Abstract
An increasing number of studies have strongly correlated the composition of the human microbiota with many human health conditions and, in several cases, have shown that manipulating the microbiota directly affects health. These insights have generated significant interest in engineering indigenous microbiota community members and nonresident probiotic bacteria as biotic diagnostics and therapeutics that can probe and improve human health. In this review, we discuss recent advances in synthetic biology to engineer commensal and probiotic lactic acid bacteria, bifidobacteria, and Bacteroides for these purposes, and we provide our perspective on the future potential of these technologies.
Keywords: probiotics, genetic tools, CRISPR, genome engineering, microbiome
1. INTRODUCTION
A microbiota is a community of microbes that naturally associate closely with a multicellular host (1). These communities of bacteria and fungi interact extensively with the host in often symbiotic or commensal relationships that can cause a disease-associated state called dysbiosis when perturbed or unbalanced (2). Most studies on these communities have used metagenomics and metatranscriptomic sequencing techniques to provide a compositional snapshot of microbial species and their expressed genes to draw correlations to health or disease states. However, experimental approaches that extend beyond sequencing are necessary in order to fully understand the mechanistic relationships among the individual microbial species and those with their host. Recent advances in synthetic biology offer a means not only to determine structure- function relationships but also to engineer the microbiota to positively influence its host. Similar efforts could also advance probiotics, microbial species that are believed to confer health benefits when administered to humans. However, the tools needed to engineer complex phenotypes have been developed for use in model laboratory microbes like Escherichia coli, and they must be extended to host-associated microbial species to advance mechanistic understanding and engineering of host-microbiota interactions. The abundance of human-associated bacteria such as lactic acid bacteria (LABs), Bacteroides, and bifidobacteria that provide health benefits make them desirable engineering targets for therapeutic applications. This review focuses on recent advances in synthetic biology and approaches to microbial engineering for health applications.
2. CURRENT UNDERSTANDING OF THE ROLE OF MICROBIOTA IN HEALTH AND DISEASE
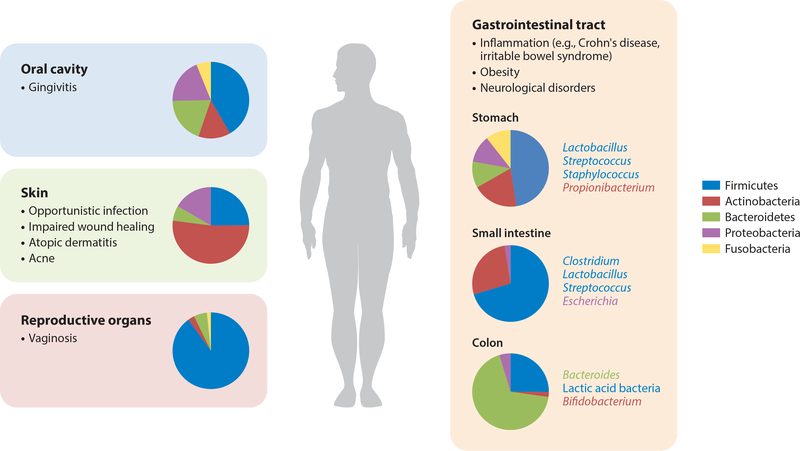
Different niches of a host are populated with distinct microbial communities that are thought to encode specialized functions (Figure 1). These niches include nearly all external and epithelial surfaces of the human body, including the skin, oral cavity, digestive tract, and reproductive system (see the sidebar titled Formation and Stability of the Microbiota) (1,3). The microbial community composition is often distinct even across niches of the same organs. For example, the composition of the microbiota associated with the duodenum differs from that associated with the ileocecal junction, despite both regions being close to one another in the digestive tract. When both the host and microbes benefit from the association, the relationship is considered symbiotic. Conversely, when the positive arrangement of the community is disrupted, dysbiosis can occur. This dysbiotic state is best understood when considering the human gut microbiota and its impact on health and disease (2). For instance, incidence of infections and subsequent treatment with broad-spectrum antibiotics have been shown to destabilize these communities and lead to dysbiosis (4). Such imbalance and/or instability of the various microbiota has been linked with disease states such as gut inflammation disorders [irritable bowel syndrome (IBS), Crohn’s disease, and others] (5, 6), obesity (7), cancer (8), and even neurological disorders (9). While the focus of most research efforts has been the bacteria associated with the gastrointestinal tract (GIT), microbiota of other organs and niches have been similarly correlated with other disease states. For instance, there are strong associations between the skin microbiota and conditions such as acne and dandruff (10, 11). Separately, dysbiosis of the oral cavity has been suggested to be a causative factor in conditions such as gingivitis and periodontitis (12, 13). Finally, bacterial vaginosis is believed to be caused by an overabundance of microbial diversity (14). The literature is replete with accounts of such correlations, which have sparked interest in fully elucidating the molecular mechanisms through which these communities of microbes interact with their human host, as well as in exploiting those insights to develop novel therapeutic interventions.
Figure 1.
Human microbiota. Microbes exist in niches throughout the human body. A healthy microbiota contributes to the overall health of the host and, conversely, when imbalanced can cause health complications. Microbial influences on disease states and the relative abundance of host-associated bacterial genera for different regions of the body are depicted. The most abundant species present in distinct locations of the digestive tract. The data presented are estimated from References 1 and 147–150.
Ingestion of commercial probiotics or prebiotics (nutrients closely associated with probiotics or microbiota) was the earliest form of microbial therapeutics with the goal of assisting or maintaining a healthy gut composition. Most probiotics are isolated from fermented foods (e.g., dairy, pickles) and are temporary members of the microbiota that transiently interact with the host and resident microbes (15). Products are often used either as single-strain formulations or as collections of naturally co-inhabiting species to benefit gut health. Most studies in this area to date have focused on correlating species composition with disease and health markers, although recent research by Garrett and colleagues (16) has elucidated the molecular mechanism through which Lactococcus lactis, a widely used probiotic, may counter gut inflammation. Supplementation with specific probiotic bacteria has also shown success in treating digestive tract disorders such as ulcerative colitis (17), IBS (5), and Crohn’s disease (18). Wholesale transfer of colonic microbiota from healthy donors to diseased individuals in a process known as fecal microbiota transplant (FMT) is significantly more effective than the use of vancomycin for the treatment of recurrent Clostridium difficile infections (CDI) (19, 20). Such findings have sparked interest in engineering these bacteria as vectors for a wide array of biomedical applications.
The efficacy of current probiotic supplementation strategy can be inconsistent and difficult to optimize due to limited knowledge of their mechanism of action. While FMTs have been successful at treating CDI, their mechanisms of action and knowledge about which constituent species constitute the bioactive component are poorly understood. In contrast, synthetic biology offers a direct way to rationally engineer probiotic organisms to directly treat disease, prevent infection, and maintain a healthy body. The inherent safety of probiotics and their ability to directly interact with a number of tissues during transit through the GIT make them highly desirable targets as a novel class of biotic therapeutics. However, a major bottleneck in the development of these technologies has been the genetic intractability of most of these largely fastidious bacteria. For these reasons, E. coli Nissle 1917 (EcN) has been the probiotic of choice for most proof-of-principle applications due to the availability of a wide array of genetic tools. Recently, however, researchers have recognized the unique advantages of using LABs, bifidobacteria, and Bacteroides, motivating the characterization and creation of new engineering tools.
3. SYNTHETIC BIOLOGY AS A MEANS TO STUDY AND ENGINEER HOST-MICROBE INTERACTIONS
Synthetic biology is a rapidly growing field that aims to design and achieve programmed cellular behavior by using natural and synthetic biological components. This type of forward engineering has created numerous biotechnological advances from chemical product biosynthesis to complex therapeutics (21, 22). Research in the field accelerated when it became possible to generate and quickly analyze large libraries of genetic components by next-generation sequencing. Notable developments include synthetic cell-to-cell communication (23), complex and large-scale genetic circuits (24), and clustered regularly interspaced short palindromic repeats (CRISPR)-based regulation (25). For the field of microbiota engineering, synthetic biology provides a means to study structure-function relationships among microbiota and engineer novel biotic therapeutics. Incorporation of synthetic genetic components broadens the abilities of the engineered microbe to sense, record, and respond to its local environment (26, 27). The success of the designed function depends on the availability of genetic tools. Currently, this toolbox is limited to model laboratory strains and a select few host-associated species. However, recent advances in expanding the genetic tools available for other non-model-host-associated microbes have opened the door for new and emerging applications for engineering synthetic biotics.
3.1. Current State of Genetic Engineering Tools
Metagenomic and 16S ribosomal RNA (rRNA) sequencing of human fecal samples has given rise to a plethora of genomic data and the means of identifying the composition of the gut microbiome (1 ). Comparing samples from healthy individuals and diseased individuals has generated hypotheses that the compositional makeup of the gut microbiota may play a major role in determining the physiological state of the host. For instance, reduced bacterial diversity in the gut has been correlated with obesity (7), and increased abundance of species in the vagina has been associated with bacterial vaginosis (28). In another study, researchers monitored the overall mass gain of mice who received a community microbiota transfer from either an obese or a lean donor twin (29). The recipient mouse who received microbiota from the lean donor twin gained less mass when fed a specific diet, suggesting that the lean donor microbiota was in some way responsible for the host’s metabolism (29).
In order to further elucidate these interactions and others, it is often desirable to culture strains individually, genetically modify them, and assess their effects on community composition and function. Unfortunately, it is estimated that most species present in the gut have not been isolated and remain unculturable using traditional laboratory techniques. Identifying conditions for the proper isolation and culturing of individual species are the first steps in genetic tool development. Recently, Lagier et al. (30) combined high-throughput culture condition testing with MALDTTOF (matrix-assisted laser desorption/ionization-time of flight) or 16S rRNA amplification and sequencing to identify unassigned species within metagenomic sequence databases. High-throughput isolation and screening will greatly increase the number of culturable species from the gut microbiota. The next steps to creating genetically tractable strains will be to (a) deliver and maintain exogenous DNA, (b) enable predictable expression of heterologous genes, and (r) edit or regulate endogenous genes. Of the identified bacterial species of the human gut microbiota, only a few currently possess the tools necessary for genetic manipulation. Nonmodel gut isolates of the genera Bacteroides, Bifidobacterium, and Lactobacillus, as well as isolates from various fermented food-associated LABs (such as pediococci, lactococci, and streptococci), have recently been genetically modified, although not to the same extent as strains of E. coli (31).
3.2. Incorporation and Maintenance of Foreign DNA
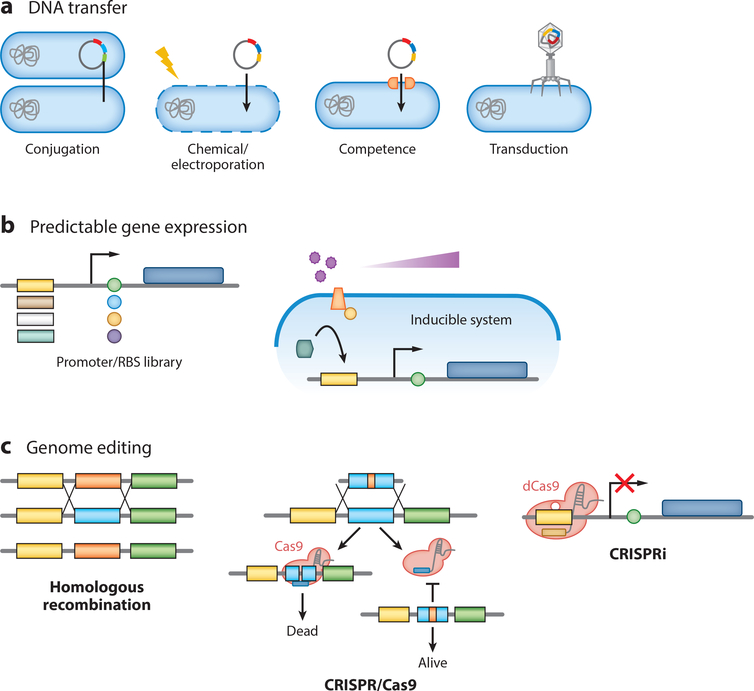
Once a bacterial strain has been isolated and deemed culturable, the next step is to stably incorporate exogenous DNA into the cell. Acceptance of foreign DNA depends on transport across the cell wall and membrane(s), avoiding rejection by the host’s defense systems, and active replication within the host. Transport into the cell has been achieved by disrupting the cellular wall and membrane (chemical disturbance, electroporation), injecting the DNA (transduction, conjugation), or using already-present machinery within the host cell (natural competence) (Figure 2a). Often, conditions for transformation need to be optimized even for different strains within the same species. Once inside the cell, the DNA is surveyed by the host cell’s defense systems, which have evolved to eliminate invading DNA. Examples of these systems are restriction-modification systems, CRISPR/Cas systems, and abortive infection systems. Restriction-modification systems are estimated to be present in almost all bacterial species and appear to pose the largest barrier to DNA transformation (31). Three distinct methods have been reported to overcome this barrier: (a) use of a readily transformable intermediate host with compatible méthylation patterns, (b) use of an engineered intermediate host expressing the methyltransferases that are predicted to be present in the target microbe, and (c) incubation of DNA with commercially available methyltransferases in vitro to match the host’s DNA méthylation patterns. This final technique has recently been used to increase the transformation efficiency of gut-associated Lactobacillus plantarum to a level comparable to that of E. coli (~109 colony-forming units per microgram of DNA) (32; for a more detailed review of these defense systems and how to overcome the barrier, see References 33 and 34). Once accepted by the host, the introduced DNA is maintained episomally as a plasmid with the aid of selectable markers and a compatible origin of replication, or by integration into the genome through recombination. The first broad-host-range plasmid replicons suitable for expression in many LABs and even E. coli are pWVOl, pSH71, andpAMβ−1 (35–39). E. coli-Bifidobacterium and E. coli—Bacteroides shuttle vectors have been also constructed as genetic tools (40–43).
Figure 2.
Current state of genetic engineering tools. Manipulation of host-associated microbes is dependent on the ability to modify their genetic content, (a) DNA can be transferred to microbes by conjugation or electric or chemical disruption of the cell membrane, inducing natural competence, or by phage transduction.(b) Expression of genes or pathways can be controlled by creation of mutant promoter and/or ribosomal binding site (RBS) libraries or by using an inducible system in which an exogenous molecule drives expression in a concentration-dependent manner, (c) Genome editing is traditionally accomplished through homologous recombination by use of selectable markers. Recent advances in the use of clustered regularly interspaced short palindromic repeats (CRISPR)-based technologies have significantly contributed to development of genome-scale engineering tools. Genome editing with Cas9 relies on the use of Cas9 as a negative selection to cleave unmodified DNA. CRISPR interference (CRISPRi) uses a mutant form of Cas9, termed dCas9, which can bind but not cleave DNA, thereby regulating gene expression by blocking transcription.
3.3. Achieving Predictable Gene Expression
Introduction of DNA into the cell allows for the expression of genes or pathways, modification of cell metabolism, or tuning of cellular regulation. Although unregulated constitutive expression of genes can be a suitable solution for some applications, it is often desirable to specifically tune expression levels (Figure 2b). One method to achieve a range of expression is to tune constitutive expression through modification of the ribosomal binding site (RBS) or Shine-Dalgarno sequence in the form of a promoter library. This approach was used to create a library of promoters with an ~104-fold range of expression in L. plantarum and Bacteroides thetaiotaomicron (44, 45). Another method is to use a one-or two-component signal transduction regulatory system for inducible gene expression. Two quorum sensing systems have been adapted for use as inducible promoter systems in LABs—a nisin lantibiotic-dependent system from L. lactis (NICE) and a sakacin P pheromone-controlled system from Lactobacillus sakei—both of which have been used extensively as part of LAB engineering (46–48). Chemical-based induction systems from E. coli [isopropyl β-D-1-thiogalactopyranoside (IPTG), D-xylose] have also been developed with some success in L. plantarum (49) and B. thetaiotaomicron (50). Importation of normative induction systems depends on the compatibility of components across species, which can sometimes be limiting. For example, the sakacin P induction system is incompatible with L. lactis, whereas the NICE system is compatible with a variety of L. lactis strains (51). By contrast, native induction systems can often overcome limitations of using heterologous systems when adapted to express heterologous genes. To this end, an endogenous mannan-mducible promoter was used as part of a synthetic regulation system for heterologous gene expression in B. thetaiotaomicron (50).
A caveat to the use of native induction systems is that they may interfere with native cell regulation or metabolism. Commonly, both native and nonnative expression systems described above are species or even strain dependent, resulting in genetic tools that are not widely applicable to other organisms. There is a great need to design systems that maintain their function when applied across different species. An example of such a system is an entirely orthogonal T7 RNA polymerase system that is decoupled from cell metabolism and has been successfully used as a portable expression system in model organisms across phyla (52). If the biological system is engineered to function in vivo, it may be desirable to design the system to function only when it is in the intended location of the body. Allain et al. (53) induced expression of heterologous proteins in LABs at high temperature (42°C) by using a temperature-sensitive constitutive promoter (Plp_0775). The result was an in vivo inducible system, which is highly desirable for many biomedical applications; unfortunately, few such systems currently exist for use in commensal organisms.
Following gene expression, it may be important to control the localization of expressed proteins, such as expression of an antigen on the cell surface to generate an immune response or an environmental sensor that is naturally present in the cytoplasmic membrane. Proteins can be directed for secretion or displayed on the surface of the host bacteria, offering even more potential applications for direct interactions with host cells or other microbiota members. This has been accomplished in LABs by encoding an N-terminal secretion signal sequence upstream of the gene of interest, which is recognized by the conserved SecY pathway machinery and directed for secretion (54). The Usp45 secretion signal is commonly used in L. lactis, whereas a variety of secretion signals have been tested in L. plantarum, and their efficiency is passenger protein dependent (55–57). Anchoring of the protein to the cell surface is accomplished by creating a fusion protein between a cell-surface anchor protein and the protein of interest. Anchor proteins exist in a variety of motifs (e.g., transmembrane, lipobox, LysM, LPxTG), all of which have been used in applications with LABs (58, 59). Unfortunately, it is very difficult to predict which secretion signal or anchor protein will yield the highest secretion efficiency or desired level of protection or exposure, respectively.
3.4. Gene- and Genome-Scale Engineering Tools
Heterologous DNA can also be transferred into a microbial cell to precisely alter the genomic content of a cell, whether to alter metabolic pathway fluxes or to analyze gene function and regulation (Figure 2c). Conventional methods for site-specific genome editing are based on homologous recombination events, which have been improved through the use of phage-based recombinases. Another advance came from multiplexed genome editing, which allows multiple loci to be altered in a high-throughput manner (60). Unfortunately, recombination has been demonstrated only in a few strains of commensal organisms (61, 62). CRISPR/Cas9, a genome-editing tool that uses RNA-guided DNA cleavage to select for cells that have undergone recombination, has been transformational for genome editing (63). This technique can be used to create large mutant libraries in species that have high transformation efficiencies and recombinase activity—a challenge currently limiting broader application of CRISPR/Cas9-based genome editing. For example, van Pijkeren & Britton (64) and Oh & van Pijkeren (65) have pioneered the development of oligonucleotide- mediated recombineering techniques in LABs, a technique that was recently improved with the use of CRISPR/Cas9. These pioneering tools in heterologous gene expression and site-specific genomic mutations will give rise to many new opportunities in the synthetic biology of LABs.
A catalytically inactive form of Cas9, dCas9, has been developed for use in guiding promoter or coding regions to interfere with transcription rather than cleave the DNA This tool is termed CRISPR interference (CRISPRi) (66). This technique has been used to control gene expression of both native and heterologous DNA in B. thetaiotaomicron, significantly enhancing the ability to engineer this commensal bacterium, which is one of the most stable inhabitants in the gut (45; for a more detailed summary of the challenges of adapting CRISPR/Cas9-based tools to other organisms, see Reference 31).
3.5. Genetic Circuits to Program Cellular Behavior
Cell behavior can be controlled by synthetic genetic regulatory circuits consisting of sensors, processors, and actuators. These components can be combined to act as more complex genetic devices, such as switches, logic gates, oscillators, and biosensors, which enable more precise cellular programming for “smart” applications (24). Currently, the use of these tools has been demonstrated only in a select set of bacteria, mainly E. coli. Danino et al. (67) developed a programmable EcN platform (termed PROP-Z) suitable for in vivo diagnostics. In one study, orally administered EcN reported the presence of hepatic tumors in rodents. PROP-Z EcN contains a luminescence expression system for ex vivo imaging, a toxin-antitoxin system for long-term plasmid maintenance, and an inducible lacZ reporter. The lacZ reporter can be used to produce a colorimetric, fluorescent, or luminescent readout. This strain selectively aggregated in organs containing métastasés and converted the coadministered luciferin-conjugate substrate into its active form, producing a compound detectable in rodent urine. Thus far, the ability of EcN to colonize tumors has been demonstrated only in mice, and its ability to colonize the human GIT is in question (68, 69). Due to the breadth of available genetic tools, EcN is an attractive, engineerable probiotic host; however, expanding the genetic tools available to other gut inhabitants, such as LABs, bifidobacteria, and Bacteroides, would create significant and unique new opportunities for microbial engineering for health applications.
4. OPPORTUNITIES FOR ENGINEERING HOST-ASSOCIATED MICROBES
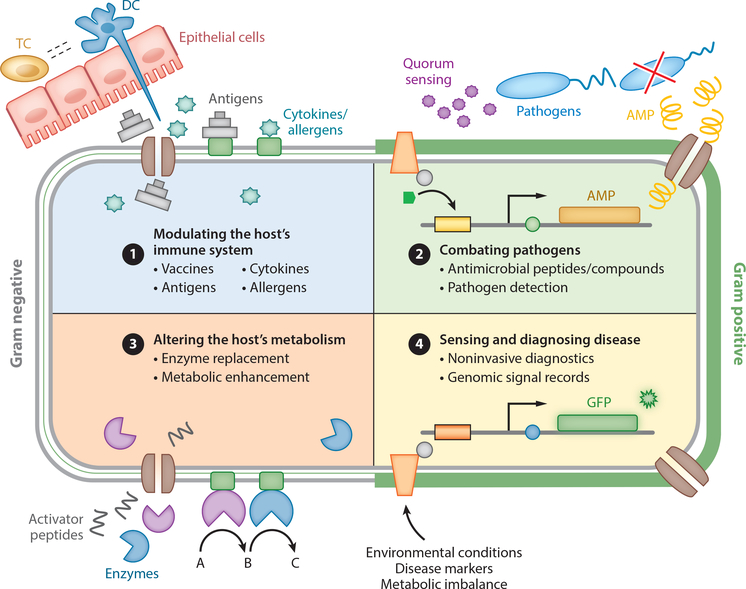
As genetic tools for manipulation of LABs, bifidobacteria, and Bacteroides continue to develop, so will the opportunities to engineer them to enhance human health. Microbes exist in diverse communities in and on almost all niches of the human body, where their importance is becoming increasingly evident. Given that microbiota have preference for specific body locations, their biogeographic preferences could be leveraged to engineer microbial therapeutics to treat diseases or maintain host health. These opportunities include developing sensors and diagnostics, preventing or treating pathogen infection, boosting the host’s immune system, enhancing metabolism, altering microbiomes, and understanding structure-function relationships between microbiota and host (Figure 3).
Figure 3.
Opportunities to engineer host-associated microbes. Both gram-positive and gram-negative members of the human microbiota can be engineered into biotic therapeutics for applications such as modulating the host immune system, combating pathogens, altering the host’s metabolism, and sensing and diagnosing disease. ① Gut-associated microbes interact extensively with the mucosal immune system and can be used to deliver vaccines, antigens, cytokines, and allergens. ② Probiotics or members of the microbiota can be engineered to produce antimicrobial compounds and peptides effective at combating pathogens. Pathogen-specific quorum sensing can be used to detect infections to actuate antimicrobial response. ③ Host metabolism can be altered through the expression of enzymes and activator peptides. ④ Microbial biosensors capable of detecting environmental conditions (pH, temperature, etc.), disease states (inflammation), or metabolic imbalance can be coupled to the expression of a reporter (fluorescent protein, enzyme) to detect and diagnose disease. Abbreviations: AMP, antimicrobial peptide; DC, dendritic cell; GFP, green fluorescent protein; TC, T cell.
4.1. Modulating the Host Immune System
Gut-associated lymphoid tissue is a prominent part of the GIT and represents almost 70% of the entire immune system. During their passage through the GIT, ingested bacteria interact directly and extensively with the host’s immune system, an interaction that affects the host’s health in many ways. These effects include inflammatory responses to bacterial metabolites and factors, alterations to the host’s immunological tolerance, and influences on abnormal immune responses at locations spatially distant from the interaction site (70). These features make ingested bacteria attractive vehicles to deliver vaccines and adjuvants as well as to rectify pathological immunological conditions. Recent demonstrations include engineering LABs, Bacteroides, and bifidobacteria to present pathogenic antigens as vaccines, allergens to desensitize the immune system, or immunomodulatory cytokines to illicit a host response (Figure 3a) (71–74). Orally administered L. lactis-secreting interleukin-27 was demonstrated to be effective in treatment of IBS in a murine model (75). The ease of delivery, ability to elicit a specific immunoglobulin A response at mucosal surfaces, and capability of mediating multivalent and adjuvant interactions are significant advantages over injectable vaccines (71). Probiotics have also successfully been used to deliver nucleic acid therapeutics to inhibit regulatory pathways controlling immune responses (76). A short hairpin RNA was successfully transferred from an engineered bacterial vector to murine intestinal cells, resulting in RNA interference-mediated downregulation of an inflammatory response protein, an overall decrease in inflammation, and an increase in survival in a colitis model (77). Christophe et al. (78) increased the internalization of plasmids in murine dendritic cells by surface-displaying the dendritic cell activator anti-DEC-205 on L. plantarum. They found that the efficacy of the therapy depended on the type of anchor used to fuse the functional single-chain variable fragment targeting the pattern recognition receptor DEC-205. Takei et al. (79) overcame the difficulty of expressing full-length antibodies in bacteria by creating chimeric epitope displaying proteins on Bifidobacterium longum. When orally delivered, the B. longum cells expressing HCV-NS3 epitopes provided vaccination against chronic hepatitis C virus infections in a murine model. These successes demonstrate the strength and utility of engaging the immune system at the intestinal mucosa using ingested microbes. However, much research still needs to be done to determine factors that contribute to efficacy, such as the ideal bacterial vehicle and identity of the surface-display anchor, which affect the strength of the host response.
4.2. Combating Pathogens
The rapid increase in microbial antibiotic resistance and formation of persister cells within biofilm matrices have led to a greater urgency to develop new and effective antimicrobial treatments. For instance, pathogenic infections often cannot be cleared and persist in different tissues of the body as chronic wound infections (80), periodontitis (81), inflammatory bowel disorder (82), and cystic fibrosis (83). Eradication of the pathogens and their protective biofilms presents unique challenges that could be addressed by engineering microbes capable of sensing pathogens; degrading biofilms; producing antimicrobial compounds; or producing narrow-spectrum antimicrobial peptides (AMPs), termed bacteriocins (Figure 3b) (84, 85). Microbes of the intestines produce an array of antimicrobial compounds as they compete for colonization and nutrients in the GIT (86–88). LABs control environmental pH through the production of lactic acid as well as the secretion of antimicrobial compounds or peptides that directly inhibit the growth of competitive species ( 15 ). For example, Lactobacillus reuteri produces the small molecule reuterin under anaerobic conditions, which has broad-spectrum activity against both gram-positive and gram-negative bacteria (89). van Pijkeren et al. (90) engineered a strain that exhibited a threefold increase in reuterin production to combat intestinal pathogens using a multiplex genome editing technology. The Kaznessis group (91) recently engineered L. lactis to detect the sex pheromone produced by Enterococcus faecalis, a multidrug-resistant opportunistic nosocomial pathogen, and initiate production of three AMPs capable of limiting the growth of E. faecalis. A pathogen-seeking engineered microbial device created by Hwang et al. (92, 93) demonstrates how synthetic biology can be applied to engineer probiotic bacteria to defend against pathogens. In these studies, the researchers outfitted EcN with receptors for quorum sensing molecules to intercept communication between pathogenic Pseudomonas aeruginosa cells and coupled it to the expression of three effector proteins. The first effector protein was a motor protein that directs movement of EcN cells toward the pathogenic signal source; the second was a biofilm-degrading enzyme (DNase); and the third was an AMP called microcin S that is effective at killing this pathogen. The use of a motor protein to direct the therapeutic bacteria to the pathogen represented a novel and effective strategy to target the therapeutic to the pathogen prior to releasing the antibiofilm and antimicrobial components. Thus, targeted delivery combined with antimicrobial synthesis enables precision medicine and offers an alternative to broad-spectrum antibiotics.
4.3. Altering Host Metabolism
Compositional changes in the human microbiota have been associated with metabolic syndromes such as obesity, type 2 diabetes, and nonalcoholic fatty liver disease. These links directly extend to the metabolome, the collective pool of metabolites within the host and the microbiota. The metabolome was previously known to be important for host-microbe interactions, such as the host benefiting from the production of microbial products like vitamins, essential amino acids, and other chemical by-products (94–96). It is now established that the microbiota produces short-chain fatty acid by-products that feed colonic epithelial cells and help maintain the epithelial barrier integrity (97). Microbiota-induced inflammation alters gut function and permeability, leading to a cascade of complications (98). Other inherited genetic disorders that arise from metabolic inadequacy (such as inborn errors of metabolism) could also be treated through supplementation with microbes engineered to provide enzyme-replacement therapy (Figure 3c). Rhimi et al. (99) demonstrated that tagatose, a bioactive metabolite that reduces blood glucose levels, can be synthesized on demand through oral administration of recombinant L. lactis and subsequent feeding with galactose. Mice treated with this recombinant system demonstrated moderate decrease in blood sugar— evidence that the secreted enzyme was converting galactose to tagatose in vivo. In a recent study, Lactobacillus gasseri was engineered to secrete glucagon-like peptide to reprogram intestinal epithelial cells into glucose-responsive insulin-producing cells (100). Bifidobacteria also have a dramatic effect on the host’s metabolism. Mice treated with Bifidobacterium pseudocatenulatum CECT 7765 showed a decrease in obesity markers when fed a high-fat diet in comparison to controls, an effect that may be due to altered expression of key genes involved in energy metabolism and lipid transport (101). A clinical trial conducted by Ojetti et al. (102) showed the ability of orally administered L. reuteri to aid in the digestion of lactose in lactose-intolerant patients lacking a native lactase enzyme in the GIT. L. reuteri was also genetically engineered for the treatment of phenylketonuria, a rare genetic disorder caused by incomplete phenylalanine metabolism leading to severe neurological complications, for which the current treatment is dietary restriction (103). Orally administered L. reuteri expressing heterologous phenylalanine ammonia lyase, an enzyme that degrades phenylalanine, was successful in reducing serum phenylalanine levels in a mouse model. In another study, L. plantarum was engineered to effectively reduce dietary oxalate, a molecule that may give rise to kidney stones, by constitutive secretion of oxalate decarboxylase in the intestines (104). Daily supplementation with this recombinant bacterium decreased the levels of urinary oxalate and reduced the risk of calcium oxalate crystal formation in mice. The ability to engineer such bacteria for enzyme replacement or supplementation offers new avenues to develop therapeutics that have been underexplored thus far.
4.4. Sensing and Diagnosing Disease
Microbes can be programmed to act as living diagnostic tools capable of sensing environmental cues and to respond by either recording information about the state of the body or outputting programmed signals for subsequent measurement (27). Biological systems across kingdoms use biosensors to sense environmental conditions (e.g., pH, temperature), the presence or absence of nutrients, and other bacteria through a form of inter- and intraspecies communication. Ingested probiotic bacteria interact with many host cells, metabolites, and other microbes during transit through the digestive tract, making them attractive targets as biosensors that could be designed to detect disease states in tissues or organs, metabolic imbalance, and the presence of pathogens (Figure 3d) (26). Environmental stresses or chemicals often elicit a cascade of cellular responses, including transcriptional changes in gene expression and metabolic pathways. These existing sensory and response proteins can be adapted or ported for use in other microbial species. Many in vivo whole-cell biosensors have already been engineered to respond to well-characterized biochemical signals (e.g., IPTG, rare sugars, tetracycline), but extension of this technology to biomedically relevant gut-associated metabolites and conditions (e.g., pH, inflammation markers) has been more challenging (45, 105, 106).
In an exceptional study conducted by Daeffler et al. (105), the researchers developed an EcN gut inflammation biosensor capable of detecting the presence of the proinflammatory molecules tetrathionate and thiosulfate. Two sensors, a thiosulfate sensor from Salmonella typhimurium and a novel tetrathionate sensor from Shewanella baltica discovered through a bioinformatics-based screen, were reprogrammed to drive expression of green fluorescent protein. The EcN cells, having been subjected to transit through an inflamed mouse GIT, were collected from fecal matter or regions of the colon and analyzed for fluorescence by flow cytometry analysis. The engineered thiosulfate microbial biosensor successfully detected inflammation in vivo; however, the tetrathionate sensor did not. The authors suggested that the concentration of tetrathionate in vivo was below the limit of detection for the sensor. Detection of clinically relevant concentrations of metabolites can be a challenge. One drawback to this approach has been the use of a protein- based reporter and the need to postprocess the samples to detect the output because the signal could have degraded over time. Anaerobic environments, such as that of the lower intestines, will affect the maturation of most fluorescent proteins due to their dependence on the presence of molecular oxygen (107, 108). Devices using an anaerobic microbial host or in anaerobic regions of the body could use an anaerobic fluorescent protein output such as iLov or FbFB (107), a luminescent protein output (108), or infrared detection (109). Avery recent solution to the problem of storing larger amounts of information over longer periods of time involves storing the information as memory in genomic DNA, although this has only been demonstrated in E. coll (110, 111). Such microbial biosensors could provide clinicians with a method to monitor patient physiology in a safe and noninvasive manner for numerous health applications.
4.5. In Situ Microbiota Engineering
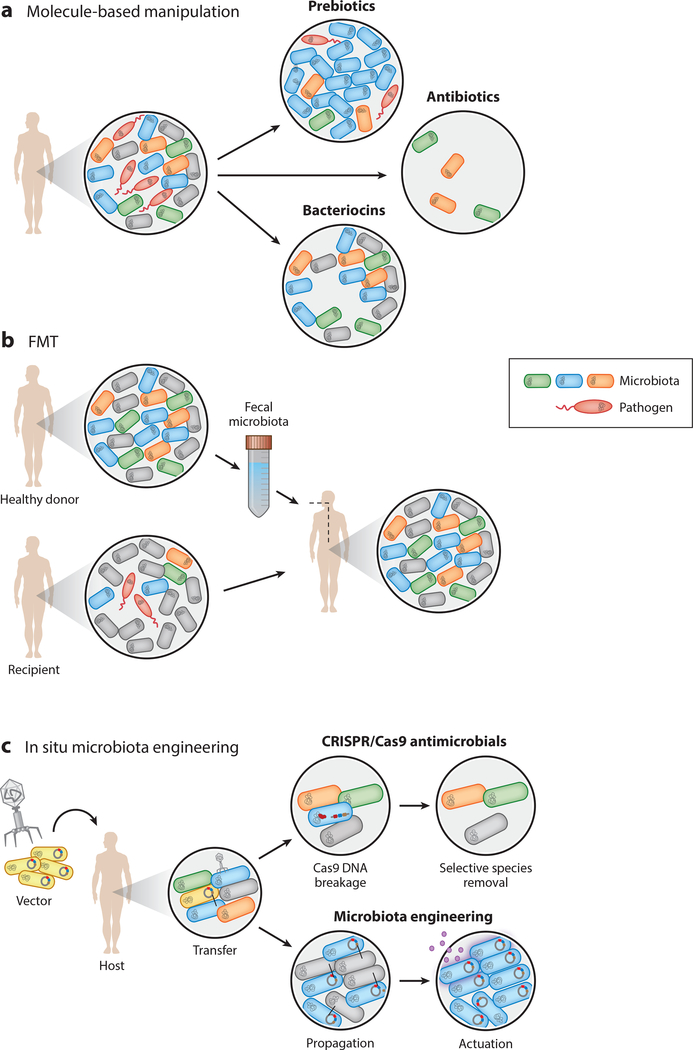
Contemporary methods to modulate the microbiota composition (chemical ingestion, prebiotics, xenobiotics, and antibiotics) (Figure 4a), probiotic supplementation, and community transplants (FMTs, designer consortia) (Figure 4b) vary widely in effectiveness and broadly affect the entire microbiota; therefore, targeted microbiota manipulation tools that can rationally alter the community composition are needed. Antibiotic administration can result in drastic, unpredictable changes in microbial composition and elimination of pathogens; however, the mechanism through which the healthy flora repopulates the gut is poorly understood (4). Conversely, probiotic and prebiotic supplementation is presumed to stimulate proliferation of beneficial microbes in the gut, but the health benefits of the microbial population distribution changes through such supplementation has not been demonstrated rigorously (112). Finally, probiotic supplementation and FMT treatments can be effective at augmenting the population, but the mechanisms through which they alter the native microbiota are poorly understood. Therefore, there is growing interest in the synthetic biology community to devise methods to rationally alter the microbiota community composition and function without performing extensive engineering work ex vivo. The goal of in situ microbiota engineering is to directly transfer synthetic genetic material from an engineered entity (e.g., a microbe or a phage) to native members of the microbiota (Figure 4c). This method is an alternative approach to the above-discussed ex vivo manipulations. A challenge shared by this approach and those discussed above is the need to deliver genetic material to alter the mi- crobiome in vivo, a considerable challenge given the current state of genetic tools available for host-associated microbes (see Section 3.1).
Figure 4.
Modulating the microbiota, (a) Molecule-mediated alteration of dysbiotic microbiota. Prebiotics promote the growth of beneficial species. Broad-spectrum antibiotics indiscriminately kill bacteria and could inadvertently lead to opportunistic infections. Bacteriocins can be used to specifically eliminate pathogens without significantly perturbing the remaining community composition,(b)In fecal matter transplants (FMTs), the fecal microbiota from a healthy donor is collected and transferred to a recipient. FMTs are effective at combating some gastrointestinal infections, (c)DNA delivery and transfer through microbes or phages can provide a means of rational in situ microbiota engineering. Clustered regularly interspaced short palindromic repeats (CRISPR) antimicrobials use targeted delivery of CRISPR/Cas machinery to create lethal double-strand breaks in the chromosomal DNA of target bacteria, leading to selective elimination. Future applications could apply in situ transfer of DNA to members of the microbiota to (re)program microbiota functions.
Recent advances in CRISPR-based tools for engineering commensal bacteria could provide the means for such engineering. One example is CRISPR-based antimicrobial therapies. These therapies rely on the delivery of Cas9 and a guide RNA to create a lethal double-strand break in the chromosome of the species to be eliminated (113–116). Thus far, this technique has been demonstrated using conjugative and phage-based DNA delivery vectors (117,118). Whereas phage transfer is generally much more efficient, transfer of exogenous DNA between the vector and target is still generally low and is limited by the phage’s host range. It is well known that genetic material is constantly being transferred between members of the microbiota; the transferred material has been termed the mobilome (119, 120). Further research needs to be done in order for us to fully understand known horizontal gene transfer (HGT) mechanisms (conjugation and natural transformation) and exploit them to transfer genetic programming on plasmids, transposons, or other DNA. Fortunately, plasmid transfer has been demonstrated in mouse gut-associated L. reuteri to Enterococcus faecium, and E.faecaiis to Lactobacillusfennentum, providing a foundation for in situ engineering technologies (121–123). Further research is necessary to discover newmobile plasmids and to identify novel nucleic acid transfer methods suited for microbiota engineering in situ (124). For a more thorough review of this topic, see Reference 120.
5. FROM BENCH TO BEDSIDE
Although there are many ongoing efforts to engineer members of the microbiota for biomedical applications, few biotic therapies have been tested in clinical settings. Demonstrating safety and efficacy in patients is paramount to the success of these biotic therapies. Challenges that need to be overcome include the creation of in vitro models that can capture interactions between host cells and microbial cells, the development in vitro and animal microbiota models that accurately capture the localization of species in the human gut, and the creation of strong biocontainment systems to prevent proliferation of engineered species upon release into the environment. Currently, in vitro cocultures of microbes and mammalian cell lines are not sufficiently well developed to provide significant new insight into the types of interactions that may be occurring in vivo. A continuous-flow “gut-on-a-chip” microfluidic device and a three-dimensional functional human intestinal model were recently developed to overcome some of the drawbacks of single-species culturing methods (125–127). Cocultures of host tissue and microbiota could be more representative of the in vivo setting and may advance our mechanistic understanding of host-microbe interactions.
Many of the applications discussed in this review used a murine model to test the effectiveness of the biotic therapies in vivo. Tests performed in mouse models can provide data on efficacy; however, there are many limitations that must be considered when translating the results to humans. Factors such as the anatomy of the digestive tract, diet, external stresses, and immune systems differ between humans and mice, and they are known to affect microbiota composition and responses to external stimuli and therapeutics (1). Additionally, it is common to use germ-free mice, which lack an endogenous microbiota, to more easily delineate host-microbe interactions (128). The utility of this approach to study single-microbe interactions is helpful to identify singular effects; however, it does not capture interactions between the engineered microbe and the community that are necessary for studies of the structure-function relationships important for microbiota engineering. In addition, germ-free mice have underdeveloped immune systems (129, 130), making it difficult to account for any potential immunological effects of microbiota engineering. Gut-associated species have evolved to survive in the conditions of their respective hosts. A colonizing LAB isolated from the human intestinal tract may express mucus-binding proteins specific to human colonic mucus and therefore may not exhibit a colonization phenotype in the mouse intestinal tract (131). Similarly, EcN is a good colonizer of the mouse GIT but cannot colonize the human intestinal tract (68, 132). These differences in fitness and colonization factors are expected to significantly affect the efficacy of biotic therapies. As new design tools and synthetic microbes are developed for increasingly complex biomedical applications, there will be a greater need for more “humanized” animal models for different disease states in which the animal contains a microbiota derived from a human patient (133–135).
Modifying the complex microbiota environment through the addition of engineering microbes can increase the risk of intestinal dysbiosis or overgrowth. Further research is required to monitor colonization and the distribution of bacterial species postadministration. Indeed, the factors that influence colonization and the residence time of microbes in the gut are still poorly understood (see the sidebar titled Colonization Factors). Another area for concern involves the release of genetically modified organisms into the environment by bowel movements. The first clinically administered genetically modified organism was engineered to have thymidine auxotrophy, which resulted in the rapid death of the engineered microbe in the absence of continuous thymidine supplementation (136). Since then, auxotrophic selection has been the primary method for biocontainment; however, it has the potential to be suppressed by mutations or HGT or circumvented by metabolism of alternate nutrients from the environment. Mandell et al. (137) developed a stronger biocontainment system in a synthetic E. coli strain. The E. coli genome was recoded to require a nonnatural amino acid for growth, creating a synthetic auxotroph incapable of being complemented with nutrients found in the environment. Containment of engineered bacteria has also been demonstrated by programming a synthetic genetic microbial kill switch (138). Biocontainment and safety are major concerns that have not yet been fully addressed in the field of microbial therapeutics and may need to be made more broadly applicable to any engineered species in the near future.
6. FUTURE OUTLOOK AND CHALLENGES
As we further elucidate the importance of a healthy microbiota to host health, it is crucial that we have a clear understanding of the contributions of the different constituents of the community. Advances in synthetic biology could be used in the future to genetically manipulate microbiota to better understand the complex structure-function relationships present in microbial communities. Such progress has allowed researchers to transform bacteria into so-called genomic tape recorders capable of observing events and storing the information to be analyzed at a later time (110). Although this technology is still in its early stages, there exists the opportunity to design bacterial recorders that can take up residence in the gut and record ecological changes. The use of such biotic recorders, along with simple and noninvasive measurement techniques, could help advance our knowledge of the events occurring within the microbiota. One could imagine designing two sets of engineered bacteria: one that perturbs the environment and one that records specific biological changes. As the development of these and other technologies advances, so will the opportunities to determine structure-function relationships between members of the human microbiota and their connection to human health.
7. CONCLUSION
Synthetic biology is a rapidly developing field with great potential for advancing studies of the role of microbiota in health and disease and for engineering constituent microbes to perform new biomedical functions. Many of the organisms present in the microbiota are considered “untamed,” with few to no genetic tools available for manipulation. The tools that do exist are often species or strain specific, a problem that must be resolved if we are to develop platforms that can be broadly applied to different microbiota that associate with the different parts of the human body. Such tools will greatly advance our understanding of how these communities affect our bodies in health and disease. The forward-engineering process of synthetic biology has already generated biotic therapeutics designed to combat infection, treat metabolic disorders, boost metabolism, alter the composition of the microbiota, and study genotype-phenotype relationships of individual host- associated species. Challenges at the frontier of synthetic microbe-based therapeutics are associated primarily with incomplete knowledge of how the microbiota and its individual members interact with one another and the host. In this area, too, synthetic biology could provide powerful tool sets for geneticists, ecologists, computational biologists, and clinicians. With that information in hand, our understanding of the importance of the human microbiota will be greatly enhanced and will make way for improved microbial therapeutics.
FORMATION AND STABILITY OF THE MICROBIOTA
Our microbiota forms at birth as the newborn passes through the vaginal canal, and rapidly develops following contact with the mother’s skin and exposure to the environment and food. Vaginally delivered babies who are breastfed have greater resistance to Clostridium diffidle and Escherichia coli infections compared with infants delivered by caesarean section and those fed formula (139). The microbial community undergoes the most dramatic changes during infancy and early childhood, when early colonization of “bad” bacteria leads to numerous complications. The composition of the microbiota steadily stabilizes after adolescence on the basis of our habitat, diet, and antibiotic use (140). Diet influences the available nutrients present in the GIT, which can favor some species over others. As diet changes, so does the composition of the gut microbiota, allowing for selective enrichment of probiotic species upon supplementation with certain indigestible carbohydrates (prebiotics) (112). Use of broad-spectrum antibiotics to treat infections significantly affects the homeostasis of the microbiota by clearing susceptible “good” bacteria, creating an imbalance in the metabolic network, and opening ecological niches for colonization by pathogens.
COLONIZATION FACTORS
Successful microbiota engineering hinges on the ability to design microbes that can integrate themselves into the community and/or reside there for a sufficient length of time so as to provide therapeutic benefit (127, 141). Colonization is generally believed to depend on the organism’s ability to adhere to and occupy a niche, compete for nutrients, survive environmental stresses, and defend this niche from other organisms (142). These abilities have evolved over long periods of time, and many systems are still unknown, making it difficult to enhance or to replicate them in noncolonizing strains. A deletion of a conserved polysaccharide utilization gene locus reduces the ability of Bacteroides fragilis to colonize the gut, suggesting that metabolic function plays a role in bacterial colonization (143). In the GIT, many bacterial members have evolved to express mucus adhesion proteins (MAPs) (144) or surface factors such as polysaccharides (145) on their cell surface, facilitating their ability to colonize. MAPs are responsible for the microbes’ ability to stick to the thick mucosal lining of the lower intestinal tract. Blocking these surface proteins with antibodies eliminates this ability to bind to mucus in vitro (146). Controllable expression of surface factors and MAPs could provide fitness to otherwise non-mucus-binding species, as well as increase the natural binding ability of colonizing strains to withstand environmental stresses or provide better protection from pathogens. Increasing colonization and fitness can help maintain homeostasis of bacterial members to exert a dampening effect on perturbations, promote host-microbe interactions at the mucosal-epithelial junction, and increase the effective duration of a biotic therapy.
SUMMARY POINTS
Synthetic biology offers a means to engineer new functionalities into host-associated microbes for biomedical benefit.
Human gut microbes and probiotics have been engineered to protect against pathogens, modulate the immune system, and alter the host’s metabolism.
Biotic sensors and recorders can be integrated into microbiota as noninvasive diagnostic tools.
Recent advances in CRISPR-based technologies could enable the rational in situ engineering of the microbiota.
Currently, efforts are limited to a small set of model bacteria (EcN, LABs, Bacteroides) due to the difficulty of genetically engineering most members of the microbiota.
Development of a standardized engineering tool set can enable engineering of nonmodel microbes within the microbiota.
FUTURE ISSUES
What are the contributions of the different individual species to different microbiota?
How can the process of developing genetic tools be streamlined to more readily engineer any member of the microbiota?
How can we design experiments and models to better predict the in vivo efficacy of biotic therapies?
How can synthetic biologists address concerns of accidental environmental release of engineered microbes or even synthetic DNA components?
How can synthetic biology navigate the challenges related to the public perception of genetically modified organisms as they relate to therapeutic synthetic microbes?
Microbiota: an ecological community of commensal, symbiotic, and pathogenic microorganisms living in or on a multicellular host
Dysbiosis: microbiota imbalance that can potentially lead to disease
Probiotics: live microorganisms that when consumed provide some benefit; often, fermented food-associated microbes and not permanent members of the microbiota that can transiently interact with the host and/or the native microbiota to impart health benefits
LABs: lactic acid bacteria
GIT: gastrointestinal tract
Fecal matter transplant (FMT): procedure in which feces containing microbiota from a healthy donor are transferred into a recipient
Escherichia coli Nissle, 1917 (EcN): E. coli species isolated from a soldier who was resistant to Shigella infection; used to treat gastrointestinal disorders
Clustered regularly interspaced short palindromic repeats (CRISPR): part of a prokaryotic immune system whose nucleases have been co-opted as genome-editing tools
Microbiome: genomic content of the collective microbiota
CRISPR interference (CRISPRi): a CRISPR-based gene-regulation technique
Horizontal gene transfer (HGT): movement of genetic material between species
Biocontainment: engineering strategy to prevent unintended release of genetically modified organisms
ACKNOWLEDGMENTS
The writing of this review was supported by funding from the National Science Foundation (MCB-1452902 to C.L.B.) and from the National Institutes of Health (1DP2HD91798–01 to N.U.N.).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review
LITERATURE CITED
- 1.Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, et al. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belizârio JE, Napolitano M. 2015. Human microbiomes and their roles in dysbiosis, common diseases, and novel therapeutic approaches. Front. Microbiol 6:1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lau JT, Whelan FJ, Herath I, Lee CH, Collins SM, et al. 2016. Capturing the diversity of the human gut microbiota through culture-enriched molecular profiling. Genome Med. 8:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Langdon A, Crook N, Dantas G. 2016. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med. 8:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Didari T, Mozaffari S, Nikfar S, Abdollahi M. 2015. Effectiveness of probiotics in irritable bowel syndrome: updated systematic review with meta-analysis. World J. Gastroenterol 21:3072–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verna EC, Lucak S. 2010. Use of probiotics in gastrointestinal disorders: what to recommend? Ther. Adv. Gastroenterol 3:307–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsai YT, Cheng PC, Pan TM. 2014. Anti-obesity effects of gut microbiota are associated with lactic acid bacteria. Appl. Microbiol. Biotechnol 98:1–10 [DOI] [PubMed] [Google Scholar]
- 8.Brennan CA, Garrett WS. 2016. Gut microbiota, inflammation, and colorectal cancer. Annu. Rev. Microbiol 70:395–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moos WH, Faller DV, Harpp DN, Kanara I, Pernokas J, et al. 2016. Microbiota and neurological disorders: a gut feeling. Biores. Open Access 5:137–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brandwein M, Steinberg D, Meshner S. 2016. Microbial biofilms and the human skin microbiome. npj Biofilms Microbiomes 2:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egert M, Simmering R, Riedel CU. 2017. The association of the skin microbiota with health, immunity, and disease. Clin. Pharmacol. Ther 102:62–69 [DOI] [PubMed] [Google Scholar]
- 12.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner ACR, et al. 2010. The human oral microbiome. J. Bacteriol 192:5002–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kilian M, Chappie ILC, Hannig M, Marsh PD, Meuric V, et al. 2016. The oral microbiome—an update for oral healthcare professionals. Br. Dent.J 221:657–66 [DOI] [PubMed] [Google Scholar]
- 14.Gopinath S, Iwasaki A. 2015. Cervicovaginal microbiota: Simple is better. Immunity 42:790–91 [DOI] [PubMed] [Google Scholar]
- 15.Sanders AIE, Gibson GR, Gill HS, Guarner F. 2007. Probiotics: their potential to impact human health Issue pap. 36, Counc. Agric. Sei. Teehnol, Ames, IA [Google Scholar]
- 16.Ballal SA, Veiga P, Fenn K, Michaud M, Kim JH, et al. 2015. Host lysozyme-mediated lysis of Lactococcus lactis facilitates delivery of colitis-attenuating superoxide dismutase to inflamed colons. PNAS 112:7803–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fedorak RN. 2010. Probiotics in the management of ulcerative colitis. Gastroenterol. Hepatol. 6:688–90 [PMC free article] [PubMed] [Google Scholar]
- 18.Prantera C 2006. Probiotics for Crohn’s disease: What have we learned? Gut 55:757–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, et al. 2013. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med 368:407–15 [DOI] [PubMed] [Google Scholar]
- 20.Petrof EO, Gloor GB, Vanner SJ, Weese SJ, Carter D, et al. 2013. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: “rePOOPulating” the gut. Microbiome 1:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ullah MW, Khattak WA, Ul-Islam M, Khan S, Park JK. 2016. Metabolic engineering of synthetic cell-free systems: strategies and applications. Biochem. Eng.J 105:391M–05 [Google Scholar]
- 22.Cameron DE, Bashor CJ, Collins JJ. 2014. A brief history of synthetic biology. Nat. Rev. Microbiol 12:381–90 [DOI] [PubMed] [Google Scholar]
- 23.Marchand N, Collins CH. 2016. Synthetic quorum sensing and cell-cell communication in gram-positive Bacillus megaterium. ACS Synth. Biol 5:597–606 [DOI] [PubMed] [Google Scholar]
- 24.Brophy JAN, Voigt CA. 2014. Principles of genetic circuit design. Nat. Methods 11:508–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu PD, Lander ES, Zhang F. 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157:1262–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park M, Tsai SL, Chen W. 2013. Microbial biosensors: engineered microorganisms as the sensing machinery. Sensors 3:5777–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rong G, Corrie SR, Clark HA. 2017. In vivo biosensing: progress and perspectives. ACS Sens. 2:327–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cribby S, Taylor M, Reid G. 2008. Vaginal microbiota and the use of probiotics. Interdiscip. Perspect. Infect. Dis 2008:256490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ridaura VΤΚ, Faith JJ, Rey FE, Cheng J, Duncan AE, et al. 2013. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341:1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lagier J-C, Khelaifia S, AIou MT, Ndongo S, Dione N, et al. 2016. Culture of previously uncultured members of the human gut microbiota by eulturomies. Nat. Microbiol 1:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Waller MC, Bober JR, Nair NU, Beisei CL. 2017. Toward a genetic tool development pipeline for host-associated bacteria. Curr. Opin. Microbiol 38:156–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spath K, Hein S, Grabherr R. 2012. Direct cloning in Lactobacillus plant arum·. Electroporation with non-methylated plasmid DNA enhances transformation efficiency and makes shuttle vectors obsolete. Microb. Cell Fact 11:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Houte S Van, Buckling A, Westra ER. 2016. Evolutionary ecology of prokaryotic immune mechanisms. Microbiol. Mol. Biol. Rev 80:745–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts RJ, Vincze T, Posfai J, Maeelis D. 2015. REBASE—a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 43 :D2 98–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kok J, Van Der Vossen JMBM, Venema G. 1984. Construction of plasmid cloning vectors for lactic streptococci which also replicate in Bacillus subtilis and Escherichia colt. Appl. Environ. Microbiol 48:726–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Vos WM. 1987. Gene cloning and expression in lactic streptococci. FEMS Microbiol. Lett 46:281–95 [Google Scholar]
- 37.Leenhouts KJ, Kok J, Venema G. 1991. Lactococcal plasmid pWVOl as an integration vector for lactococci. Appl. Environ. Microbiol 57:2562–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang F, O’Toole PW. 2009. Genetic tools for investigating the biology of commensal laetobaeilli. Front. Biosci 14:3111–27 [DOI] [PubMed] [Google Scholar]
- 39.Landete JM. 2017. A review of food-grade vectors in lactic acid bacteria: from the laboratory to their application. Crit. Rev. Biotechnol 37:296–308 [DOI] [PubMed] [Google Scholar]
- 40.Missieh R, Sgorbati B, LeBlane DJ. 1994. Transformation of Bifidobacterium longum with pRM2, a constructed Escherichia coli-B. longum shuttle vector. Plasmid 32:208–11 [DOI] [PubMed] [Google Scholar]
- 41.Matsumura H, Takeuchi A, Kano Y. 1997. Construction of Escherichia coli-Bifidobacterium longum shuttle vector transforming B. longum 105-A and 108-A. Biosci. Biotechnol. Biochem 61:1211–12 [DOI] [PubMed] [Google Scholar]
- 42.Alvarez-Martin P, Florez AB, Margolles A, Del Solar G, Mayo B. 2008. Improved cloning vectors for bifidobacteria, based on the Bifidobacterium catenulatum pBCl replicon. Appl. Environ. Microbiol 74:4656–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garrigues-Jeanjean N, Wittmer A, Ouriet M, Duval-Iflah Y. 1999. Transfer of the shuttle vector pRRI207 between Escherichia colt and Bacteroides spp. in vitro and in vivo in the digestive tract of ax- enic mice and in gnotoxenic mice inoculated with a human microflora. FEMS Microbiol. Ecol 29:33–43 [Google Scholar]
- 44.Tauer C, Heini S, Egger E, Heiss S, Grabherr R. 2014. Tuning constitutive recombinant gene expression in Lactobacillus plantarum. Microb. Cell Fact 13:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mimee M, Tucker AC, Voigt CA, Lu TK. 2015. Programming a human commensal bacterium, Bacteroides thetaiotaomicron, to sense and respond to stimuli in the murine gut microbiota. Cell Syst. 1:62–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mierau I, Kleerebezem M. 2005. 10 Years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl. Microbiol. Biotechnol 68:705–17 [DOI] [PubMed] [Google Scholar]
- 47.Kaswurm V, Nguyen T-T, Maischberger T, Kulbe KD, Michlmayr H. 2013. Evaluation of the food grade expression systems NICE and pSIP for the production of 2,5-diketo-D-glueonie acid reductase from Corynebacterium glutamicum. AMB Express 3:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mathiesen G, Sorvig E, Blatny J, Naterstad K, Axelsson L, Eijsink VGH. 2004. High-level gene expression in Lactobacillus plantarum using a pheromone-regulated bacteriocin promoter. Lett. Appl. Microbiol 39:137–43 [DOI] [PubMed] [Google Scholar]
- 49.Heiss S, Hörmann A, Tauer C, Sonnleitner M, Egger E, et al. 2016. Evaluation of novel inducible promoter/repressor systems for recombinant protein expression in Lactobacillus plantarum. Microb. Cell Fact 15:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Horn N, Carvalho AL, Overweg K, Wegmann U, Carding S R, Stentz R. 2016. A novel tightly regulated gene expression system for the human intestinal symbiont Bacteroides thetaiotaomicron. Front. Microbiol 7:1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bosma EF, Forster J, Nielsen AT. 2017. Lactobacilli and pedioeoeei as versatile cell factories—evaluation of strain properties and genetic tools. Biotechnol. Adv 35:419–42 [DOI] [PubMed] [Google Scholar]
- 52.Kushwaha M, Salis HM. 2015. A portable expression resource for engineering cross-species genetic circuits and pathways. Nat. Commun 6:7832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Allain T, Mansour NM, Bahr MMA, Martin R, Florent I, et al. 2 016. A new laetobaeilli in vivo expression system for the production and delivery of heterologous proteins at mucosal surfaces. FEMS Microbiol. Lett 363:fnw ll7. [DOI] [PubMed] [Google Scholar]
- 54.Schneewind O, Missiakas D. 2014. Sec-seeretion and sortase-mediated anchoring of proteins in grampositive bacteria. Biochim. Biophys. Acta 1843:1687–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dieye Y, Usai S, Clier F, Grass A, Piard J. 2001. Design of a protein-targeting system for lactic acid bacteria. J. Bacteriol 183:4157–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Le Loir Y, Nouaille S, Commissaire J, Brétigny L, Langella P. 2001. Signal peptide and propeptide optimization for heterologous protein secretion in Lactococcus lactis. Appl. Environ. Microbiol 67:4119–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mathiesen G, Sveen A, Piard J-C, Axelsson L, Eijsink VGH. 2008. Heterologous protein secretion by Lactobacillusplantarum using homologous signal peptides. J. Appl. Microbiol 105:215–26 [DOI] [PubMed] [Google Scholar]
- 58.Zadravec P, Strakelj B, Berlec A. 2015. Heterologous surface display on lactic acid bacteria: non-GMO alternative? Bioengineered 6:179–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Michon C, Langella P, Eijsink VGH, Mathiesen G, Chatel JM. 2016. Display of recombinant proteins at the surface of lactic acid bacteria: strategies and applications. Microb. Cell Fact 15:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dalia AB, McDonough E, Camilli A. 2014. Multiplex genome editing by natural transformation. PNAS 111:8937–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Pijkeren JP, Britton RA. 2012. High efficiency recombineering in lactic acid bacteria. Nucleic Acids Res. 40:e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lambert JM, Bongers RS, Kleerebezem M. 2007. Cre-lox- based system for multiple gene deletions and selectable-marker removal in Lactobacillus plantarum. Appl. Environ. Microbiol 73:1126–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiang W, Bikard D, Cox D, Zhang F, Marraffin LA. 2013. CRISPR-assisted editing of bacterial genomes. Nat Biotechnol. 31:233–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Pijkeren JP, Britton RA. 2014. Precision genome engineering in lactic acid bacteria. Microb. Cell Fact 13(Suppl. 1):S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oh JH, van Pijkeren JP. 2014. CRISPR-Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res 42:el31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, Qi LS. 2013. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc 8:2180–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Danino T, Prindle A, Kwong GA, Skalak M, Li H, et al. 2015. Programmable probiotics for detection of cancer in urine. Sei. Transi. Med 7:289ra84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ou B, Yang Y, Tham WL, Chen L, Guo J. 2016. Genetic engineering of probiotic Escherichia colt Nissle 1917 for clinical application. Appl. Microbiol. Biotechnol 100:8693–99 [DOI] [PubMed] [Google Scholar]
- 69.Stritzker J, Weibel S, Hill PJ, Oelsehlaeger TA, Goebel W, Szalay AA. 2007. Tumor-specific colonization, tissue distribution, and gene induction by probiotic Escherichia colt Nissle 1917 in live mice. Int.J. Med. Microbiol 297:151–62 [DOI] [PubMed] [Google Scholar]
- 70.Gasteiger G, D’osualdo A, Schubert DA, Weber A, Braseia EM, Hartl D. 2017. Cellular innate immunity: an old game with new players. J. Innate Immun 9:111–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wyszynska A, Kobierecka P, Bardowski J, Jagusztyn-Krynicka EK. 2015. Lactic acid bacteria—20 years exploring their potential as live vectors for mucosal vaccination. Appl. Microbiol. Biotechnol 99:2967–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tarahomjoo S 2012. Development of vaccine delivery vehicles based on lactic acid bacteria. Mol. Biotechnol 51:183–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wells JM, Mereenier A. 2008. Mucosal delivery of therapeutic and prophylactic molecules using lactic acid bacteria. Nat. Rev. Microbiol 6:349–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cronin M, Morrissey D, Rajendran S, El Mashad SM, van Sinderen D, et al. 2010. Orally administered bifidobacteria as vehicles for delivery of agents to systemic tumors. Mol. Ther 18:13 97–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hanson ML, Hixon JA, Li W, Felber BK, Anver MR, et al. 2014. Oral delivery of IL-27 recombinant bacteria attenuates immune colitis in mice. Gastroenterology 146:210–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Meister G, Tuschi T. 2004. Mechanisms of gene silencing by double-stranded RNA. Nature 431:343–49 [DOI] [PubMed] [Google Scholar]
- 77.Spisni E, Valerii MC, De Fazio L, Cavazza E, Borsetti F, et al. 2015. Cyclooxygenase-2 silencing for the treatment of colitis: a combined in vivo strategy based on RNA interference and engineered Escherichia colt. Mol. Ther 23:27 8–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Christophe M, Kuczkowska K, Langella P, Eijsink VGH, Mathiesen G, Chatel J-M. 2015. Surface display of an anti-DEC-205 single chain Fv fragment in Lactobacillus plantarum increases internalization and plasmid transfer to dendritic cells in vitro and in vivo. Microb. Cell Fact 14:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Takei S, Omoto C, Kitagawa K, Morishita N, Katayama T, et al. 2014. Oral administration of genetically modified Bifidobacterium displaying HCV-NS3 multi-epitope fusion protein could induce an HCV-NS3-speeifie systemic immune response in mice. Vaccine 32:3066–74 [DOI] [PubMed] [Google Scholar]
- 80.Frykberg RG, Banks J. 2015. Challenges in the treatment of chronic wounds. Adv. Wound Care 4:560–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim J, Amar S. 2006. Periodontal disease and systemic conditions: a bidirectional relationship. Odontology 94:10–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bai A-P, Ouyang Q. 2006. Probiotics and inflammatory bowel diseases. Postgrad. Med. J 82:376–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ratjen FA. 2009. Cystic fibrosis: pathogenesis and future treatment strategies. Respir. Care 54:595–605 [DOI] [PubMed] [Google Scholar]
- 84.Brogden KA. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol 3:238–50 [DOI] [PubMed] [Google Scholar]
- 85.Geldart K, Borrero J, Kaznessis YN. 2015. Chloride-inducible expression vector for delivery of antimicrobial peptides targeting antibiotic-resistant Enterococcus faecium. Appl. Environ. Microbiol 81:3889–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brurberg MB, Nes IF, Eijsink VG. 1997. Pheromone-induced production of antimicrobial peptides in Lactobacillus. Mol. Microbiol 26:347–60 [DOI] [PubMed] [Google Scholar]
- 87.Jones SE, Versalovie J. 2009. Probiotic Lactobacillus reuteri biofilms produce antimicrobial and anti-inflammatory factors. BMC Microbiol. 9:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Roelofs KG, Comstock LE. 2016. Molecules necessary for gut colonization and mediate competition in vivo. mBio 7:e0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Spinier JK, Taweeehotipatr M, Rognerud CL, Ou CN, Tumwasorn S, Versalovie J. 2008. Human- derived probiotic Lactobacillus reuteri demonstrate antimicrobial activities targeting diverse enteric bacterial pathogens. Anaerobe 14:166–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.van Pijkeren J-P, Neoh KM, Sirias D, Findley AS, Britton RA. 2012. Exploring optimization parameters to increase ssDNA recombineering in Lactococcus lactis and Lactobacillus reuteri. Bioengineered 3:209–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Borrero J, Chen Y, Dunny GM, Kaznessis YN. 2015. Modified lactic acid bacteria detect and inhibit multirésistant enterococci. ACS Synth. Biol 4:299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hwang IΥ, Tan MH, Koh E, Ho CL, Poh CL, Chang MW. 2014. Reprogramming microbes to be pathogen-seeking killers. ACS Synth. Biol 3:228–37 [DOI] [PubMed] [Google Scholar]
- 93.Hwang IΥ, Koh E, Wong A, March JC, Bentley WE, et al. 2017. Engineered probiotic Escherichia coli can eliminate and prevent Pseudomonas aeruginosa gut infection in animal models. Nat. Commun 8:15028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Joyce SA, MacSharry J, Casey PG, Kinsella M, Murphy EF, et al. 2014. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. PNAS 111:7421–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wikofif WR, Anfora AT, Liu J, Schultz PG, Lesley SA, et al. 2009. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. PNAS 106:3698–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sanz Y, Olivares M, Moya-Pérez A, Agostoni C. 2015. Understanding the role of gut microbiome in metabolic disease risk. Pediatr. Res 77:236–44 [DOI] [PubMed] [Google Scholar]
- 97.Krishnan S, Alden N, Lee K. 2015. Pathways and functions of gut microbiota metabolism impacting host physiology. Curr. Opin. Biotechnol 36:137–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Le Barz M,Anhê FF,Varin TV, Desjardins Y, Levy E, et al. 2015. Probiotics as complementary treatment for metabolic disorders. Diabetes Metab. J 39:291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rhimi M, Bermudez-Humaran LG, Huang Y, Boudebbouze S, Gaci N, et al. 2015. The secreted L-arabinose isomerase displays anti-hyperglycemic effects in mice. Microb. Cell Fact 14:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Duan FF, Liu JH, March JC. 2015. Engineered commensal bacteria reprogram intestinal cells into glucose-responsive insulin-secreting cells for the treatment of diabetes. Diabetes 64:1794–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mauricio MD, Serna E, Fernândez-Murga ML, Portero J, Aldasoro M, et al. 2017. Bifidobacterium pseudocatenulatum CECT 7765 supplementation restores altered vascular function in an experimental model of obese mice. Int. J. Med. Sei 14:444–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ojetti V, Gigante G, Gabrielli M, Ainora AIE, Mannoeei A, et al. 2010. The effect of oral supplementation with Lactobacillus reuteri or tilactase in lactose intolerant patients: randomized trial. Eur. Rev. Med. Pharmacol. Sei 14:163–70 [PubMed] [Google Scholar]
- 103.Dürrer KE, Allen AIS, Hunt von Herbing I. 2017. Genetically engineered probiotic for the treatment of phenylketonuria (PKU); assessment of a novel treatment in vitro and in the PAHenu2 mouse model of PKU. PLOS ONE 12:e0176286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sasikumar P, Gomathi S, Anbazhagan K, Abhishek A, Paul E, et al. 2014. Recombinant Lactobacillus plantarum expressing and secreting heterologous oxalate decarboxylase prevents renal calcium oxalate stone deposition in experimental rats. J. Biomed. Sei 21:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.DaefHer KN, Galley JD, Sheth RU, Ortiz-Velez LC, Bibb CO, et al. 2017. Engineering bacterial thiosulfate and tetrathionate sensors for detecting gut inflammation. Mol. Syst. Biol 13:923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Benbouziane B, Ribelles P, Aubry C, Martin R, Kharrat P, et al. 2013. Development of a stress-inducible controlled expression (SICE) system in Lactococcus lactis for the production and delivery of therapeutic molecules at mucosal surfaces. J. Biotechnol 168:120–29 [DOI] [PubMed] [Google Scholar]
- 107.Landete JM, Medina M, Arqués JL. 2016. Fluorescent reporter systems for tracking probiotic lactic acid bacteria and bifidobacteria. World J. Microbiol. Biotechnol 32:119. [DOI] [PubMed] [Google Scholar]
- 108.Karimi S, Ahl D, Vagesjö E, Holm L, Phillipson M, et al. 2016. In vivo and in vitro detection of luminescent and fluorescent Lactobacillus reuteri and application of red fluorescent mCherry for assessing plasmid persistence. PLOS ONE 1l:e0151969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Berlec A, Zavrsnik J, Butinar M, Turk B, Strukelj B. 2015. In vivo imaging of Lactococcus lactis, Lactobacillus plantarum and Escherichia colt expressing infrared fluorescent protein in mice. Microb. Cell Fact 14:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Farzadfard F, Lu TK. 2014. Synthetic biology. Genomieally encoded analog memory with precise in vivo DNA writing in living cell populations. Science 346:1256272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bonnet J, Subsoontorn P, Endy D. 2012. Rewritable digital data storage in live cells via engineered control of recombination directionality. PNAS 109:8884–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pandey KR, Naik SR, Vakil BV. 2015. Probiotics, prebioties and synbioties—a review. J. Food Sei. Technol 52:7577–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Beisei CL, Gomaa AA, Barrangou R. 2014. A CRISPRdesign for next-generation antimicrobials. Genome Biol. 15:516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bikard D, Euler CW, Jiang W, Nussenzweig PM, Goldberg GW, et al. 2014. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat. Biotechnol 32:1146–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gomaa AA, Klumpe HE, Luo ML, Selle K, Barrangou R, Beisei CL. 2014. Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. mBio 5:e00928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Citorik RJ, Mimee M, Lu TK. 2014. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat. Biotechnol 32:1141–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Park JY, Moon BY, Park JW, Thornton JA, Park ΥΉ, Seo KS. 2017. Genetic engineering of a temperate phage-based delivery system for CRISPR/Cas9 antimicrobials against Staphylococcus aureus. Sei. Rep 7:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Goren M, Yosef I, Qimron U. 2017. Sensitizing pathogens to antibiotics using the CRISPR-Cas system. Drug Resist Update 30:1–6 [DOI] [PubMed] [Google Scholar]
- 119.Brito IL, Yilmaz S, Huang K, Xu L, Jupiter SD, et al. 2016. Mobile genes in the human microbiome are structured from global to individual scales. Nature 535:435–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sheth RU, Cabral V, Chen SP, Wang HH. 2016. Manipulating bacterial communities by in situ microbiome engineering. Trends Genet. 32:189–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.T annoek GW. 1987. Conjugal transfer of plasmid pAM ß 1 in Lactobacillus reuteri and between laetobaeilli and Enterococcus faecalis. Appl. Environ. Microbiol 53:2693–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.McConnell MA, Mercer AA, Tannoek GW. 1991. Transfer of plasmid pAM βl between members of the normal microflora inhabiting the murine digestive tract and modification of the plasmid in a Lactobacillus reuteri host. Microb. Ecol. Health Dis 4:343–55 [Google Scholar]
- 123.Netherwood T, Bowden R, Harrison P, O’Donnell G, Parker DS, Gilbert HJ. 1999. Gene transfer in the gastrointestinal tract. Appl. Environ. Microbiol 65:5139–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shintani M, Sanchez ZK, Kimbara K. 2015. Genomics of microbial plasmids: classification and identification based on replication and transfer systems and host taxonomy. Front. Microbiol 6:242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chen Y, Lin Y, Davis KM, Wang Q, Rnjak-Kovacina J, et al. 2015. Robust bioengineered 3D functional human intestinal epithelium. Sci. Rep 5:13708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Arnold JW, Roach J, Azcarate-Peril MA. 2016. Emerging technologies for gut microbiome research. Trends Microbiol 24:887–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yaung SJ, Church GM, Wang HH. 2014. Recent progress in engineering human-associated miero- biomes. Methods Mol. Biol 1151:3–25 [DOI] [PubMed] [Google Scholar]
- 128.Hazebrouck S, Oozeer R, Adel-Patient K, Langella P, Rabot S, et al. 2006. Constitutive delivery of bovine β-laetoglobulin to the digestive tracts of gnotobiotic mice by engineered Lactobacillus casei. Appl. Environ. Microbiol 72:7460–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rooks MG, Garrett WS. 2016. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol 16:341–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fiebiger U, Bereswill S, Heimesaat MM. 2016. Dissecting the interplay between intestinal microbiota and host immunity in health and disease: lessons learned from germfree and gnotobiotic animal models. Eur.J. Microbiol. Immunol 6:253–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.MacKenzie DA, Jeffers F, Parker ML, Vibert-Vallet A, Bongaerts RJ, et al. 2010. Strain-specific diversity of mucus-binding proteins in the adhesion and aggregation properties of Lactobacillus reuteri. Microbiology 156:3368–78 [DOI] [PubMed] [Google Scholar]
- 132.Jourovâ L, Anzenbaeher P, Liskovâ B, Matuskovâ Z, Hermanovâ P, et al. 2017. Colonization by non-pa thogenic bacteria alters mRNA expression of cytochromes P450 in originally germ-free mice. Folia Microbiol. 62:463–69 [DOI] [PubMed] [Google Scholar]
- 133.Ito R, Takahashi T, Katano I, Ito M. 2012. Current advances in humanized mouse models. Cell. Mol. Immunol 9:208–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nguyen TLA, Vieira-Silva S, Liston A, Raes J. 2015. How informative is the mouse for human gut microbiota research? Dis. Model. Mech 8:1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Arrieta M, Walter J, Finlay BB. 2016. Forum human microbiota-associated mice: a model with challenges. Cell Host Microbe 19:575–78 [DOI] [PubMed] [Google Scholar]
- 136.Steidler L, Neirynek S, Huyghebaert N, Snoeck V, Vermeire A, et al. 2003. Biological containment of genetically modified Lactococcus lactis for intestinal delivery of human interleukin 10. Nat. Biotechnol 21:785–89 [DOI] [PubMed] [Google Scholar]
- 137.Mandell DJ, Lajoie MJ, Mee MT, Takeuchi R, Kuznetsov G, et al. 2015. Biocontainment of genetically modified organisms by synthetic protein design. Nature 518:55–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chan CTY, Lee JW, Cameron DE, Bashor CJ, Collins JJ. 2015. “Deadman” and “Passcode” microbial kill switches for bacterial containment. Nat. Chem. Biol 12:82–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Penders J, Thijs C, Vink C, Stelma FF, Snijders B, et al. 2006. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118:511–21 [DOI] [PubMed] [Google Scholar]
- 140.O’Connor EM. 2013. The role of gut microbiota in nutritional status. Curr. Opin. Clin. Nutr. Metab. Care 16:509–16 [DOI] [PubMed] [Google Scholar]
- 141.Kali A 2015. Human microbiome engineering: the future and beyond. J. Clin. Diagn. Res 9:1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Krumbeck JA, Marsteller NL, Frese SA, Peterson DA, Ramer-Tait AE, et al. 2016. Characterization of the ecological role of genes mediating acid resistance in Lactobacillus reuteri during colonization of the gastrointestinal tract. Environ. Microbiol 18:2172–84 [DOI] [PubMed] [Google Scholar]
- 143.Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. 2013. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature 501:426–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Etzold S, Kober OI, Mackenzie DA, Tailford LE, Gunning AP, et al. 2014. Structural basis for adaptation of lactobacilli to gastrointestinal mucus. Environ. Microbiol 16:888–903 [DOI] [PubMed] [Google Scholar]
- 145.Coyne MJ, Chatzidaki-Livanis M, Paoletti LC, Comstock LE. 2008. Role of glyean synthesis in colonization of the mammalian gut by the bacterial symbiont Bacteroides fragilis. PNAS 105:13099–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Etzold S, Mackenzie DA, Jeffers F, Walshaw J, Roos S, et al. 2014. Structural and molecular insights into novel surface-exposed mucus adhesins from Lactobacillus reuteri human strains. Mol. Microbiol 92:543–56 [DOI] [PubMed] [Google Scholar]
- 147.Angelakis E, Armougom F, Carrière F, Bachar D, Laugier R, et al. 2015. A metagenomic investigation of the duodenal microbiota reveals links with obesity. PLOS ONE 10:e0137784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, et al. 2006. Molecular analysis of the bacterial microbiota in the human stomach. PNAS 103:732–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shin NR, Whon TW, Bae JW. 2015. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 33:496–503 [DOI] [PubMed] [Google Scholar]
- 150.Sankar SA, Lagier J-C, Pontarotti P, Raoult D, Fournier P-E. 2015. The human gut microbiome, a taxonomic conundrum. Syst. Appl. Microbiol 38:276–86 [DOI] [PubMed] [Google Scholar]