Abstract
With the advent of three-dimensional (3D) imaging technologies such as electron tomography, serial-block-face scanning electron microscopy and confocal microscopy, the scientific community has unprecedented access to large datasets at sub-micrometer resolution that characterize the architectural remodeling that accompanies changes in cardiomyocyte function in health and disease. However, these datasets have been under-utilized for investigating the role of cellular architecture remodeling in cardiomyocyte function. The purpose of this protocol is to outline how to create an accurate finite element model of a cardiomyocyte using high resolution electron microscopy and confocal microscopy images. A detailed and accurate model of cellular architecture has significant potential to provide new insights into cardiomyocyte biology, more than experiments alone can garner. The power of this method lies in its ability to computationally fuse information from two disparate imaging modalities of cardiomyocyte ultrastructure to develop one unified and detailed model of the cardiomyocyte. This protocol outlines steps to integrate electron tomography and confocal microscopy images of adult male Wistar (name for a specific breed of albino rat) rat cardiomyocytes to develop a half-sarcomere finite element model of the cardiomyocyte. The procedure generates a 3D finite element model that contains an accurate, high-resolution depiction (on the order of ~35 nm) of the distribution of mitochondria, myofibrils and ryanodine receptor clusters that release the necessary calcium for cardiomyocyte contraction from the sarcoplasmic reticular network (SR) into the myofibril and cytosolic compartment. The model generated here as an illustration does not incorporate details of the transverse-tubule architecture or the sarcoplasmic reticular network and is therefore a minimal model of the cardiomyocyte. Nevertheless, the model can already be applied in simulation-based investigations into the role of cell structure in calcium signaling and mitochondrial bioenergetics, which is illustrated and discussed using two case studies that are presented following the detailed protocol.
Keywords: Bioengineering, Issue 134, Cardiomyocyte, cardiac ultrastructure, electron microscopy, confocal microscopy, computational modelling, finite element modelling, systems biology, calcium signaling, mitochondria, bioenergetics
Introduction
Excitation-contraction coupling (ECC) in the heart refers to the important and intricate coupling between electrical excitation of the cardiomyocyte membrane and the subsequent mechanical contraction of the cell during each heartbeat. Mathematical models have played a key role in developing a quantitative understanding of the interlinked biochemical processes that regulate the action potential1, cytosolic calcium signaling2, bioenergetics3, and subsequent contractile force generation. Such models have also successfully predicted changes to the heartbeat when one or several of these biochemical processes undergo alterations4,5. The highly-organized ultrastructure of the cardiomyocyte has increasingly been recognized to play a critical role in the normal contractile function of the cell and the whole heart. Indeed, changes to the morphology and organization of components of cardiac ultrastructure occur in parallel to biochemical changes in disease conditions such as hypertrophy6, heart failure7, and diabetic cardiomyopathy8. Whether these structural changes are minor, adaptive, or pathological responses to the changing biochemical conditions is still largely unknown9. The inherently tight coupling between form and function in biology means that experimental studies alone cannot provide deeper insights than correlations between structural remodeling and cardiomyocyte function. A new generation of mathematical models that can incorporate the structural assembly of the sub-cellular components, along with the well-studied biochemical processes, are necessary to develop a comprehensive, quantitative understanding of the relationship between structure, biochemistry, and contractile force in cardiomyocytes. This protocol describes methods that can be used to generate structurally accurate finite element models of cardiomyocytes that can be used for such investigations.
The last decade has seen significant advances in 3D electron microscopy10, confocal11, and super-resolution microscopy12 that provide unprecedented, high-resolution insights into the nano-scale and micro-scale assembly of the sub-cellular components of the cardiomyocyte. Recently, these datasets have been used to generate computational models of cardiomyocyte ultrastructure13,14,15,16. These models use a well-established engineering simulation method, called the finite element method17, to create finite element computational meshes over which biochemical processes and cardiomyocyte contractions can be simulated. However, these models are limited by the resolution and detail that a microscopy method can provide in an image dataset. For example, electron microscopy can generate nanometer-level detail of cell structure, but it is difficult to identify specific proteins within the image that would be necessary to create a model. On the other hand, super-resolution optical microscopy can provide high contrast images at resolutions on the order of 50 nm of only a select few molecular components of the cell. Only by integrating complementary information from these imaging modalities can one realistically explore the sensitivity of function to changes in structure. Correlative light and electron microscopy is still not a routine procedure and it would still suffer the limitation that only a limited number of components could be stained in the immunofluorescence view and correlated with the electron microscopy view.
This protocol presents a novel approach18 that uses statistical methods19 to analyze and computationally fuse light microscopy information on the spatial distribution of ion-channels with electron microscopy information on other cardiac ultrastructure components, such as myofibrils and mitochondria. This produces a finite element model that can be used with biophysical models of biochemical processes to study the role of cardiomyocyte sub-cellular organization on the biochemical processes that regulate cardiomyocyte contraction. For example, this protocol could be used to create models from healthy and streptozotocin-induced diabetic cardiac myocytes to study the effect of structural remodeling on cardiac cell function that is observed in diabetic animal models8. An additional advantage of the statistical nature of the presented method is also illustrated in the protocol: the method can generate multiple instances of finite element geometries that closely mimic the experimentally observed variations in cell structure.
As an overview, the protocol steps include: (i) preparation of cardiac tissue for electron microscopy to generate 3D images with sufficient resolution and contrast; (ii) reconstruction and segmentation of 3D image stacks from electron microscopy data using a 3D electron microscopy reconstruction and image analysis software called IMOD20; (iii) using iso2mesh21 to generate a finite element mesh using the segmented data as input; (iv) using the novel algorithm and codes to map the distribution of ion channels onto the finite element mesh.
The premise of the approach to each step is outlined within the protocol, and representative results are provided in the accompanying figures. An overview is outlined specifying how the generated spatially detailed models can be used to study the spatial dynamics of calcium during ECC, as well as mitochondrial bioenergetics. Some of the current limitations of the protocol are discussed, as well as new developments that are underway to overcome them and further advance a quantitative understanding of the role of cell structure to cardiac systems biology. How these methods could be generalized to create finite element models of other cell types is also addressed.
Users of this protocol may skip step 1 and the reconstruction part of step 2 if they have access to a pre-existing electron microscopy image stack. Users who intend to acquire their data in collaboration with more experienced electron microscopists may wish to discuss and compare the fixation and staining procedures in step 1 with the expert to determine an optimal protocol for acquisition.
Protocol
All methods described here have been approved by the University of Auckland Animal Ethics Committee and the University of California San Diego Institutional Animal Care and Use Committee, where the tissue protocol was originally developed.
1. Experimental Preparation
Prepare stock solutions of 0.15 M and 0.3 M sodium cacodylate buffers at pH 7.4 according to Table 1.
- Prepare glutaraldehyde fixative (2% paraformaldehyde (PFA) + 2.5% glutaraldehyde + 0.2% tannic acid in 0.15 M sodium cacodylate buffer at pH 7.4) using components listed in Table 1.
- Dissolve 2 g of PFA powder in 20 mL of distilled water at 60 °C on a hotplate with constant stirring.
- Add 1 M NaOH solution until the solution is clear.
- Wait until the temperature of the solution is below 40 °C, then add 10 mL of glutaraldehyde and 20 mL of 0.3 M sodium cacodylate.
- Add 0.2 g of tannic acid and dissolve it in the solution.
- Add 50 mL of 0.15 M sodium cacodylate.
- Store three or four 20 mL scintillation vials of fixative in fridge at 4 °C, or on ice if used on the same day.
Prepare Tyrode's solution with 20 mM 2,3- butanedione monoxime (BDM), according to the recipe in Table 1.
- Construct a Langendorff apparatus inside a fume cupboard.
- Clamp two 100 mL plastic syringe tubes to two different retort stands, approximately 70 cm high from the stand base.
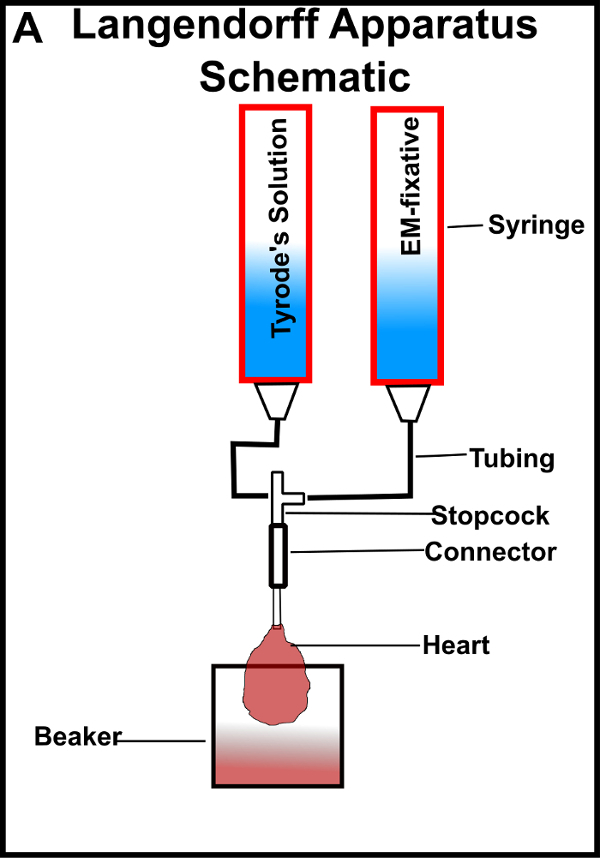
- Connect the plastic syringe tubes and a connector tube using 3 mm-inner-diameter tubing and a three-way stopcock as shown in Figure 1.
- Fit a 3 mm outer-diameter cannula to the bottom of the connector tubing, where the heart will be tied for perfusion fixation.
- Fill the syringe tubes with Tyrode's solution and fixative solutions at 37 °C.
- Remove air bubbles within the tubing by passing the syringe solutions through them.
2. Acquire Electron Microscopy Data of Cardiomyocyte Ultrastructure
- Prepare the animal for excision and chemical fixation of the heart. NOTE: This protocol details the steps to process adult male Wistar (name for a specific breed of albino rat) rat cardiac tissue. The protocol may require modifications when the cardiac tissue is sourced from other species.
- Anesthetize the animal by treating the rat (200-250 g by body weight) with 0.5 mL of 250 U/mL heparin via intraperitoneal injection. Wait for 10 min, then treat the rat with intraperitoneal injection of pentobarbital (210 mg/kg of body weight).
- Confirm a proper degree of anesthesia by testing the toe pinch reflex.
- Euthanize the animal by cervical dislocation.
- Isolate the ascending aorta and cannulate. Ensure that the tip of the cannula sits just above the semi-lunar valve, where the coronary arteries branch off. Tie a thread tightly around the ascending aorta.
- Connect the cannula to the gravity-driven Langendorff apparatus operated at ~70 cm above the base of the system (Figure 1).
- Twist the stopcock on the Langendorff system to perfuse the heart with Tyrode's solution, including 20 mM BDM (a myosin inhibitor) for 2-3 min.
- Twist the stopcock to begin perfusion with fixative of 2% paraformaldehyde, 2.5% glutaraldehyde, and 0.2% tannic acid in 0.15 M sodium cacodylate at 37 °C for 10 min. If successful, the heart will turn pale brown and become stiff and rubbery.
- Using a razor blade, dissect tissue blocks from the left ventricular (lateral) free wall and obtain tissue blocks approximately 1 mm3 in size.
- Store the samples in pre-cooled 20 mL scintillation vials of the same fixative on ice for 2 h. Store as many samples as possible in the vials, and ensure that the samples are submerged in the solution. CAUTION: It is important to keep samples cold until after the 100% dehydration step below.
- Further fixation and staining of samples for electron microscopy.
- Pour 100 mL of 0.15 M sodium cacodylate buffer in a glass beaker and cool it on ice.
- Prepare equal volumes of 4% osmium tetroxide and 0.3 M sodium cacodylate, followed by the addition of 0.08 g of potassium ferrocyanide to make a heavy metal stain solution containing 2% potassium ferrocyanide in 2% osmium tetroxide and 0.15 M sodium cacodylate. Cool the solution on ice.
- Pipette the fixative out and replace it with enough cold 0.15 M sodium cacodylate buffer to submerge samples in the vials. Place the vials on ice for 5 min.
- Repeat step 2.2.3 four more times with 0.15 M sodium cacodylate to remove excess fixative. CAUTION: Do not use glass pipettes because tiny shards can get into the sample and can ruin diamond knives during tissue block sectioning. It is important to pre-cool all solutions on ice before adding them to the sample. It is also important that the sample remains covered by solution all time (very brief periods of partial coverage lasting no more than a few seconds may be briefly tolerated during solution exchanges). When washing or replacing solutions, add the new solution quickly after removing the previous solution. Keep the scintillation vials on ice between washes. Note that this step is not time sensitive, the tissue blocks can be washed slightly longer, if needed.
- Replace sodium cacodylate buffer with ice-cold heavy metal stain solution and store it on ice overnight. Ensure that the ferrocyanide is well mixed by shaking the vial by hand; the solution should turn black. CAUTION: Work in the fume hood when performing this step because osmium is toxic.
- Dilute 4% aqueous uranyl acetate (UA) stock solution (Table 1) to 2% using equal volumes of stock UA solution and double distilled water, and cool it down on ice.
- Replace heavy metal stain with ice-cooled 0.15 M sodium cacodylate buffer, and let the sample incubate for 5 min on ice.
- Repeat step 2.2.7 four more times on ice to rinse off any excess heavy metal stain solution.
- Rinse the samples four times, with a 2-min wait-period in between in purified water (double-distilled is sufficient).
- Replace purified water with ice-cold 2% UA and let the sample incubate for 60-120 min on ice.
- Cool down five 20 mL scintillation vials of ethanol (enough to submerge samples) on ice that will be used in the dehydration step. The six vials have increasing percentages of ethanol in distilled water: 20%, 50%, 70%, 90%, and 100%.
- Pour pure acetone into another 20 mL scintillation vial and cool on ice along with the ethanol vials.
- Rinse samples in purified water four times with 2 min waiting periods on ice to wash excess UA.
- Dehydrate samples in ethanol.
- Dehydrate in cold ethanol series by successively replacing solution within the sample vial as follows, on ice: 20% ethanol for 10 min; 50% ethanol for 10 min; 70% ethanol for 10 min; 90% ethanol for 10 min; then twice in 100% ethanol for 10 min.
- Transition sample to room temperature by replacing the final 100% ethanol with cooled pure acetone and place the sample vial on a room-temperature lab bench for 10 min.
- Perform one more 10 min wash in pure acetone at room temperature to remove condensation that is observable within the vials. While incubating, make up the acetone/resin solutions.
- Replace the pure acetone in the vials with 50:50 pure acetone and epoxy resin (with proportions for epoxy resin components as stated by the manufacturer). Use all components from the same batch. Leave the resin-filled sample vials overnight on a rotor.
- Embed samples in resin.
- Replace 50:50 resin with 75% resin (25% acetone) and incubate for 3 h, followed by further incubation/infiltration in 100% resin for 4 h at room temperature.
- Replace the 100% resin with fresh 100% resin, and leave the sample vials overnight for deeper penetration of the resin into the tissue samples.
- Take samples out of the vials and place them in containers that are oven-friendly and have a flat base; aluminum or silver foil baking cups can be used for this purpose, for example.
- Gently pour (to avoid displacement of the samples) fresh resin over the samples (thus embedding samples in resin) and polymerize the resin and samples in an oven at 60 °C for 48 h.
- Re-orient resin blocks to image cells in cross-section.
- Obtain 1 µm thick test sections from the resin blocks using an ultramicrotome with a glass knife to assess cell orientation24.
- Stain the thick test sections for 20 s with 1% toluene blue and 1% borax.
- Examine muscle cell orientation under a bright field microscope.
- Image tissue sections using electron tomography.
- Obtain semi-thin sections (~300 nm) of the reoriented tissue blocks using a diamond knife and transfer the sections onto copper grids25.
- Stain the thick sections with 2% UA and Sato lead25.
- Apply colloidal gold particles on both sides of the sections25.
- Obtain sets of single or dual-axis tilt series of projected images from -70 ° to +70 °25.
Use IMOD20 to reconstruct the 3D image stack (a series of 2D images that provide the 3D information of a single electron tomography section) of the cell. This reconstructed stack will be segmented in the next step.
3. Segment Myofibrils and Mitochondria Regions from the EM 3D Image Dataset
Execute the IMOD program "3dmod".
Within the 3dmod graphical user interface, enter the address of the ".rec" or ".mrc" file that contains the 3D reconstructed image dataset of the cell (generated in step 2.7) into the entry box labeled "Image file(s):", then press "OK".
Under the "File" menu, select New Model, then save the file with an appropriate name using the "Save Model as"… menu item under "File". NOTE: An object in IMOD can contain a collection of segmented components that make up the "object". By default, a new model contains a new object with the id "#1".
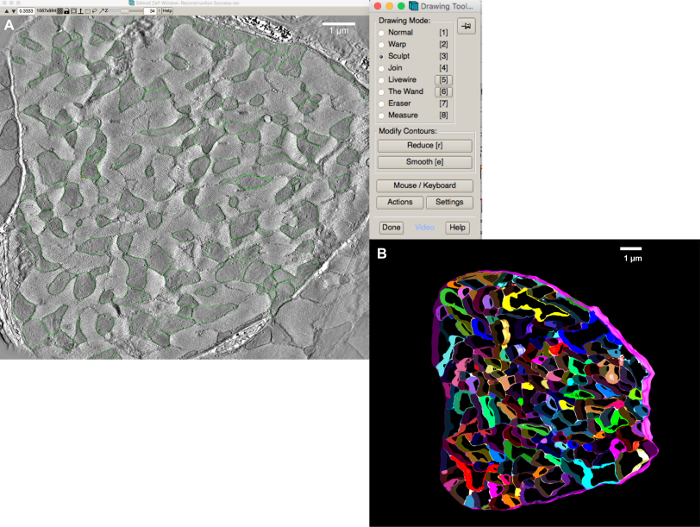
Under the "Special" menu, select "Drawing Tools". This will open a new tool bar similar to that shown in Figure 3A. NOTE: There are several tools that can be used to create contours around the different organelle boundaries. More help on ways to use these tools can be found in the IMOD20 documentation.
Choose the "Sculpt" option in the "Drawing Tools" menu, and move the mouse over to the image window; a circular contour centered on the mouse pointer will appear.
By holding the middle mouse button down over a mitochondrion (the darker regions of the image files, as illustrated by demarcations with green contour lines in Figure 3A), drag the perimeter of the circle contour into the shape of its boundary.
Once contouring of the mitochondrion boundary is complete, release the middle button and repeat steps 3.6 for each mitochondrion in the data. Each contour will automatically be recognized by IMOD as a new contour within the same object.
Under the "Edit | Object "menu, select "New" to create a new object. This will automatically increment the total number of objects by one and assign this number to the new object.
Repeat steps 3.6 and 3.7 to segment and save myofibril contours.
Repeat step 3.8, followed by step 3.6, to segment the cell boundary as well.
Save the model file under the "File" menu.
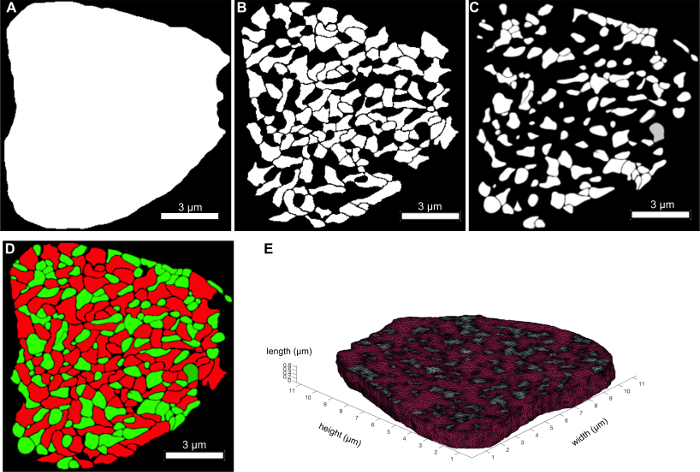
- Execute the following commands in a command-line window to convert each object grouping into a binary mask as illustrated in Figure 4A-C.
- Extract a specific object from the model "imodextract object modelfile outputmodelfile" where "object" is the number of the object to be extracted.
- Create a mask for that object: imodmop -mask 255 outputmodelfile imagefile outputmask.mrc
- Convert the file into a tif stack: mrc2tif -s outputmask.mrc outputmask.tif
4. Create a Finite Element Mesh from the Segmented Components
Iso2mesh is a freely available MATLAB program to convert TIFF image stacks into volumetric tetrahedral finite element meshes. Download and add iso2mesh to the MATLAB path from iso2mesh.sourceforge.net.
Download the source codes and data to simulate RyR clusters on the mesh from the github website https://github.com/CellSMB/RyR-simulator.
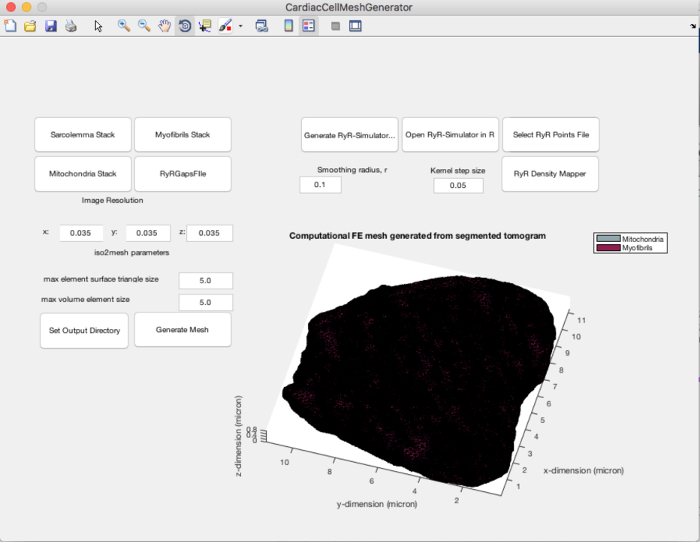
Start the CardiacCellMeshGenerator MATLAB application (Figure 5).
Load the different organelle component masks into MATLAB using the three push buttons on the upper left hand side of the GUI.
- Create another binary image stack that demarcates gaps between myofibrils and mitochondria as shown in Figure 6A.
- Open ImageJ.
- Using the File | Open dialog, load the myofibrils and mitochondria tiff stacks into the program.
- Initiate the image addition plugin by selecting "Process | Calculator Plus".
- Select the myofibril image stack as i1, the mitochondria image stack as i2, and choose the "Add" operator. Click "OK".
- After a new image stack representing the result of 4.5.4 appears, select "Edit | Invert" to produce an image stack similar to Figure 6A.
Load the file containing the binary image stack of the gaps between myofibrils and mitochondria by pushing the "RyRGapsFile" button on the CardiacCellMeshGenerator program.
Push "Generate Mesh" on the GUI. This will trigger the iso2mesh command v2m with the 'cgalmesh' option to generate a tetrahedral mesh similar to Figure 4E. Three files will be output at the end of this step: an .ele file, a .face file, and a .node file that will contain the listing of nodes that make up the elements, the nodes that make up the faces, and the coordinates of the nodes, respectively.
5. Mathematically Map the Spatially Varying Density of Ion-channels of Interest onto the Finite Element Mesh.
- Generate the necessary inputs for the RyR-Simulator by pushing the button labelled Generate RyR-Simulator inputs on the GUI. NOTE: The button will trigger a function generateRyRsimulatorInputs.m, which uses the following information from step 4 to generate the inputs: (1) outDir: the location to output files that are necessary for RyR cluster simulation. (2) imres: the pixel resolution in the three directions of the image stacks. (3) myofibril_file: the file containing the binary image stack like that shown in Figure 4B. (4) sarcolemma_file: the file containing the binary image stack like that shown in Figure 4A. (5) ryrgaps_file: the file containing the binary image stack like that shown in Figure 5A.
- After this function executes, check that the following files have been created within the directory specified as the outDir path:
- d_axial_micron.txt, which represents the axial distance between the position of the z-disc and the remainder of the pixels in the image stack.
- d_radial_micron.txt, which represents the Euclidean distance (excluding the axial component) from each pixel in the set of possible RyR cluster locations to the pixels on the z-disc plane.
- W_micron.txt, which represents the list of spatial coordinates of all the available positions for RyR clusters to be present.
- The remaining 3 files in the folder contain the suffix "_pixel" rather than "_micron" to denote that the values within these files have been written out in pixel coordinate form.
- Simulate RyR cluster distributions on the binary image stack of myofibrils.
- Push the button labelled "Open RyR-simulator in R" to initiate the R program.
- On the R-gui, select "File | Open" and find the file "settings.R" within the RyR-Simulator package (RyR-Simulator/source/settings.R).
- Also open the file ryr-simulator-parallel.R (located in RyR-Simulator/source/ryr-simulator-parallel.R). Environments like Rstudio (https://www.rstudio.com) or a plain command line interface can be used, e.g. using the R64 command in a shell or command window.
- Change the parameters in settings.R file as detailed below:
- Set Path2 to the folder address that contains files listed in 5.1.2 for an experimentally acquired confocal image stack of RyR clusters and myofibrils. NOTE: The github repository folder input-files/master-cell/ already contains files that were generated for a previously collected image stack.
- Set Path4 to a folder address where the files that were generated in step 5.1.2 are stored.
- Set Path3 point to a folder where the user wants the simulated RyR cluster locations to be saved.
- Set N to the number of RyR clusters to be simulated in the model (typically in the range of 200 to 300).
- Set etol, a tolerance setting, for the difference between the experimentally measured spatial distribution of RyR clusters and the model simulated spatial distribution of RyR clusters.
- Set numIter to limit the number of attempts that the RyR-Simulator should take to find a simulated RyR cluster pattern that satisfies the etol value. NOTE: Values for etol and numIter have been set to typical values within the settings.R file.
- Set numPatterns to the number of different RyR cluster patterns that the user wants to simulate (typically, it is useful practice simulating 99 patterns for statistical confidence).
- Set numCores to enable the use of several CPU threads (cores) for faster parallel processing with R to simulate the point patterns.
- Check that the following packages are installed using the package installer gui in R: snow, doSNOW, doparallel, foreach, iterators, and rgl.
- Execute the simulator by entering the following command in the R command window: source('path to ryr-simulator-parallel.R',chdir=TRUE) NOTE: The output of the RyR-Simulator program is a list of .txt files (a numPatterns file will be generated) that contain coordinate lists in N rows and 3 columns, which represent the x, y, and z coordinates of the N simulated RyR clusters.
- Map points as spatial densities onto a computational model using the CardiacCellMeshGenerator.
- By selecting the button labelled "Select RyR points file", choose a simulated RyR cluster distribution text file from those that were output by the RyR-Simulator.
- Execute "RyR Density Mapper" on the GUI in MATLAB. This will map the spatial locations of the simulated RyR clusters in the .txt file in step 5.3.1 onto the finite element mesh that was generated in step 4.8 using a method called a spherical kernel intensity estimator method26. NOTE: The output from this step is a file with .txt extension that contains a list of values for the numeric density of the ion-channel per unit spherical volume at each of the computational mesh nodes.
Representative Results
Figure 2 through Figure 7 provide representative results of several key steps in this protocol: (i) visualizing and reorienting tissue blocks for cross-sectional electron microscopy views; (ii) generating a 3D electron microscopy image stack; (iii) segmenting sub-cellular ultrastructure for organelles of interest; (iv) generation of a finite element mesh using iso2mesh; (v) simulating a realistic distribution of RyR clusters on the mesh; (vi) followed by mapping this spatial distribution as a spatial density over the computational mesh.
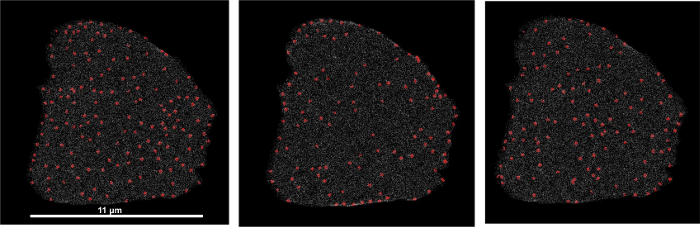
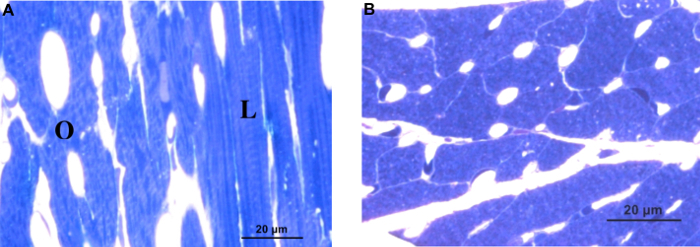
Figure 2 displays typical bright field images that show how cells appear when oriented longitudinally (Figure 2A, labelled L), obliquely (Figure 2A, labelled O) and cross-sectionally (Figure 2B) with respect to the cutting plane. Oblique and longitudinally oriented samples exhibit striations, while cross-sectionally oriented samples do not exhibit striations. The capillaries also appear more circular in cross-sectional views than in oblique views.
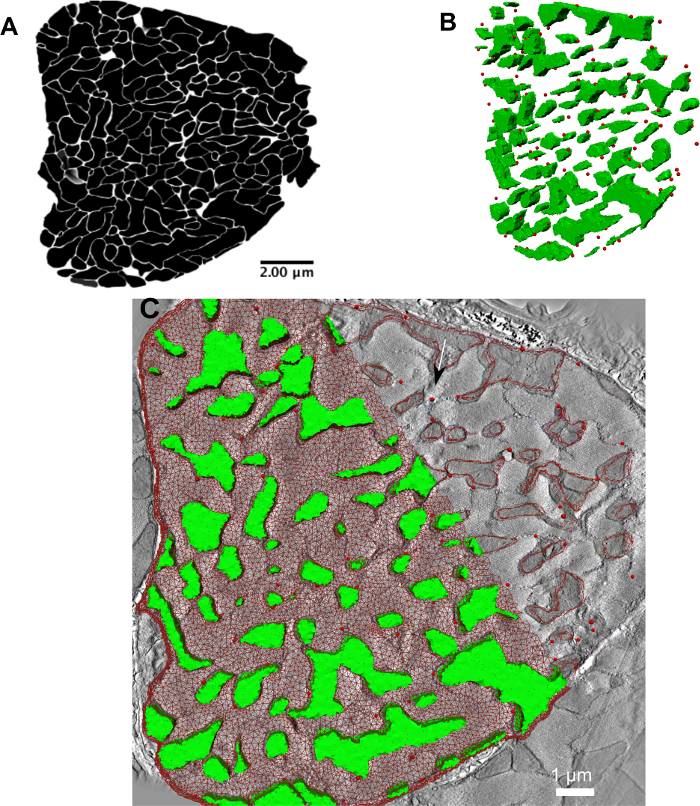
Figure 3A shows a representative image of a good quality tomogram stack that can be acquired when the tissue preparation protocol in step 1 is followed. Care must be taken when performing 3D reconstruction from the microscope image tilt series. Incorrect tracking of colloidal gold particles, or lack of sufficient gold particles can affect the quality — image contrast and image focus — of the tomogram considerably. This affects the ability to segment different structures significantly. Care and experience are necessary to ensure that the tissue blocks are stained sufficiently and that there is an even distribution of colloidal gold particles through the tissue volume. If in doubt, it may be useful to consult with a specialist laboratory or facility at the local institution. If model generation is attempted with low contrast or incorrectly reconstructed data, models may still be generated but the correspondence of modelling results with the properties of the biological systems to be simulated may be poor. Figure 3B shows a representative image of what a segmented model of myofibrils and mitochondria looks like.
Figure 4E shows a 3D rendering of the tetrahedral mesh that is produced by iso2mesh. As long as the original image stack and segmentation tasks are of good quality, this step is fairly robust. Figure 6B and Figure 6C show a typical RyR cluster distribution (in red) within the 3D cellular topology. At this stage, the mesh itself is not the input in the RyR simulator. An image stack of the cell boundary and the gaps between myofibrils and mitochondria — generated from the segmented images in Figure 4 — form the major inputs into the RyR simulator. Care must be taken to segment the boundaries of myofibrils and mitochondria as accurately as possible; inaccurate segmentations would result in an inaccurate representation of the cell topology in the data, which will consequently affect RyR placement within the mesh. This would then affect the spatio-temporal dynamics of calcium signaling in the cardiomyocyte (Application 1).
Figure 7 shows 3 instances of simulated RyR clusters on the same mesh topology after using the spherical kernel intensity estimator algorithm. Notice the variation in organization of the RyR clusters. These variations have been measured to be within the variability that was found in the experimental data18. Care must be taken in choosing the kernel sphere radius and the step size over which the kernel samples the RyR cluster density. These two factors affect how sharply or diffusely a representation of the density of RyR clusters will be depicted on the mesh. An analysis of the effect of different values for these parameters will help to narrow down on the parameter choices for a reasonable mesh.
The resulting mesh of RyR clusters, myofibrils, and mitochondria is a minimal model of the cardiomyocyte structure. This mesh has been applied, as well as variations of it that were derived from other cellular structure data, to study calcium dynamics and bioenergetics. An overview of these two applications is outlined below to illustrate the power of finite element modelling of cell structure in giving insights on structure-function relationships.
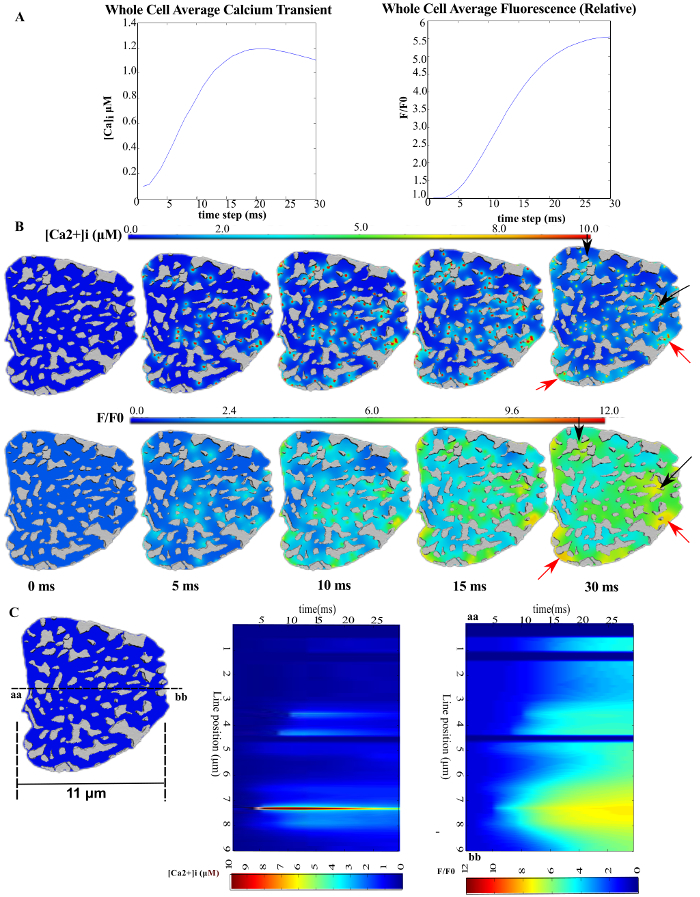
Application 118: Spatiotemporal dynamics of calcium release into cardiomyocytes. This model has been used to examine the role of heterogeneous distribution of RyR clusters, myofibrils, and mitochondria on calcium signaling. Figure 8 shows the spatiotemporal dynamics of cytosolic calcium during the rising phase of the transient when simulated on the mesh topology generated from this protocol. Figure 8B shows the heterogeneous free calcium and fluorophore-bound-calcium concentration over time. The arrows point to small spots of high calcium due to a higher density of RyR clusters or mitochondria in those regions. Figure 8C shows simulated line-scan images that can be compared to experimental line-scan images of calcium dynamics that are typically collected using live confocal microscopy. The model simulation provides a much higher resolution view of the spatial heterogeneity of calcium than an experimentally acquired image. Furthermore, due to the model being derived from structural microscopy data, the number of clusters that must remain inactivated (~4-6) after electrical activation for heterogeneities to occur in the confocal line-scan image could be precisely determined. Similar heterogeneities are observed in experimentally acquired line-scan images of calcium dynamics in animal models of heart failure18
Application 227: Spatiotemporal dynamics of mitochondrial bioenergetics in cardiomyocytes. An overarching aim of this work is to study the relationship between calcium dynamics, mitochondrial bioenergetics, and muscle contraction. As a first step, simulations of mitochondrial oxidative phosphorylation in isolation within the cardiomyocyte have been conducted. As such, the distribution of RyR clusters is not needed, and one can simply use the mesh at step 3.4. As a more complex application, the bioenergetics (application 2) could be coupled with calcium dynamics (application 1) in a model that uses all components considered in mesh and model construction, i.e. mitochondria, myofibrils, and ryanodine receptors.
Figure 9 represents the results from these spatial metabolism simulations over different regions of a mesh generated using step 3.4. Figure 9A depicts the oxygen concentration throughout the cell cross-section, while Figure 9B shows the concentration of ADP only in the myofibril compartment of the cell. Figure 9C depicts the distribution of mitochondrial membrane potential across the mitochondrial network. These figures reveal the non-uniform distribution of mitochondria and myofibrils in the cell cross-section. They also reveal the inhomogeneous metabolic landscape resulting from this non-uniform ultrastructure. This demonstrates the power of microscopy data based finite element simulations for giving insights on structure-function relationships that are otherwise difficult to obtain through experimental methods alone.
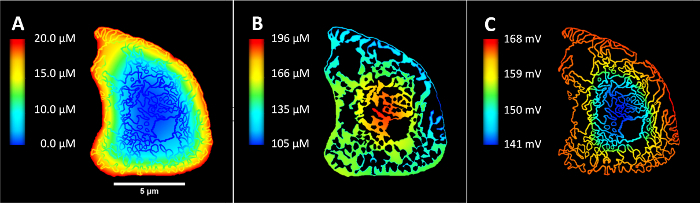
Figure 1: Langendorff preparation of the heart for chemical fixation. (A) Schematic of Langendorff perfusion apparatus. The stopcock contains a valve to switch between Tyrode's and fixative solutions. The beaker at the base is used to catch solutions that perfuse through the heart. Please click here to view a larger version of this figure.
Figure 2: Examining cell orientation under bright field microscopy. (A) Bright field view of toluene blue stained thick section of cardiac tissue illustrating cells that are longitudinally oriented (L) and obliquely oriented (O) with respect to the imaging plane; note the striations in the longitudinally oriented cell, for example. (B) Bright field view of a tissue section that has the transverse, cross-section of the cell aligned with the imaging plane. Please click here to view a larger version of this figure.
Figure 3: Segmentation of the electron microscopy data. (A) Snapshot of manual segmentation of mitochondria using the sculpt tool. (B) A 3D view of the complete manually segmented mitochondria, myofibrils (in a variety of colors), and sarcolemma (in pink). Please click here to view a larger version of this figure.
Figure 4: Converting binary image stacks into 3D tetrahedral mesh. (A-C) Binary masks of sarcolemma, myofibrils and mitochondria, respectively. (D) Merged view of the binary masks of myofibrils (in red) and mitochondria (in green). (E) Representative tetrahedral mesh that was generated from the image stack in (D) using iso2mesh. Please click here to view a larger version of this figure.
Figure 5: CardiacCellMeshGenerator. A screenshot of the graphical user interface that accompanies this protocol publication for users to perform steps 4 and 5. Please click here to view a larger version of this figure.
Figure 6: Simulating a realistic RyR cluster distribution onto a segmented electron microscopy image stack of cardiac ultrastructure. (A) A binary image stack depicting the gaps between myofibrils and mitochondria, which represent the set of all pixels where RyR clusters could be found. (B) A 3D rendering (image not drawn to scale) of one of the simulated RyR cluster distributions (shown as red spheres) within the 3D geometry depicted in the 3D image stack; the green surfaces represent the mitochondria in the image stack. (C) A higher-magnification (thus a small portion is cut out of the cell cross-section at the boundaries) view of the overlay of the RyR clusters from step 4 and the computational mesh from step 3 onto one slice of the 3D electron tomography image stack. (B) and (C) have been modified from Rajagopal et al.18 Please click here to view a larger version of this figure.
Figure 7: Spatial distribution of numeric density of RyR clusters from three simulated RyR cluster patterns. All mesh nodes are color coded from white for numeric density of 0.0 RyR clusters/µm3 to red for a maximum numeric density of 0.99 RyR clusters/µm3. Please click here to view a larger version of this figure.
Figure 8: Spatial heterogeneity in [Ca2+]i during the rising phase of the Ca2+ transient. (A) The average cytosolic freely diffusing [Ca2+]i (left) and Fluo-4 bound Ca2+, [F4Ca]i (right). (B) and (C) show time-lapse snapshots of freely diffusing Ca2+ and F4Ca at the z-disc, transverse plane; black arrows mark regions of high [Ca2+]i due to mitochondrial clustering; red arrows mark regions of high [Ca2+]i due to several RyR clusters in close proximity. (C) Simulated line scans of [Ca2+]i (middle) [F4Ca]i (right). The line position is shown on the cell cross-section on the left. The horizontal dimension of the cell cross-section has been provided as an indicator of the spatial scale in the cross-sections of the image. This figure has been modified from Rajagopal et al.18 Please click here to view a larger version of this figure.
Figure 9: Spatial distribution of metabolites and mitochondrial membrane potential generated from the spatial simulation of cardiac energy metabolism. (A) Oxygen concentration throughout the cell cross-section in units of µM, (B) Concentration of ADP only in the myofibril part of the cell in units of µM. (C) Distribution of mitochondrial membrane potential across the mitochondrial network in units of mV. Please click here to view a larger version of this figure.
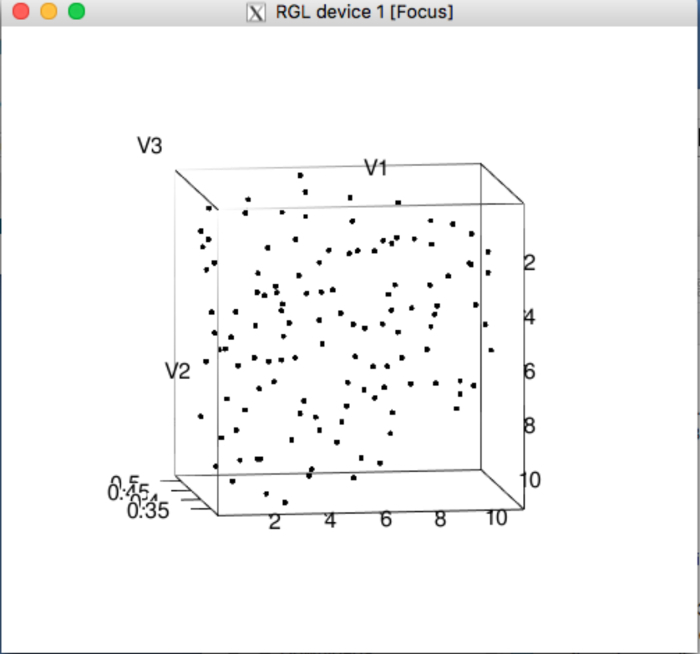
Figure 10: Visualization of a simulated RyR cluster point pattern on the R-statistics package. This window appears at the conclusion of the RyR clusters simulations, depicting the first of the simulated point patterns. This can be used as an indicator that the RyR-simulator has concluded as well, and may interrogate the quality of the simulated patterns. Please click here to view a larger version of this figure.
| Chemicals | Quantities |
| 0.2 N HCl | |
| 1 N HCl | 5 mL |
| Distilled water | 20 mL |
| 0.15 M Sodium cacodylate buffer stock | |
| Sodium cacodylate | 32.1 g [3H2O] |
| 0.2 N HCl | 25 mL |
| Make up 1 L with distilled water | |
| pH 7.4 | |
| 0.3 M Sodium cacodylate buffer stock | |
| Sodium cacodylate | 64.2 g [3H2O] |
| 0.2 N HCl | 25 mL |
| Make up 1 L with distilled water | |
| pH 7.4 | |
| Tyrode Solution Composition | |
| NaCl | 139 mM |
| KCl | 3 mM |
| CaCl2 | 3 mM |
| MgCl2 | 1 mM |
| Glucose | 12 mM |
| NaHCO3 | 17 mM |
| Probenecid | 1 mM |
| BDM | 20 mM |
| NaCl/KCl/NaHCO3 1 L Solution | |
| NaCl (Molecular Weight 58.44 g/mol) | 81.2 g |
| KCl (Molecular Weight 74.55 g/mol) | 2.24 g |
| NaHCO3 (Molecular Weight 84.01 g/mol) | 14.28 g |
| Dissolve in 1 L distilled water | |
| MgCl2/CaCl2 1 L solution | |
| CaCl2 (2H2O) (Molecular Weight 147 g/mol) | 1.764 g |
| MgCl2 (6H2O) (203.31 g/mol) | 0.814 g |
| Dissolve in 1 L distilled water | |
| 1 M glucose 200 mL solution | |
| Dextrose (Molecular Weight 180.16 g/mol) | 36.03 g |
| Dissolve in 200 mL double distilled water | |
| Tyrode Solution 500 mL solution | |
| NaCl/KCl/NaHCO3 | 50 mL |
| Glucose | 6 mL |
| Double distilled water | 400 mL |
| Bubble oxygen in solution for 5 min | |
| 2% PFA+ 2.5% Glutaraldehyde +0.2% tanic acid fixative in 0.15 M sodium cacodylate | |
| Paraformaldehyde powder (PFA) | 2 g |
| Tanic acid | 0.2 g |
| Distilled water | 20 mL |
| 25% Glutaraldehyde | 10 mL |
| 0.3 M sodium cacodylate | 20 mL |
| 0.15 M sodium cacodylate | 50 mL |
| 1 N NaOH | 4 g NaOH/100 mL distilled water |
| 4% Uranyl Acetate Stock Solution | |
| Uranyl Acetate | 4 g |
| Near boiling double distilled water | 96 mL |
| Dissolve UA in near-bioling water using a stirrer in a fumehood. | |
| Filter UA through a Whatman #1 filter into 200 mL brown bottle for light protection | |
| Cap tightly and label, store at 4 °C |
Table 1: Reagents and quantities for tissue preparation for electron microscopy.
Discussion
The above protocol outlines key steps to generate a novel finite element geometric model of cardiomyocyte ultrastructure. The method enables computational fusion of different microscopy (or, in principle, other data) modalities to develop a more comprehensive computational model of cardiomyocyte dynamics that includes details of spatial cell architecture. There is currently no other protocol available to create such a model of a cardiomyocyte.
Step 1 outlines a protocol for perfusion fixation. Alternatively, the heart could be excised, dissected into smaller pieces, and immersed in fixative. However, this process has to be done quickly and can have varied results, depending on the size of the samples. In the authors' experience, perfusion ensures thorough infiltration of the fixative into the tissue and cells. The authors have also previously perfused the heart with Krebs-Henseliet solution to make the heart beat before fixation. This enabled measurements on the working heart before fixation.
The electron microscopy processing steps in Step 2 are typical for electron tomography. The processing solution quantities, particularly of the staining solutions, and timings may differ when using other imaging techniques such as serial-block-face scanning electron microscopy or focused ion-beam microscopy28. The acquired electron microscopy images must be segmented into different organelle regions, such as mitochondria, myofibrils, and transverse tubules. This can be conducted manually or in an automated fashion. The contrast in the image stack is partly dependent on the success of the staining protocol, but some of this contrast is lost when performing tomography. This protocol describes manual steps for segmenting the different structures, although new methods to perform automatic segmentation29 will be more favorable in general to improve data throughput.
The most important qualities to examine in the electron microscopy dataset are the image contrast and structure preservation. The staining recipe and the resin embedding mixtures in this protocol have been refined for high contrast between myofibrils, mitochondria, and the z-discs of the cardiac cell. For example, the authors have previously used calcium chloride in place of tannic acid in step 1.2 to demarcate the sarcoplasmic reticular network. This helped with identifying the outer boundaries of myofibril bundles during organelle segmentation. Additional staining chemical components and times may also be used for visualizing membrane structures, such as the sarcoplasmic reticulum or transverse-axial tubules30.
The images must also be examined for structural degradation. Poor quality fixation or electron microscopy processing can result in severe loss of mitochondrial cristae, high proportion of dislodged or swollen mitochondria, and disintegration of portions of the cell membrane (sarcolemma). A closer and careful examination may also show loss of the t-tubular and SR membranes, but are less obvious to the untrained eye. Solutions to this issue include: (i) ensuring thorough perfusion; (ii) dissecting the tissue into small samples that also ensure deep penetration of the fixative and stains; (iii) ensuring that electron microscopy processing is done in cold solutions or on ice wherever specified; and (iv) ensuring that the timings for staining and dehydration (dehydration in particular) are strictly followed.
A typical cardiomyocyte is ~20 µm in cross-section diameter and ~100 µm in length. This makes imaging entire heart cells a significant challenge. Furthermore, the changing orientation of muscle fibers within the heart31 further complicates the acquisition of an image volume whose axes are aligned with the transverse and longitudinal axes of a cell of interest. Image analysis programs such as IMOD enable synthetic re-alignment of the data into a desired orientation. Recent advances in serial block-face scanning electron microscopy28 and associated image processing algorithms29 also help resolve these issues. Nevertheless, careful examination of thick sections under a light microscope and subsequent re-orientation of the block (step 2.5) will increase the likelihood of imaging cells with reasonable alignment of the longitudinal and transverse axes with the imaging axes. This will ensure that most of the cell volume will be captured within the field of view.
Protocol steps 3, 4 and 5 require software listed in the Table of Materials to be installed. IMOD, the R-statistics package, iso2mesh and FiJi (a version of ImageJ for microscopy data) have extensive documentation and tutorials on how to use them. CardiacCellMeshGenerator is a MATLAB application that has been developed to make it easier for the user to build the model in a simple workflow. Users can download the entire source code, and application package from the github RyR-simulator repository link (https://github.com/CellSMB/RyR-simulator/tree/JoVE). The application, CardiacCellMeshGenerator.miappinstall, can be installed in MATLAB as an app. The inputs for the mesh generated in this protocol can be found inside RyR-simulator/input-files/target-tomo-cell/, while the additional inputs for the RyR-cluster simulator can be found inside RyR-simulator/input-files/master-cell/. Before initiating the app, users should make sure that the installed iso2mesh folder and its sub folders have been added to the MATLAB path. Similarly, ensure that the R-packages necessary (Table of Materials) for the cluster simulator have also been installed within R.
A progress bar has been included in the app when generating the mesh, when generating the inputs for the RyR cluster simulator, and when mapping RyR cluster densities on the mesh using the RyR density mapper; the app will render the 3D mesh in the GUI when the mesh generation step has concluded. Upon completing the RyR cluster simulations on the R user interface, a graphical window will appear (Figure 10), depicting one of the simulated 3D RyR cluster point patterns.
The "Generate Mesh" button on the GUI triggers the iso2mesh function to generate a tetrahedral mesh. The model resolution can be adjusted by setting the "iso2mesh parameters" on the GUI that adjust the maximum tetrahedral element volume and edge length. The program currently distinguishes elements that make up the myofibril region and the mitochondria regions; this feature will be extended to include other components of the cell in the future.
Users can use the 3D rendering of the mesh and the RyR clusters to troubleshoot these steps. The RyR cluster simulation settings, etol and numIter, inside settings.R affect the accuracy of the RyR cluster simulations; these two parameters determine when an RyR cluster pattern simulation terminates. These parameters can be adjusted upon inspection of the distribution of the RyR clusters in the R window (Figure 10) to ensure that: (i) all clusters are close to the z-discs; and (ii) the clusters are spread across the entire cross-section rather than aggregating in any one region. An ad hoc sensitivity analysis of the simulations to these two parameters can be conducted to determine an appropriate combination of etol and numIter.
The spherical kernel intensity estimator in step 5 uses a sphere of chosen diameter as a kernel, which is passed over the volume of interest (the 3D spatial model in this context). These parameters can be adjusted for this smoothing algorithm on the GUI in Figure 5: (1) r_sphere defines the radius of spherical kernel smoothing; (2) hval defines the iterative spatial step size over which the spherical kernel must be iteratively passed over the volume.
A simple approach to determine whether the spatial density parameters are sufficient is to inspect the distribution of the RyR cluster density values as shown in Figure 7. The density values should be distributed such that the neighborhood of high density values is a similar size to the size of an RyR cluster in microscopy data.
The current protocol produces a finite element model that only contains the realistic distribution of myofibrils, mitochondria, and RyR clusters. An extension to this step would be to simulate the distribution of other ion-channels that play a role in ECC, such as sarco-endoplasmic reticular calcium pumps (SERCA2a), sodium-calcium exchangers (NCX) and the sarcolemmal calcium pumps. The key to these extensions is an analysis of the spatial distribution of the ion channels from immunofluorescence data. For instance, NCX channels have been measured to be distributed uniformly over the t-tubular membranes at an average spacing of 0.67 µm, but their relationship to RyR clusters is not so clear32. On the other hand, SERCA2a targeting antibodies have previously been used to mark the SR33, but the spatial density of SERCA2a has not been reported in the literature. Therefore, at this stage, a uniform distribution of SERCA 2A over the SR network and NCX over the t-tubules may be assumed with caution.
As demonstrated in the Representative Results, the resultant finite element mesh can be used with finite element solvers34 to simulate biochemical processes such as calcium signaling18 and bioenergetics27 as reaction-diffusion processes. To this end, ion-channels are represented as reaction sites at nodes within the mesh topology. The reaction sites act as sources or sinks of different chemical species to represent the flow of chemical species through these ion channels into different compartments of the cell. The output mesh and RyR density files from this protocol can be used with any other integrator or within any other environment, provided that the software has the feature to recognize these text files. The text files can also be manipulated by scripts or third-party software to satisfy input file format requirements of the environment of choice. The first application on calcium dynamics only demonstrates the utility of this model for the calcium upstroke but this does not indicate that the mesh cannot be used for longer time scales. The mesh and cluster densities can be used to simulate multiple calcium cycles, with the maintenance of homeostasis and stability only dependent on the finite element solver that is used.
A longer-term aim is to improve this pipeline of model building to make it easy to create structurally accurate finite element models of cardiomyocytes and other cell types. An automated segmentation algorithm has been developed recently, which can be applied to serial-block-face scanning electron microscopy data of cardiomyocytes to create a model of the myofibrils and mitochondria distribution in the entire cell with minimal user input29. This method will be extended to segment the t-tubule and SR membrane networks in future. A more generalized version of the RyR-simulator algorithm that will enable users to analyze and simulate co-locations between ion-channel and organelles, as well as between different ion-channel types, in a high throughput fashion is also under development. A seamless protocol for building a complete model of the cell would enable scientists to perform model-based hypotheses tests on the relationship between cell structure and cell function more routinely than is currently possible with experimental approaches alone.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
This work was supported by the Royal Society of New Zealand Marsden Fast Start Grant 11-UOA-184, the Human Frontiers Science Program Research grant RGP0027/2013 and the Australian Research Council Discovery Project Grant DP170101358.
References
- Noble D, Rudy Y. Models of cardiac ventricular action potentials: iterative interaction between experiment and simulation. Phil. Trans. R. Soc. A: Math., Phys. and Eng. Sci. 2001;359(1783):1127–1142. [Google Scholar]
- Williams GSB, Smith GD, Sobie EA, Jafri MS. Models of cardiac excitation-contraction coupling in ventricular myocytes. Math. Biosci. 2010;226(1):1–15. doi: 10.1016/j.mbs.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard DA, Vendelin M. Systems biology of the mitochondrion. Am. J. Phys. - Cell Phys. 2006;291(6):C1101–C1103. doi: 10.1152/ajpcell.00481.2006. [DOI] [PubMed] [Google Scholar]
- Crampin EJ, Smith NP. A Dynamic Model of Excitation-Contraction Coupling during Acidosis in Cardiac Ventricular Myocytes. Biophys. J. 2006;90(9):3074–3090. doi: 10.1529/biophysj.105.070557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Louch WE, et al. Calcium Dynamics in the Ventricular Myocytes of SERCA2 Knockout Mice: A Modeling Study. Biophys. J. 2011;100(2):322–331. doi: 10.1016/j.bpj.2010.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu I, Minamino T. Physiological and pathological cardiac hypertrophy. J. Mol. Cell. Cardiol. 2016;97:245–262. doi: 10.1016/j.yjmcc.2016.06.001. [DOI] [PubMed] [Google Scholar]
- Wei S, Guo A, et al. T-tubule remodeling during transition from hypertrophy to heart failure. Circ. Res. 2010;107(4):520–531. doi: 10.1161/CIRCRESAHA.109.212324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz J, Ghosh S, et al. Changes in mitochondrial morphology and organization can enhance energy supply from mitochondrial oxidative phosphorylation in diabetic cardiomyopathy. Am. J. Phys. - Cell Phys. 2017;312(2):C190–C197. doi: 10.1152/ajpcell.00298.2016. [DOI] [PubMed] [Google Scholar]
- González A, Ravassa S, Beaumont J, López B, Díez J. New Targets to Treat the Structural Remodeling of the Myocardium. J. Am. Coll. Cardiol. 2011;58(18):1833–1843. doi: 10.1016/j.jacc.2011.06.058. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Martone ME, Yu Z, Thor A, Doi M. Three-dimensional electron microscopy reveals new details of membrane systems for Ca2+ signaling in the heart. J. Cell Sci. 2009. [DOI] [PMC free article] [PubMed]
- Soeller C, Crossman D, Gilbert R, Cannell MB. Analysis of ryanodine receptor clusters in rat and human cardiac myocytes. Proc. Natl. Acad. Sci. 2007;104(38):14958–14963. doi: 10.1073/pnas.0703016104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeller C, Baddeley D. Super-resolution imaging of EC coupling protein distribution in the heart. J. Mol. Cell. Cardiol. 2013;58(1):32–40. doi: 10.1016/j.yjmcc.2012.11.004. [DOI] [PubMed] [Google Scholar]
- Yu Z, Holst MJ, et al. Three-dimensional geometric modeling of membrane-bound organelles in ventricular myocytes: bridging the gap between microscopic imaging and mathematical simulation. J. Struct. Biol. 2008;164(3):304–313. doi: 10.1016/j.jsb.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake J, Edwards AG, et al. Modelling cardiac calcium sparks in a three-dimensional reconstruction of a calcium release unit. J. Physiol. 2012;590(18):4403–4422. doi: 10.1113/jphysiol.2012.227926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeller C, Jayasinghe ID, Li P, Holden AV, Cannell MB. Three-dimensional high-resolution imaging of cardiac proteins to construct models of intracellular Ca2+ signalling in rat ventricular myocytes. Exp. Physiol. 2009;94(5):496–508. doi: 10.1113/expphysiol.2008.043976. [DOI] [PubMed] [Google Scholar]
- Kekenes-Huskey PM, Cheng Y, Hake JE. Modeling effects of L-type Ca2+ current and Na+-Ca2+ exchanger on Ca2+ trigger flux in rabbit myocytes with realistic t-tubule geometries. Front. in Physiol. 2012;3:1–14. doi: 10.3389/fphys.2012.00351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zienkiewicz OC, Taylor RL. The finite element method. Vol. 1. Butterworth-Heinemann; 2000. [Google Scholar]
- Rajagopal V, Bass G, et al. Examination of the effects of heterogeneous organization of RyR clusters, myofibrils and mitochondria on Ca2+ release patterns in cardiomyocytes. PLoS Comp. Biol. 2015;11(9):e1004417. doi: 10.1371/journal.pcbi.1004417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illian J, Penttinen A, Stoyan H, Stoyan D. Statistical Analysis and Modelling of Spatial Point Patterns. Statistical Analysis and Modelling of Spatial Point Patterns. Chichester, UK: John Wiley & Sons; 2008. pp. 1–534. [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer Visualization of Three-Dimensional Image Data Using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Fang Q, Boas DA. Tetrahedral mesh generation from volumetric binary and grayscale images. Proc. ISBI. 2009. pp. 1142–1145.
- Aune DJ, Herr SE, Menick DR. Induction and assessment of ischemia-reperfusion injury in langendorff perfused rat hearts. J. Vis. Exp. 2015. p. e52908. [DOI] [PMC free article] [PubMed]
- Judd J, Lovas J, Huang GN. Isolation, culture and transduction of adult mouse cardiomyocytes. J. Vis. Exp. 2016. [DOI] [PMC free article] [PubMed]
- Hagler HK. Ultramicrotomy for biological electron microscopy. Electron Microscopy: Methods and Protocols. 2007;369(Chapter 5):67–96. doi: 10.1007/978-1-59745-294-6_5. [DOI] [PubMed] [Google Scholar]
- He W, He Y. Electron tomography for organelles, cells, and tissues. Electron Microscopy: Methods and Protocols. 2014;1117(20):445–483. doi: 10.1007/978-1-62703-776-1_20. [DOI] [PubMed] [Google Scholar]
- Diggle P, Marron JS. Equivalence of smoothing parameter selectors in density and intensity estimation. J. Am. Stat. Assoc. 1988;83(403):793–800. [Google Scholar]
- Ghosh S, Crampin EJ, Hanssen E, Rajagopal V. A computational study of the role of mitochondrial organization on cardiac bioenergetics. Proc. EMBC. 2017. pp. 2696–2699. [DOI] [PubMed]
- Pinali C, Kitmitto A. Serial block face scanning electron microscopy for the study of cardiac muscle ultrastructure at nanoscale resolutions. J. Mol. Cell. Cardiol. 2014;76:1–11. doi: 10.1016/j.yjmcc.2014.08.010. [DOI] [PubMed] [Google Scholar]
- Hussain A, Hanssen E, Rajagopal V. A Semi-Automated Workflow for Segmenting Contents of Single Cardiac Cells from Serial-Block-Face Scanning Electron Microscopy Data. Microsc Microanal. 2017;23(S1):240–241. [Google Scholar]
- Pinali C, Bennett H, Davenport JB, Trafford AW, Kitmitto A. Three-dimensional reconstruction of cardiac sarcoplasmic reticulum reveals a continuous network linking transverse-tubules: this organization is perturbed in heart failure. Circ. Res. 2013;113(11):1219–1230. doi: 10.1161/CIRCRESAHA.113.301348. [DOI] [PubMed] [Google Scholar]
- LeGrice IJ, Hunter PJ, Smaill BH. Laminar structure of the heart: a mathematical model. Am. J. Physiol. 1997;272(5 Pt 2):H2466–H2476. doi: 10.1152/ajpheart.1997.272.5.H2466. [DOI] [PubMed] [Google Scholar]
- Jayasinghe ID, Cannell MB, Soeller C. Organization of ryanodine receptors, transverse tubules, and sodium-calcium exchanger in rat myocytes. Biophys. J. 2009;97(10):2664–2673. doi: 10.1016/j.bpj.2009.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasinghe ID, Crossman DJ, Soeller C, Cannell MB. Comparison of the organization of t-tubules, sarcoplasmic reticulum and ryanodine receptors in rat and human ventricular myocardium. Clinic. Exp. Pharmacol. P. 2012;39(5):469–476. doi: 10.1111/j.1440-1681.2011.05578.x. [DOI] [PubMed] [Google Scholar]
- Bradley C, Bowery A, et al. OpenCMISS: a multi-physics & multi-scale computational infrastructure for the VPH/Physiome project. Prog. Biophys. Mol. Bio. 2011;107(1):32–47. doi: 10.1016/j.pbiomolbio.2011.06.015. [DOI] [PubMed] [Google Scholar]


