical characteristics and risk factors in patients infected with MBLs.
Materials and Methods:
Study was conducted by the Department of Microbiology, Jawaharlal Nehru Medical College from February 2014 to December 2015. Bacterial culture, identification, and antibiotic susceptibility were carried out according to standard guidelines. MBL production was detected both phenotypically (Modified Hodge test [MHT], imipenem-ethylene diamine tetraacetic acid double disk potentiation test [IMP-EDTA DDPT], IMP-EDTA combined disk synergy test [IMP-EDTA CDST]), and genotypically (blaNDM-1, blaVIM and blaIMP).
Results:
Among 116 carbapenem-resistant Gram-negative Bacilli (CRGNB), Citrobacter species 28 (24.1%) was the most common pathogen. Phenotypically, MHT, IMP-EDTA DDPT, and IMP-EDTA CDST detected MBL production in 105 (90.5%), 96 (81%), and 87 (75%) CRGNB, respectively. BlaNDM-1 genes were detected in 6 6 (56.8%) isolates, however, very few blaVIM (16, 15.2%) and blaIMP (1, 1.2%) were identified. Considering polymerase chain reaction (PCR) as the gold standard, it was observed that IMP-EDTA CDST was most specific (78.3%) while MHT was most sensitive (97.4%). Results of blaNDM-1 gene by PCR were further confirmed by sequencing (Triyat genomics, Nagpur). All the 11 representative strains were confirmed to be NDM-1 gene. Major risk factors in patients infected with MBLs were in-dwelling devices (68%), prolonged hospital stay (72%) and prior antibiotic treatment (86%). However, on tracing their outcome, it was interesting to note that mortality was relatively low 5 (4.3%).
Conclusion:
The present study shows a rising trend of blaNDM-1 in CRGNB, an ominous sign heralding the post antibiotic era. It is essential to assess the prevalence of various MBLs so that infection control measures can be reinforced. We recommend three phenotypic tests in tandem for the detection of MBL. While phenotypic tests are easy and cost-effective to perform, quick, effective molecular diagnostic techniques can tailor treatment guidelines to optimize patient's management.
Keywords: BlaIMP, blaNDM1, blaVIM, carbapenem resistant Gram-negative bacilli, polymerase chain reaction, phenotypic tests
INTRODUCTION
Carbapenemases represent the most versatile family of beta-lactamases, with the spectrum unrivalled by the other beta-lactam-hydrolysing enzymes.[1] Although known as carbapenemases, many of these enzymes recognize almost all hydrolysable beta-lactams and are resilient against inhibition by all commercially available beta-lactamase inhibitors.[2,3] This resilience is often accompanied by resistance to other classes of antibiotics. Carbapenem-resistant Gram-negative bacilli (CRGNB) represents difficult-to-treat infections in hospitalized patients and is associated with high mortality.[4] Carbapenem antibiotics have been reserved as drugs of the last resort for salvage treatment for infections caused by multidrug-resistant Gram-negative bacteria. Thus, resistance to carbapenems becomes a real threat to the survival of seriously ill patients, with overall mortality exceeding 50%.[5] It is estimated that we are on the edge of a worldwide carbapenemase-pandemic. This is a prospective study performed on a significant number of strains to establish the prevalence of different carbapenemase genes in fermenting and nonfermenting Gram-negative bacilli isolated from patients admitted in wards or attending the outpatient department (OPD) of a tertiary care hospital in Northern India. We compared different methods for detection of metallo-β-lactamase (MBL) so as to establish the most specific test. Clinical characteristics and risk factors associated with patients infected with MBLs were assessed.
MATERIALS AND METHODS
The present study was conducted in patients admitted to the wards or attending the OPD of Jawaharlal Nehru Medical College Hospital, AMU, Aligarh in the Department of Microbiology from February 2014 to December 2015. This study was done after approval from the Institutional Ethics Committee of JNMC, and the procedures followed in the study were in accordance with its guidelines. Clinical specimens including pus, urine, and fluid submitted to the bacteriology were investigated. Urinary tract infection (UTI) was defined as positive quantitative urine culture (>105 microorganisms/ml) with a maximum of two isolated microbial species in patients with symptoms suggestive of UTI. Wound infection was defined as per Southampton grading. Majority of the cases belonged to Grade III and IV. Fluid samples were obtained from intercostal tube drainage, peritoneal fluids, and pleural fluids. Fluid cultures were interpretated according to Bergey's Manual of Systematic Bacteriology Vol 3, 2009. The samples were obtained with proper aseptic techniques and transported to the laboratory within 1 h of their collection. The organisms were identified on the basis of cultural characteristics, morphology, and biochemical tests (Bailey and Scotts, 2007,[6] Mackie and McCartney, 2007[7]). Antibiotic susceptibility testing was performed by Kirby-Bauer disc diffusion technique on Mueller-Hinton agar according to Clinical and Laboratory Standards Institute (CLSI) 2010, M 100-S20[8] guidelines. However, certain other drugs apart from CLSI 2010 guidelines were also tested. Antimicrobial discs used were as follows:
For enterobacteriaceae (pus and fluids)
Amikacin (30 μg), gentamicin (10 μg), levofloxacin (5 μg), ofloxacin (5 μg), ceftriaxone (30 μg), cefoperazone (75 μg), cefotaxime (30 μg), cefixime (5 μg), cefepime (30 μg), ceftazidine (30 μg), cepodoxime (10 μg), cefoperazone/sulbactam. (75/10 μg), ceftazidine/clavulanicacid (30/10 μg), ceftriaxone/sulbactam (30/15 μg), cefotaxime/clavulanicacid (30/10 μg), ceftazidine/tazobactam (80/10 μg), piperacillin (100 μg), piperacillin/tazobactam (100/10 μg), fosfomycin (200 μg), tobramycin (10 μg), sparfloxacin (5 μg), ertapenem (10 μg), faropenem (5 μg), and imipenem (IMP) (10 μg).
For nonfermenting Gram-negative bacilli
Amikacin (30 μg), gentamicin (10 μg), levofloxacin (5 μg), ceftriaxone (30 μg), cefoperazone (75 μg), cefepime (30 μg), nitrofurantoin (300 μg), ceftazidine (30 μg), cefotaxime (30 μg), cepodoxime (10 μg), cefoperazone/sulbactam (75/10 μg), ceftriaxone/sulbactam (30/15 μg), ceftazidine/clavulanicacid (30/10 μg), ticarcillin (75 μg), piperacillin (100 μg), piperacillin/tazobactam (100/10 μg), tobramycin (10 μg), sparfloxacin (5 μg), IMP (10 μg), colistin (10 μg), and Polymyxin B (300 units).
For urinary tract infection, the drugs used were as follows
Amikacin (30 μg), gentamicin (10 μg), levofloxacin (5 μg), ceftriaxone (30 μg), cefoperazone (75 μg), cefoperazone/sulbactam (75/10 μg), cefixime (5 μg), cefepime (30 μg), nitrofurantoin (300 μg), ceftazidine (30 μg), ceftazidine/tazobactam (80/10 μg), piperacillin (100 μg), piperacillin/tazobactam (100/10 μg), tobramycin (10 μg), sparfloxacin (5 μg), ertapenem (10 μg), faropenem (5 μg), and imipenem (10 μg).
Consecutive, nonduplicate, and CRGNB isolates were screened phenotypically for the detection of MBL production. Confirmation of MBL production was done by polymerase chain reaction (PCR). Detailed history and investigations of all these patients were noted and followed up for outcome.
Phenotypic detection of metallo-β-lactamase production
Modified Hodge test[9](MHT), IMP-ethylene diamine tetraacetic acid-Double Disk Synergy test[9] (IMP-EDTA DDST), and IMP-EDTA-combined disk synergy test[10] (IMP-EDTA CDST) were done for phenotypic detection of MBL.
Genotypic detection of metallo-β-lactamase genes (BlaNDM-1, BlaVIM, BlaIMP)
Genotypic detection of MBL genes was performed only on those isolates which were phenotypically identified as MBL producers. Thus, 116 isolates were selected for detailed molecular characterization. PCR amplification was carried out using the 2x PCR master mix (Fermentas, Thermo scientific, USA) on a Gradient Thermo Cycler named Le cycler of LABNIC, USA, with primers targeting MBL genes as given below.
Molecular detection of blaNDM-1, blaVIM, and blaIMP was performed using PCR according to methods described previously (Nordmann et al., 2011[11] for blaNDM-1, Manoharan et al., 2010[12] for blaVIM and blaIMP) to amplify the 621, 382, and 587 base pair fragments, respectively [Table 1]. The amplified PCR products were electrophoresed on a 1.5% agarose gel stained with ethidium bromide and visualized under ultraviolet light.
Table 1.
Primers used in polymerase chain reaction for various MBL genes in the study

Sequencing
DNA sequencing was performed at TRIYAT SCIENTIFIC CO.(Nagpur, India) using the ABI Prism® Big Dye® Terminator version 3.1 Cycle Sequencing Kit (Applied Biosystems, USA), for forward and reverse primers of NDM-1 gene on 3730xl Genetic Analyzer (Applied Biosystems, USA), separately.
Phylogenetic analysis
Phylogenetic analysis was performed using the software MEGA, version 4, after multiple alignments of data using CLUSTAL X. Pairwise evolutionary distances were estimated by Kimura's two-parameter method, and a phylogenetic tree was constructed by the neighbor-joining method. The reliability of the topologies was estimated by performing bootstrap analysis (1000 replicates). All phylogenetic analysis was performed only for NDM-1 gene.
All CRGNB positive patients were kept isolated from the other patients, and all the hospital staff were advised to practice transmission-based precautions and other universal precautions with more conscious attention to hand hygiene.
RESULTS
A total of 116 consecutive, nonduplicate CRGNB were included in the study, out of which 85 (73.2%) were isolated from pus, 17 (14.6%) from urine and 14 (12.1%) from fluid. Citrobacter species 28 (24.1%) predominated followed by Escherichia coli 26 (22.4%), Pseudomonas aeruginosa 24 (20.6%), Serratia species 17 (14.6%), Klebsiella species 9 (7.7%), Acinetobacter species 5 (4.3%), Proteus species 4 (3.4%), Providencia species 2 (1.7), and Aeromonas species 1 (0.86%). Fractures 29 (29.5%), G. I. tract surgical site infections 27 (27.5%), and skin and soft-tissue infections 13 (13.2%) were the predominant clinical conditions associated with patients who were admitted in the hospital.
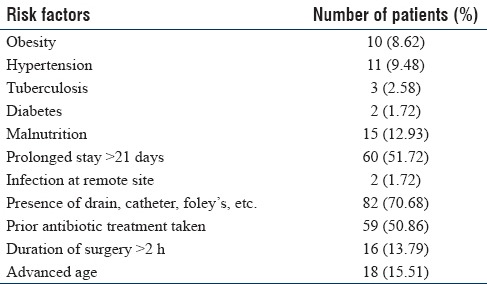
Table 2 describes the risk factors associated with hospitalized patients. It was observed that 82 (70.6%) patients had indwelling devices such as intravenous catheter and urinary catheters, 60 (51.7%) patients had prolonged stay in hospital >21 days and 59 (50.8%) patients were on prior antibiotic treatment. Risk factors associated with advanced age, surgery for >2 h duration, and malnutrition were lower 18 (15.5%), 15 (12.9%) and 16 (13.7%), respectively.
Table 2.
Risk factors in hospitalized patients for acquiring carbapenem-resistant Gram-negative bacilli (n=116)
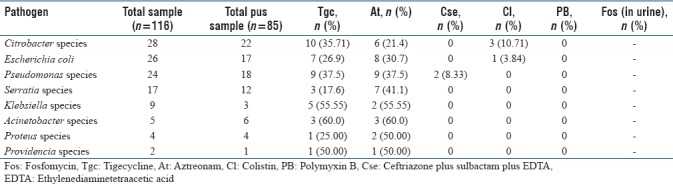
In the current study, we observed that all the study isolates (urine, pus, and fluids) were resistant to amikacin, gentamicin, levofloxacin, ceftriaxone, cefoperazone, cefepime, nitrofurantoin, ceftazidime, cefotaxime, cefpodoxime, cefoperazone/sulbactam, ceftriaxone/sulbactam, ceftazidime/clavulanic acid, ticarcillin, piperacillin, tobramycin, sparfloxacin, piperacillin-tazobactam, and carbapenems (IMP, ertapenem, doripenem, and meropenem). Among higher drugs- polymyxin B (100%), ceftriaxone plus sulbactam plus EDTA (CSE) (96.5%), and colistin (90.5%) were the only drugs found to be active against all CRGNB clinical isolates. High resistance was observed to aztreonam (51.7%), tigecycline (40.1%) and fosfomycin (35.2%). Antibiotic resistance pattern of the isolates against polymyxin B, ceftriaxone plus sulbactam plus EDTA (CSE), colistin, aztreonam, tigecycline, fosfomycin among pus, urine, and fluids are depicted in Tables 3–5, respectively.
Table 3.
Antimicrobial resistance pattern of carbapenem resistant Gram-negative bacilli among urine samples
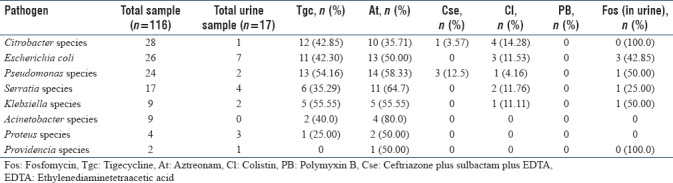
Table 5.
Antimicrobial resistance pattern of carbapenem resistant Gram-negative bacilli among fluid samples

Table 4.
Antimicrobial resistance pattern of carbapenem resistant Gram-negative bacilli among pus samples
MHT, IMP-EDTA CDST, and IMP-EDTA DDST phenotypically detected 105 (90.5%), 87 (75%) and 93 (81%) MBL producers, respectively. Sensitivity and specificity of MHT, IMP-EDTA DST, IMP-EDTA DDST was calculated with PCR as the gold standard. Sensitivity of MHT, IMP-EDTA DST, and IMP-EDTA DDST was 97.41%, 84.81%, 84.81%, whereas specificity was 29.73%, 59.49%, and 78.38%, respectively.
Only those isolates which were screened phenotypically as MBL producers were subjected to molecular detection of MBL genes. Of the 116 isolates, MBL genes were detected in 79 (68.1%) strains. Table 2 shows the prevalence of NDM-1, VIM, and IMP-1 genes in clinical specimen. NDM-1 was found to be the most prevalent MBL gene among all specimens 66 (83.5%), followed by VIM [12 (15.1%)], and IPM-1 (1 [1.2%]) [Table 6]. It was interesting to observe that all MBL producers in urine samples were NDM-1. On the other hand, maximum VIM gene positivity was seen in pus 10 (17.8%). IMP-1 was detected in ascitic fluid. Figures 1–3 show positive results of PCR for blaNDM, blaIMP, and blaVIM genes, respectively.
Table 6.
Prevalence of MBL genes in clinical specimen

Figure 1.
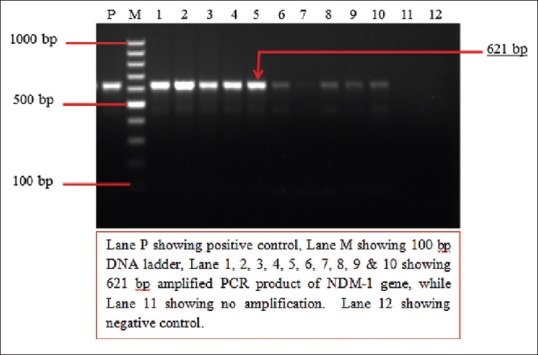
Two percent agarose gel electrophoresis showing results of polymerase chain reaction for the detection of blaNDM-1 gene
Figure 3.

Two percent agarose gel electrophoresis showing results of polymerase chain reaction for the detection of blaIMP gene
Figure 2.

Two percent agarose gel electrophoresis showing results of polymerase chain reaction for the detection of blaVIM gene
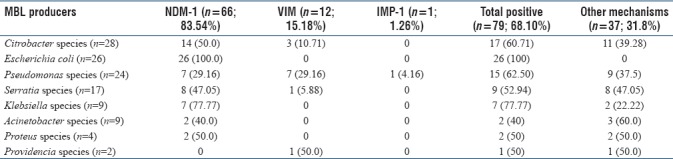
Table 7 shows the distribution of NDM-1, VIM, and IPM genes among various MBL producers. All E. coli 26 (100%) were positive for NDM-1 gene, followed by Klebsiella species 7 (77.7%), Citrobacter species 14 (50%), and Serratia species 8 (47%). Majority of VIM was observed in P. aeruginosa 7 (29.1%) followed by Citrobacter species 3 (10.7%). IMP was detected only in one isolate of P. aeruginosa.
Table 7.
Prevalence of NDM-1, VIM and IMP genes in various MBL producers
Results of blaNDM-1 gene were further confirmed by DNA sequencing for all the 11 representative strains (DNA extraction No. 90, 97, 103, 110, 111,115, 116, 119,139, 159, and176). After analyzing the nucleotide sequences of our study strains for NDM-1 gene, we prepared a phylogenetic tree with the help of MEGA 4 software. We observed that our most of the study isolates were showing the proximity with the reference sequences of NDM-1 gene from India as shown in Figure 4 while some patients were exhibiting slight relatedness with other reference NDM strains.
Figure 4.

Phylogenetic analysis of NDM-1 strains
DISCUSSION
The predominant CRGNB in ward patients were Citrobacter species (25.2%),E. coli (24.2%), and P. aeruginosa (18.2%) followed by Serratia species (13.1%) and Klebsiella species (8.1%), whereas in outpatients, the predominant CRGNB were P. aeruginosa (35.2%), Serratia species (23.5%), Citrobacter species (17.6%) and followed by E. coli (11.7%). Overwhelmingly, most isolates were from inpatients. Wankhede et al., 2011[13] reported that P. aeruginosa (23.7%), Acinetobacter spp. (18.4%), K. pneumoniae (8.3%), and E. coli (5%) were the most common MBL producers.
This study highlights that the drugs of choice for treating CRGNB are polymyxin B which was active against all isolates, ceftriazone-sulbactam plus EDTA, and colistin with 96.56% and 90.5% sensitivity, respectively. Unfortunately, high resistance was observed for aztreonam, tigecycline (40.1%), and fosfomycin. Thus, in our center, use of the latter drugs is discouraged. Behera et al., 2008[14] reported that MBL-positive isolates are usually resistant to all β-lactam antibiotics, aminoglycosides, tetracycline, and fluoroquinolones. However, they remain sensitive to polymyxin B. Nordmann et al.,2011[11] reported that therapeutic options against serious infections due to NDM-1 producers are limited to tigecycline and polymyxins although the former may not reach desired serum levels to treat systemic infections.
In this study, the 116 CRGNB isolates were subjected to phenotypic MBL detection. It was observed that 90.5% isolates were MBL positive by MHT. According to Cury et al.,2012,[15] MHT was positive in 35.5% of Enterobacteriaceae isolates that were not susceptible to ertapenem 71% of those isolates showed true-positive results, and 29% had inconclusive results. Phenotypic detection by IPM-EDTA-CDST showed that 75% isolates were MBL positive. Similar result was observed in a study from Vellore.[16] Phenotypic detection by IMP-EDTA DDST showed that 81% isolates were MBL positive. From our study, it was observed that among these standard phenotypic detection tests the least sensitive was CDST (75%), and MHT was the most sensitive (90.5%).
The high prevalence of CRE phenotype of Citrobacter and Serratia spp. may be due to hyperproduction of AmpC. This suggests that hyperproduction of AmpC results in a positive MHT. We suggest that in case of Citrobacter and Serratia spp., a positive phenotypic test need not suggest MBL production. They should be further subjected to genotypic detection.
The present study demonstrates that blaNDM-1 (56.8%) was the most prevalent MBL gene followed by blaVIM 10.3% and blaIMP (4.1%). Surprisingly, all MBL producers in urine samples were blaNDM-1. In our study, all E. coli (100%) were blaNDM-1 producers followed by Klebsiella species (77.7%) and Citrobacter species (50%). Amudhan et al., 2011[17] too reported that blaNDM-1 was the most prevalent MBL (57.6%) gene in their study. In other Indian studies, the prevalence of blaNDM-1 producers among carbapenem resistant Enterobacteriaceae ranged between 31.2% and 91.6%.[12,18] Maximum blaVIM gene positivity was seen in pus 17.8% while blaIMP was found in ascitic fluid. Common blaVIM producing bacterial isolates in pus were P. aeruginosa 100% and Citrobacter species 66.6%. However, in fluids, Serratia species (100%) predominated followed by Citrobacter species (33.3%). BlaVIM was not detected in E. coli and Klebsiella species. However, 1 out of 2 isolates of Providencia species was positive for blaVIM. Further, spread of carbapenemase-producing P. aeruginosa strains would represent a significant threat for the future of β-lactams. Confirmation of blaIMP gene among our clinical isolates showed that only one P. aeruginosa species (4.16%) give positive result. The prevalence of MBL genotypes varies from one country to another. Two genotypes, namely, VIM and IMP are most common in Asian countries.
On comparing phenotypic and genotypic detection methods for MBL, only 68.1% isolates were confirmed as MBL producers by PCR. In our study, IPM-EDTA CDST correlated best with PCR results and thus is the most specific phenotypic test for MBL detection. However, an interspecies variation was also observed. For Citrobacter species, IPM-EDTA DDST with 78.5% correlation most closely matched with PCR 60.7% while in case of Escherichia coli, MHT (96.1%) was almost equivalent to PCR (100%). However, for Pseudomonas species, Serratia species and Klebsiella species, IPM-EDTA DST most closely mirrored with PCR results.
We suggest that a combination of MHT and IPM-EDTA DDST should be put up for more specific identification of MBL. Debasrita et al., 2011[19] too compared phenotypic and genotypic methods. They observed that 26% bacteria were false positive by phenotypic method. They suggested that three phenotypic methods should be positive before labeling bacteria MBL producing. However, gold standard remains PCR. Thus, amplification clearly highlighted that phenotypic tests wrongly identify some CRGNB as MBL producers.
Thus, 47.1% strains of Serratia species, 39.29% of Citrobacter species, and 37.5% of Pseudomonas species were not MBL producers. In all probability, these strains were Amp C hyper-producers which can be effectively treated with higher doses of IMP or doripenem. Other causes may be hyperproduction of CTX-M or presence of other MBLs such as KPC or OXA. Porin defects or overexpression of efflux pumps also may play a significant role.[11] Manoharan et al., 2015[12] reported that there are other resistance mechanisms involved apart from MBLs such as permeability mutations through the loss of porins or the upregulation of efflux systems, which may be missed by the E test or the MBL PCR.
On analysis of the phylogenetic tree, NDM gene sequences from our patients were found to be evolutionarily close to NDM-1 reference sequences. Most of the study subjects were found to be infected with NDM-1. As per our finding, other studies also support the dominance of NDM-1 strains in India.[20,21]
In our study, 77.2% patients improved clinically and discharged, 7.1% did not show any improvement while 4.04% died. As the majority of patients recovered (P < 0.001), it appears that MBL carrying bacteria may have low virulence, or on the other hand, the patients are immunocompetent as the cases were drawn from wards and not Intensive Care Units (ICUs). Since this was a study in patients admitted in wards (not seriously ill patients), their robust immune response may also have played a significant role in recovery. However, recovery did take time as is manifested by the increased duration of hospital stay (P < 0.001). Thus, in a non ICU patient, morbidity due to MBL infection increased; however, mortality was significantly low. It may be premature to think that NDM-1 is a resistant but a less virulent pathogen, although some clinical facts point toward this possibility.[22,23]
CONCLUSION
Development of quick, effective molecular diagnostic techniques for identification and epidemiological surveillance of resistance genes can significantly improve treatment protocols, which are currently guideline based. Such improved strategies can effectively intervene to prescribe on a case-to-case basis, in other words tailoring it to the requirements of the individual patient.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Queenan AM, Bush K. Carbapenemases: The versatile beta-lactamases. Clin Microbiol Rev. 2007;20:440–58. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Livermore DM, Woodford N. Carbapenemases: A problem in waiting? Curr Opin Microbiol. 2000;3:489–95. doi: 10.1016/s1369-5274(00)00128-4. [DOI] [PubMed] [Google Scholar]
- 3.Nordmann P, Boulanger AE, Poirel L. NDM-4 metallo-β-lactamase with increased carbapenemase activity from Escherichia coli. Antimicrob Agents Chemother. 2012;56:2184–6. doi: 10.1128/AAC.05961-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: An evolving crisis of global dimensions. Clin Microbiol Rev. 2012;25:682–707. doi: 10.1128/CMR.05035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paterson DL, Doi Y. A step closer to extreme drug resistance (XDR) in gram-negative bacilli. Clin Infect Dis. 2007;45:1179–81. doi: 10.1086/522287. [DOI] [PubMed] [Google Scholar]
- 6.Forbes BA, Sahm DF, Weissfeld AS. 12th ed. St. Louis, Missouri: Mosby Elseiver; 2007. International. Bailey and Scott's Diagnostic Microbiology. [Google Scholar]
- 7.Collee GJ, Marmion PB, Fraser GA. 14th ed. London, England: Churchill Livinstone; 2007. Simmons A Mackie & McCartney Practical Medical Microbiology. [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Seventeenth Informational Supplement: Approved Standards M100-S20. 2010 [Google Scholar]
- 9.Lee K, Chong Y, Shin HB, Kim YA, Yong D, Yum JH, et al. Modified Hodge and EDTA-disk synergy tests to screen metallo-beta-lactamase-producing strains of pseudomonas and acinetobacter species. Clin Microbiol Infect. 2001;7:88–91. doi: 10.1046/j.1469-0691.2001.00204.x. [DOI] [PubMed] [Google Scholar]
- 10.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, et al. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53:5046–54. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011;17:1791–8. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manoharan A, Chatterjee S, Mathai D, SARI Study Group Detection and characterization of metallo beta lactamases producing Pseudomonas aeruginosa. Indian J Med Microbiol. 2010;28:241–4. doi: 10.4103/0255-0857.66486. [DOI] [PubMed] [Google Scholar]
- 13.Wankhede SV, Iyer VS, Bharadwaj RS. The study of MBL producers in Gram negative isolates from ICUs and wards. Indian J Basic Appl Med Res. 2011;1:38–46. [Google Scholar]
- 14.Behera B, Mathur P, Das A, Kapil A, Sharma V. An evaluation of four different phenotypic techniques for detection of metallo-beta-lactamase producing Pseudomonas aeruginosa. Indian J Med Microbiol. 2008;26:233–7. doi: 10.4103/0255-0857.39587. [DOI] [PubMed] [Google Scholar]
- 15.Cury AP, Andreazzi D, Maffucci M, Caiaffa-Junior HH, Rossi F. The modified Hodge test is a useful tool for ruling out Klebsiella pneumoniae carbapenemase. Clinics (Sao Paulo) 2012;67:1427–31. doi: 10.6061/clinics/2012(12)13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jesudason MV, Kandathil AJ, Balaji V. Comparison of two methods to detect carbapenemase & metallo-beta- lactamase production in clinical isolates. Indian J Med Res. 2005;121:780–3. [PubMed] [Google Scholar]
- 17.Amudhan MS, Sekar U, Kamalanathan A, Balaraman S. Bla(IMP) and bla(VIM) mediated carbapenem resistance in Pseudomonas and Acinetobacter species in India. J Infect Dev Ctries. 2012;6:757–62. doi: 10.3855/jidc.2268. [DOI] [PubMed] [Google Scholar]
- 18.Athanassios T, Spyros P, Neil W, Gioia PS, Douboyas J, Livermore DM. Outbreak of Infections Caused by Pseudomonas aeruginosa Producing VIM-1 Carbapenemase in Greece. J Clin Microbiol. 2000;38:1290–2. doi: 10.1128/jcm.38.3.1290-1292.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Debasrita C, Basu S, Das S. Study on some Gram negative multi drug resistant bacteria and their molecular characterization research article. Asian J Pharm Clin Res. 2011;4:108–12. [Google Scholar]
- 20.Bharadwaj R, Joshi S, Dohe V, Gaikwad V, Kulkarni G, Shouche Y, et al. Prevalence of New Delhi metallo-β-lactamase (NDM-1)-positive bacteria in a tertiary care centre in Pune, India. Int J Antimicrob Agents. 2012;39:265–6. doi: 10.1016/j.ijantimicag.2011.09.027. [DOI] [PubMed] [Google Scholar]
- 21.Mohan B, Hallur V, Singh G, Sandhu HK, Appannanavar SB, Taneja N, et al. Occurrence of blaNDM-1 & absence of blaKPC genes encoding carbapenem resistance in uropathogens from a tertiary care centre from North India. Indian J Med Res. 2015;142:336–43. doi: 10.4103/0971-5916.166601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kus JV, Tadros M, Simor A, Low DE, McGeer AJ, Willey BM, et al. New Delhi metallo-β-lactamase-1: Local acquisition in Ontario, Canada, and challenges in detection. CMAJ. 2011;183:1257–61. doi: 10.1503/cmaj.110477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen TL, Fung CP, Lee SD. Spontaneous eradication of a NDM-1 positive Klebsiella pneumoniae that colonized the intestine of an asymptomatic carrier. J Chin Med Assoc. 2011;74:104. doi: 10.1016/j.jcma.2011.01.022. [DOI] [PubMed] [Google Scholar]


