Abstract
Simo decoction (SMD), as a traditional medicine, is widely used in the treatment of gastrointestinal dysmotility in China. In this study, a combined method of liquid chromatography quadrupole time-of-flight mass spectrometry (LC-Q-TOF-MS) and ultrafiltration high-performance liquid chromatography molecular docking (UF-HPLC-MD) was efficiently employed to identify and screen bioactive ingredients in SMD. Ninety-four major constituents were identified or tentatively characterized by comparing their retention times and mass spectra with standards or literature data by using LC-Q-TOF-MS, and the ascription of those compounds were classified for the first time. Among them, 13 bioactive ingredients, including norisoboldine, eriocitrin, neoeriocitrin, narirutin, hesperidin, naringin, neohesperidin, hesperitin-7-O-glucoside, linderane, poncirin, costunolide, nobiletin, and tangeretin, were primarily identified as the human serum albumin (HSA) ligands at a range of docking scores from −29.7 to −40.6 kJ/mol by UF-HPLC-MD. The results indicate the systematic identification and screening of HSA ligands from Simo decoction guided by LC-Q-TOF-MS and UF-HPLC-MD represents a feasible and efficient method that could be extended for the identification and screening of other bioactive ingredients from natural medicines.
Keywords: quadrupole time-of-flight mass spectrometry, ultrafiltration, HSA ligands, molecular docking, Simo decoction
1. Introduction
As a traditional Chinese medicine prescription, Simo decoction (SMD) is composed of Semen arecae, Radix linderae, Radix aucklandiae, and Aurantii fructus. It has been used abundantly to regulate gastrointestinal function and bloating in clinical applications for a thousand years [1,2]. Literature shows that SMD combined some methods e.g., chewing gum or acupuncture, could enhance bowel function recovery, prevent postoperative ileus, and shorten hospital stay in postoperative patients [3,4]. The positive effects may be due to its participation of the regulation of gastrointestinal hormones of the digestive system, and promotion of gastrointestinal motility by promoting contraction of smooth muscle [5,6]. Despite many clinical treatments having been applied, arecoline, norisobodine, naringin, hesperidin, neohesperidin, and narirutin have been identified as the main effective components [7,8]. Besides, some compounds e.g., narirutin, naringin, hesperidin, neohesperidin, and nobiletin were detected in the plasma of rats [9]. However, chemical compounds of SMD were still not completely identified and systematically classified, and the bioactive ingredients should be further investigated in detail.
The degree to which a drug is protein-bound in plasma has a marked effect on its toxicological, pharmacological, and pharmacokinetic parameters. It is widely believed that only the free concentration, rather than the total drug concentration, can elicit pharmacological responses [10,11]. Human serum albumin (HSA) is the most abundant protein in the circumstance of blood circulation, playing a crucial role of the protein to transport and transmit many endogenous and exogenous constituents such as fatty acids, hormones, and drugs [12,13]. The binding affinity of HSA with drugs is connected to the efficiency of clinical treatment. Therefore, the binding affinity of HSA and drug is an essential parameter that should be carefully analyzed in drug studies [14]. In vitro means have been frequently applied to select HSA ligands from purified extracts of medicinal plants. However, trials based on active compounds need sophisticated and multiple isolation steps which are labor-intensive, time-consuming, and expensive [15]. With the advance of the analytical techniques for active ingredients in complex systems, one method based on ultrafiltration coupled with liquid chromatography mass spectrometry (UF-LC-MS) is considered to investigate the combination between HSA and bioactive compounds [11,16]. Due to its low sample consumption, reuse of receptors (e.g., HSA, enzymes), and obviated need for immobilization, bioactive ingredients have been high-throughput screened and identified via the UF-LC-MS technique [17]. This method enables an efficient separation of the binder–receptor complexes from unbound ingredients [18]. Besides, the binding affinity of the bioactive could be calculated by comparing the ultrafiltration chromatogram and reference chromatogram, by yielding the ratios of the unbound and total amount of single component [11,19]. The structure types could also be obtained by the MS/MS system. In addition, molecular docking has also been employed as a crucial tool to select bioactive components, and has exhibited efficient screening ability from multiple targets with a substantial degree of accuracy, time-saving, and cost-effectiveness in drug discovery [20,21]. It could therefore be an appropriate assistant in the ultrafiltration screening method.
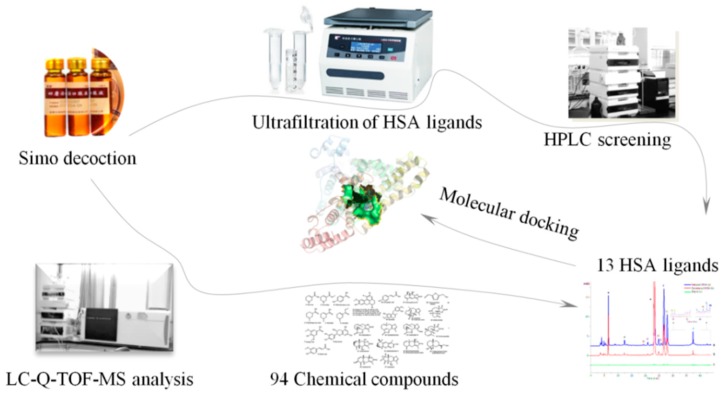
Inspired by the applications mentioned above, a simplified and efficient strategy on the strength of liquid chromatography quadrupole time-of-flight mass spectrometry (LC-Q-TOF-MS) and ultrafiltration high-performance liquid chromatography molecular docking (UF-HPLC-MD) to investigate the bioactive ingredients in SMD was developed, as depicted in Figure 1. To the best of our knowledge, this is the first time that LC-Q-TOF-MS and UF-HPLC-MD have been integrated in the identification and screening of major bioactive components from SMD. The LC-Q-TOF-MS technique could improve the fast detection of chemical compounds, while UF-HPLC-MD supports an approach for the recognition of bioactive ligands of HSA, predicting their binding sites and illustrating more information about the interaction mechanisms between receptor and active ligands [22]. The present study illustrates and explains the practical application of the bioactive compounds of SMD for the clinical treatment of gastrointestinal diseases.
Figure 1.
Strategy based on liquid chromatography quadrupole time-of-flight mass spectrometry (LC-Q-TOF-MS) and ultrafiltration high-performance liquid chromatography molecular docking (UF-HPLC-MD) method to identify and screen the bioactive ingredients in Simo decoction (SMD). HSA = human serum albumin; HPLC = high-performance liquid chromatography.
2. Materials and Methods
2.1. Chemicals and Reagents
SMD, used for gastrointestinal dysmotility in China (approval number: guo-yao-zhun-zi Z20025044; specification: 10 mL/division), was obtained from Hunan Hansen Pharmaceutical Company, Ltd. (Yi Yang, China). HSA was acquired from Sigma Chemical Co. (St. Louis, MO, USA), standards, including arecaidine, arecoline, norisoboldine, linderane, costunolide, dehydrocostus lactone, synephrine, rutin, limonin, eriocitrin, narirutin, naringin, hesperidin, heohesperidin, poncirin, naringenin, hesperetin, nobiletin, tangeretin, and sorbic acid with a purity of over 98%, were purchased from Yuan-ye Bio-Technology Co., Ltd. (Shanghai, China). The formic acid, acetonitrile, and methanol used for HPLC analysis were chromatographic grade and purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China).
2.2. HPLC Conditions
An Agilent 1260 HPLC system (Agilent Technologies, Palo Alto, CA, USA), equipped with a quat pump, an automatic sampler with a 20 μL sample loop, a thermostat of column, a diode array detector (DAD), and an Agilent ChemStation (Agilent Technologies, Palo Alto, CA, USA) had been employed to analyze samples. A Waters-XTerra™ C18 column (250 mm × 4.6 mm, 5 μm, Waters Corp., Milford, MA, USA) was performed for the chromatographic separation of SMD.
2.3. Q-TOF-MS Apparatus
Identification of mass spectrum was employed on an accurate mass spectrometer of Agilent 6530 Q-TOF-MS (Agilent Technologies, Palo Alto, CA, USA). Chromatographic separation was employed on an Agilent-ZORBAX SB-C18 column (250 mm × 4.6 mm, 5 μm, Agilent Technologies, Palo Alto, CA, USA), and the effluent of the HPLC mobile phase was split and guided into the electrospray ionization (ESI) source. Parameter conditions were performed as following: capillary voltage, 3500 V; nebulizer pressure, 50 psi; nozzle voltage, 1000 V; flow rate of drying gas, 6 L/min; temperature of sheath gas, 350 °C; flow rate of sheath gas, 11 L/min; skimmer voltage, 65 V; OCT1 RF Vpp, 750 V; fragmentor voltage, 135 V. The spectra data were recorded in the range of m/z 100–1000 Da in a centroid pattern of full-scan MS analysis mode. The MS/MS data of the selected compounds were obtained by regulating diverse collision energy (18–45 eV).
2.4. Sample Preparations
The SMD for ultrafiltration and LC-MS were filtered through a 0.22 μm membrane, then diluted to 1:10 V/V with a buffer solution of ammonium acetate buffer solution (ABS; 10 mM, pH 7.4) before experiments. The HSA (600 µM) was dissolved in ABS and prepared as the work solution
2.5. UF-HPLC-Based Binding Assay
The procedure of screening was manipulated according to the approach of previous research and consisted of three steps: incubation, washing, and dissociation [11,23]. Briefly, 100 µL of tested SMD solution was incubated with 200 µL HSA (600 µM) and 200 µL buffer solution for 20 min at 37 °C. Meanwhile, denatured HSA solution (boiled for 15 min in a water bath) was used as the negative control in the same manner. The incubated solutions were then filtered through ultrafiltration devices (Millipore Corp., Billerica, MA, USA) with a 30 kDa molecule weight cut-off membrane (Millipore AmiconUltra-0.5 mL, item: UFC503096) and centrifuged at 14,000× g to separate the non-specific ingredients from the HSA-ligand complexes for 15 min at room temperature. The residues were then washed with 200 µL of buffer solution by centrifugation to remove the unbound components three times. The ligands showing specific binding to HSA were then released from the mixtures by elution with 400 µL 50% methanol (pH = 3) for 20 min, and then centrifuged at 14,000× g for 15 min at room temperature, a process which was repeated twice. The dissociated filtrates were combined and added to 1000 µL by 50% methanol and then directly analyzed.
2.6. Molecular Docking Study
To further study the coactions of the bioactive ligands with HSA, a molecular docking study which could conjecture the interactions of ligands within the constraint of receptors binding sites was performed in silico. In the prediction, the initial three-dimensional structure of the HSA was acquired from the Protein Data Bank (PDB, ID: 1E7I). The binders and water molecules were removed from the crystal structure of HSA by using PyMOL (Schrödinger LLC, New York, NY, USA) [11]. The 3D structures of the ligands were drawn and converted using ChemBioDraw Ultra and ChemBio 3D Ultra (Cambridgesoft Corp., Waltham, MA, USA) [11].
The AutoDock Vina [24] was employed for the docking simulation of these ligands. The docking steps were performed according to the protocol described by Ma et al. [25], with some modifications. Due to the various molecule sizes, in the first round of docking, each grid computation was calculated covering all amino acid residues of HSA to recognize the binding sites, and the simulation was then performed with flexible docking of all molecules in HSA. The grid was then concentrated on the center of Sudlow’s site I (60 Å × 60 Å × 60 Å, 0.375 Å, central coordinates x = 30.938, y = 13.241, and z = 7.960) and Sudlow’s site II (60 Å × 60 Å × 60 Å, 0.375 Å, central coordinates x = 9.491, y = 5.575, and z = 18.576), respectively, to find the appropriate binding sites [11]. The calculation of docking score was repeated three times for each ligand. Finally, PyMOL was used to present the docking results.
3. Results and Discussion
3.1. Optimization of HPLC Conditions
Because of the complicated compositions of four main traditional Chinese medicines, the adequate separation of the aimed constituents is a challenging and essential procedure for HPLC analysis [23]. The SMD was rich in flavonoids, alkaloids, and lactone compounds, and therefore, in the HPLC analytical procedures, the separation conditions containing the mobile phase system, column detection wavelength (nearly higher absorption), temperature, and so on should be investigated. Acid is known to improve separation for constituents with hydroxyl groups by reducing the tailing of the chromatographic peaks. Therefore, formic acid was added to the mobile phase composed of solvents A (0.1% formic acid in water) and B (acetonitrile) [26], and a flow rate of gradient elution was elected at 0.7 mL/min. In consideration of the variety of constituents in SMD in previous pre-experiments, the solvent gradient of the mobile phase was finally optimized as follows: 15% B for 0–5 min, 15–20% B for 5–15 min, 20–25% B for 15–30 min, 25–65% B for 30–42 min, 65–90% B for 42–45 min. The programmed wavelength was selected at 284 nm via comparison of the higher absorption of the main compounds. The column temperature was maintained at 30 °C and the volume of injection was 5 µL.
3.2. Identification of Constituents in SMD
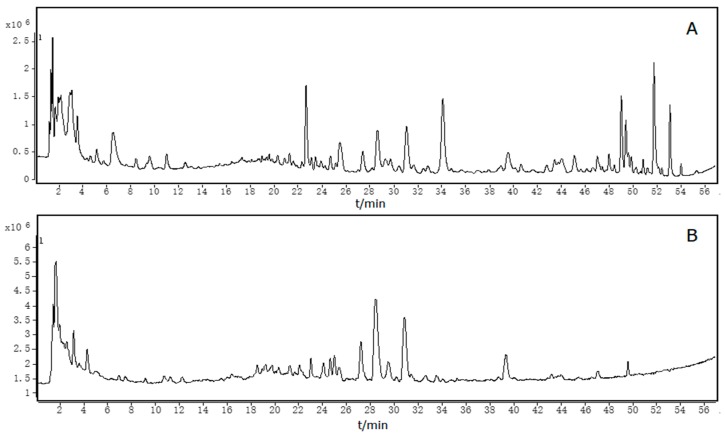
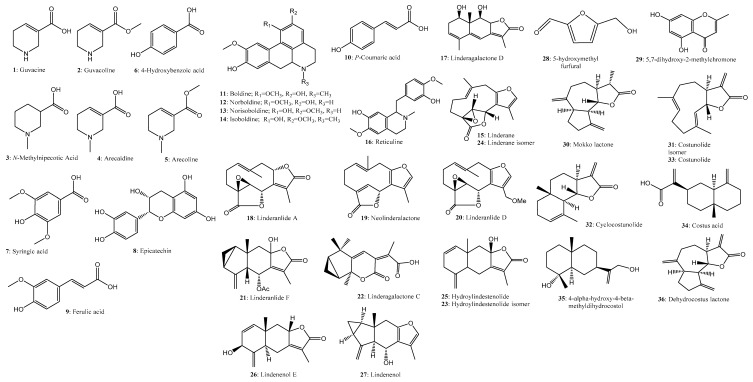
As many as 94 compounds were identified as the main constituents by ESI-Q-TOF-MS in the positive and negative ion mode (Figure 2), and their origin was classified according to the chemical information of single herb and literatures (Table 1). Compounds 4, 5, 13, 15, 33, 36, 38, 41, 46, 47, 53, 55, 57, 59, 67, 71, 73, 82, and 86 were unambiguously identified as arecaidine [27,28], arecoline [27,28], norisoboldine [29], linderane [30,31,32], costunolide [33,34], dehydrocostus lactone [33,34], synephrine [35], rutin [36], limonin [37,38,39], eriocitrin [36,40], narirutin [36,41], naringin [36,41], hesperidin [36,41], neohesperidin [36,41], poncirin [36,41], naringenin [36], hesperetin [36], nobiletin [36,42], and tangeretin [36,42], respectively, by comparison of the retention time, absorption wavelengths, and m/z values with the standards and values reported in the literature. The remaining compounds could be tentatively assigned by comparing the fragmentation patterns, the accurate mass data (absolute value of error < 5 ppm), and the formula predictor software (Table 1). The chemical structures of these compounds were drawn clearly as shown in Figure 3.
Figure 2.
Total ion chromatography of Simo decoction (SMD) in positive (A) and negative (B) modes.
Table 1.
Identification of constituents from Simo decoction (SMD) by liquid chromatography quadrupole time-of-flight mass spectrometry (LC-Q-TOF-MS) analysis in positive and negative ion modes.
| No. | TR (min) | ESI+ (m/z) | ESI− (m/z) | Fragment Ions (Positive/Negative) | MW (Mea.) | MW (MFG) | Formula | Compound | Ref. | Error (ppm) b |
|---|---|---|---|---|---|---|---|---|---|---|
| Areca catechu | ||||||||||
| 01 | 1.899 | 128.0704 | 109.0289 | 127.0631 | 127.0633 | C6H9NO2 | Guvacine | [27] | 1.61 | |
| 02 | 2.113 | 142.0860 | / | 141.0787 | 141.0790 | C7H11NO2 | Guvacoline | [27] | 1.96 | |
| 03 | 3.001 | 144.1021 | / | 143.0948 | 143.0946 | C7H13NO2 | N-Methylnipecotic Acid | [28] | −1.06 | |
| 04 | 5.163 | 142.0864 | 124.0252, 109.0289 | 141.0791 | 141.0790 | C7H11NO2 | Arecaidine a | [27,28] | −0.80 | |
| 05 | 7.021 | 156.1018 | 127.0410 | 155.0946 | 155.0946 | C8H13NO2 | Arecoline a | [27,28] | 0.24 | |
| 06 | 16.247 | 137.0240 | 138.0312 | 138.0317 | C7H6O3 | 4-Hydroxybenzoic acid | [43] | 3.23 | ||
| 07 | 17.508 | 199.0597 | 198.0525 | 198.0528 | C9H10O5 | Syringic acid | [44] | 1.62 | ||
| 08 | 19.379 | 291.0866 | 290.0793 | 290.0790 | C15H14O6 | Epicatechin | [44] | −0.93 | ||
| 09 | 25.337 | 193.0500 | 194.0572 | 194.0579 | C10H10O4 | Ferulic acid | [43] | 3.4 | ||
| Radix linderae | ||||||||||
| 10 | 5.722 | 165.0545 | 164.0472 | 164.0473 | C9H8O3 | p-Coumaric acid | [29] | 0.6 | ||
| 11 | 20.782 | 328.1546 | 327.1474 | 327.1471 | C19H21NO4 | Boldine | [29,31] | −0.91 | ||
| 12 | 21.558 | 314.1388 | 297.1125, 265.0839, 237.0743 | 313.1315 | 313.1314 | C18H19NO4 | Norboldine | [29,31] | −0.26 | |
| 13 | 22.677 | 314.1387 | 297.1141, 265.0787, 237.0619 | 313.1314 | 313.1314 | C18H19NO4 | Norisoboldine a | [29] | −0.12 | |
| 14 | 23.466 | 328.1543 | 297.1110, 265.0859, 237.0627 | 327.1470 | 327.1471 | C19H21NO4 | Isoboldine | / | 0.03 | |
| 15 | 24.093 | 261.1116 | 243.1018, 173.0132 | 260.1043 | 260.1049 | C15H16O4 | Linderane a | [30,31,32] | 2.07 | |
| 16 | 24.698 | 330.1698 | 330.1691, 299.1472, 192.0682 | 329.1625 | 329.1627 | C19H23NO4 | Reticuline | [29,31] | 0.57 | |
| 17 | 25.234 | 263.1270 | 262.1197 | 262.1205 | C15H18O4 | Linderagalactone D | [30,31] | 3.13 | ||
| 18 | 35.746 | 277.1068 | 276.0995 | 276.0998 | C15H16O5 | Linderanlide A | [32] | 1.01 | ||
| 19 | 40.231 | 245.1169 | 244.1096 | 244.1099 | C15H16O3 | Neolinderalactone | [30,31] | 1.50 | ||
| 20 | 40.590 | 291.1223 | 290.1150 | 290.1154 | C16H18O5 | Linderanlide D | [32] | 1.51 | ||
| 21 | 44.323 | 305.1376 | 304.1303 | 304.1311 | C17H20O5 | Linderanlide F | [32] | 2.54 | ||
| 22 | 46.623 | 263.1279 | 262.1207 | 262.1205 | C15H18O4 | Linderagalactone C | [30] | −0.57 | ||
| 23 | 47.462 | 247.1326 | 246.1254 | 246.1256 | C15H18O3 | Hydroylindestenolide isomer | [29] | 0.92 | ||
| 24 | 49.032 | 261.1117 | 260.1044 | 260.1049 | C15H16O4 | Linderane isomer | / | 1.64 | ||
| 25 | 50.118 | 247.1325 | 246.1252 | 246.1256 | C15H18O3 | Hydroylindestenolide | [30,31] | 1.53 | ||
| 26 | 50.126 | 247.1329 | 246.1256 | 246.1256 | C15H18O3 | Lindenenol E | [29] | 0.08 | ||
| 27 | 54.009 | 231.1380 | 230.1307 | 230.1307 | C15H18O2 | Lindenenol | [31] | −0.01 | ||
| Radix aucklandiae | ||||||||||
| 28 | 11.014 | 127.0388 | / | 126.0315 | 126.0317 | C6H6O3 | 5-HydroxymethylFurfual | [45] | 1.24 | |
| 29 | 25.283 | 193.0490 | 192.0417 | 192.0423 | C10H8O4 | 5,7-dihydroxy-2-methylchromone | [45] | 2.89 | ||
| 30 | 44.022 | 233.1534 | 232.1461 | 232.1463 | C15H20O2 | Mokko lactone | [46] | 0.91 | ||
| 31 | 46.101 | 233.1532 | 232.1459 | 232.1463 | C15H20O2 | Costunolide isomer | / | 1.84 | ||
| 32 | 47.095 | 233.1536 | 232.1464 | 232.1463 | C15H20O2 | Cyclocostunolide | [46] | −0.16 | ||
| 33 | 49.408 | 233.1530 | 187.1475, 121.0516 | 232.1457 | 232.1463 | C15H20O2 | Costunolide a | [33,34] | 2.75 | |
| 34 | 49.895 | 235.1691 | 234.1618 | 234.1620 | C15H22O2 | Costus acid | [46] | 0.71 | ||
| 35 | 51.221 | 239.2003 | 238.1930 | 238.1933 | C15H26O2 | 4-α-hydroxy-4-β-methyldihydrocostol | [46] | 1.2 | ||
| 36 | 54.001 | 231.1373 | 230.1301 | 230.1307 | C15H18O2 | Dehydrocostus lactone a | [33,34] | 2.69 | ||
| Aurantii fructus | ||||||||||
| 37 | 3.201 | 191.0189 | 192.0262 | 192.0270 | C6H8O7 | Citric acid | [37,47] | 4.06 | ||
| 38 | 3.802 | 168.1017 | 167.0944 | 167.0946 | C9H13NO2 | Synephrine a | [35] | 1.45 | ||
| 39 | 9.369 | 268.1035 | 267.0962 | 267.0968 | C10H13N5O4 | Adenosine | [37] | 2.03 | ||
| 40 | 17.108 | 196.0967 | 195.0894 | 195.0895 | C10H13NO3 | N-Acetylnorsynephrine | / | 0.8 | ||
| 41 | 20.439 | 611.1590 | 465.0874, 303.0511 | 610.1518 | 610.1534 | C27H30O16 | Quercetin-3-O-rutinoside (Rutin) a | [36] | 2.53 | |
| 42 | 21.337 | 595.1659 | 593.1500 | 594.1587 | 594.1585 | C27H30O15 | Isovitexin-7-O-glucoside (Saponarin) | [48] | −0.31 | |
| 43 | 21.903 | 625.1766 | 623.1615 | 301.0723 | 624.1693 | 624.1690 | C28H32O16 | Diosmetin-6,8-di-C-glucoside | [47] | −0.46 |
| 44 | 22.234 | 741.2245 | 579.1833, 417.1323, 271.0756 | 742.2318 | 742.2320 | C33H42O19 | Naringenin-7-O-triglycoside | [36,40] | 0.36 | |
| 45 | 22.377 | 625.1761 | 624.1688 | 624.1690 | C28H32O16 | Diosmetin 6,8-di-C-glucoside (isomer) | [47] | 0.41 | ||
| 46 | 23.947 | 471.2007 | 470.1935 | 470.1941 | C26H30O8 | Limonin a | [37,38,39] | 1.28 | ||
| 47 | 24.235 | 597.1813 | 595.1663 | 435.1278, 417.1185, 331.1826, 289.0702 | 596.1740 | 596.1741 | C27H32O15 | Eriodictyol-7-O-rutinoside (Eriocitrin) a | [36] | 0.12 |
| 48 | 24.460 | 481.1683 | 480.1610 | 480.1632 | C23H28O11 | Paeoniflorin, Albiflorin | [40] | 4.44 | ||
| 49 | 24.603 | 649.2501 | 650.2573 | 650.2575 | C32H42O14 | Limonin-17-β-d-glucoside | [39,49] | 0.16 | ||
| 50 | 25.124 | 597.1807 | 595.1656 | 451.1287, 289.0699 | 596.1735 | 596.1741 | C27H32O15 | Eriodictyol-7-O-neohesperidoside (Neoeriocitrin) | [36,37] | 1.06 |
| 51 | 26.946 | 461.1067 | 462.1140 | 462.1162 | C22H22O11 | Diosmetin-7-O-glucoside | [35] | 4.85 | ||
| 52 | 26.986 | 595.1653 | 463.1303, 287.0559 | 594.1579 | 594.1585 | C27H30O15 | Luteolin-7-O-rutinoside (Veronicastroside) | / | 0.9 | |
| 53 | 27.421 | 581.1853 | 579.1705 | 435.1274, 273.0757 | 580.1781 | 580.1792 | C27H32O14 | Naringenin-7-O-rutinoside (Narirutin) a | [36,41] | 1.97 |
| 54 | 27.989 | 625.2107 | 643.1461, 267.1224 | 624.2034 | 624.2054 | C29H36O15 | Magnoloside A | [37,47] | 3.31 | |
| 55 | 28.694 | 581.1857 | 579.1687 | 435.1278, 419.1330, 273.0754, 153.0186 | 580.1785 | 580.1792 | C27H32O14 | naringenin-7-O-neohesperidoside (Naringin) a | [36,41] | 1.26 |
| 56 | 29.032 | 435.1274 | 273.0757 | 434.1201 | 434.1213 | C21H20O10 | Naringenin-7-O-glucoside | 2.71 | ||
| 57 | 29.692 | 611.1965 | 609.1803 | 465.1432, 303.0858, 273.0757 | 610.1891 | 610.1898 | C28H34O15 | Hesperetin-7-O-rutinoside (Hesperidin) a | [36,41] | 1.13 |
| 58 | 30.385 | 579.1708 | 577.1549 | 433.1323, 271.0596 | 578.1636 | 578.1636 | C27H30O14 | Apigenin-7-O-rutinoside (Isorhoifolin) | [40] | −0.01 |
| 59 | 31.051 | 611.1962 | 609.1811 | 465.1434, 303.0862, 153.0188 | 610.1889 | 610.1898 | C28H34O15 | Hesperetin-7-O-neohesperidoside (Neohesperidin) a | [36,41] | 1.37 |
| 60 | 31.121 | 465.1395 | 331.1881, 303.0861, 155.0372, 121.0216 | 464.1322 | 464.1319 | C22H24O11 | Hesperitin-7-O-glucoside | [37] | −0.73 | |
| 61 | 31.638 | 609.1819 | 463.1409, 301.0723 | 608.1747 | 608.1741 | C28H32O15 | Diosmetin-7-O-rutinoside (Diosmin) | −0.88 | ||
| 62 | 32.515 | 609.1806 | 463.1411, 301.0723 | 608.1734 | 608.1741 | C28H32O15 | Diosmetin-7-O-neohesperidoside (Neodiosmin) | [47] | 1.12 | |
| 63 | 32.531 | 693.2756 | 694.2829 | 694.2837 | C34H46O15 | Nominin-17-β-d-glucoside | [39,49] | 1.12 | ||
| 64 | 32.787 | 651.1541 | 652.1614 | 652.1639 | C29H32O17 | Obacunoic acid-17-β-d-glucoside | [39] | 3.92 | ||
| 65 | 33.509 | 711.2850 | 712.2923 | 712.2942 | C34H48O16 | Nomilinic acid 17-O-β-d-glucoside | [39,49] | 2.74 | ||
| 66 | 34.361 | 261.1120 | 260.1047 | 260.1049 | C15H16O4 | Meranzin hydrate | [50] | 0.67 | ||
| 67 | 39.559 | 595.2016 | 593.1875 | 449.1505, 287.0917 | 594.1944 | 594.1949 | C28H34O14 | Isosakuranetin-7-O-neohesperidoside, (Poncirin) a | [36,41] | 0.77 |
| 68 | 39.546 | 287.0913 | 286.0840 | 286.0841 | C16H14O5 | Oxypeucedanin | [48] | 0.48 | ||
| 69 | 41.776 | 697.1975 | 696.1901 | 696.1902 | C31H36O18 | Isovitexin-7-O-xylocoside 2″-O-arabinoside | [40] | 0.05 | ||
| 70 | 42.803 | 728.3970 | 727.3896 | 727.3905 | C36H53N7O9 | Citrusin III | [35,51,52] | 1.15 | ||
| 71 | 43.149 | 271.0609 | 272.0682 | 272.0685 | C15H12O5 | Naringenin a | [36] | 1.18 | ||
| 72 | 44.264 | 725.2283 | 724.2210 | 724.2215 | C33H40O18 | Melitidin | [53] | 0.65 | ||
| 73 | 45.375 | 301.0714 | 302.0787 | 302.0790 | C16H14O6 | Hesperetin a | [36] | 1.27 | ||
| 74 | 47.003 | 704.3968 | 703.3895 | 703.3905 | C34H53N7O9 | Citrusin I | [52] | 1.40 | ||
| 75 | 47.187 | 329.1023 | 314.0762, 299.0543 | 328.0950 | 328.0947 | C18H16O6 | Monohydroxytrimethoxyflavone | [54] | −1.08 | |
| 76 | 47.976 | 355.1533 | 354.1460 | 354.1467 | C21H22O5 | Epoxybergamottin or Cnidicin | [55] | 2.01 | ||
| 77 | 48.440 | 359.1119 | 344.0877, 326.0771 | 358.1046 | 358.1053 | C19H18O7 | 5-Hydroxy-6,7,3′,4′-tetramethoxy-flavone | [54] | 1.74 | |
| 78 | 49.028 | 261.1117 | 260.1044 | 260.1049 | C15H16O4 | Meranzin, IsoMeranzin | [50] | 1.64 | ||
| 79 | 49.634 | 471.2005 | 470.1932 | 470.1941 | C26H30O8 | Limonin isomer | [37,38] | 1.76 | ||
| 80 | 50.227 | 373.1276 | 358.1024, 343.0811 | 372.1204 | 372.1209 | C20H20O7 | 5,7,8,3′,4′-Pentamethoxyflavone (Isosinensetin) | [36,54] | 1.44 | |
| 81 | 50.853 | 373.1278 | 358.1036, 343.0812 | 372.1205 | 372.1209 | C20H20O7 | 5,6,7,3′,4′-Pentamethoxyflavone (Sinensetin) | [54] | 1.04 | |
| 82 | 51.721 | 403.1385 | 388.1025, 373.1253 | 402.1312 | 402.1315 | C21H22O8 | 5,6,7,8,3′,4′-Hexamethoxyflavone (Nobiletin) a | [36,42] | 0.74 | |
| 83 | 51.847 | 433.1485 | 403.1021, 388.0773 | 432.1413 | 432.1420 | C22H24O9 | 3′,4′,3,5,6,7,8-Heptamethox-yflavone | [36] | 1.79 | |
| 84 | 52.030 | 343.1174 | 328.0927, 285.0749 | 342.1101 | 342.1103 | C19H18O6 | 5,6,8,4′-Tetramethoxyflavone | [54] | 0.63 | |
| 85 | 52.381 | 343.1175 | 328.0919, 313.0705 | 342.1102 | 342.1103 | C19H18O6 | 4′,5,7,8-Tetramethoxyflavone | [54] | 0.36 | |
| 86 | 53.099 | 373.1281 | 358.1007, 343.1182 | 372.1208 | 372.1209 | C20H20O7 | 5,6,7,8,4′-Pentamethoxyflavone, (Tangeretin) a | [36,42] | 0.25 | |
| Common Compounds | ||||||||||
| 87 | 1.711 | 116.0705 | 115.0632 | 115.0633 | C5H9NO2 | Proline | [38] | 0.83 | ||
| 88 | 2.849 | 118.0865 | 117.0792 | 117.0790 | C5H11NO2 | Valine | [38] | −1.83 | ||
| 89 | 4.328 | 132.1016 | 131.0943 | 131.0946 | C6H13NO2 | Isoleucine | [38] | 2.32 | ||
| 90 | 4.678 | 132.1019 | 131.0947 | 131.0946 | C6H13NO2 | Leucine | [38] | −0.32 | ||
| 91 | 5.813 | 182.0810 | 181.0737 | 181.0739 | C9H11NO3 | tyrosine | [38] | 0.83 | ||
| 92 | 9.561 | 166.0859 | 165.0786 | 165.0790 | C9H11NO2 | Phenylalanine | [38] | 2.36 | ||
| 93 | 17.316 | 205.0969 | 204.0896 | 204.0899 | C11H12N2O2 | Tryptophan | [38] | 1.34 | ||
| 94 | 25.552 | 113.0597 | 112.0524 | 112.0524 | C6H8O2 | Sorbic acid a | / | 0.25 | ||
a Compound identified with standards; b The error (ppm < 5 ppm) was obtained via the accurate mass data and formula predictor software of TOF mass spectrometer.MW (Mea.) = Molecular weight (measured); MW (MFG) = Molecular weight (molecular formula generated).
Figure 3.
The compounds were identified or preliminarily assigned from Simo decoction (SMD) based on the time-of flight-mass spectrometer.
For example, compound 12 had [M + H]+ ions m/z at 314.1372 yielding the product ions m/z at 297.1125 [M + H-17]+, 265.0839 [M + H-17-32]+, and 237.0743 [M + H-17-32-28]+, and compound 14 generated [M + H]+ ions m/z at 328.1534 yielding the similar product ions m/z at 297.1110 [M + H-31]+, 265.0859 [M + H-31-32]+, and 237.0627 [M + H-31-32-28]+, both of which had coincident ions with compound 13 and were tentatively assigned as norboldine [29,31] and isoboldine [29,31], respectively. Similarly, compound 16 was initiatively detected as reticuline according to the ions at 330.1691 [M + H]+, 299.1472 [M + H-31]+, and 192.0682 [M + H-138]+ [29,31].
The sugar parts in O-glycosylflavone, such as neohesperidose (1→2) and rutinose (1→6) could be distinguished because neohesperidose in glycosides could yield a stronger abundance of parent nuclei contrasted with rutinose-contained glycosides, and thus could be identified by their characteristic fragmentation behaviors [36]. For example, ion m/z at 273 of naringin was higher than naritutin, and ion m/z at 303 of neohesperidin was higher than hesperidin in ESI+ mode. Compounds 47 and 50 had the same [M + H]+ ions at m/z 597 and molecular formula of C27H32O15. Compound 50 presented product ions m/z at 451 [M + H-146]+ and 289 [M + H-146-162]+, exhibited the same ions with compound 47, and could yield a much higher abundance of fragment ions at m/z 289 when compared with that of compound 47 (eriocitrin), suggesting that it contained neohesperidose (1→2), and it was accurately identified as neoeriocitrin. Compound 60 showed [M + H]+ ion at m/z 465.1387 and [M − H]− ion at m/z 463.1252, and produced the parent ion at m/z 303.0861 [M + H]+, with this compound being preliminarily identified as hesperitin-7-O-glucoside. Compound 44 of m/z 741.2245 [M − H]− produced product ions at 579.1833 [M − H-162]–, 417.1323 [M − H-2 × 162]–, and 271.0756 [M − H-3 × 162]– that was identified as naringenin-7-O-triglycoside [36,40]. The remaining ingredients were similarly analyzed and classified by referring to the original medical plants of SMD as shown in Table 1.
3.3. Optimization of Screening Conditions
Working factors of pH and temperature influenced the activity of HSA, time of incubation influenced the binding degree of binders, eluting steps removed the disturbance of unbound compounds, and dissolution reagent was necessary for the dissociation of HSA-drug complexes. Optimum conditions in the study were established by referring to related studies [56,57,58] which performed single factor experiments in previous work. This study was performed at 37 °C (physiological temperature) with pH 7.4 (the plasma condition) to provide optimal reaction conditions. HSA concentration (from 100 μM to 800 μM), incubation time (from 0 min to 50 min), eluting time, and dissolution reagent (methanol of diverse concentration and pH) were improved before experiments. The results showed that each bioactive ligand could be obtained with the best binding affinity when the concentration of HSA was 600 μM to avoid competitive binding, and the incubation time was set at 20 min. ABS was employed as an eluting solution in triplicate, and a 50% methanol solution (pH 3) was optimally chosen to dissociate HSA-drug complexes.
3.4. Screening Bioactive HSA Ligands from SMD
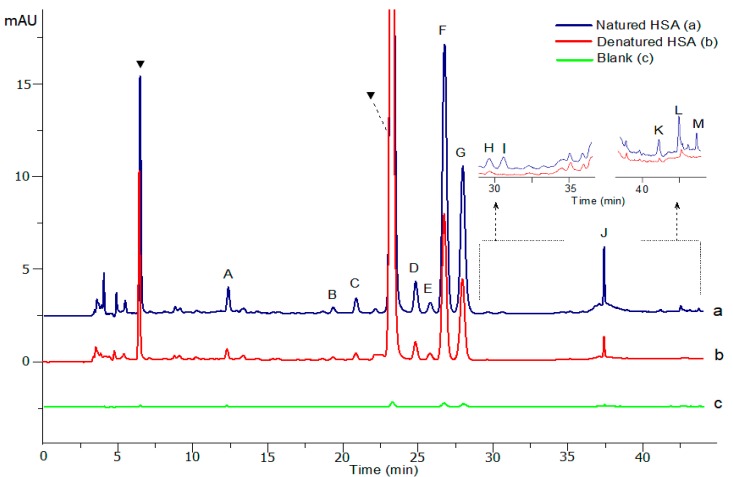
Using the above screening method, 15 binders were identified as “tight-binding” ligands (Figure 4). However, not all the binders bound to the HSA are specific ligands because some are just ‘‘frequent hitters’’, unselectively clogging the protein by hydrophobic interaction without any specific interactions [59], even though the washing procedures were performed. To distinguish between specific ligands and ‘‘frequent hitters’’, the method of ultrafiltration and dissociation could be efficiently combined. In short, if one compound in the complicated sample is able to interact with a specific target receptor, the peak area of the bound constituent will significantly increase in the total ion chromatogram after dissociation from the drug-protein complexes. In this way, the UF-HPLC assay could rapidly screen and identify the ligand-receptor complexes from unbound or nonspecific binding compounds, by directly comparing the chromatogram peak areas between natured and denatured HSA after ultrafiltration, as shown in (a) and (b) in Figure 4.
Figure 4.
The high-performance liquid chromatography (HPLC) chromatograms for screening of the human serum albumin (HSA) ligands in Simo decoction (SMD) after ultrafiltration and dissociation procedures. The blue solid line represents HPLC profile of SMD sample mixed with natured HSA, and the red and green lines represent HPLC profiles of SMD sample mixed with denatured HSA and buffer solution, respectively. Bioactive ligands (A–M) were identified as norisoboldine (A), eriocitrin (B), neoeriocitrin (C), narirutin (D), hesperidin (E), narigin (F), neohesperidin (G), hesperidin-7-O-glucoside (H), linderane (I), neoponcirin (J), costunolide (K), nobiletin (L), and tangeretin (M), respectively. Triangle (▼) represents a compound with a high response but no specific binding named “frequent hitters”.
Based on the variation of the chromatographic proportion before and after incubation with natured and denatured HSA, the real reduced peak areas can be used to determine the degree of affinity between the ligand and the enzyme. The binding degree (BD) was calculated as follows:
| (1) |
where As, An, and Ad represent the peak areas obtained from the SMD sample, natured and denatured HSA after dissociation, respectively. The results in Table 2 suggest that flavonoids and alkaloids could exert good affinity activity to HSA. Finally, 13 bioactive ingredients (A–M), containing norisoboldine, eriocitrin, neoeriocitrin, narirutin, hesperidin, naringin, neohesperidin, hesperitin-7-O-gulcoside, linderane, poncirin, costunolide, nobiletin, and tangeretin, were preliminarily identified as the specific HSA ligands.
Table 2.
Binding affinity (%) and docking score (kj/mol) of the ligands in Simo decoction (SMD) with human serum albumin (HSA).
| No. | Ligand | Binding Affinity | Docking Score | |
|---|---|---|---|---|
| Site I | Site II | |||
| A | Norisoboldine | 26.1 | −34.7 | −36.1 |
| B | Eriocitrin | 14.2 | −39.7 | −30.5 |
| C | Neoeriocitrin | 15.3 | −38.9 | −31.8 |
| D | Narirutin | 15.5 | −40.6 | −31.4 |
| E | Hesperidin | 11.6 | −39.3 | −30.1 |
| F | Naringin | 13.9 | −39.7 | −33.9 |
| G | Neohesperidin | 12.8 | −39.3 | −30.1 |
| H | Hesperitin-7-O-glucoside | 9.8 | −36.4 | −35.5 |
| I | Linderane | 22.5 | −34.3 | −36.0 |
| J | Poncirin | 16.7 | −38.9 | −31.0 |
| K | Costunolide | 19.6 | −33.5 | −35.9 |
| L | Nobiletin | 14.7 | −32.2 | −33.5 |
| M | Tangeretin | 12.9 | −31.4 | −34.7 |
| Drugs a | Warfarin | − | −33.5 | − |
| Ibuprofen | − | − | −32.2 | |
a The drugs warfarin and ibuprofen were specific ligands for site I and site II, respectively.
3.5. Repeatability of Ultrafiltration
Due to the potential for nonspecific binders to the HSA to lead to erroneous calculation, the repeatability of the bioactive ingredients in the SMD during ultrafiltration was studied. The repeatability showed the degree of affinity of these specific ingredients in SMD exhibiting large variety from 9.8–26.1% as shown in Table 2, which might be caused by their structure types and proportions. Besides, the bioactivities of the targets might be affected by not only the binding properties but also the drug-like properties [56], and the complicated components exhibited competitive relationships. Therefore, the binding degree in complex compounds might be different from that of single compounds. The relative standard deviation (RSD) of binding degrees of these binders was below 12.7%, indicating that each of the bioactive ligands could interact well with HSA.
3.6. Analysis of Molecular Docking
In order to expound how bioactive drugs conjugate with HSA, a molecular docking simulation was used for further illustration in the active sites and binding degree of the ligands on HSA. The interactions of drugs with HSA typically occur at two major hydrophobic sites, known as Sudlow’s site I (subdomain IIA) and site II (subdomain IIIA), which are located in subdomains [57]. Many studies have verified that warfarin and ibuprofen were specific binders for site I and site II, respectively. In this study, the mode of specific docking (grid in site I and site II) showed more specific affinity than the full grid mode, indicating that these components were appropriate ligands of HSA. The simulation scores of the drugs to the two binding sites are listed in Table 2.
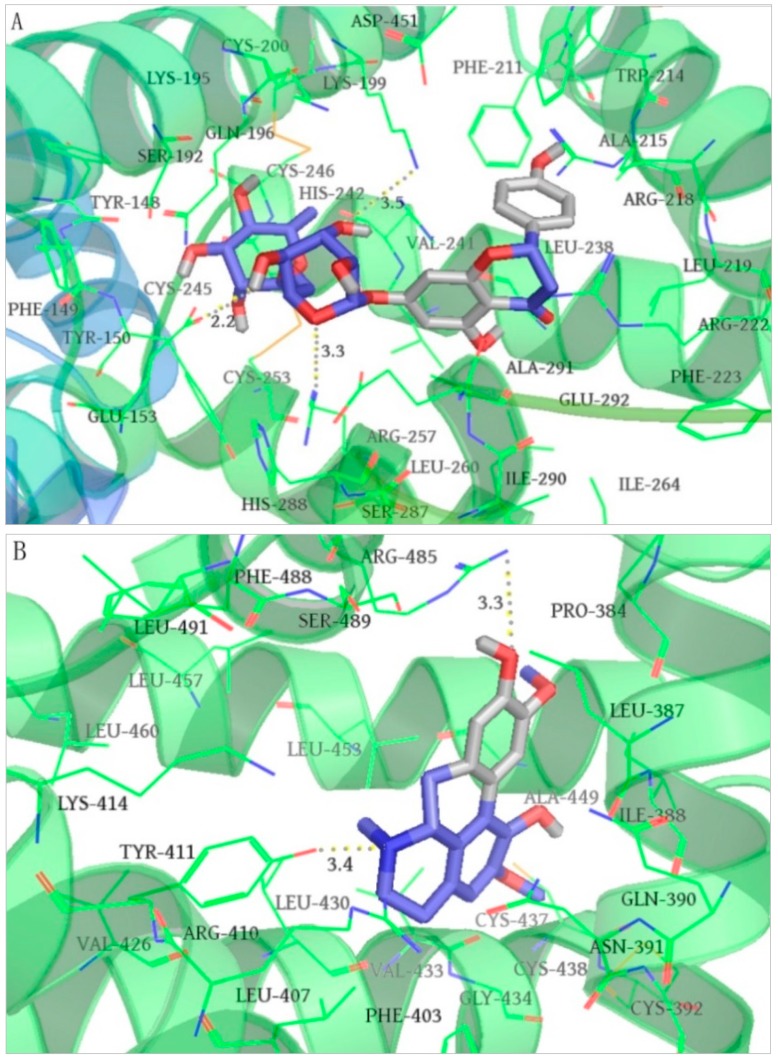
To further investigate the interactions between ligands and binding sites, the ligands, narirutin and norisoboldine, with the highest scores of binding site I and site II, respectively, were analyzed. As shown in Figure 5A, narirutin easily inserted into site I with a docking score of −40.6 kJ/mol and was mainly surrounded by 30 amino acid residues within a range of 4 Å. These residues are believed to be important in the binding affinity. Three hydrogen bonds (dash lines) were formed on the 7-rutinose with Glu153, Lys199, and Arg257, respectively. Hydrophobic bonds were generated on the mother nucleus of the flavanone surrounded with 14 amino acid residues, as follows: Phe211, Trp214, Ala215, Arg218, Leu219, Arg222, Phe223, Leu238, Val241, Arg257, Leu260, Ile264, Ile290, and Ala291. As shown in Figure 5B, norisoboldine efficiently bound with site II and was mainly surrounded by 25 amino acid residues within a range of 4 Å. Two hydrogen bonds were formed on the 6-N and 9-C (OH) with Tyr411 and Arg458, respectively. Pro384, Leu387, Ile388, Phe403, Leu407, Val426, Leu430, Val 433, Ala449, Leu453, Leu457, Leu460, Phe488, and Leu491 surrounded the molecule forming hydrophobic bonds with a docking score at −36.1 kJ//mol.
Figure 5.
Molecular docking of narirutin docked to site I (A) and norisoboldine docked to site II (B) of human serum albumin (HSA), respectively (Ligands were shown in stick form and gray dashed lines were hydrogen bonds. The figure was prepared with PyMol. The interactions between bioactive ligands and binding sites were detailed in the article).
It was found that the interactions between HSA and the flavonoids were dependent on the structures of the flavonoids. The glycoside of flavonoid was very important for the affinity degree, which mainly formed hydrogen bonds, while the A, B, and C rings supported hydrophobic bonds. In addition, in site I, as the number of methoxyl groups decreased, the affinity degree might increase. In site II, the docking scores of polymethoxy flavonoids were higher than at site I. This phenomenon was consistent with the tendency of site I to bind bulky heterocyclic anionic compounds and site II to aromatic carboxylates [57]. Moreover, the effect of a hydroxyl on glycosyl was found to be less than that of a hydroxyl on the parent nucleus, perhaps because of the large area of steric hindrance [58]. Although the current simulation studies could be considered efficient and reasonable, we also expect to further apply more advanced methods, such as fluorescence or X-ray, to explain the mechanisms of interactions between the bioactive ingredients and the related receptors, as well as the establishment of animal models to illustrate the metabolic pathways of effective constituents to clarify the pharmacological effects of SMD in future research.
4. Conclusions
In the current study, we established a simplified and effective strategy based on LC-Q-TOF-MS and UF-HPLC-MD for the identification of complicated ingredients and the screening of bioactive HSA ligands from SMD. A total of 94 compounds were identified or tentatively speculated by LC-Q-TOF-MS. Among them, nine compounds were derived from Semen arecae, 28 compounds were derived from Radix linderae, nine compounds were derived from Radix aucklandiae, and 40 compounds were derived from Aurantii fructus, in addition to the speculation of a further eight common compounds (e.g., amino acids). Flavonoids were abundant in these identified compounds in SMD (Table 1). In addition, HSA binders from SMD were screened by the established UF-HPLC-MD method. A total of 13 bioactive ingredients was primarily illustrated as the specific HSA ligands in SMD which may be the main medicinal components. Molecular docking was employed for further illustration in the active site and binding degree of bioactive ligands on HSA.
SMD is widely used in the clinical treatment of gastrointestinal dynamic disorder, and these results provide reliable data to support the pharmacological research of SMD in the future. They also provide a reference for the reasonable combination of SMD with other methods or drugs in the treatment of gastrointestinal dysmotility. In addition, and compared with the conventional bioassay approach, the proposed strategy enables the rapid illustration of the identification and screening of bioactive components from complex mixtures.
Author Contributions
Y.H., H.X. and D.L. conceived and designed the experiments; Y.H., P.C., and W.W. performed the experiments and designed the figures; Y.H., W.W., S.Y., and Q.T. analyzed and helped in data interpretation; Y.H., H.X., and D.L. wrote the paper.
Funding
This research was funded by the Program of International Science & Technology Cooperation of Ministry of Science and Technology (No. 2013DFA31790, No. 2013DFG32060), the Project of Hunan Science and Technology Innovation (No. S2017SFXYZ0116), the Major Projects of Hunan Provincial Science and Technology Department (No. 2015SK1001), the Project of Hunan Postgraduate Research and Innovation (No. CX2016B311), and the China Agriculture Research System (No. CARS-21).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of the compounds are available from the authors.
References
- 1.Chen S.P., Wang X.P. Effect of Simotang oral liquid on anal exhaust in patients after abdominal gynecological operation. Chin. J. Integr. Med. 2006;12:221–223. doi: 10.1007/BF02836528. [DOI] [PubMed] [Google Scholar]
- 2.Chong T., Jin H.B., Zhang J.D., Tan Y. Clinical effects of the treatment of gastrointestinal dysfunction after stable thoracolumbar fractures with Simo decoction oral liquid. Zhongguo Gu Shang. 2010;23:595–597. [PubMed] [Google Scholar]
- 3.You X.M., Mo X.S., Ma L., Zhong J.H., Qin H.G., Lu Z., Xiang B.D., Wu F.X., Zhao X.H., Tang J., et al. Randomized Clinical Trial Comparing Efficacy of Simo Decoction and Acupuncture or Chewing Gum Alone on Postoperative Ileus in Patients With Hepatocellular Carcinoma After Hepatectomy. Medicine. 2015;94:e1968. doi: 10.1097/MD.0000000000001968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Y., Zuo H.Q., Li Z., Qin Y.Z., Mo X.W., Huang M.W., Lai H., Wu L.C., Chen J.S. Comparison of efficacy of simo decoction and acupuncture or chewing gum alone on postoperative ileus in colorectal cancer resection: A randomized trial. Sci. Rep. 2017;7:37826. doi: 10.1038/srep37826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai G.X., Liu B.Y., Yi J., Chen X.M., Liu F.L. Simotang enhances gastrointestinal motility, motilin and cholecystokinin expression in chronically stressed mice. World J. Gastroenterol. 2011;17:1594–1599. doi: 10.3748/wjg.v17.i12.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dai C., Liu N., Chen W., Qian W., Hou X. Simo decoction promotes contraction of antral circular smooth muscle mainly via muscarinic M3 receptor. J. Ethnopharmacol. 2012;144:270–276. doi: 10.1016/j.jep.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 7.Yi Y.N., Cheng X.M., Liu L.A., Hu G.Y., Wang Z.T., Deng Y.D., Huang K.L., Cai G.X., Wang C.H. Simultaneous determination of synephrine, arecoline, and norisoboldine in Chinese patent medicine Si-Mo-Tang oral liquid preparation by strong cation exchange high performance liquid chromatography. Pharm. Biol. 2012;50:832–838. doi: 10.3109/13880209.2011.637505. [DOI] [PubMed] [Google Scholar]
- 8.Yi Y.N., Cheng X.M., Liu L.A., Hu G.Y., Cai G.X., Deng Y.D., Huang K.L., Wang C.H. HPLC Fingerprint with Multi-components Analysis for Quality Consistency Evaluation of Traditional Chinese Medicine Si-Mo-Tang Oral Liquid Preparation. Chem. Res. Chin. Univ. 2011;27:756–763. [Google Scholar]
- 9.Zhang X., Han L., Liu J., Xu Q., Guo Y., Zheng W., Wang J., Huang X., Ren P. Pharmacokinetic Study of 7 Compounds Following Oral Administration of Fructus Aurantii to Depressive Rats. Front. Pharmacol. 2018;9:131. doi: 10.3389/fphar.2018.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berezhkovskiy L.M. On the calculation of the concentration dependence of drug binding to plasma proteins with multiple binding sites of different affinities: Determination of the possible variation of the unbound drug fraction and calculation of the number of binding sites of the protein. J. Pharm. Sci. 2007;96:249–257. doi: 10.1002/jps.20777. [DOI] [PubMed] [Google Scholar]
- 11.Zhu J., Yi X., Huang P., Chen S., Wu Y. Drug-protein binding of Danhong injection and the potential influence of drug combination with aspirin: Insight by ultrafiltration LC-MS and molecular modeling. J. Pharm. Biomed. Anal. 2017;134:100–107. doi: 10.1016/j.jpba.2016.11.028. [DOI] [PubMed] [Google Scholar]
- 12.He X.M., Carter D.C. Atomic structure and chemistry of human serum albumin. Nature. 1992;358:209–215. doi: 10.1038/358209a0. [DOI] [PubMed] [Google Scholar]
- 13.Gokara M., Sudhamalla B., Amooru D.G., Subramanyam R. Molecular interaction studies of trimethoxy flavone with human serum albumin. PLoS ONE. 2010;5:e8834. doi: 10.1371/journal.pone.0008834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vuignier K., Schappler J., Veuthey J.L., Carrupt P.A., Martel S. Drug-protein binding: A critical review of analytical tools. Anal. Bioanal. Chem. 2010;398:53–66. doi: 10.1007/s00216-010-3737-1. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y., Liu S., Liu Z. Screening and determination of potential xanthine oxidase inhibitors from Radix Salviae Miltiorrhizae using ultrafiltration liquid chromatography-mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2013;923–924:48–53. doi: 10.1016/j.jchromb.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Li S., Li S., Tang Y., Liu C., Chen L., Zhang Y. Ultrafiltration-LC-MS combined with semi-preparative HPLC for the simultaneous screening and isolation of lactate dehydrogenase inhibitors from Belamcanda chinensis. J. Sep. Sci. 2016;39:4533–4543. doi: 10.1002/jssc.201600703. [DOI] [PubMed] [Google Scholar]
- 17.Zhou H., Xing J., Liu S., Song F., Cai Z., Pi Z., Liu Z., Liu S. Screening and determination for potential alpha-glucosidase inhibitors from leaves of Acanthopanax senticosus harms by using UF-LC/MS and ESI-MS(n) Phytochem. Anal. 2012;23:315–323. doi: 10.1002/pca.1360. [DOI] [PubMed] [Google Scholar]
- 18.Chen G.L., Tian Y.Q., Wu J.L., Li N., Guo M.Q. Antiproliferative activities of Amaryllidaceae alkaloids from Lycoris radiata targeting DNA topoisomerase I. Sci. Rep. 2016;6:38284. doi: 10.1038/srep38284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin S., Ren Y., Fu X., Shen J., Chen X., Wang Q., Bi X., Liu W., Li L., Liang G., et al. Multiple ligand detection and affinity measurement by ultrafiltration and mass spectrometry analysis applied to fragment mixture screening. Anal. Chim. Acta. 2015;886:98–106. doi: 10.1016/j.aca.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 20.Ferreira L.G., Dos Santos R.N., Oliva G., Andricopulo A.D. Molecular docking and structure-based drug design strategies. Molecules. 2015;20:13384–13421. doi: 10.3390/molecules200713384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma X.H., Shi Z., Tan C., Jiang Y., Go M.L., Low B.C., Chen Y.Z. In-silico approaches to multi-target drug discovery: Computer aided multi-target drug design, multi-target virtual screening. Pharm. Res. 2010;27:739–749. doi: 10.1007/s11095-010-0065-2. [DOI] [PubMed] [Google Scholar]
- 22.Scotti L., Mendonca Junior F.J., Ishiki H.M., Ribeiro F.F., Singla R.K., Barbosa Filho J.M., Da Silva M.S., Scotti M.T. Docking Studies for Multi-Target Drugs. Curr. Drug Targets. 2017;18:592–604. doi: 10.2174/1389450116666150825111818. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y., Peng M., Liu L., Shi S., Peng S. Screening, identification, and potential interaction of active compounds from Eucommia ulmodies leaves binding with bovine serum albumin. J. Agric. Food Chem. 2012;60:3119–3125. doi: 10.1021/jf205135w. [DOI] [PubMed] [Google Scholar]
- 24.Trott O., Olson A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma X., He J., Yan J., Wang Q., Li H. Comparative analysis the binding affinity of mycophenolic sodium and meprednisone with human serum albumin: Insight by NMR relaxation data and docking simulation. Chem. Biol. Interact. 2016;248:52–59. doi: 10.1016/j.cbi.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 26.Qing Z.X., Zhao H., Tang Q., Mo C.M., Huang P., Cheng P., Yang P., Yang X.Y., Liu X.B., Zheng Y.J., et al. Systematic identification of flavonols, flavonol glycosides, triterpene and siraitic acid glycosides from Siraitia grosvenorii using high-performance liquid chromatography/quadrupole-time-of-flight mass spectrometry combined with a screening strategy. J. Pharm. Biomed. Anal. 2017;138:240–248. doi: 10.1016/j.jpba.2017.01.059. [DOI] [PubMed] [Google Scholar]
- 27.Srimany A., George C., Naik H.R., Pinto D.G., Chandrakumar N., Pradeep T. Developmental patterning and segregation of alkaloids in areca nut (seed of Areca catechu) revealed by magnetic resonance and mass spectrometry imaging. Phytochemistry. 2016;125:35–42. doi: 10.1016/j.phytochem.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 28.Lee H.H., Chen L.Y., Wang H.L., Chen B.H. Quantification of Salivary Arecoline, Arecaidine and N-Methylnipecotic Acid Levels in Volunteers by Liquid Chromatography-Tandem Mass Spectrometry. J. Anal. Toxicol. 2015;39:714–719. doi: 10.1093/jat/bkv077. [DOI] [PubMed] [Google Scholar]
- 29.Deng G.M., Xiang B., Xiao X.Q., Ge J.W., Chen Z., Yang L.P., Wei F. Study on Chemical Constituents of Lindera aggregate by GC-MS and UPLC-ESI-MS/MS. Zhong Yao Cai. 2016;39:2229–2236. [Google Scholar]
- 30.Wu Y., Zheng Y., Liu X., Han Z., Ren Y., Gan L., Zhou C., Luan L. Separation and quantitative determination of sesquiterpene lactones in Lindera aggregata (Wu-yao) by ultra-performance LC-MS/MS. J. Sep. Sci. 2010;33:1072–1078. doi: 10.1002/jssc.200900768. [DOI] [PubMed] [Google Scholar]
- 31.Wu Y.J., Zheng Y.L., Luan L.J., Liu X.S., Han Z., Ren Y.P., Gan L.S., Zhou C.X. Development of the fingerprint for the quality of Radix Linderae through ultra-pressure liquid chromatography-photodiode array detection/electrospray ionization mass spectrometry. J. Sep. Sci. 2010;33:2734–2742. doi: 10.1002/jssc.201000243. [DOI] [PubMed] [Google Scholar]
- 32.Qiang Y., Yang Z.D., Yang J.L., Gao K. Sesquiterpenoids from the root tubers of Lindera aggregata. Planta Med. 2011;77:1610–1616. doi: 10.1055/s-0030-1270922. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J., Hu X., Gao W., Qu Z., Guo H., Liu Z., Liu C. Pharmacokinetic study on costunolide and dehydrocostuslactone after oral administration of traditional medicine Aucklandia lappa Decne. by LC/MS/MS. J. Ethnopharmacol. 2014;151:191–197. doi: 10.1016/j.jep.2013.10.024. [DOI] [PubMed] [Google Scholar]
- 34.Zhang J., Gao W., Liu Z., Zhang Z. Identification and Simultaneous Determination of Twelve Active Components in the Methanol Extract of Traditional Medicine Weichang’an Pill by HPLC-DAD-ESI-MS/MS. Iran. J. Pharm. Res. 2013;12:15–24. [PMC free article] [PubMed] [Google Scholar]
- 35.Ye X., Cao D., Zhao X., Song F., Huang Q., Fan G., Wu F. Chemical fingerprint and metabolic profile analysis of Citrus reticulate ‘Chachi’ decoction by HPLC-PDA-IT-MS(n) and HPLC-Quadrupole-Orbitrap-MS method. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2014;970:108–120. doi: 10.1016/j.jchromb.2014.06.035. [DOI] [PubMed] [Google Scholar]
- 36.Lin Z., Wang H., Xu Y., Dong J., Hashi Y., Chen S. Identification of antioxidants in Fructus aurantii and its quality evaluation using a new on-line combination of analytical techniques. Food Chem. 2012;134:1181–1191. doi: 10.1016/j.foodchem.2012.02.161. [DOI] [PubMed] [Google Scholar]
- 37.Wang M., Li Y., Huang Y., Tian Y., Xu F., Zhang Z. Chemomic and chemometric approach based on ultra-fast liquid chromatography with ion trap time-of-flight mass spectrometry to reveal the difference in the chemical composition between Da-Cheng-Qi decoction and its three constitutional herbal medicines. J. Sep. Sci. 2014;37:1148–1154. doi: 10.1002/jssc.201301378. [DOI] [PubMed] [Google Scholar]
- 38.Wang S., Tu H., Wan J., Chen W., Liu X., Luo J., Xu J., Zhang H. Spatio-temporal distribution and natural variation of metabolites in citrus fruits. Food Chem. 2016;199:8–17. doi: 10.1016/j.foodchem.2015.11.113. [DOI] [PubMed] [Google Scholar]
- 39.Tian Q., Schwartz S.J. Mass spectrometry and tandem mass spectrometry of citrus limonoids. Anal. Chem. 2003;75:5451–5460. doi: 10.1021/ac030115w. [DOI] [PubMed] [Google Scholar]
- 40.Zhou W.J., Song J.Z., Fu W.W., Tan H.S., Bian Z.X., Xu H.X. Chemical comparison of two dosage forms of Hemp Seed Pills by UHPLC-Q-ToF-MS/MS and multivariate statistical techniques. J. Pharm. Biomed. Anal. 2013;84:59–68. doi: 10.1016/j.jpba.2013.05.034. [DOI] [PubMed] [Google Scholar]
- 41.Liu X., Gu Z., Guo Y., Liu J., Ma M., Chen B., Wang L. Rapid analysis of Aurantii Fructus Immaturus (Zhishi) using paper spray ionization mass spectrometry. J. Pharm. Biomed. Anal. 2017;137:204–212. doi: 10.1016/j.jpba.2017.01.034. [DOI] [PubMed] [Google Scholar]
- 42.Chen H.F., Zhang W.G., Yuan J.B., Li Y.G., Yang S.L., Yang W.L. Simultaneous quantification of polymethoxylated flavones and coumarins in Fructus aurantii and Fructus aurantii immaturus using HPLC-ESI-MS/MS. J. Pharm. Biomed. Anal. 2012;59:90–95. doi: 10.1016/j.jpba.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 43.Lee J., Chan B.L., Mitchell A.E. Identification/quantification of free and bound phenolic acids in peel and pulp of apples (Malus domestica) using high resolution mass spectrometry (HRMS) Food Chem. 2017;215:301–310. doi: 10.1016/j.foodchem.2016.07.166. [DOI] [PubMed] [Google Scholar]
- 44.Zhang W.M., Huang W.Y., Chen W.X., Han L., Zhang H.D. Optimization of extraction conditions of areca seed polyphenols and evaluation of their antioxidant activities. Molecules. 2014;19:16416–16427. doi: 10.3390/molecules191016416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang T., Wang H., Du G., Chen R. Study on chemical constituents from roots of Saussurea lappa. Zhongguo Zhong Yao Za Zhi. 2009;34:1223–1224. [PubMed] [Google Scholar]
- 46.Wei H., He C., Peng Y., Ma G., Xiao P. Chemical constituents of Dolomiaea souliei. Chin. J. Chin. Mater. Med. 2012;37:1249–1253. doi: 10.1007/s10600-014-0985-x. [DOI] [PubMed] [Google Scholar]
- 47.Brito A., Ramirez J.E., Areche C., Sepulveda B., Simirgiotis M.J. HPLC-UV-MS profiles of phenolic compounds and antioxidant activity of fruits from three citrus species consumed in Northern Chile. Molecules. 2014;19:17400–17421. doi: 10.3390/molecules191117400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Durand-Hulak M., Dugrand A., Duval T., Bidel L.P., Jay-Allemand C., Froelicher Y., Bourgaud F., Fanciullino A.L. Mapping the genetic and tissular diversity of 64 phenolic compounds in Citrus species using a UPLC-MS approach. Ann. Bot. 2015;115:861–877. doi: 10.1093/aob/mcv012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raman G., Cho M., Brodbelt J.S., Patil B.S. Isolation and purification of closely related Citrus limonoid glucosides by flash chromatography. Phytochem. Anal. 2005;16:155–160. doi: 10.1002/pca.835. [DOI] [PubMed] [Google Scholar]
- 50.Mencherini T., Campone L., Piccinelli A.L., Mesa M.G., Sanchez D.M., Aquino R.P., Rastrelli L. HPLC-PDA-MS and NMR characterization of a hydroalcoholic extract of Citrus aurantium L. var. amara peel with antiedematogenic activity. J. Agric. Food Chem. 2013;61:1686–1693. doi: 10.1021/jf302815t. [DOI] [PubMed] [Google Scholar]
- 51.Kuroyanagi M., Ishii H., Kawahara N., Sugimoto H., Yamada H., Okihara K., Shirota O. Flavonoid glycosides and limonoids from Citrus molasses. J. Nat. Med. 2008;62:107–111. doi: 10.1007/s11418-007-0198-8. [DOI] [PubMed] [Google Scholar]
- 52.Matsubara Y., Yusa T., Sawabe A., Iizuka Y., Takekuma S., Yoshida Y. Structures of new cyclic peptides in young unshiu (Citrus unshiu Marcov.), orange (Citrus sinensis Osbeck.) and amanatsu (Citrus natsudaidai) peelings. Agric. Biol. Chem. 1991;55:2923–2929. [PubMed] [Google Scholar]
- 53.Di Donna L., De Luca G., Mazzotti F., Napoli A., Salerno R., Taverna D., Sindona G. Statin-like principles of bergamot fruit (Citrus bergamia): Isolation of 3-hydroxymethylglutaryl flavonoid glycosides. J. Nat. Prod. 2009;72:1352–1354. doi: 10.1021/np900096w. [DOI] [PubMed] [Google Scholar]
- 54.Xing T.T., Zhao X.J., Zhang Y.D., Li Y.F. Fast Separation and Sensitive Quantitation of Polymethoxylated Flavonoids in the Peels of Citrus Using UPLC-Q.-TOF-MS. J. Agric. Food Chem. 2017;65:2615–2627. doi: 10.1021/acs.jafc.6b05821. [DOI] [PubMed] [Google Scholar]
- 55.Dugrand A., Olry A., Duval T., Hehn A., Froelicher Y., Bourgaud F. Coumarin and furanocoumarin quantitation in citrus peel via ultraperformance liquid chromatography coupled with mass spectrometry (UPLC-MS) J. Agric. Food Chem. 2013;61:10677–10684. doi: 10.1021/jf402763t. [DOI] [PubMed] [Google Scholar]
- 56.Wang Z., Kwon S.H., Hwang S.H., Kang Y.H., Lee J.Y., Lim S.S. Competitive binding experiments can reduce the false positive results of affinity-based ultrafiltration-HPLC: A case study for identification of potent xanthine oxidase inhibitors from Perilla frutescens extract. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2017;1048:30–37. doi: 10.1016/j.jchromb.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 57.Anguizola J., Debolt E., Suresh D., Hage D.S. Chromatographic analysis of the effects of fatty acids and glycation on binding by probes for Sudlow sites I and II to human serum albumin. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2016;1021:175–181. doi: 10.1016/j.jchromb.2015.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fu L., Sun Y., Ding L., Wang Y., Gao Z., Wu Z., Wang S., Li W., Bi Y. Mechanism evaluation of the interactions between flavonoids and bovine serum albumin based on multi-spectroscopy, molecular docking and Q-TOF HR-MS analyses. Food Chem. 2016;203:150–157. doi: 10.1016/j.foodchem.2016.01.105. [DOI] [PubMed] [Google Scholar]
- 59.Deng S., Xia L., Xiao H. Screening of alpha-glucosidase inhibitors from green tea extracts using immobilized enzymes affinity capture combined with UHPLC-QTOF MS analysis. Chem. Commun. 2014;50:2582–2584. doi: 10.1039/c3cc49681c. [DOI] [PubMed] [Google Scholar]