Abstract
The Protein Kinase C (PKC) enzymes have long been established as critical for synaptic plasticity. However, it is unknown whether Ca2+-dependent PKC isozymes are activated in dendritic spines during plasticity, and if so, how this synaptic activity is encoded by PKC. Here, using newly-developed, isozyme-specific sensors, we demonstrate that classic isozymes are activated to varying degrees and with unique kinetics. PKCα is activated robustly and rapidly in stimulated spines and is the only isozyme required for structural plasticity. This specificity, depends on a PDZ-binding domain present only in PKCα. The activation of PKCα during plasticity requires both NMDAR Ca2+-flux and autocrine BDNF-TrkB signaling, two pathways that differ vastly in their spatiotemporal scales of signaling. Our results suggest that by integrating these signals, PKCα combines a measure of recent, nearby synaptic plasticity with local synaptic input, enabling complex cellular computations such as heterosynaptic facilitation of plasticity necessary for efficient hippocampal-dependent learning.
Introduction:
The integration of signals across various time and space scales is essential for effective adaption to a changing environment. This is particularly apparent in neurons, which receive input from thousands of other neurons at discrete synaptic sites with various temporal patterns. During learning, the strength and structure of synapses are modulated in a synapse-specific manner through complex signaling cascades that transduce compartmentalized Ca2+ transients into long-lasting biochemical changes 1,2. However, the threshold for inducing plasticity at a single synapse is also regulated by the history of neuronal activity across many synapses 3. For example, potentiation at one dendritic spine can facilitate potentiation of nearby spines receiving subsequent input 4–6. This heterosynaptic facilitation of plasticity has been suggested to contribute to the clustered patterns of inputs and plasticity observed in vivo 7–9. Therefore, the refinement of neuronal circuits during development and learning depends critically on the integration of biochemical signals that differ markedly in their spatiotemporal scales.
The protein kinase C (PKC) family has long-been implicated as essential in synaptic plasticity 10–16. Of particular interest is the classic subfamily of PKC isozymes (PKCα, PKCβ, PKCγ) which are widely expressed in many brain regions, including in hippocampal CA1 pyramidal neurons 17–20. Moreover, this subfamily requires both Ca2+ and lipid signals to be activated and thus, during plasticity, may play a role in integrating multiple signals across time and space domains. However, due to the complexity of the PKC family of isozymes and a lack of specific tools, analysis of PKC during plasticity has primarily been limited to loss-of-function/ gain-of-function studies. Thus, our understanding of PKC in plasticity has remained unclear. Specifically, it is unknown if classic PKCs are activated in spines during plasticity of hippocampal synapses. Moreover, if they are activated, the mechanisms, spatiotemporal activity patterns, and functional role of activation are unknown.
Here, we develop highly-sensitive, isozyme-specific sensors for each of the classic PKCs in order to analyze Ca2+-dependent PKC function in plasticity. We find that classic PKCs have distinct spatiotemporal profiles of activation, and that among the classic PKCs, only PKCα is involved in synaptic plasticity. This high degree of functional isozyme specificity relies on a PDZ-binding-domain present only in PKCα. Further, we delineate the relationship between PKC and known signaling pathways of plasticity and demonstrate the critical role of PKCα in plasticity and spatial learning. Finally, we reveal a mechanism of signal integration by PKCα, which combines a measure of local, current synaptic input with a measure of the recent history of nearby synaptic plasticity. The integration of these signals, which have spatiotemporal scales that differ by orders of magnitude, supports heterosynaptic facilitation of plasticity.
Results:
Development of sensors to probe classic PKC isozyme activity during synaptic plasticity
We set out to address the precise subcellular regulation of specific PKC isozyme activity with high temporal resolution during synaptic plasticity. Since previously developed PKC FRET-based sensors 21–23 lack sufficient sensitivity and isozyme specificity to delineate the functionally unique roles of PKC isozymes during plasticity (see supplementary note), we developed novel, isozyme-specific, FRET-based sensors. Moreover, sensors were optimized for 2-photon fluorescence lifetime imaging (2pFLIM): a quantitative, fast, and robust readout of FRET well-suited to monitor protein activity in light-scattering brain tissue 24. For each of the three classic PKC isozymes (PKCα, PKCβ, PKCγ), two different sensor approaches were designed based on well-characterized properties of PKC activation: translocation to the plasma membrane, and binding to downstream substrate 25. We named these sensors ITRACK (Isozyme-specific TRAnslocation of C Kinase, Figure 1A) and IDOCKS (Isozyme-specific Docking Of C Kinase Substrate, Figure 1B) respectively. For both sensor designs, specific PKC isozymes were tagged with a donor fluorophore, monomeric eGFP, and expressed together with an acceptor construct. For ITRACK, the acceptor construct consisted of the acceptor fluorophore, mCherry, which was targeted to the plasma membrane by a K-Ras CAAX domain 26. With PKC activation, the eGFP-tagged PKC isozyme translocates to the mCherry labeled membrane, increasing FRET between eGFP and mCherry. This translocation could be detected as a decrease in the fluorescence lifetime of eGFP. For the IDOCKS sensor, the acceptor construct consisted of the pseudosubstrate region from the regulatory domain of PKCα/β (aa 19–36), which was tagged on the N-terminus by two mCherry fluorophores. Activation of eGFP-PKC allows binding of the mCherry tagged substrate to the kinase domain, increasing FRET and decreasing fluorescence lifetime of eGFP.
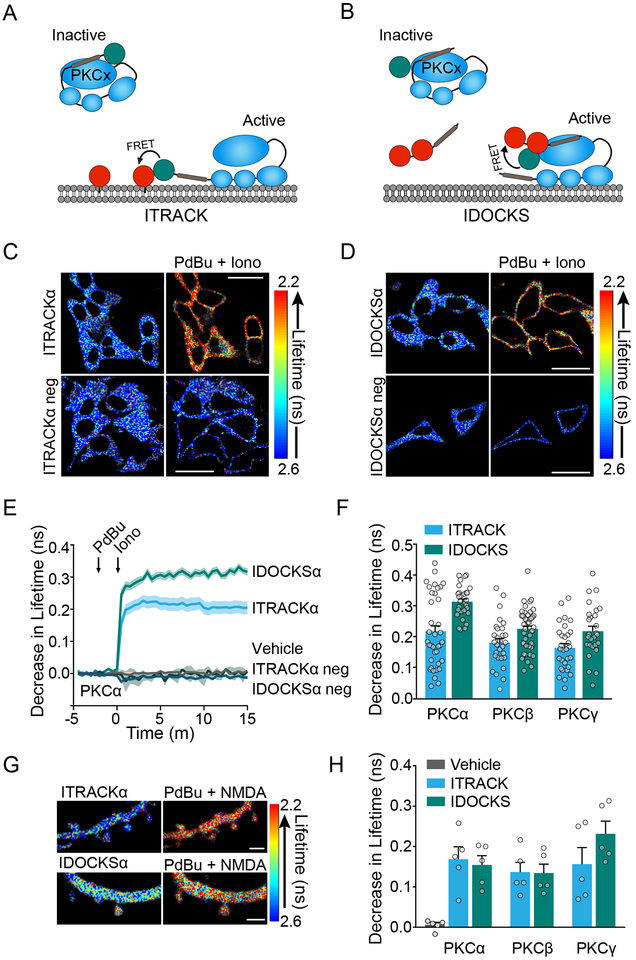
Fig. 1: Development of FLIM-FRET sensors for isozyme-specific PKC activity.
A) ITRACK sensor design. Donor: Classic PKC isozyme is tagged with mEGFP. Acceptor: mCh is targeted to membrane with K-CAAX domain.
B) IDOCKS sensor design. Donor: Classic PKC isozyme is tagged with mEGFP. Acceptor: The pseudosubstrate domain from PKCα/β is tagged by 2mCh fluorophores.
C) Representative fluorescent lifetime images of HeLa cells expressing ITRACKα or ITRACKα neg (see Fig. S1A) before and after PKC activation by PdBu (1 μM) and ionomycin (1 μM). Warmer colors indicate a decrease in lifetime and an increase in PKCα activity (Quantification in (E)). Scale bar = 20 μm.
D) Same as in C but cells express IDOCKSα or IDOCKSα neg (Quantification in (E)).
E) Mean time course of the change in fluorescence lifetime of PKCα sensors upon stimulation with PdBU (1 μM) and ionomycin (1 μM) or vehicle (n [experiments/cells]: Veh=6/30, ITRACKα= 7/37, ITRACKα neg= 5/28, IDOCKSα= 6/29, IDOCKSα neg= 6/36, shaded regions indicate SEM).
F) Quantification of drug-induced fluorescence lifetime changes of ITRACK and IDOCKS for PKCα (shown in E), PKCβ and PKCγ in HeLa cells as indicated (n [experiments/cells]: ITRACKβ= 6/29, IDOCKSβ=7/44, ITRACKγ= 6/30, IDOCKSγ= 6/27, Bars indicate mean and SEM, symbols indicate individual cells).
G) Representative fluorescent lifetime images of secondary apical dendrites of CA1 hippocampal neurons expressing ITRACKα or IDOCKSα before or 2 mins after stimulation with PdBu (1 μM) and NMDA (20 μM). Scale bar = 2 μm.
H) Quantification of drug-induced fluorescence lifetime changes of ITRACK and IDOCKS for PKCα, PKCβ, and PKCγ in neurons as indicated (n [experiments/neurons] =5/5 for all conditions, Bars indicate mean and SEM, symbols indicate individual neurons).
ITRACK and IDOCKS sensors for each of three classic PKC isozymes were expressed separately in HeLa cells and tested for sensitivity and specificity. Classic PKC isozymes were activated by sequential application of phorbol ester (PdBu 1 μM), a DAG analog, and the Ca2+ ionophore ionomycin (1 μM). Upon addition of both drugs, a robust decrease in the lifetime of both the ITRACK and IDOCKS sensors was seen (PKCα shown Figure 1C-E). Control sensors, in which the acceptor fluorophore was no longer localized to the membrane (ITRACKneg, Figure S1A) or in which a single point mutation (R27E) was introduced to disrupt pseudosubstrate domain docking to active PKC (IDOCKSneg, Figure S1B), were unable to report PKC activation (Figure 1C-E). This indicates that the sensors report translocation and substrate docking as designed. Similar results were obtained for sensors of all classic isozymes, demonstrating that each of the isozyme-specific sensors have similar sensitivity (Figure 1F).
Sensors were then introduced into organotypic hippocampal mouse brain slices through biolistic transfection and dendrites of CA1 neurons were imaged through 2pFLIM. After baseline measurements of sensors, PKC isozymes were activated by co-application of PdBu (1 μM) and NMDA (20 μM). Drug application, but not vehicle, induced a robust decrease in fluorescence lifetime in spines and dendrites of all sensors (Figure 1G). Similarly to the results in HeLa cells, sensors for all classic isozymes exhibited similar sensitivity to strong drug-induced activation (Figure 1H). Thus, our PKC sensors have sensitivity sufficient for imaging in single dendritic spines and can probe the activity of each of the classic PKC isozymes.
Activity of classic PKC isozymes during plasticity of dendritic spines
Using these new sensors, we investigated the activity of classic PKC isozymes during the induction of single-spine structural long-term potentiation (sLTP): a robust model of functional synaptic plasticity and learning 27,28. sLTP was induced in single spines on secondary or tertiary apical dendrites of CA1 pyramidal neurons in organotypic hippocampal slices by a 0.5 Hz train of 2-photon glutamate uncaging pulses in zero extracellular Mg2+. This stimulation, as previously shown, led to a sustained increase in size and functional strength of the stimulated spine 28. The activity of each PKC isozyme was monitored by ITRACK (Figure 2A) in the stimulated spine and surrounding dendrite during sLTP with 128 ms temporal resolution. With each glutamate uncaging pulse, classic PKCs showed rapid activation (~200 ms) and decay (~1 s) in the stimulated spine (PKCα: Figure 2B-D, Video S1, PKCβ, PKCγ: Figure S1C). PKC activity was compartmentalized in the stimulated spine and a short stretch of dendrite immediately below the stimulated spine (~1 μm), suggesting a spine-specific, spatially-confined role in plasticity (Figure 2F, G). To quantify the decay kinetics of PKC activation, we averaged the activity in response to each glutamate uncaging pulse (uncaging triggered average, Figure 2E). This revealed differential activity profiles of the three classic PKC isozymes: PKCα had a larger peak amplitude and longer decay constant than PKCβ and PKCγ (Figure 2E), and thus the largest averaged activation among the classic isozymes (PKCα: 5.66 ± 0.64, PKCβ: 2.3 ± 0.11, PKCγ: 1.43 ± 0.12 % s, Figure S1C inset).
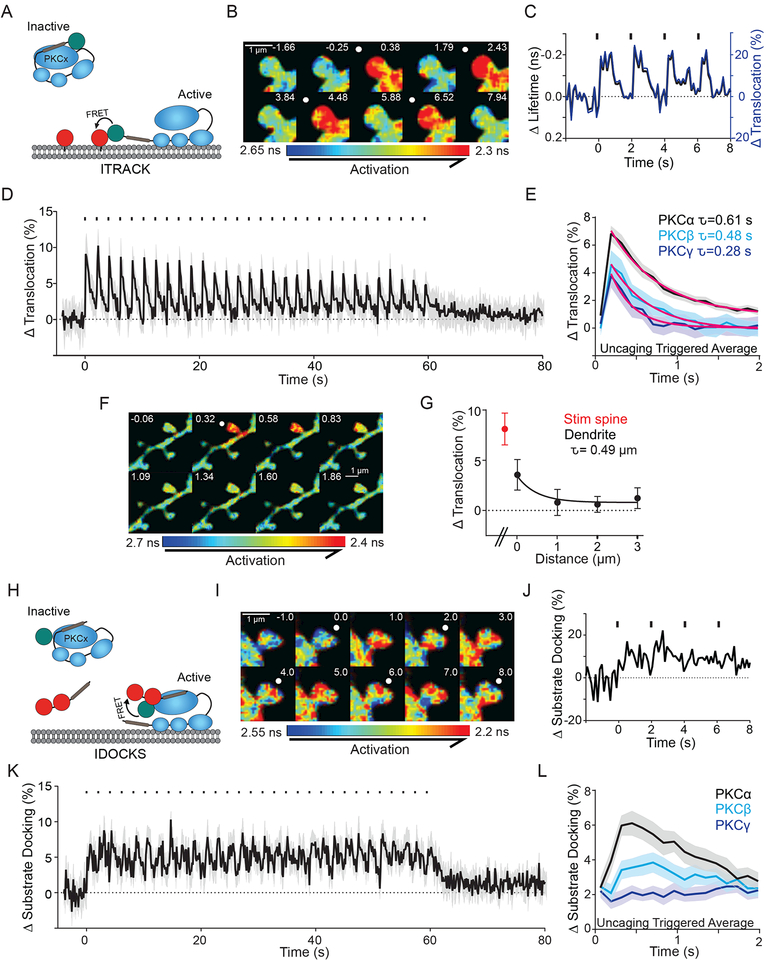
Fig. 2: Structural plasticity induces robust, compartmentalized and rapid PKCα activity.
A) ITRACK sensor design.
B, C) Fluorescence lifetime images (B) and quantification (C) of ITRACKα in a single spine undergoing sLTP. White spots (B) and black dashes (C) indicate glutamate uncaging. Fluorescence lifetime data (black) can be quantified as percent translocation (blue) by calculating the percent of the ITRACKα donor (eGFP tagged PKCα) that is undergoing FRET with the ITRACKα acceptor (membrane targeted mCh).
D) Mean time course of PKCα activity measured by ITRACKα during sLTP. n [neurons/spines] = 32/103. Shaded area represents SEM.
E) Mean response of ITRACK to each glutamate uncaging pulse (uncaging triggered average, shaded area is SEM) and decay time constants for PKCα (τ= 0.61 ± 0.07 s, n [neurons/spines] =32/103), PKCβ (τ= 0.48 ± 0.11 s, n [neurons/spines] =13/31), and PKCγ (τ= 0.28 ± 0.04 s, n [neurons/spines] = 10/27). Extra Sum of squares F test- Yo and K (1/τ) are not shared between groups p<0.0001.
F) Uncaging triggered average of fluorescence lifetime images of ITRACKα during sLTP. White spot indicates location of glutamate uncaging.
G) Spatial profile of mean peak PKCα activity in the stimulated spine (red), and the adjacent stretch of dendrite (n [neurons/ spines] = 4/17). Spatial decay constant calculated using non-linear regression. Error bars represent SEM.
H) IDOCKS sensor design.
I, J) Fluorescence lifetime images (I) and quantification (J) of IDOCKSα of a single spine undergoing sLTP
K) Average time course of PKCα as measured by IDOCKSα during sLTP (n [neurons/spines] = 6/28. Mean and SEM (shaded) are shown).
L) Uncaging triggered average (mean and SEM (shaded)) measured by IDOCKS of PKCα (n [neurons/spines] = 6/28), PKCβ (n [neurons/spines] = 6/24), and PKCγ (n [neurons/= 8/29).
To test that PKC is not only translocating to membrane, but also binding downstream substrate on this timescale, we measured PKC activity using IDOCKS (Figure 2H). Similar to the activity profile measured using ITRACK, IDOCKS reported rapid activation of PKC isozymes in response to the induction of plasticity (PKCα: Figure 2I-K, PKCβ, PKCγ: Figure S1D). Compared to ITRACK, IDOCKS displayed slightly slower kinetics of both the rising phase and decay of PKC activity. It did not fully return to baseline before the next glutamate uncaging pulse was triggered, instead reaching a plateau until the uncaging train was complete (Figure 2K, S1D). This is reflected in the uncaging triggered average as a higher initial value (Figure 2L). It should be noted that the pseudosubstrate used in IDOCKS cannot be phosphorylated and thus decay kinetics reported by the sensor may be slower than endogenous substrates. Consistent with measurements by ITRACK, PKCα was activated to a greater extent than PKCβ or PKCγ (Figure 2L, S1D).
Several control experiments validate these measurements of PKC activity during plasticity. Experiments performed without MNI-glutamate or that used negative control sensors ITRACKneg or IDOCKSneg showed no change in PKC activity during sLTP induction (Figure S1E, F). The magnitude of sensor response did not correlate with expression level of the sensor (Figure S2A) or the ratio of its components (Figure S2B). Basal sensor signals were also independent of spine size and surface area to volume ratio of spines or dendrites (Figure S2C, D). The expression of the ITRACK and IDOCKS sensor, particularly the expression of the pseudosubstrate used in the IDOCKS sensor, has the potential to interfere with PKCα localization or signaling. We therefore monitored the diffusion of PKCα in spines by measuring the decay of fluorescence after photoactivation of a photoactivatable-GFP (paGFP) tagged PKCα. Photoactivated paGFP-PKCα diffused out of the spine with a time constant of ~5 sec, consistent with cytosolic distribution of PKCα, although it was significantly slower than paGFP alone (Figure S2E). Co-expression of ITRACK acceptor (CAAX) or IDOCKS acceptor (pseudosubstrate) did not change the diffusion of PKCα (Figure S2F), suggesting that they have little effect on the localization of PKCα. Moreover, sLTP of stimulated spines expressing either ITRACK or IDOCKS was consistent with spines expressing eGFP alone (Figure S2G) demonstrating that any effects of sensor overexpression on endogenous signaling are minimal. Finally, ITRACK and IDOCKS, as well as a variant of the ITRACK sensor, which used an alternative membrane targeting domain (H-Ras derived CAAX), all yielded similar results, demonstrating that our sensors are reporting physiologic PKC activity that is minimally modulated by sensor design (Figure S2H).
PKCα is the only classic PKC isozyme required for structural plasticity
Our measurements reveal that classic PKC isozymes are only active during the induction phase of sLTP. We therefore tested the temporal requirement of PKC activity in plasticity by applying a broad-spectrum PKC inhibitor, Gö6983 29, before or after sLTP induction. Although Gö6983 effectively blocked sLTP when applied 10 min before stimulation, plasticity was not attenuated when it was applied immediately after the induction protocol (Figure 3A-C, Figure S3A). Consistent with our sensor measurements, these data suggest that PKC activity is required as an early signal during the induction of sLTP.
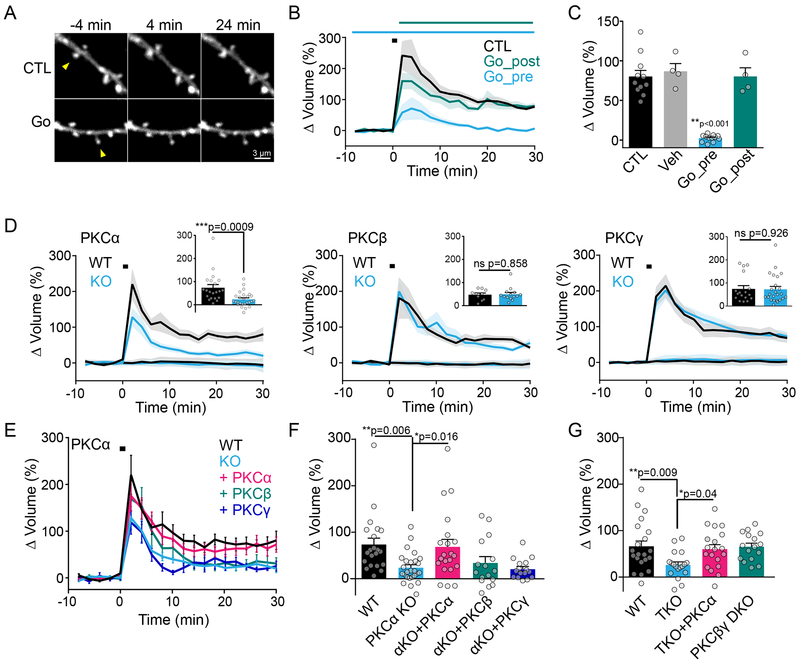
Fig. 3: Classic isozyme PKCα, but not PKCβ or PKCγ, regulates structural plasticity.
A) Representative intensity images (eGFP) of uncaging-induced sLTP of single spines (arrow) in the absence (CTL) or with pretreatment of PKC inhibitor Gö6983 (Gö, 1 μM).
B) Average time courses of sLTP induced by glutamate uncaging (black square) in CTL or Gö6983 treated slices. Gö6983 was applied 10+ mins before (Gö_pre) or immediately after (Gö_post) the uncaging stimulation (n [neurons/spines]: CTL=11/11, Gö_pre= 12/12, Gö_post=4/4). Two-way ANOVA significant by drug application (F (2, 480) = 45.35, p<0.001). Mean and SEM (shaded) shown.
C) Quantification of sustained sLTP (25–30 min) in B (n [neurons/spines]: Veh= 4/4). One-way ANOVA with Sidak’s multiple comparison test Gö_pre vs all other groups. Mean and SEM shown.
D) Average time courses of sLTP induced by glutamate uncaging (black square) in neurons from WT and KO littermates of classic PKC isozymes alpha (Mean and SEM (shaded) shown, n[neurons/spines]: WT=10/21, KO= 12/26, Two-way ANOVA significant by genotype for PKCalpha (F (1, 920) = 97.41, p<0.001), beta ns (n[neurons/spines]: WT= 7/10, KO=9/14) and gamma ns (n[neurons/spines]: WT=8/17, KO=10/23) as indicated. Insets: quantification of mean sustained sLTP (25–30 min, error bars represent SEM). Unpaired, two-tailed t-test shown.
E) Average time courses of sLTP of spines from PKCα WT and KO littermates, as shown in D, overlaid with sLTP of PKCα KO neurons overexpressing eGFP tagged PKCα, PKCβ or PKCγ. Mean and SEM shown (shaded). Two-way ANOVA significant by group (F (4, 1900) = 32.59, p<0.0001).
F) Quantification of mean sustained sLTP (25–30 min) in E (n [neurons/spines]: WT = 10/21, KO = 12/26, KO+PKCα = 9/21, KO+PKCβ = 6/16, KO+PKCγ = 6/16, One way ANOVA with Sidak’s multiple comparison test shown).
G) Quantification of mean sustained sLTP (25–30 min) in neurons from WT mice, PKCα,β,γ triple KO mice (TKO), TKO mice overexpressing eGFP-tagged PKCα, or PKCβ,γ double KO mice (DKO) (n[neurons/spines]: WT=8/21, TKO=7/18, TKO +PKCα=7/17, βγDKO=5/16). Error bars represent SEM. One way ANOVA with Sidak’s multiple comparison.
Of the three Ca2+- sensitive PKC isozymes, PKCα activity was particularly robust during structural plasticity. Consistently, hippocampal CA1 neurons from PKCα, but not PKCβ or PKCγ knockout mice, showed a deficit in sustained sLTP (Figure 3D, quantified at 25–30 min). This deficit could be fully rescued by acute, sparse and postsynaptic expression of eGFP-tagged PKCα, but not by PKCβ or PKCγ (Figure 3E, F). These data reveal exquisite specificity of signaling between isozymes even in the context of knockout and overexpression (Figure S2B) and that the eGFP-tagged PKC used in our sensors retains its enzymaticaction30–32. The inability of PKCβ and PKCγ to rescue the plasticity deficits of PKCα KO suggests that compensation by other isozymes is not masking a loss-of-function phenotype. To investigate this further, we bred triple-null mice (TKO), which lacked all classic PKC isozymes. TKO mice showed impairments in sLTP similar to PKCα KO mice, which could also be fully rescued by acute, sparse postsynaptic overexpression of eGFP-PKCα (Figure 3G, S2C, D). Finally, a double knockout (DKO) of PKCβ and PKCγ exhibit no deficits in sLTP (Figure 3G). These data suggest that PKCα is the only Ca2+-sensitive PKC isoform involved in sLTP.
Isozyme specificity of PKCα during sLTP is defined by a C-terminal PDZ binding domain.
Classic PKC isozymes have high sequence homology and poor in-vitro substrate specificity, suggesting that isozyme specificity of function may be due to selective protein-protein interactions. One domain that is unique to PKCα is a 4 amino acid (QSAV) C-terminal PDZ binding motif (Figure 4A) 33. In order to determine whether this domain contributes to the isozyme specificity of kinase activation during the induction of plasticity, we used ITRACKα FLIM sensors which lacked this domain (Figure 4B, PKCα ΔQSAV) and ITRACKγ sensors in which this domain was added to the C-terminus (Figure 4B, PKCγ+QSAV) to monitor the activation of WT and mutated PKC in response to glutamate uncaging. The amplitude of PKCα ΔQSAV translocation was reduced to a level similar to PKCγ WT. Moreover PKCγ+QSAV was activated more robustly, to a similar level as PKCα WT (Figure 4C, D).
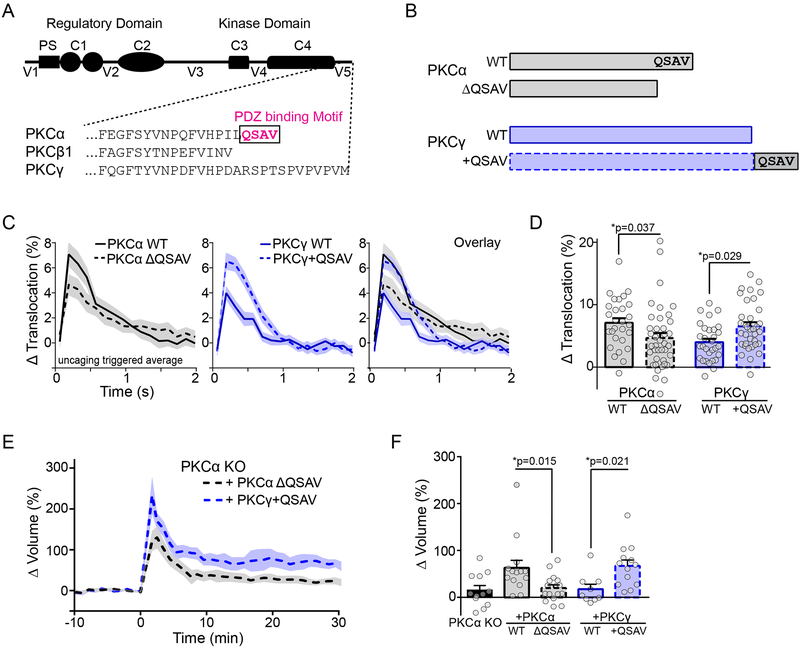
Fig. 4: Isozyme-specific PKCα activity and function is defined by a C-terminal PDZ binding domain.
A) Schematic of classic PKC isozyme domain structure. PS: pseudosubstrate, C1: DAG binding domain, C2: Calcium binding domains, C3: ATP binding site, C4: substrate binding domain. V1–V5: regions of lower homology between isozymes. Amino acid sequence of C-terminal portion of V5 for three classic isozymes shown. The PDZ binding motif is indicated.
B) Schematic of eGFP-tagged PKC constructs used in ITRACK sensors for panels D-G. Donor constructs consisted of eGFP tagged PKCα (WT) or a deletion mutant with no PDZ binding domain PKCα (ΔQSAV) and PKCγ (WT) or a chimeric PKCγ with the PDZ binding domain from PKCα added to the C terminus (+QSAV).
C) Uncaging triggered average measured by ITRACK (mean and SEM (shaded) shown, n [neurons/spines] = PKCα WT: 5/28, PKCα ΔQSAV: 8/39, PKCγ WT: 4/30, PKCγ +QSAV: 6/35).
D) Quantification of peak translocation in (C, mean and SEM). Two way ANOVA significant for presence of the QSAV domain (F (1, 128) = 11.81, p=0.0008)). Sidak’s multiple comparison test.
E)Average time courses of sLTP of spines from PKCα KO animals overexpressing eGFP-tagged PKCα ΔQSAV (n [neurons/spines] = 6/17) or PKCα KO neurons overexpressing eGFP tagged PKCγ +QSAV (n=4/13). Mean and SEM (shaded) shown.
F) Quantification of mean sustained sLTP (25–30 min) in neurons from PKCα KO mice overexpressing eGFP alone (PKCα KO: n [neurons/spines] = 4/12), eGFP-tagged PKCα WT (n= 5/14), PKCα ΔQSAV (shown in E), PKCγ WT (n=3/10), PKCγ +QSAV (shown in E). Error bars represent SEM. Two-way ANOVA significant by presence of QSAV domain (F (1, 49) = 14.63, p=0.0004). Sidak’s multiple comparison test.
We then expressed these mutant constructs in PKCα KO neurons and monitored sLTP to whether this domain determined the functional isozyme specificity of PKCα in sLTP. PKCα ΔQSAV was unable to rescue the sLTP deficits of PKCα KO animals, similar to expression of other PKC isozymes (Figure 4E, F). On the other hand, expression of PKCγ+QSAV fully rescued the PKCα KO sLTP deficit (Figure 4E, F). This suggests that functionally, the presence or absence of the PDZ binding domain at the C terminus of PKCα is sufficient to account for its isozyme-specific role of in sLTP.
PKCα is required for functional plasticity and spatial learning
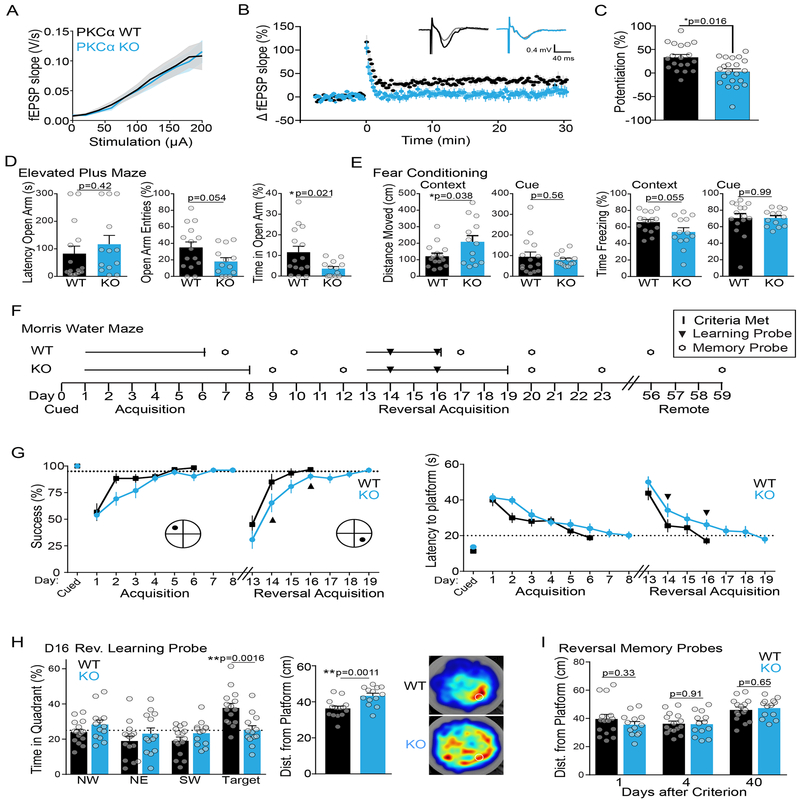
In order to evaluate whether PKCα plays a central role in other measures of synaptic plasticity, we performed electrophysiological LTP measurements in PKCα KO animals. While PKCα KO mice showed no apparent difference in basal transmission (Figure 5A), LTP induction at Schaeffer Collateral synapses by high frequency electrical stimulation was severely impaired compared to non-transgenic littermates (3 × 100 Hz 1s trains stimulation, Figure 5B, C).
Fig. 5: Functional and Behavioral characterization of plasticity in PKCα KO animals.
A) Electrophysiological recordings showing average input/output curve of PKCα KO (n [animals/experiments] =8/20) and WT (n=8/20) littermate mice. Mean and SEM shown.
B) Time course of change in extracellularly recorded EPSP slope and average representative EPSP traces (inset) before and after LTP induction in CA1 pyramidal neurons from PKCα KO (n[animals/experiments]=8/22) and WT (n=8/19) littermate mice. Mean and SEM shown. Two way ANOVA significant by genotype (F (2, 8818) = 463.7 p<0.0001).
C) Quantification of average potentiation (40–60 min) of neurons in (B). Mean and SEM shown. Two tailed unpaired t-test.
D) Anxiety related measurements (mean and SEM) of performance of male PKCα KO (n [animals] = 13) and WT (n = 15) littermate mice in elevated plus maze. Mean and SEM shown. Two tailed unpaired t-tests.
E) Distance moved and percent time freezing of PKCα KO and WT littermates in cued and contextual fear conditioning. Mean and SEM shown. Two tailed unpaired t-tests.
F) Schematic of training and testing protocol for the Morris water maze (MWM).
G) Acquisition curves of success and latency in finding the hidden platform in MWM task during training of PKCα KO (n [animals] = 13) and WT (n = 15) littermate mice (Two-way repeated measures ANOVA for success finding platform significant by day (F (10, 260) = 26.67, p<0.0001) and genotype (F (1, 26) = 8.302, p=0.0078), and for latency to find platform significant by day (F (9, 234) = 19.5, p<0.0001) and genotype (F (1, 26) = 5.11, p=0.0324). Animals were trained each day until they reached a group average performance criterion (dotted lines: success of finding platform > 95% and latency to platform < 20 s). Mean and SEM shown. Black dot indicates location of platform. Black triangles indicate probe tests to assess learning.
H) Average quadrant time, distance from platform location and heat maps for WT (n [animals] = 15) and PKCα KO (n=13) mice in the first 30s of probe test after day 3 of reversal training (D16). Mean and SEM shown. Quadrant time analyzed by two-way repeated measures ANOVA which showed significant interaction of location and genotype (F (3, 104) = 5.97, p=0.0009). Sidak’s multiple comparison test of genotypes by location was significant in the target quadrant. Distance from platform location analyzed by unpaired two-tailed t-test.
I) Average distance from the trained platform location of PKCα WT (n [animals] = 15) and KO mice (n=13) during memory probe tests at one, four and forty days after reaching performance criterion of reversal training. Mean and SEM shown. Unpaired two-tailed t tests (ns).
We next assessed whether PKCα KO animals had behavioral phenotypes that might be associated with deficits in synaptic plasticity. WT and PKCα KO littermates (male, n=15, 13 respectively, 9–16 weeks) showed no differences in locomotion or anxiety as assessed through an open field test (Figure S4A), working memory as assessed through a spontaneous alternation task (Figure S4B), or nociception as assessed through a hot plate test (Figure S4C). PKCα KO animals showed a mild anxiety phenotype in the elevated plus maze as they spent significantly less time and had fewer entries into the open arm (Figure 5D, unpaired t-test). In a cued and contextual fear conditioning task, WT and KO animals showed similar behavior in response to the conditioned cue (Figure 5E, cue). However, PKCα KO animals showed a tendency to freeze less than their WT counterparts and moved significantly more in the conditioned context (Figure 5E, context). These results suggested a possible impairment in hippocampal dependent spatial learning or memory.
Hippocampal dependent spatial learning and memory were assessed in more detail through the Morris Water Maze task (Figure 5F). Both WT and KO animals performed equally well on visual platform training as well as on day 1 of training in the hidden platform task (Figure 5G). However, KO animals showed delays in learning acquisition, taking two days longer to reach a predefined group criterion of performance (Figure 5G). Once learning criteria were met, however, PKCα WT and KO animals showed similar performance on memory probe tests at 1 and 4 days post training. To further investigate a potential learning deficit, MWM training was repeated with the location of the platform reversed to the opposite quadrant. Probe tests were included on days two and four of the reversal training. In these probe tests, PKCα KO animals showed impaired learning of the platform location compared to their WT littermates (Figure 5H). Quadrant time analysis revealed that by reversal day 4, WT animals spent significantly longer in the target quadrant, whereas KO animals spent equal time in all four quadrants (Figure 5H, left). The average distance from the platform location was also significantly less for WT than KO animals (Figure 5H, middle). These differences are reflected in the group-average heat maps of WT and KO animals (Figure 5H, right). Consistent with slower learning, KO animals also took three days longer than WT animals to reach the group performance criterion (Figure 5G). However, again, there was no difference between WT and KO animals in memory probe tests at 1, 4 or 40 days after reaching criterion (Figure 5I). These results suggest that PKCα KO animals have a deficit in learning, consistent with their deficits in structural and functional spine plasticity.
NMDAR and TrkB activation converge to activate PKCα during spine plasticity
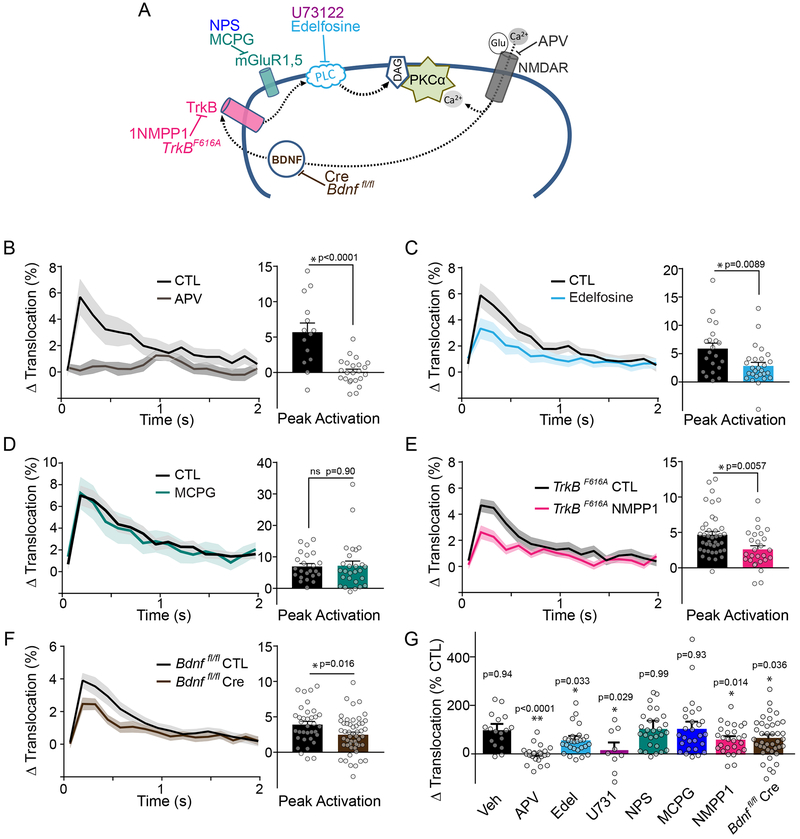
Having determined a critical role for PKCα in synaptic plasticity and learning, we next investigated the upstream signals that activate PKCα during plasticity. To do so, we measured PKCα activity in response to plasticity-inducing stimuli before and after pharmacological manipulation (Figure 6A). First, we found that inhibiting NMDA receptors with AP5 (50 μM) blocked the induction of sLTP as well as PKCα activation as measured by ITRACK (Figure 6B, G, S5A), suggesting that the Ca2+ flux through NMDARs is necessary for PKCα activation. Classic PKCs require both Ca2+ and lipid signals (diacylglycerol (DAG)) to be activated. We, therefore, measured PKCα translocation and substrate binding before and after application of edelfosine (50 μM), an inhibitor of phosphatidylinositol-directed phospholipase C (PLC), which produces DAG. Edelfosine significantly impaired PKCα activation and structural plasticity (Figure 6C, G, S5A, B), suggesting DAG production contributes to PKCα activation during plasticity. This finding was confirmed through the use of a second PLC inhibitor, U73122 (10 μM), which impaired PKCα translocation (Figure 6G, S5C). One potential signaling pathway upstream of PLC and activated by glutamate is the group I metabotropic glutamate receptors (mGluRs), which signal through Gαq to activate PLC. However, inhibiting mGluR with competitive or non-competitive antagonists S-MCPG (250 μM, Figure 6D, G, S5A) and NPS 2390 (20 μM, Figure 6G, S5A, D), showed no effects on PKCα translocation or structural plasticity.
Fig. 6: NMDAR and BDNF/TrkB activation converge to activate PKCα.
A) Schematic of pharmacological agents used to inhibit various components of signaling pathway.
B-E) Uncaging triggered average of PKCα activity (measured with ITRACKα) beforeand after drug application. Mean and SEM (shaded) shown. B) APV (50 μM n [neurons/spines] = 6/26, CTL 6/15). C) Edelfosine (50μM n [neurons/spines] =7/29, CTL 7/23). D) MCPG (250 μM n [neurons/spines] =7/23, CTL 7/23). E) 1NMPP1 (TrkBF616A mice; 1 μM 1NMPP1 n [neurons/spines] =7/27, CTL= 11/40, Veh= 4/17). Significance of two-tailed, unpaired t-tests are shown.
F) Average change in uncaging triggered average of PKCα activity (measured with ITRACKα) in neurons from Bdnf fl/fl mice expressing ITRACKα with Cre (n [neurons/spines] = 14/49) or without Cre (CTL n [neurons/spines] =10/35). Mean and SEM (shaded) shown. Two way ANOVA significance by Cre treatment.
G) Quantification of mean percent inhibition by indicated treatments of PKCα activation (same data as in B-F, Figure S5C, D). The peak response of ITRACKα (t = 0.192 s) in each treatment was normalized to its own control experiment (NPS 20 μM n [neurons/spines] =7/28, U73122 10 μM n [neurons/spines] =3/9). Error bars reflect combined standard error of the mean of treatment and control group. Significance of two-tailed one sample t-test against 100% is shown.
We next tested the involvement of the BDNF/TrkB pathway, a canonical plasticity promoting signaling pathway that activates PLC34,35. To selectively inhibit TrkB during the induction of plasticity, we took advantage of TrkBF616A knock-in mice36. These mice have normal TrkB function that can be selectively inhibited by the pharmacologic agent 1NMPP1. Blockade of TrkB during stimulation inhibited both PKCα translocation and substrate docking as well as sLTP to a similar extent as inhibition of PLC (Figure 6E, G, S5A, E). It has been recently shown that, during the induction of sLTP, post-synaptic BDNF can be rapidly released in response to each glutamate uncaging pulse from the stimulated spine to activate TrkB in the same spine and recruit PLC 37. To determine if this autonomous signaling mechanism contributes to PKCα activation during plasticity, we knocked out BDNF expression in single neurons using floxed BDNF knockout mice (Bdnf fl/fl) and sparse biolistic expression of Cre recombinase. Single-cell postsynaptic knockout of BDNF impaired PKCα activity during spine plasticity (Figure 6F, G), demonstrating autocrine BDNF/TrkB signaling is upstream of PKCα activity. We cannot exclude, however, that other sources of BDNF (e.g. presynaptic) might also contribute to postsynaptic TrkB activation during synaptic stimulation. Taken together, these results suggest that PKCα is activated by the convergence of two pathways in the stimulated spine: activation of NMDARs and autocrine BDNF-TrkB activation, which provide Ca2+ and DAG, respectively.
Integration of spatiotemporally distinct signals by PKCα
The timescale of PKCα activity (activated within ~0.1 s and decayed within ~1s) is one of the fastest signaling proteins measured during plasticity. PKCα activity decays on a timescale slightly longer than that of one of its upstream activators, Ca2+ (~0.1 s) 38, but much faster than its other upstream activator, TrkB (minutes)37. Moreover, PKCα activity is confined to the stimulated spine, similar to Ca2+. TrkB activity, on the other hand, is activated in a more widespread manner including within non-stimulated spines along a ~ 10 μm stretch of dendrite 6,37. Thus, while BDNF-TrkB signaling regulates PKCα activation, the temporal and spatial profile of PKCα appears to be shaped almost entirely by Ca2+ transients. This suggests that PKCα can integrate information about the history of nearby synaptic activity over minutes (long-lasting, spreading TrkB activity), while maintaining the spatiotemporal specificity of ongoing synaptic activity (compartmentalized, transient Ca2+ signals). These properties suggest that PKCα may be particularly well suited to mediate heterosynaptic facilitation of spine plasticity, whereby induction of plasticity at an individual spine can lower the threshold for the induction of plasticity in nearby spines receiving subsequent input 4–6.
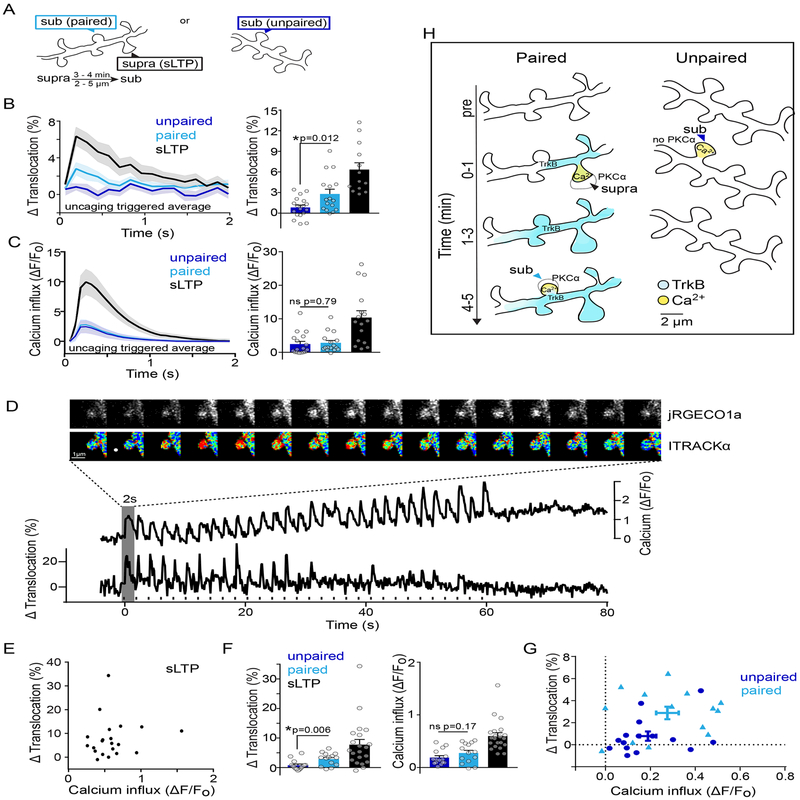
To test this, we monitored PKCα activation in dendritic spines that were given a weak, subthreshold uncaging stimulation either alone or several minutes after a nearby plasticity-inducing stimulation (Figure 7A). In contrast to sLTP-inducing stimulation, which induces both Ca2+ influx and long lasting TrkB activity, subthreshold stimulations given alone induce very little BDNF release and TrkB activity and smaller Ca2+ transients6,37. This subthreshold stimulation induced neither PKCα translocation measured by ITRACK (Figure 7B: unpaired) nor sLTP 6. However, when the same subthreshold stimulation was given 3–4 mins after a nearby spine (< 9 μm) had received a potentiating stimulation, PKCα translocated to a greater extent (Figure 7B: paired) and sLTP was induced 6. Measurements of Ca2+ activity by GCaMP6f 39 in spines receiving subthreshold stimuli given alone or after a plasticity-inducing stimulation did not differ in their amplitude, eliminating the possibility that facilitation of PKCα activation was a reflection of changes in Ca2+ dynamics (Figure 7C: paired vs unpaired).
Fig. 7: Integration of multiple upstream signals by PKCα induces heterosynaptic-facilitated plasticity.
A) Schematic of experimental design. A subthreshold stimulus that was unable to induce plasticity was given alone (unpaired) or after a nearby suprathreshold stimulus (paired).
B) Uncaging triggered average and peak translocation of PKCα after unpaired (n [neurons/spines] = 5/18) or paired stimulation (n [neurons/spines] = 5/16). Mean and SEM are shown, symbols represent individual spines. Two-tailed unpaired t-test.
C) Uncaging triggered average and peak activation of Ca2+ measured by GCaMP6f after unpaired (n [neurons/spines] = 7/16) or paired stimulation (n [neurons/spines] = 7/16). Mean and SEM are shown, symbols represent individual spines. One Way ANOVA with Sidak’s multiple comparison.
D) Simultaneous monitoring of calcium influx and PKCα translocation in a single dendritic spine undergoing sLTP. Intensity images of jRGECO1a and fluorescence lifetime images of ITRACKα and respective timecourses. Rise in Ca2+ baseline is due to volume increase of stimulated spine. White dot and black dashes indicate glutamate uncaging.
E) Relationship of peak PKCα translocation and Ca2+ influx calculated from the uncaging triggered average in single spines undergoing sLTP (n[neurons/spines] = 11/20). Pearson correlation is ns (p=0.597, r2=0.015, linear regression line shown is constrained to 0,0 intercept).
F) Peak translocation of PKCα and Ca2+ after unpaired (n [neurons/spines] = 8/16) or paired stimulation (n [neurons/spines] = 8/15). Bars represent mean and SEM, symbols individual spines. Two-tailed unpaired t-test.
G) Relationship of peak PKCα translocation and Ca2+ influx calculated from the uncaging triggered average of individual spines (points) given a subthreshold stimulation (unpaired-dark blue) or a subthreshold stimulation shortly after an sLTP inducing stimulation in a nearby spine (paired-light blue). Mean and Standard error of the mean are shown. There is no significant correlation for unpaired (p=0.247, r2=0.101) or paired (p=0.572, r2 =0.027) spines. Linear regression lines drawn are constrained to 0,0 intercept.
H) Model of integration of TrkB and Ca2+ signals to induce PKCα activation in paired and unpaired subthreshold stimulations.
To more closely examine the relationship between PKCα translocation and calcium dynamics within single spines, we simultaneously measured PKC translocation and calcium influx in single spines. To do this, we used the red-shifted Ca2+sensor jRGECO1a40 and modified the ITRACKα sensor by replacing mCherry on the acceptor construct with ShadowY, a dark YFP variant 41. With this approach, we were able to successfully measure PKCα translocation and calcium dynamics simultaneously in single spines (Figure 7D). In spines undergoing sLTP, the magnitude of PKCα translocation and calcium influx were not correlated, but rather showed high variability (Figure 7E), consistent with PKCα’s dependence on the integration of calcium with other upstream signals such as DAG. In agreement with our previous data subthreshold stimulation when given alone induced very little PKCα translocation, but when paired with previous induction of sLTP in a nearby spine, induced significantly more PKCα translocation in the absence of changes in calcium influx (Figure 7F). Similar to spines given a sLTP stimulus, single spine analysis of paired spines showed that the magnitude of PKCα activation was not significantly correlated to the magnitude of calcium influx, nor was PKCα activation significantly correlated with distance between the paired spine and the nearby spine that had previously undergone sLTP (Figure S6 , within ~9 μm range). However, paired spines showed an overall shift in the threshold for sensor activation, such that PKCα translocation was overall greater for similar amounts of calcium influx than spines that received this subthreshold stimulation alone (Figure 7G). We suggest that long-lasting, more widespread TrkB activity induced by sLTP in a nearby spine, enables PKCα activation by lower levels of NMDAR-dependent Ca2+-influx to induce spine plasticity (Figure 7H). Taken together, these data suggest that PKCα integrates a measure of recent nearby synaptic history with local synaptic input to facilitate plasticity in dendritic regions of clustered synaptic activity.
Discussion
In this study, we clarify long-standing questions about the classic PKC family in plasticity through the development of novel, highly-sensitive isozyme-specific biosensors. These sensors demonstrate that classic PKC activity is rapid, transient, and compartmentalized (< 1 μm) during the induction of sLTP. The activation of PKC occurs within ~0.1 s and decays over 0.3 – 0.8 s. The decay of PKC is much more rapid than the other major Ca2+-dependent signaling kinase involved in synaptic plasticity, CaMKII42. Thus, unlike CaMKII, PKC has little capability to integrate Ca2+ signals over seconds during the induction of sLTP 42. Since both kinases are necessary for sLTP, they likely act in parallel pathways with different kinetics. This mechanism may be important for decoding Ca2+ signals at different temporal scales.
Amongst the closely related classic PKC isozymes, our results revealed a high-degree of specificity in signaling. Deletion of neither PKCβ nor PKCγ led to plasticity deficits and neither could compensate for the loss of PKCα in plasticity, even when overexpressed. This specificity of signaling was reflected in the extent of activation in response to glutamate uncaging pulses and could be defined by the presence or absence of the C-terminal PDZ binding domain unique to PKCα. This domain, which was also sufficient for the functional specificity of PKCα in sLTP, has been reported to bind with PICK1 (protein interacting with C-kinase 1), a PDZ- and BAR domain-containing protein implicated in bidirectional plasticity 33,43. This domain likely regulates PKCα activation by localizing it to subdomains enriched in upstream signaling molecules (e.g. calcium nanodomains and/or DAG enriched lipid domains), as well regulating its access to downstream substrates via scaffolding/effector proteins44.
Previous analysis has demonstrated that PKCβ KO animals, while displaying normal LTP and hippocampal-dependent learning, showed impaired fear learning45. PKCγ KO animals, on the other hand, had impaired long term potentiation (LTP)46,47, that could be rescued by the prior induction of long-term depression, suggesting that PKCγ may play a role in modulating hippocampal-dependent plasticity (e.g. homeostatic regulation). Taken together, while PKCα appears to be the primary classic isozyme involved in the induction of hippocampal plasticity and hippocampus-dependent learning, other isozymes are likely engaged during various physiologic neuromodulatory states such as attention, emotional salience or stress.
The convergence of canonical plasticity promoting NMDARs and BDNF/TrkB-mediated signaling on PKCα is consistent with a central role for PKCα in the mechanisms that underlie synaptic plasticity. While these upstream signals are required for PKCα activation, we cannot rule out the presence of additional upstream regulation of PKCα. In particular, phosphorylation of PKC isozymes by mTOR complex 2 (mTORC2) and 3-Phosphoinositide-dependent kinase 1 (PDK1) are required for PKCα stability and activation48. While these phosphorylation events are generally constitutive, recent evidence suggests acute regulation in some cases 48; a potentially interesting avenue for future study given the role of mTORC2 in synaptic plasticity 49.
The integration of multiscale signals by PKCα is one mechanism through which synapse-specificity of plasticity and the clustered plasticity models may be coordinated. Evidence suggests that plasticity mechanisms are allocated across multiple temporal and spatial scales, thus mechanisms of signal integration across domains is necessary50. In addition to playing a role in synaptic plasticity, PKC isozymes have been implicated in numerous cellular and disease related functions in neuronal and non-neuronal cells, including cell division, proliferation, apoptosis, and differentiation. The integration of signals that differ in their time and space scales may be a more general mechanism employed by the classic PKC family to enable complex information processing for various cellular functions. As disentangling specific functions of the closely related PKC isozymes is a common need across disciplines, we anticipate that the tools developed here will be useful for broad cell biology applications.
Materials and Methods:
DNA constructs:
ITRACK: Donor plasmids were made using site independent PIPE to clone PKCα (Bos taurus), PKCβI (rat), and PKCγ (mouse) into cmv promotor containing mEGFP C1 vectors such that mEGFP was fused to the N-terminus of PKCα and PKCγ, and the C-terminus of PKCβ (N-terminal tagging of PKCβ showed poor expression). PKCβI was chosen for sensor design instead of PKC βII due to having a larger reported membrane translocation after high frequency electrical stimulation 51. The acceptor plasmid, mCh-CAAX, consisted of mCherry followed by a 14 aa linker and a K-Ras derived lipid targeting motif (DGKKKKKKSKTKCVIM) driven by CMV promoter. Negative controls for ITRACK consisted of the same donor construct and a control acceptor construct in which a stop codon was introduced before the lipid targeting domain or an acceptor construct that was targeted to the membrane with an H-Ras derived lipid targeting domain (DESGPGCMSCKCVLS).IDOCKS:Donor plasmids consisted of PKCα, PKCβII, or PKCγ which were tagged on their C termini with mEGFP driven by CMV promotor. The acceptor construct, 2mCh-PS, consisted of two copies of mCherry separated by an eight amino acid linker followed immediately by the 18 amino acid pseudosubstrate region from PKCα/β (RFARKGALRQKNVHEVKN) driven by the CMV promoter. The negative control IDOCKS consisted of the same acceptor construct with a single point mutation R→E (RFARKGALEQKNVHEVKN). mEGFP-PKCαΔQSAV was made by introducing a single point mutation into the mEGFP-PKCα/C1 to introduce a stop codon before the last 4 amino acids (QSAV) of PKCα. mEGFP-PKCγ+QSAV was made by inserting the nucleotide sequence encoding QSAV immediately before the C-terminal stop codon. Photoactivatable GFP constructs were made by replacing mEGFP with photoactivatable mEGFP in mEGFP-PKCα/C1 plasmids. pGP-CMV-GCaMP6f was a gift from Douglas Kim (Addgene plasmid # 40755 pGP-CMV-GCaMP6f was a gift from Douglas Kim (Addgene plasmid # 40755). pGP-CMV-NES-jRGECO1a was a gift from Douglas Kim (Addgene plasmid # 61563). CMV-ShadowY was a gift from Hideji Murakoshi (Addgene plasmid # 104621). All cloning procedures were performed using standard PCR reactions and PIPE or Gibson Assembly (NEB). Single point mutations were introduced using Q5 site directed mutagenesis kit using PCR (NEB).
HeLa cell maintenance, transfection and imaging:
HeLa cells (ATCC CCL-2) were cultured in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum at 37 °C in 5% CO2 and transfected with various plasmids using Lipofectamine 2000 (Invitrogen). For HeLa cells ITRACK was transfected at a ratio of donor plasmid to acceptor plasmid of 1:3. For IDOCKS, the ratio of the plasmids encoding the donor and the acceptor was 1:2. Imaging was performed 24–48h following transfection in a HEPES buffered aCSF solution with 2 mM CaCl2 and 2 mM MgCl2 by 2pFLIM as described below. When indicated, cells were stimulated with 1 μM PdBu (Tocris) and then 2.5 minutes later 1 μM Ionomycin (Tocris) or DMSO (0.02%) vehicle.
Animals:
All experimental procedures were approved by the Max Planck Florida Institute for Neuroscience Animal Care and Use Committee, in accordance with guidelines by the US National Institutes of Health. P3–P8 mouse pups from both sexes were used for organotypic slices for imaging studies. P30–P50 mice of both sexes were used for acute slices for electrophysiological studies. 2– 4 month old male mice were used for behavioral studies.
‒Nontransgenic animals, C57Bl/6N Crl, were received from Charles River Lab.
‒PKCα KO 129/sv and PKCβ KO 129/sv animals were developed by Dr. Michael Leitges as previously described52,53. Animals were crossed to C57Bl/6N Crl and are on a mixed background. For all experiments WT littermates were used as controls for KO animals.
‒PKCγ KO animals were developed by Dr. Susumu Tonegawa as previously described46 and received from Jackson Labs B6;129P2-Prkcgtm1Stl/J Stock No: 002466. Animals were crossed with C57Bl/6N Crl and are on a mixed background. For all experiments WT littermates were used as controls for KO animals.
‒Double (β, γ) knockout and triple (α, β, γ) KO animals were bred by crossing single KO animals. Breeding pairs were KO on one or two genes and Het on the remaining. A parallel WT control line was bred to match offspring by strain and generation.
‒TrkBF616A C57bl/6 mutant mice were developed and provided by Dr. David Ginty as previously described36.
‒BDNF fl/fl C57bl/6 mice were developed and provided by Dr. Luis Parada as previously described54.
Organotypic hippocampal slice cultures and transfection:
Organotypic hippocampal slices were prepared from wildtype or transgenic postnatal 3–8 day old pups of both sexes as previously described55. In brief, pups were euthanized by deep anesthesia by isofluorane followed by decapitation. After removing the brain, hippocampi were dissected and 350 μm thick coronal hippocampal slices were cut using a McIlwain tissue chopper (Ted Pella, Inc) and plated on hydrophilic PTFE membranes (Millicell, Millipore). Slices were maintained in culture medium containing MEM medium (Life Technologies), 20% horse serum, 1mM L-Glutamine, 1mM CaCl2, 2mM MgSO4, 12.9mM D-Glucose, 5.2mM NaHCO3, 30mM Hepes, 0.075% Ascorbic Acid, 1μg/ml Insulin. Medium was exchanged every other day. After 7–10 days in culture, CA1 pyramidal neurons were transfected with biolistic gene transfer 56 (Bio-Rad) using 1.0 or 1.6 μm gold beads (8–12 mg) coated with plasmids containing DNA of interest in the following amounts. mEGFP: 15 μg ITRACK or ITRACKneg: 15 μg donor (mEGFP- PKCα, PKCβ-mEGFP, or mEGFP-PKCγ) 40 μg acceptor (mCh-CAAX or mCh-CAAX_neg). IDOCKS or IDOCKSneg: 10 μg donor (PKCα-mEGFP, PKCB-mEGFP, or PKCγ-mEGFP) 20 μg Acceptor (2mCh-PS or 2mCh-PSmut). Cre Recombinase: 10 μg Cre with or without ITRACK DNA. Photoactivatable constructs and PDZ binding domain mutant constructs were transfected in the same ratios as ITRACK and IDOCKS. Secondary or tertiary apical dendrites in the stratum radiatum of transfected CA1 pyramidal neurons were imaged 3–7 days after transfection (for PKC sensors) by 2pFLIM as described below.
2-photon microscopy and 2pFLIM:
PKC isozyme activity was measured using 2pFLIM. For quantification of spine volume change, we monitored the fluorescence intensity change of mEGFP in the spines using regular two-photon microscopy that was simultaneous with lifetime measurements. Intensity measurements as well as 2pFLIM imaging in HeLa cells and slices was performed using custom 2p microscope as previously described 57. Briefly, mEGFP and mCh were excited with a Ti:sapphire laser (Chameleon, Coherent) at a wavelength of 920 nm and a power of 1.4–1.6 mW measured below the objective. The fluorescence was collected with an objective (60×, 1.0 n.a, Olympus), divided with a dichroic mirror (565 nm) and detected with two separated photoelectron multiplier tubes placed after wavelength filters (Chroma, 510/70–2p for green and 620/90–2p for red). Photoelectron multiplier tubes with low transfer time spread (H7422–40p; Hamamatsu) were used for both red and green channels. Fluorescence images were acquired with PCI-6110 (National instrument) and displayed on ScanImage 3.8 on Matlab 58. Fluorescence lifetime images were obtained using a time-correlated single photon counting board (SPC-150; Becker and Hickl) controlled with custom software in Matlab 24. Intensity images for analysis of sLTP volume change were collected by 128×128 pixels as a z stack of three slices with 1 μm separation and averaging 5 frames/slice. Spine volume change was calculated as the background subtracted F - F0 where F0 was the average fluorescence intensity before stimulation. In many cases of sLTP volume imaging parallel automated imaging of 4 spines per neuron was done using a custom-built interface in MATLAB by employing algorithms for autofocusing and drift correction to maintain position and focus59–61. A 2 min stagger of uncaging events was incorporated to avoid data loss during uncaging events. FLIM imaging was collected by 32×32 pixels at 128 ms/frame, equal to a 7.8 Hz temporal resolution. Frames were not averaged. For spatial analysis of FLIM data, images were collected by 64×64 pixels at 128ms/frame. Frames were not averaged.
Two-photon glutamate uncaging:
Structural plasticity of dendritic spines was induced using a Ti: Sapphire laser tuned at a wavelength of 720 nm to uncage 4-Methoxy-7-nitroindolinyl-caged-L-glutamate (MNI-caged glutamate, Tocris) in a small region ~0.5 μm from the spine with a 30 pulse, 0.5 Hz train. The power of the laser was set to 2.7 mW measured at the objective. For plasticity inducing experiments the width of the uncaging pulses was set depending on depth of the stimulated spine (4 ms for spines less than 10 μm deep, 6 ms for spines between 10–20 μm deep and 8 ms for spines 20–50 μm deep). Spines deeper than 50 μm were not selected for uncaging experiments. Experiments were performed in Mg2+ fee artificial cerebral spinal fluid (ACSF; 127 mM NaCl, 2.5 mM KCl, 4 mM CaCl2, 25 mM NaHCO3, 1.25 mM NaH2PO4 and 25 mM glucose) containing 1 μM tetrodotoxin (TTX) and 4 mM MNI-caged L-glutamate aerated with 95% O2 and 5% CO2. Experiments were performed at room temperature (24–26°C). For heterosynaptic facilitation experiments, spines at a depth between 5 μm and 25 μm were selected. Unpaired spines were depth matched within 5 μm to paired spines on a given neuron. Subthreshold stimulations were given by changing the uncaging laser dwell time to 1ms. Suprathreshold stimulations had a 4 or 6 or 8 ms dwell time depending on depth as described above. ‘Unpaired’ subthreshold stimuli were given first, spread across different dendrites (~4 per neuron). Following, ‘paired’ subthreshold stimulation was given 3–4 min after sLTP was induced in a spine <10 μm away.
Calcium imaging and simultaneous imaging of calcium and FLIM sensors:
For calcium imaging in response to glutamate uncaging organotypic slices were transfected with CMV driven GCaMP6f and cmv-driven mCyRFP62. Intensity was monitored with a time resolution of 64ms. Upon induction of plasticity, spine volume increases so GCaMP6f was normalized to increases in mCyRFP intensity due to this volume change. For simultaneous FLIM and calcium imaging, fluorescence lifetime measurements, together with intensity images, were taken as described above. Calcium imaging was measured by intensity in channel 2 using jRGECO1a excited with 1100 nm, 120fs pulsed, 80MHz, two-photon INSIGHT laser (Spectra-Physics). For these experiments the ITRACK acceptor construct needed to be modified to avoid contamination of the calcium imaging channel. Therefore the mCh fluorophore was replaced with ShadowY, a dark YFP based fluorophore. Slices were transfected via biolistics with the following mix of constructs: sY-ITRACKα containing 15 μg donor (mEGFP- PKCα) 40 μg acceptor (ShadowY-CAAX) and 15ug of jRGECO1a. Care was taken to avoid imaging of neurons with particularly high shadow Y expression which could be detected by its short lifetime. For analysis of paired spines, volume change of the paired spine was monitored to ensure successful induction of heterosynaptic plasticity before further analysis of PKCα activity or calcium influx was done. FLIM data was analyzed as described below. Intensity changes in jRGECO1a were normalized for increases in spine volume due to sLTP.
2pFLIM analysis:
To measure the fraction of donor that was undergoing FRET with acceptor (% Translocation or % Substrate Docking), we fit a fluorescence lifetime curve summing all pixels over a whole image with a double exponential function convolved with the Gaussian pulse response function:
where τAD is the fluorescence lifetime of donor bound with acceptor, PD and PAD are the fraction of free donor and donor undergoing FRET with acceptor, respectively, and H(t) is a fluorescence lifetime curve with a single exponential function convolved with the Gaussian pulse response function:
in which τD is the fluorescence lifetime of the free donor, τG is the width of the Gaussian pulse response function, F0 is the peak fluorescence before convolution and t0 is the time offset, and erfc is the complementary error function.
We obtained τD from single exponential fits (PAD = 0) to fluorescence lifetime curves of mEGFP-PKCα expressed in HeLa cells (2.65 ns), and then obtained τAD by double exponential fits to fluorescence lifetime curves of the sensors (ITRACK or IDOCKS) in HeLa cells which were stimulated with strong PKC activators (1.33 ns). For experimental data, we fixed τD and τAD to these values to obtain stable fitting. To generate the fluorescence lifetime image, we calculated the mean photon arrival time, <t>, in each pixel as:
Then, the mean photon arrival time is related to the mean fluorescence lifetime, <τ>, by an offset arrival time, to, which is obtained by fitting the whole image:
The fraction of PKC isozyme that participated in FRET was calculated from obtained <τ> as:
Change in translocation or substrate docking were calculated as PAD – PAD0 where PAD0 was the average PAD before stimulation. Data with lifetime fluctuations in the baseline that were greater than .1 ns were excluded before further analysis. Lifetime drift was corrected in analysis.
Pharmacology Experiments:
All pharmacological experiments were performed in a paired manner for each neuron. Structural plasticity was induced in spines before and after drug application in the same neuron. 1NMPP1 (Santa Cruz, WD 1 μM) was prepared as a 10 mM stock in DMSO and then diluted to 500x in a solubilisation buffer (0.9% NaCl and 2.5% Tween-20). U73122 and U73343 (Tocris, μM) WD 10 (Gö6983 (Tocris, 1 μM WD), Phorbol 12,13-dibutyrate (PdBu, Tocris, WD 1 μM), Edelfosine (Tocris, WD 1 μM), NMDA (Tocris, WD 20 μM), MNI Caged Glu (Tocris, 4 mM), APV (Sigma, WD 50 μM), S-MCPG (Tocris, WD 250 μM), NPS 2390 (Tocris, WD 20 μM) were stored as recommended by supplier and diluted into aCSF to the working dilutions (WD) listed.
Electrophysiology:
Acute slice preparation: PKCα WT or KO littermate mice (P30–P50) were sedated by isoflurane inhalation, and perfused intracardially with a chilled choline chloride solution. Brain was removed and placed in the same choline chloride solution composed of 124 mM Choline Chloride, 2.5 mM KCl, 26 mM NaHCO3, 3.3 mM MgCl2, 1.2 mM NaH2PO4, 10 mM Glucose and 0.5 mM CaCl2, pH 7.4 equilibrated with 95%O2/5%CO2. Coronal slices (400 μm) containing the hippocampus were cut using a vibratome (Leica) and maintained in a submerged chamber at 32 °C for 1h and then at room temperature in oxygenated ACSF.
Extracellular Recordings and LTP protocol: Slices were perfused with oxygenated ACSF containing 2 mM CaCl2, 2 mM MgCl2 and 100 μM picrotoxin. One glass electrode (resistance ~4 MΩ) containing the same ACSF solution was placed in the dendritic layer of CA1 area (~100–200 μm away from the soma) while stimulating Schaffer Collateral fibers with current square pulses (0.1 ms) using a concentric bipolar stimulation electrode (FHC). The initial slope of the EPSP was monitored with a custom software (MatLab). The stimulation strength was set to ~50% saturation. LTP was induced by applying 3 sets of high frequency stimuli (100 Hz, 1 s) with 20 s intervals. All data was analyzed with an in-house program written with Matlab. Data is presented as mean ± SEM.
Animal Behavior:
General Methods:
Adult (2–4 months) male constitutive PKCα KO 129/sv53 and non-transgenic littermate mice from HET/HET breeding were used in behavioral experiments. All animal experiments were performed in a double-blinded manner. ~70dB background white noise was used during all tests with the exception of water maze and cued fear conditioning testing. One hour prior to testing, mice were moved from their holding room to a waiting room in which the light level was set to that of the test apparatus for each respective assay in order to ensure the mice were adjusted to the lighting level of each test. Mice were habituated to the testing facility and to being handled by the experimenter for 3 days prior to the start of the test battery. Tasks below were performed in the order they are described.
Elevated plus maze:
Mice were placed in the center of the plus maze (Med Associates, St. Albans, VT) and allowed to explore the maze for 5 minutes. Time spent in open and closed arms, number of arm entries, latency to initially enter an open arm and total distance moved were recorded using EthoVision XT (Noldus Information Technology Inc., Leesburg, VA). Uniformity in lighting was confirmed across the maze, and the maze was cleaned with 1% Micro-90 before each trial.
Open field:
The open field consisted of a 17×17 in. square acrylic open field chamber. Prior to testing, uniformity of light across the arena was confirmed using a light intensity meter, and the chambers were cleaned with 1% Micro-90 before and between trials. Mice were placed into the center of the chamber to begin testing, and activity was recorded for 30 min using EthoVision XT (Noldus Information Technology Inc., Leesburg, VA).
Spontaneous alternation:
Mice received 2 tests, each separated by three days. Two “T”-shaped mazes (Med Associates, St. Albans, VT) were used, each turned in a different configuration. Automatic doors were installed at the entry of each arm that were controlled by EthoVision XT (Noldus Information Technology Inc., Leesburg, VA). Spontaneous alternation trials were performed as described previously 63. The mouse was placed in a start and allowed to enter and choose an arm to enter (free choice trial). Once a choice was made the mouse was detained in that arm automatically by a door for a period of 10 seconds. The mouse was then immediately placed back in the start box for a second free choice trial (no delay). The second test was performed in the same way except that the mouse was delayed in the start box for 60s before the second trial (delay trial) Groups were balanced for maze, test day, and delay. Alternation success was calculated for each test. If a mouse did not leave the start box to enter the maze after 60s, it was gently nudged with a cotton swab. The maze was cleaned with 70% ethanol between trials and between mice.
Morris water maze:
The Morris water maze was performed as described previously 64,65. The water maze consisted of a 1.4 m diameter white tank with a 10 cm diameter platform submerged approximately 1 cm below the surface of the water. The water was made opaque using non-toxic white washable paint that made the platform invisible during trials. The temperature of the water was kept at 22–24°C. Visual cues were placed in the testing room around the tank for spatial reference. Prior to water maze training, mice received a ‘cued’ visual platform test where the spatial cues surrounding the tank were removed and the platform was elevated above the surface of the water and marked with a cue so it could clearly be discerned. Mice were given four trials, and the platform location was varied over trials. For the hidden platform test, mice were given four acquisition trials per day until they reached a pre-determined criteria of an average latency (by group) of 20s to find the platform and at least 95% success in finding the platform. The start location was varied for each trial and mice were allowed 60 seconds to find the platform. Mice were left on the platform for 15 seconds before removing them from the water maze. If a mouse did not find the platform within 60 seconds, it was placed on or guided to the platform and kept there for 15 seconds. Mice were dried after each trial and placed into cages located atop heating pads to prevent hypothermia. Daily acquisition trials were averaged for analysis. 24 and 96 hours after reaching criteria, mice were given a probe trial during which the platform was not present, and a novel start location was used for these trials. Activity and performance was tracked using EthoVision XT (Noldus Information Technology, Leesburg, VA). Total time spent in each quadrant, total number of entries into the target quadrant, total number of platform crossings, latency to first platform crossing, and average distance to the platform center were recorded for a 60 second probe trial.
Following the 2nd probe trial, mice were tested in a reversal learning paradigm where the platform was moved to the opposite quadrant location as before and mice were trained to find the platform in this new location. Again, mice were trained until they reached criteria. Mice received probe trials throughout the reversal learning phase of the test every 2 days (i.e., on reversal learning acquisition days 2, 4, etc., until criteria was reached) and then at 24 and 96 hours after reaching criteria for the new platform location. Mice also received a remote probe trial 40 days post-reversal acquisition.
Fear conditioning:
The fear conditioning chambers (Noldus Information Technology, Leesburg, VA) have been previously described 66. Each chamber was cleaned with 70% ethanol prior to each trial. White light was used inside the chamber for training and testing, and white noise (approximately 75dB) was played in the room during training and context tests to mask any unintended noise that might add to the context. The tray below the shock grid floor was lined with Techboard Ultra paper (Shepherd Specialty Papers) and was changed out after each trial. Mice were placed individually into each chamber and allowed to explore for 2.5 min. Mice then received a series of three 30-second 85dB white noise tone presentations co-terminating with a one-second 0.75 mA foot shock, each occurrence being separated by 60 and 90 seconds, respectively. 30 s following the last foot shock, the mice were removed from the chambers and returned to their home cages. 24 h later, mice were tested for contextual fear conditioning by placing them back into the chambers for 5 min. Percent of time spent freezing (immobility except for breathing) and activity were recorded. At least one hour after context testing, the mice were tested for cued fear conditioning by placing them into the chambers after disguising them with inserts that changed the look, dimensions and feel of the chamber and with orange extract placed behind the inserts to change the smell of the chamber. 70% isopropanol was used instead of ethanol to clean the chambers, as well, and white noise was not used during the trials. Mice were given three minutes to explore the “new” context before being presented with the 85dB white noise tone for three minutes. Again, activity and freezing during the pre-tone and tone phases was recorded.
Hot plate:
The hot plate (Ugo Basile, Collegeville, PA) temperature was automatically controlled and set to 54°C. In addition, the hot plate had a removable clear acrylic cylinder surrounding the plate area. Mice were individually placed onto the hot plate, and latency to show a nociceptive response was manually recorded with a button press. The hot plate and acrylic cylinder were cleaned with 70% ethanol in between animals and verified that was dried before placing a new animal. A nociceptive response was defined as lifting or licking of the rear paws or jumping and served as a correlation of pain sensitivity or analgesia. The maximum test time was 30 s, although all mice showed a nociceptive response prior to reaching the cutoff time.
Experimental Design and Statistical analysis:
All values are presented as mean ± SEM unless otherwise noted. Number of independent measurements (n) is indicated in figure legends. No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications6,38. For in-vitro studies, neurons were assigned to different groups to randomly interleave control and experimental groups from slices prepared from the same animals. Experiments from control and experimental groups were randomly interleaved during experimentation with the exception that each block of experiments began with one neuron from the control group to ensure technical success of experiments. Data distribution was assumed to be normal but this was not formally tested. Unpaired two-tailed student’s t test was used for comparing two independent samples. One way ANOVA followed by multiple comparison tests was used for comparing more than two independent samples. Two-way ANOVA followed by multiple comparison test was used to compare grouped data sets. If data were paired, repeated measures ANOVA was used as indicated. Šídák-Bonferroni (Sidak) multiple comparison test was used to compare specific relevant groups. Dunnett’s post-test was used when comparing all groups to a control group. For linear regression decay of activity was fit with least squared regression. Extra sum of squares F test was used to compare Y0 and K (1/τ) values of different groups. Correlation analysis was done by computing Pearson correlation coefficients. All t-tests and one-way ANOVAs included formal testing for differences in variance (e.g F test or Bartlett’s test to compare variances) and appropriate corrections were made and are indicated in instances where this was relevant. Statistical tests and p values are noted in each figure legend and were computed using GraphPad Prism 7.03 for Windows, GraphPad Software, La Jolla California USA, www.graphpad.com. For in-vitro experiments, data collection and analysis were not performed blind to the conditions of the experiments. Neurons in which there were obvious signs of poor health that developed during the experiment such as beading of processes were excluded before analysis. For in-vivo behavioural experiments, data and analysis were performed in a double blind manner. Additional information can be found in the Life Sciences Reporting Summary.
Supplementary Material
Acknowledgments
Acknowledgements: We would like to thank A.F. Brantley and the Scripps Florida behavior core who performed animal behavioral studies, Luis Parada (Memorial Sloan Kettering) for Bdnf fl/fl mice, David Ginty (Harvard) for TrkBF616A mice, M. Dowdy and the MPFI ARC for animal care, members of the Yasuda laboratory, L. Yan and D. Kloetzer. This work was funded by F32MH101954 (LAC), R01MH080047 (RY), 1DP1NS096787 (RY) and the Max Planck Florida Institute for Neuroscience.
Footnotes
Accession Codes: DNA sequences and plasmids used for the PKC sensors developed can be found at Addgene with the following accession codes. Xxxxxx
The authors declare no competing financial or non-financial interests as defined by Nature Research.
Code and Data Availability Statement:
The datasets generated during and/or the code used to analyze the data in the current study are available from the corresponding author on reasonable request.
References and Notes:
- 1.Nicoll RA & Roche KW Long-term potentiation: peeling the onion. Neuropharmacology 74, 18–22, doi: 10.1016/j.neuropharm.2013.02.010 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nishiyama J & Yasuda R Biochemical Computation for Spine Structural Plasticity. Neuron 87, 63–75, doi: 10.1016/j.neuron.2015.05.043 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hulme SR, Jones OD & Abraham WC Emerging roles of metaplasticity in behaviour and disease. Trends Neurosci 36, 353–362, doi: 10.1016/j.tins.2013.03.007 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Harvey CD & Svoboda K Locally dynamic synaptic learning rules in pyramidal neuron dendrites. Nature 450, 1195–1200, doi: 10.1038/nature06416 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Govindarajan A, Israely I, Huang SY & Tonegawa S The dendritic branch is the preferred integrative unit for protein synthesis-dependent LTP. Neuron 69, 132–146, doi: 10.1016/j.neuron.2010.12.008 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hedrick NG et al. Rho GTPase complementation underlies BDNF-dependent homo- and heterosynaptic plasticity. Nature 538, 104–108, doi:10.1038/nature19784 10.1038/nature19784http://www.nature.com/nature/journal/v538/n7623/abs/nature19784.html#supplementary-informationhttp://www.nature.com/nature/journal/v538/n7623/abs/nature19784.html#supplementary-information (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makino H & Malinow R Compartmentalized versus global synaptic plasticity on dendrites controlled by experience. Neuron 72, 1001–1011, doi: 10.1016/j.neuron.2011.09.036 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu M, Yu X, Lu J & Zuo Y Repetitive motor learning induces coordinated formation of clustered dendritic spines in vivo. Nature 483, 92–95, doi:http://www.nature.com/nature/journal/v483/n7387/abs/nature10844.html#supplementary-information (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi N et al. Locally Synchronized Synaptic Inputs. Science 335, 353–356, doi: 10.1126/science.1210362 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Olds JL, Anderson ML, McPhie DL, Staten LD & Alkon DL Imaging of memory-specific changes in the distribution of protein kinase C in the hippocampus. Science 245, 866–869 (1989). [DOI] [PubMed] [Google Scholar]
- 11.Pastalkova E et al. Storage of spatial information by the maintenance mechanism of LTP. Science 313, 1141–1144, doi: 10.1126/science.1128657 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Hongpaisan J & Alkon DL A structural basis for enhancement of long-term associative memory in single dendritic spines regulated by PKC. Proceedings of the National Academy of Sciences of the United States of America 104, 19571–19576, doi: 10.1073/pnas.0709311104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malinow R, Schulman H & Tsien RW Inhibition of postsynaptic PKC or CaMKII blocks induction but not expression of LTP. Science 245, 862–866 (1989). [DOI] [PubMed] [Google Scholar]
- 14.Hu GY et al. Protein kinase C injection into hippocampal pyramidal cells elicits features of long term potentiation. Nature 328, 426–429, doi: 10.1038/328426a0 (1987). [DOI] [PubMed] [Google Scholar]
- 15.Malinow R, Madison DV & Tsien RW Persistent protein kinase activity underlying long-term potentiation. Nature 335, 820–824, doi: 10.1038/335820a0 (1988). [DOI] [PubMed] [Google Scholar]
- 16.Routtenberg A et al. Phorbol ester promotes growth of synaptic plasticity. Brain research 378, 374–378 (1986). [DOI] [PubMed] [Google Scholar]
- 17.Huang FL, Yoshida Y, Nakabayashi H, Young WS 3rd & Huang KP Immunocytochemical localization of protein kinase C isozymes in rat brain. The Journal of neuroscience : the official journal of the Society for Neuroscience 8, 4734–4744 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark EA, Leach KL, Trojanowski JQ & Lee VM Characterization and differential distribution of the three major human protein kinase C isozymes (PKC alpha, PKC beta, and PKC gamma) of the central nervous system in normal and Alzheimer’s disease brains. Laboratory investigation; a journal of technical methods and pathology 64, 35–44 (1991). [PubMed] [Google Scholar]
- 19.Kose A, Ito A, Saito N & Tanaka C Electron microscopic localization of gamma- and beta II-subspecies of protein kinase C in rat hippocampus. Brain research 518, 209–217 (1990). [DOI] [PubMed] [Google Scholar]
- 20.Ito A et al. Immunocytochemical localization of the alpha subspecies of protein kinase C in rat brain. Proceedings of the National Academy of Sciences of the United States of America 87, 3195–3199 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schleifenbaum A, Stier G, Gasch A, Sattler M & Schultz C Genetically encoded FRET probe for PKC activity based on pleckstrin. Journal of the American Chemical Society 126, 11786–11787, doi: 10.1021/ja0460155 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Violin JD, Zhang J, Tsien RY & Newton AC A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. The Journal of cell biology 161, 899–909, doi: 10.1083/jcb.200302125 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Braun DC, Garfield SH & Blumberg PM Analysis by Fluorescence Resonance Energy Transfer of the Interaction between Ligands and Protein Kinase Cδ in the Intact Cell. Journal of Biological Chemistry 280, 8164–8171, doi: 10.1074/jbc.M413896200 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Yasuda R Imaging spatiotemporal dynamics of neuronal signaling using fluorescence resonance energy transfer and fluorescence lifetime imaging microscopy. Current opinion in neurobiology 16, 551–561, doi: 10.1016/j.conb.2006.08.012 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Steinberg SF Structural basis of protein kinase C isoform function. Physiological reviews 88, 1341–1378, doi: 10.1152/physrev.00034.2007 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Apolloni A, Prior IA, Lindsay M, Parton RG & Hancock JF H-ras but not K-ras traffics to the plasma membrane through the exocytic pathway. Molecular and cellular biology 20, 2475–2487 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayashi-Takagi A et al. Labelling and optical erasure of synaptic memory traces in the motor cortex. Nature 525, 333–338, doi: 10.1038/nature15257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuzaki M, Honkura N, Ellis-Davies GC & Kasai H Structural basis of long-term potentiation in single dendritic spines. Nature 429, 761–766, doi: 10.1038/nature02617 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gschwendt M et al. Inhibition of protein kinase C mu by various inhibitors. Differentiation from protein kinase c isoenzymes. FEBS letters 392, 77–80 (1996). [DOI] [PubMed] [Google Scholar]
- 30.Sakai N et al. Direct visualization of the translocation of the gamma-subspecies of protein kinase C in living cells using fusion proteins with green fluorescent protein. The Journal of cell biology 139, 1465–1476 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner S, Harteneck C, Hucho F & Buchner K Analysis of the subcellular distribution of protein kinase Calpha using PKC-GFP fusion proteins. Experimental cell research 258, 204–214, doi: 10.1006/excr.2000.4925 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Feng X et al. Visualization of dynamic trafficking of a protein kinase C betaII/green fluorescent protein conjugate reveals differences in G protein-coupled receptor activation and desensitization. The Journal of biological chemistry 273, 10755–10762 (1998). [DOI] [PubMed] [Google Scholar]
- 33.Staudinger J, Lu J & Olson EN Specific Interaction of the PDZ Domain Protein PICK1 with the COOH Terminus of Protein Kinase C-α. Journal of Biological Chemistry 272, 32019–32024, doi: 10.1074/jbc.272.51.32019 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Minichiello L et al. Mechanism of TrkB-mediated hippocampal long-term potentiation. Neuron 36, 121–137 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Korte M et al. Hippocampal long-term potentiation is impaired in mice lacking brain-derived neurotrophic factor. Proceedings of the National Academy of Sciences of the United States of America 92, 8856–8860 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X et al. A chemical-genetic approach to studying neurotrophin signaling. Neuron 46, 13–21, doi: 10.1016/j.neuron.2005.03.009 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Harward SC et al. Autocrine BDNF–TrkB signalling within a single dendritic spine. Nature, doi:10.1038/nature19766 10.1038/nature19766http://www.nature.com/nature/journal/vaop/ncurrent/abs/nature19766.html#supplementary-informationhttp://www.nature.com/nature/journal/vaop/ncurrent/abs/nature19766.html#supplementary-information (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhai S, Ark ED, Parra-Bueno P & Yasuda R Long-distance integration of nuclear ERK signaling triggered by activation of a few dendritic spines. Science 342, 1107–1111, doi: 10.1126/science.1245622 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen T-W et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295, doi:10.1038/nature12354 10.1038/nature12354https://www.nature.com/articles/nature12354#supplementary-informationhttps://www.nature.com/articles/nature12354#supplementary-information (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dana H et al. Sensitive red protein calcium indicators for imaging neural activity. eLife 5, doi: 10.7554/eLife.12727 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murakoshi H & Shibata ACE ShadowY: a dark yellow fluorescent protein for FLIM-based FRET measurement. Scientific reports 7, 6791, doi: 10.1038/s41598-017-07002-4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee SJ, Escobedo-Lozoya Y, Szatmari EM & Yasuda R Activation of CaMKII in single dendritic spines during long-term potentiation. Nature 458, 299–304, doi: 10.1038/nature07842 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Terashima A et al. An Essential Role for PICK1 in NMDA Receptor-Dependent Bidirectional Synaptic Plasticity. Neuron 57, 872–882, doi: 10.1016/j.neuron.2008.01.028 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woolfrey KM & Dell’Acqua ML Coordination of Protein Phosphorylation and Dephosphorylation in Synaptic Plasticity. The Journal of biological chemistry 290, 28604–28612, doi: 10.1074/jbc.R115.657262 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weeber EJ et al. A role for the beta isoform of protein kinase C in fear conditioning. The Journal of neuroscience : the official journal of the Society for Neuroscience 20, 5906–5914 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abeliovich A et al. Modified hippocampal long-term potentiation in PKC gamma-mutant mice. Cell 75, 1253–1262 (1993). [DOI] [PubMed] [Google Scholar]
- 47.Abeliovich A et al. PKC gamma mutant mice exhibit mild deficits in spatial and contextual learning. Cell 75, 1263–1271 (1993). [DOI] [PubMed] [Google Scholar]
- 48.Freeley M, Kelleher D & Long A Regulation of Protein Kinase C function by phosphorylation on conserved and non-conserved sites. Cellular Signalling 23, 753–762, doi: 10.1016/j.cellsig.2010.10.013 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Hoeffer CA & Klann E mTOR Signaling: At the Crossroads of Plasticity, Memory, and Disease. Trends in neurosciences 33, 67, doi: 10.1016/j.tins.2009.11.003 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kastellakis G, Cai DJ, Mednick SC, Silva AJ & Poirazi P Synaptic clustering within dendrites: an emerging theory of memory formation. Progress in neurobiology 126, 19–35, doi: 10.1016/j.pneurobio.2014.12.002 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Method References
- 51.Sacktor TC et al. Persistent activation of the zeta isoform of protein kinase C in the maintenance of long-term potentiation. Proceedings of the National Academy of Sciences of the United States of America 90, 8342–8346 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leitges M et al. Immunodeficiency in protein kinase cbeta-deficient mice. Science 273, 788–791 (1996). [DOI] [PubMed] [Google Scholar]
- 53.Leitges M et al. Knockout of PKC alpha enhances insulin signaling through PI3K. Mol Endocrinol 16, 847–858, doi: 10.1210/mend.16.4.0809 (2002). [DOI] [PubMed] [Google Scholar]
- 54.He XP et al. Conditional deletion of TrkB but not BDNF prevents epileptogenesis in the kindling model. Neuron 43, 31–42, doi: 10.1016/j.neuron.2004.06.019 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Stoppini L, Buchs PA & Muller D A simple method for organotypic cultures of nervous tissue. Journal of neuroscience methods 37, 173–182 (1991). [DOI] [PubMed] [Google Scholar]
- 56.O’Brien JA & Lummis SC Biolistic transfection of neuronal cultures using a hand-held gene gun. Nature protocols 1, 977–981, doi: 10.1038/nprot.2006.145 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murakoshi H, Wang H & Yasuda R Local, persistent activation of Rho GTPases during plasticity of single dendritic spines. Nature 472, 100–104, doi: 10.1038/nature09823 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pologruto TA, Sabatini BL & Svoboda K ScanImage: flexible software for operating laser scanning microscopes. Biomedical engineering online 2, 13, doi: 10.1186/1475-925X-2-13 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smirnov MS, Evans PR, Garrett TR, Yan L & Yasuda R Automated remote focusing, drift correction, and photostimulation to evaluate structural plasticity in dendritic spines. bioRxiv, doi: 10.1101/083006 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sugar JD, Cummings AW, Jacobs BW, and Robinson David B.. A Free Matlab Script for Spatial Drift Correction. Microscopy Today 22, 40–47 (2014). [Google Scholar]
- 61.Geusebroek JM, Cornelissen F, Smeulders AW & Geerts H Robust autofocusing in microscopy. Cytometry 39, 1–9 (2000). [PubMed] [Google Scholar]
- 62.Laviv T et al. Simultaneous dual-color fluorescence lifetime imaging with novel red-shifted fluorescent proteins. Nature methods 13, 989–992, doi: 10.1038/nmeth.4046 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deacon RM & Rawlins JN T-maze alternation in the rodent. Nature protocols 1, 7–12, doi: 10.1038/nprot.2006.2 (2006). [DOI] [PubMed] [Google Scholar]
- 64.Vorhees CV & Williams MT Morris water maze: procedures for assessing spatial and related forms of learning and memory. Nature protocols 1, 848–858, doi: 10.1038/nprot.2006.116 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wenk GL Assessment of spatial memory using the radial arm maze and Morris water maze. Current protocols in neuroscience / editorial board, Crawley Jacqueline N. ... [et al. ] Chapter 8, Unit 8 5A, doi: 10.1002/0471142301.ns0805as26 (2004). [DOI] [PubMed] [Google Scholar]
- 66.Pham J, Cabrera SM, Sanchis-Segura C & Wood MA Automated scoring of fear-related behavior using EthoVision software. Journal of neuroscience methods 178, 323–326, doi: 10.1016/j.jneumeth.2008.12.021 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.