Abstract
Pharmacogenomics (PGx) studies are to identify genetic variants that may affect drug efficacy and toxicity. A machine understandable drug- gene relationship knowledge is important for many computational PGx studies and for personalised medicine. A comprehensive and accurate PGx-specific gene lexicon is important for automatic drug-gene relationship extraction from the scientific literature, rich knowledge source for PGx studies. In this study, we present a bootstrapping learning technique to rank 33,310 human genes with respect to their relevance to drug response. The algorithm uses only one seed PGx gene to iteratively extract and rank co-occurred genes using 20 million MEDLINE abstracts. Our ranking method is able to accurately rank PGx-specific genes highly among all human genes. Compared to randomly ranked genes (precision: 0.032, recall: 0.013, F1: 0.018), the algorithm has achieved significantly better performance (precision: 0.861, recall: 0.548, F1: 0.662) in ranking the top 2.5% of genes.
Keywords: pharmacogenomics, text mining, NLP, natural language processing, personalised medicine
1. Introduction
Pharmacogenomics is the study of how human genetic variations affect an individual’s response to drugs, with a focus on drug metabolism, absorption and distribution (Evans and McLeod, 2003). Understanding how the genetic variants correspond to various drug responses is an essential step of personalised medicine (Swen et al., 2007; Weiss, 2008; Davis et al., 2009). The success of personalised drug treatment largely depends on the availability of accurate, comprehensive knowledge bases for machine-understandable drug-gene relationships. The volume of published biomedical research, and thus the corpora of putative pharmacogenomics knowledge, is growing fast. As of 2012, the MEDLINE database contains over 21 million biomedical literature citations (see http://www.ncbi.nlm.nih.gov/pubmed). Clearly, with the current rate of growth in this corpus, it has become increasingly likely that important knowledge connecting drugs, genes and diseases will be routinely missed. For these reasons, there is a need to systematically acquire structured pharmacogenomics knowledge from the literature. Substantial manual curation efforts have been used to achieve this end. Currently, the Pharmacogenomics Knowledge Base (PharmGKB) is the largest manually created resource of information on how variation in human genetics leads to variation in drug response (Klein et al., 2001). However, extracting biomedical information from the published literature manually and transforming it into machine-understandable knowledge is a difficult task because biomedical terminologies and knowledge are huge, dynamic and highly complicated.
Developing automatic approaches to extracting PGx-specific drug-gene relationships from free text is an active research area. Both statistical and Natural Language Processing (NLP) methods have been used (Garten et al., 2010). Chang and Altman (2004) extracted drug-gene pairs from the literature based on co-occurrence statistics and used machinelearning algorithms to build models that can classify the extracted relationships. Garten and Altman (2009) developed Pharmspresso, a text-mining tool for extracting PGxspecific concepts and relationships from full text. Guided by curated drug-gene relationships in PharmGKB, Theobald et al. (2009) constructed n-way Bayesian networks based on conditional probability tables extracted from co-occurrence statistics over the entire MEDLINE corpus, and produced a broad-coverage analysis of the relationships between these biological entities. Hansen et al. (2009) recently described an algorithm that uses existing biomedical knowledge about drug structure and indications to rank 12,460 genes in the genome on the basis of their potential relevance to specific drugs. Wu et al. (2012) showed that the Latent Dirichlet Allocation (LDA) model can be used to effectively rank candidate gene-drug relations based on MEDLINE cooccurrence. Recently, NLP-based methods have been in the mainstream of PGx-specific drug-gene relationship extraction. Ahlers et al. (2007) developed an NLP system (Enhanced SemRep) to extract pharmacogenomics relationships in MEDLINE citations. Coulet et al. (2010) have developed an NLP technique to build a PGx ontology from 17 million MEDLINE abstracts using syntactic dependency structure. Recently, we have developed a conditional approach to extract PGx-specific drug-gene pairs from MEDLINE (Xu and Wang, 2012). Our approach used known drug-gene pairs as prior knowledge to implicitly classify sentences before relationship extraction.
Semantic relationships between drugs and genes, such as drug-gene target relationships and drug-gene metabolising (PGx) relationships, are complicated. Automatically, extracting drug-gene relationships from free text and distinguishing them by PGx relevance is a challenging task. A high-quality PGx-specific lexicon is critical for automatic PGx-specific drug-gene relationship extraction tasks (Figure 1). Currently, there are over 33,310 human genes (accessed in October 2010), most of which are not PGx-related. Therefore, there is a need to identify PGx-specific genes amongst these for use in drug-gene relationship extraction. In this study, starting with all known human genes and 20 million MEDLINE abstracts, we have developed a bootstrapping, learning approach to iteratively extract and rank genes according to their relevance to drug pharmacogenomics. This technique is based on the assumption that PGx genes are often mentioned together in MEDLINE abstracts. If we start with a known PGx gene such as ‘cytochrome P450, family 2, subfamily C, polypeptide 9’ (CYP2C9), it is likely that genes mentioned with it are also PGx genes with a likelihood that decreases as the co-occurrence distance grows.
Figure 1.
Drug-gene relationship extraction input: drug and gene lexicons
For instance, consider the following sentence: “Those genes are ATP-binding cassette subfamily B member 1 (ABCB1), the noradrenaline, dopamine and serotonin transporters (SLC6A2, SLC6A3 and SLC6A4), cyclic AMP-responsive element binding protein 1 (CREB1), corticotropin-releasing hormone receptor 1 (CRHR1) and neurotrophic tyrosine kinase type 2 receptor (NTRK2)” (Dong et al., 2009). There are a total seven genes (ABCB1, CREB1, CRHR1, NTRK2, SLC6A2, SLC6A3 and SLC6A4) and three drugs (noradrenaline, dopamine and serotonin) mentioned. Among these seven genes, only four genes (ABCB1, SLC6A2, SLC6A3 and SLC6A4) are related to drug metabolism and disposition. If a PGx-specific gene lexicon is used to extract drug-gene pairs from this sentence, the false positive rate would be greatly decreased from that of an extraction using a lexicon comprised of all human genes because non-PGx genes such as CREB1, CRHR1 and NTRK2 would be excluded. While a high-quality drug lexicon is available (e.g. the FDA drug list), there is no comprehensive and accurate list of PGxspecific genes. Although HGNC (HUGO Gene Nomenclature Committee) maintains a comprehensive list of 33,310 human genes, there are no criteria for classification that incorporate the gene’s function in drug disposition (see http://www.genenames.org/).
PharmGKB is the largest pharmacogenomics knowledge base and there are 10,898 drug-gene pairs drawn from 918 drugs and 2388 genes (accessed in October 2010). However, the genes in PharmGKB are a mixture of non-PGx-specific genes such as BRCA1 (‘breast cancer 1, early onset’), TP53 (‘tumour protein p53’), IL6 (‘Interleukin 6’), VDR (‘vitamin D (1, 25-dihydroxyvitamin D3) receptor’) and AR (‘androgen receptor’), and PGx-specific genes such as CYP2C9 and VKROC1 (‘vitamin K epoxide reductase complex, subunit 1’). Correspondingly, the drug-gene pairs in PharmGKB are also a mixture of non-PGX-specific pairs (clodronate-VDR and levonorgestrel-AR) and PGxspecific pairs (warfarin-CYP2C9 and warfarin-VKORC1). For example, two drug-gene pairs, caffeine-BRCA1 and cisplatin-BRCA1, are incorrectly assigned semantic subtype ‘PD’ (Pharmacodynamics). In addition, total 15 interleukin ligand or receptor genes (e.g. L2, IL4, IL4R, IL5, IL6) are associated with 34 drug-gene pairs (e.g. warfarin-IL6, salbutamol-IL6R and cetuximab-IL8) and these drug-gene pairs are incorrectly assigned the semantic subtypes ‘PD’ or ‘PK’ (Pharmaokenetics). If we can correctly classify genes such as BRCA1, TP53 and IL6 as non-PGx-specific, the precision of drug-gene semantic type classification in PharmGKB can be improved.
The manual examination of gene names in order to find PGx genes will be a difficult task. For instance, it is difficult to tell if gene CALU (a calumenin gene) is a PGx gene by its name alone. However, when gene CALU appears together with other PGx genes in a specific context, it becomes easier to make this determination. For example, consider the following sentence: “Compound genetic profiles comprising VKORC1, CALU and CYP2C9 improve categorisation of individual warfarin dose” (Vecsler et al., 2006). From this sentence alone and without further knowledge about CALU, we can determine CALU is a PGx gene based on its co-occurrence with two other typical PGx genes VKORC1 and CYP2C9. There is research into ranking genes based on co-citation metrics. Jesson et al. (2001) created a weighted gene-to-gene co-citation network (PubGene) for 13,712 human genes by analysis of over 10 million MEDLINE records. In this network, nodes represent each gene or protein and the edges connecting them represent the number of articles in which each gene or protein pair is co-cited (Jenssen et al., 2001). PubGene ranks genes using link structure analysis with the purpose of helping researchers to retrieve information on genes and proteins. Morrison et al. (2005) developed the GeneRank algorithm by combining gene expression information with a network structure derived from the Gene Ontology (GO) or expression profile correlations. However, none of these studies rank genes based on their role in drug metabolism, absorption and distribution.
2. Approach
Our method is based on the assumption that genes with a similar function tend to be mentioned together in the biomedical research literature. Intuitively, a gene that is cocited with many other PGx genes is likely to be assigned a high probability of being PGx-related. In our recent studies, we have developed and evaluated semi-supervised, bootstrapping approaches to extracting and ranking disease entities (Xu et al., 2008), drug entities (Xu et al., 2009a; Xu et al., 2009b), and binary is a relationships from MEDLINE abstracts (Xu et al., 2009a; Xu et al., 2009b). In this study, we demonstrate that these bootstrapping techniques can be extended to extract and rank PGx genes. This iterative algorithm runs as follows: a first iteration begins with a known PGx gene such as CYP2C9 (the seed gene) and finds other PGx genes that co-occur. For example, using CYP2C9, we can extract 13 additional PGx genes from the following sentence: “The aim of this study was to explore the frequencies of polymorphisms in drugmetabolising enzymes (CYP1A1, CYP2C9, CYP2C19, CYP3A4, CYP2D6, CYP3A5, DPYD, UGT1A1, GSTM1, GSTP1, GSTT1) and drug transporters (ABCB1[MDR1] and ABCC2[MRP2])” (Bosch et al., 2006). In the next iteration, those genes extracted in previous iterations are used as seeds. As a result, we can extract additional genes that do not co-occur with the initial seed gene CYP2C9. For instance, using GSTT1 (‘glutathione S-transferase theta 1’) from the previous iteration, we can extract additional glutathione S-transferase genes such as GSTO1 and GSTO2 from the following sentence: “Genotyping of GSTM1 and GSTT1 genes was carried out by a multiplex PCR; GSTA1, GSTO1, GSTO2, GSTP1 polymorphisms were determined using the PCR-RFLP method” (Piacentini et al., 2010). Neither GSTO1 nor GSTO2 appear together with the initial seed CYP2C9 in MEDLINE. Similarly, using ABCB10, we can extract a whole set of ATC-binding cassette transporter genes such as ABCB10, ABCB2, ABCB3, ABCB8, ABCB9, TAP1 and TAP2 from sentence “In the human genome, the five adenosine triphosphate (ATP)-binding cassette (ABC) half transporters ABCB2 (TAP1), ABCB3 (TAP2), ABCB9 (TAP-like), and in part, also ABCB8 and ABCB10 are closely related with regard to their structural and functional properties” (Herget and Tampé, 2007). Since this is a semi-supervised approach without human intervention, false positives can be introduced. The criterion of co-occurrence with a known PGx gene is not by itself decisive regarding whether or not the gene in question is indeed a PGx gene. For example, 12 genes co-occur with PGx gene CYP3A4 in the sentence that follows: “The expression levels of 13 of these genes, ALPK2, ASAP1, CEACAM5, CYP3A4, ENAH, ERBB2, HHIPL2, LTB4R, MMP9, PERLD1, PNMT, PTPRA, and OSMR, were validated in a total of 118 gastric samples using either the qRT-PCR” (Kapucuoglu et al., 2003). None of these 12 genes are PGx genes. Therefore, it is important to rank genes in a manner that takes all co-occurring genes into account. In this study, we developed a ranking algorithm to prioritise PGx-specific genes.
3. Data and methods
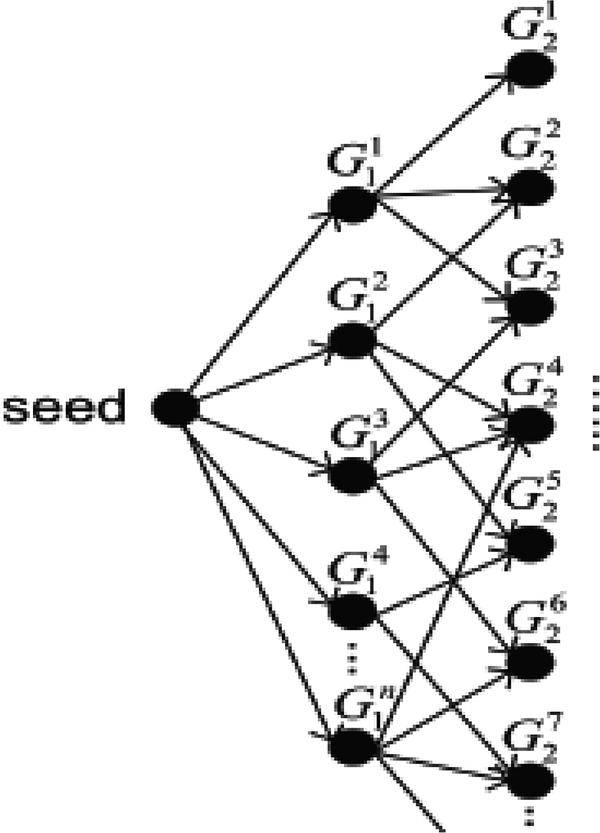
We used 20 million MEDLINE abstracts (roughly 100 million sentences) published from 1965 to 2010 as our text corpus for PGx gene extraction and ranking. We used the publicly available information retrieval library Lucene (see http://lucene.apache.org/java/docs/index.html) to create an index of the 100 million MEDLINE sentences. The 33,310 gene symbols downloaded from HGNC were used as the universe of human genes (accessed in October 2010). We downloaded the drug-gene pairs from PharmGKB, which contains a total of 10,898 drug-gene pairs, 918 drugs and 2388 genes (accessed in October 2010). We applied our algorithm in ranking the 2388 genes available in PharmGKB. The algorithm started with a known PGx gene CYP2C9 as seed and looped over a procedure consisting of a gene extraction step and a gene-ranking step. In the gene extraction step, the seed or extracted gene(s) from the previous step were used as search queries to the Lucene search engine in order to find all the sentences in which these genes appear. The genes that co-occur were then extracted from returned sentences. The process stopped after two iterations. This stopping point was determined by a manual examination that revealed a lack of any new PGx genes in subsequent iterations. Figure 2 shows the iterative gene extraction process.
Figure 2.
Iterative approach in finding PGx genes starting from a single seed
The ranking score of a gene being a PGx-specific gene at a given iteration is given as following:
The term on the left is the ranking score of gene Gi being a PGx-specific gene in iteration N, and the term within the summation on the right is the ranking score of co-occurring gene Gj being a PGx-specific gene in iteration N-1 weighted by the number of times Gi and Gj co-ocurring. The ranking algorithm is similar to topic sensitive page ranking algorithm (Haveliwala, 2003). Starting from a seed gene, the ranking algorithm iteratively propagates its confidence score to its co-occurred genes. The ranking score of each gene is the sum of scores of its co-occurring genes weighted by co-occurrence count. The confidence score of the seed gene was given a score of 1.0. For example, starting with the seed gene CYP2C9, we extracted four additional genes G1, G2, G3 and G4 after first iteration, with the co-occurrence (with seed) counts 100, 20, 10, 1, respectively. Then the weight of G1 after first iteration is (100/131) × 1.0. At the second iteration, the ranking scores will be calculated again based on the score vector from the first iteration.
Since there is no gold standard list of PGx-specific genes for precision and recall evaluation, it is hard to evaluate the precision and recall in a totally unbiased way. The genes in PharmGKB drag–gene pairs are not necessarily PGx–specific. For example, when searching for drug tamoxifen at PharmGKB (see http://www.pharmgkb.org/drag/PA451581#tabview=tab5&subtab=31), we got its related genes such as ABCB1, BRCA1, CYP2B6, ESR1 and ESR2, which is a mix of PGx genes (ABCB1 and CYP2B6) and non-PGx–specific genes (BRCA1, ESR1 and ESR2). Therefore for precision evaluation, we manually examined a randomly selected 20% of the extracted 13,581 genes. For each gene, we examined its name and the MEDLINE sentences where it appears. Since it is often impossible to decide whether a gene is PGx-specific or not by examining its symbol or name, the actual context where a gene appears is necessary for deciding whether or not a gene is PGx-specific. Three evaluators with graduate level of biomedical background conducted the evaluation. A gene was assigned true positive only if all three evaluators agreed. Precision was calculated as the proportion of PGx genes amongst the ranked genes. We used the java.util.Random class to generated random numbers between 1 and the size of the input list and selected 20% of input genes for manual examination. Due to the manual curation effort, we only did random sampling once, which is one of the limitations of our study. It could generate a biased data set, but the chance will be very small since the sampling method is random and not biased towards any particular genes and our ranking method. The manual examination process may introduce biase towards to specific evaluators, but not towards the algorithm.
For recall evaluation for extracting and ranking HGNC genes, we used all 79 CYP (Cytochrome P450 enzymes) genes selected from 33,310 HGNC genes as the gold standard. The CYP genes account for 70%−80% of enzymes involved in drug metabolism. Our intuition is that if our algorithm can find all or most of CYP genes after two iterations and also ranked all the CYP genes high in the list, it indicates that the algorithm has good recall in both gene extraction and ranking. For recall evaluation for extracting and ranking HGNC genes, we used all 79 CYP genes selected from 33,310 HGNC genes as the gold standard. The recall is estimated as the proportion of all 79 CYP genes that appear amongst the ranked genes. The assumption is that these 79 CYP genes are uniformly distributed among all the PGx genes. Similarly, we used the 50 CYP genes (out of 2388 genes) in PharmGKB for recall measure of our algorithms in extracting and ranking PharmGKB genes. Since both precision and recall are important for our task, F1 measure was used to evaluate the overall performance of the extraction and ranking process. The F1 measure is the harmonic mean of precision and recall:
4. Results
4.1. Performance of the gene extraction and ranking algorithms in ranking HGNC genes
Using CYP2C9 as the seed gene, we extracted 13,581 genes after two iterations. Table 1 shows the precision, recall and F1 measure of the ranked 13,581 genes in terms of their score of being PGx genes. As shown in Table 1, the algorithm was able to extract 75 out of the total 79 CYP genes (a recall of 0.949) after two iterations. Additionally, most of the extracted CYP genes were ranked high on the list. Thus, this demonstrates that both the extraction and ranking algorithms have high recalls. For example, there were 41 CYP genes amongst the 136 genes in the 1.0% rank (a recall of 0.519) and 60 CYP genes in the 340 genes in the 2.5% rank (a recall of 0.759). Among the 79 CYP genes, six CYP genes (CYP2D8P1, CYP2C17, CYP2B7P1, CYP2D7P1, CYP20A1 and CYP2G2P) appeared only once in MEDLINE, and our algorithm was able to extract them and ranked them high on the list. Most of the high-ranking genes were PGx genes with a precision of 0.807 for the 1.0% rank, and the precisions dropped rapidly to 0.037 for the whole gene list, while recall did not increase significantly (column 3 and 4 in Table 1). The precision and recall of a rank determined from a random ordering were significantly lower (column 6 and 7 in Table 1). In summary, we have shown that (a) the bootstrapping gene extraction algorithm using a single seed has high recall (0.949) and is able to extract most of the PGx-specific genes after two iterations, (b) the gene-ranking algorithm has achieved both high precision and high recall by ranking most PGx-specific genes high on the list.
Table 1.
Precision, recall and F1 of the ranked 13,581 HGNC genes after two iterations using seed CYP2C9
| Ranked | Randomly Ranked | ||||||
|---|---|---|---|---|---|---|---|
| Cut-off (%) | Count | Precision | Recall | F1 | Precision | Recall | F1 |
| 1.0 | 136 | 0.807 | 0.519 | 0.632 | 0.015 | 0.000 | 0.000 |
| 2.5 | 340 | 0.587 | 0.759 | 0.662 | 0.032 | 0.013 | 0.018 |
| 5.0 | 680 | 0.402 | 0.861 | 0.548 | 0.041 | 0.051 | 0.045 |
| 7.5 | 1018 | 0.312 | 0.886 | 0.462 | 0.038 | 0.089 | 0.053 |
| 10 | 1358 | 0.250 | 0.899 | 0.392 | 0.035 | 0.101 | 0.052 |
| 20 | 2716 | 0.142 | 0.911 | 0.246 | 0.035 | 0.253 | 0.061 |
| 30 | 4074 | 0.103 | 0.924 | 0.185 | 0.037 | 0.304 | 0.065 |
| 40 | 5432 | 0.081 | 0.937 | 0.150 | 0.036 | 0.380 | 0.066 |
| 50 | 6790 | 0.067 | 0.937 | 0.126 | 0.037 | 0.405 | 0.068 |
| 60 | 8148 | 0.059 | 0.949 | 0.111 | 0.038 | 0.494 | 0.070 |
| 70 | 9506 | 0.051 | 0.949 | 0.097 | 0.038 | 0.608 | 0.071 |
| 80 | 10864 | 0.046 | 0.949 | 0.088 | 0.038 | 0.734 | 0.072 |
| 90 | 12223 | 0.041 | 0.949 | 0.079 | 0.038 | 0.810 | 0.072 |
| 100 | 13581 | 0.037 | 0.949 | 0.071 | 0.037 | 0.949 | 0.071 |
4.2. Performance of the gene extraction and ranking algorithms in ranking PharmGKB genes
Next we showed that our algorithm was able to accurately rank genes available in the PharmGKB drug-gene relationship database, in which there were a total 2388 genes (of which 50 are CYP genes). In a manner similar to the last experiment, these 50 CYP genes were used to estimate recalls of extraction and ranking algorithms. After two iterations, we extracted 2093 genes (out of 2388 genes) using CYP2C9 as the seed. Based on our manual curation of a random sample of 20% of the extracted PharmGKB genes, we estimated that only 13.0% of the extracted 2093 PharmGKB genes were PGx genes (Table 2). Our algorithm successfully ranked these genes highly according to their relevance to PGx. The precision dropped rapidly from 100% at the 1.0% rank to 13.0% for the whole list. Compared to the randomly ranked gene list (column 6 and 7 in Table 2), our ranking algorithm was able to rank most PGx genes high on the list (column 3 and 4). For the recall estimate, 94% of the 50 CYP genes were ranked among the 418 in the 20% rank. This ranked gene list may be used to refine or augment the drug-gene relationship classification in the PharmGKB.
Table 2.
Precision, recall and F1 of the ranked 2093 PharmGKB genes after two iterations using seed CYP2C9
| Ranked | Randomly Ranked | ||||||
|---|---|---|---|---|---|---|---|
| Cutoff (%) | Count | Precision | Recall | F1 | Precision | Recall | F1 |
| 1.0 | 21 | 1.000 | 0.300 | 0.462 | 0.150 | 0.040 | 0.063 |
| 2.5 | 52 | 1.000 | 0.500 | 0.667 | 0.135 | 0.040 | 0.062 |
| 5.0 | 104 | 0.875 | 0.580 | 0.698 | 0.154 | 0.080 | 0.105 |
| 7.5 | 157 | 0.821 | 0.780 | 0.800 | 0.135 | 0.100 | 0.115 |
| 10 | 209 | 0.722 | 0.840 | 0.777 | 0.129 | 0.180 | 0.150 |
| 20 | 418 | 0.474 | 0.940 | 0.630 | 0.122 | 0.200 | 0.152 |
| 30 | 628 | 0.354 | 0.960 | 0.517 | 0.129 | 0.340 | 0.187 |
| 40 | 837 | 0.287 | 0.960 | 0.442 | 0.130 | 0.420 | 0.193 |
| 50 | 1047 | 0.231 | 0.980 | 0.374 | 0.130 | 0.520 | 0.208 |
| 60 | 1256 | 0.200 | 0.980 | 0.332 | 0.131 | 0.620 | 0.216 |
| 70 | 1465 | 0.177 | 0.980 | 0.300 | 0.132 | 0.740 | 0.224 |
| 80 | 1674 | 0.160 | 0.980 | 0.275 | 0.133 | 0.820 | 0.229 |
| 90 | 1883 | 0.144 | 0.980 | 0.251 | 0.133 | 0.920 | 0.232 |
| 100 | 2093 | 0.130 | 0.980 | 0.229 | 0.130 | 0.980 | 0.229 |
4.3. Effects of seeds on the gene extraction and ranking algorithms
We investigated the effects of seeds in subsequent gene extraction and ranking processes. We compared five seed genes: CYP2C9, CYP2D6 (‘cytochrome P450, family 2, subfamily D, polypeptide 6’), VKORC1, BRCA1 and EGFR (‘epidermal growth factor receptor’). Among these five seed genes, CYP2C9, CYP2D6 and VKORC1 are typical PGx genes. Both CYP2C9 and CYP2D6 are cytochrome P450 family genes involved in drug metabolism, and VKORC1 is a typical PGx gene involved in vitamin K metabolism. Neither BRCA1 nor EFGR is PGx gene. BRCA1 is a tumour suppressor gene and its mutation is related to breast cancer. EGFR is a proto-oncogene involved in many cancers. We applied our extraction and ranking algorithms using each of the five genes as seed. Table 3 shows the top ten-ranked genes for each seed after two iterations. As shown in Table 3, all top ten-ranked genes for the three PGx seed genes (CYP2C9, CYP2D6 and VKORC1) are true PGx genes, while none of the top-ranked genes using seed genes BRCA1 and EGFR are PGx genes. Interestingly, most of the top ten genes for seed EGFR, which is oncogene, are also oncogenes, such as BRAF, PIK3CA, NRAS, HRAS, PTPN11, RAF1 and KRAS. Similarly, for the tumour suppressor gene BRCA1, which encodes a nuclear phosphoprotein that plays a role in maintaining genomic stability, most of the top-ranked genes are also tumour suppressor genes, such as FANCD1, FANCD2, PALB2, RAD51, CHEK2, BRIP1, BCCIP and TP53. This demonstrated that out algorithm was able to find genes similar to a given seed. The seed can be a PGx gene, oncogene, tumour suppressor gene or other type of gene. The top-ranked genes for the three PGx seed genes were all typical PGx genes, which demonstrated that our algorithm may not be sensitive to specific seed choices as long as the seed is a typical PGx gene (e.g. CYP2C9, CYP2D6, CYP2C19, CYP3A4 and VKORC1). The final PGx genes extracted using different PGx gene seeds may converge after two iterations. To approve this, we need to manually examine the extracted gene lists for different seed genes.
Table 3.
Top ten-ranked genes for five seeds: three PGx genes (CYP2C9, CYP2D6 and VKORC1), one tumour suppressor gene (BRCA1) and one oncogene (EGFR).
| Rank | Seed1 (CYP2C9) |
Seed2 (CYP2D6) |
Seed3 (VKORC1) |
Seed4 (BRCA1) |
Seed5 (EGFR) |
|---|---|---|---|---|---|
| 1 | CYP2C9 | CYP2D6 | VKORC1 | BRCA1 | EGFR |
| 2 | CYP2D6 | CYP2C9 | CYP2C19 | FANCD1 | BRAF |
| 3 | CYP3A4 | CYP2C19 | CYP2C8 | PALB2 | PIK3CA |
| 4 | CYP1A2 | VKROC1 | CYP3A4 | RAD51 | NRAS |
| 5 | CYP2C19 | CYP3A4 | CYP2D6 | CHEK2 | HRAS |
| 6 | CYP2B6 | CYP1A2 | CYP1A2 | BRIP1 | PTPN11 |
| 7 | CYP3A5 | CYP2C8 | CYP2B6 | FANCD2 | RAF1 |
| 8 | CYP2C8 | CYP3A5 | CYP2C18 | BRCA3 | SOS1 |
| 9 | GGCX | CYP2B6 | CYP2A6 | BCCIP | KRAS |
| 10 | CYP2A6 | CYP1A1 | CYP2E1 | TP53 | MAP2K1 |
We then used the Jaccard index to measure the similarity of top-ranked gene lists for these five seed genes in order to investigate the effect of seed genes on PGx gene extraction and ranking. The Jaccard coefficient measures the similarity between two sets, and is defined as the size of the intersection divided by the size of the union of two sets. As shown in Table 4, there were considerable overlaps in top-ranked genes when PGxspecific seeds were used (columns 2 and 3). On the other hand, there was a little overlap when the gene lists derived using PGx-specific seeds were compared to those using non- PGx-specific seeds (columns 4 and 5). For instance, while 67.2% of the top 500 genes were the same for seeds CYP2C9 and CYP2D9, only 2.7% of the top 500 genes were the same for seeds CYP2C9 and EGFR. There were no overlaps in the top-ranked 100 genes between CYP2C9 and BRCA1 or EGFR. Both CYP2C9 and CYP2D6 are cytochrome P450 family genes and the top-ranked gene lists for these two seeds are more similar (67.2% for top 500 ranked genes) than those between CYP2C9 and VKORC1 (48.1% for top 500 ranked genes).
Table 4.
Jaccard similarity among top ranked genes for seeds: CYP2C9, CYP2D6, VKORC1, BRCA1 and EGFR.
| Rank | CYP2C9/CYP2D6 | CYP2C9/VKORC1 | CYP2C9/BRCA1 | CYP2C9/EGFR |
|---|---|---|---|---|
| 10 | 66.7 | 53.8 | 0.0 | 0.0 |
| 25 | 61.3 | 56.2 | 0.0 | 0.0 |
| 50 | 75.4 | 51.5 | 0.0 | 0.0 |
| 100 | 65.3 | 46.0 | 0.0 | 0.0 |
| 200 | 71.7 | 40.8 | 1.8 | 0.5 |
| 300 | 68.5 | 56.9 | 4.0 | 0.8 |
| 400 | 68.4 | 47.1 | 5.5 | 2.2 |
| 500 | 67.2 | 48.1 | 7.4 | 2.7 |
5. Discussion
We have developed a bootstrapping learning to iteratively search for and rank PGxspecific genes among 33,310 human genes and 2388 PharmGKB genes. Our algorithm is able to accurately find and rank PGx-specific genes highly on the input gene lists. We also demonstrated that this gene searching and ranking algorithm is domain independent and can be used to ranking other gene types, such as oncogenes, or tumour suppressor genes. This algorithm can be used to create domain-specific lexicons for other types of biomedical relationship extraction. However, it has several limitations and requires further improvements in precision and recall. First, the precision is not perfect and some non-PGx genes are ranked high on the list. The highest precision is 0.807 for genes in the 1.0% rank as shown in Table 1. For example, a total of 12 genes co-occur with the highly ranked PGx gene CYP3A4 in the following sentence: “The expression levels of 13 of these genes, ALPK2, ASAP1, CEACAM5, CYP3A4, ENAH, ERBB2, HHIPL2, LTB4R, MMP9, PERLD1, PNMT, PTPRA, and OSMR, were validated in a total of 118 gastric samples using either the qRT-PCR or TRAC assay” (Junnila et al., 2010). Based on our algorithm, all 12 genes are ranked highly since they co-occur with a highly ranked PGx gene. Another example is the gene MKNK, which is a MAP kinase interacting serine/threonine kinase 1 gene. The MKNK gene symbol appears in MEDLINE only twice, once with the highly ranked PGx gene CYP2C8. However, the MEDLINE abstract where the gene MKNK co-occurs with CYP2C8 is not related to PGx studies. We can improve the precision of our gene extraction and ranking algorithm using text classification techniques to first classify MEDLINE abstracts into PGx-related or non- PGx-related. Our ranking algorithm implicitly uses the knowledge about the gene entities themselves to classify MEDLINE sentences and their gene occurrences based on known PGx genes. For example, if a gene co-occurs with a PGx gene in a sentence, then it is likely that the sentence is related to a PGx study and the gene is a PGx-specific gene. On the other hand, text classifiers can learn text features surrounding gene entities, not entities themselves to classify sentences. We can improve our algorithm by training a text classifier using manually curated PGx articles available in PharmGKB and incorporating the text classification score into our ranking algorithm to reduce false positive rate. We expect that incorporating both text features and the gene ranking score will result in a higher precision algorithm. The effect of a text classifier on precision and recall will largely depend on how good the text classifier is in classifying a text document and how the text classification scores with the gene ranking score are combined.
Another limitation in our ranking algorithm is that it ranks common, ambiguous genes highly. For example, the gene symbol SD is a valid gene symbol for ‘segregation distorter homolog’. But in most MEDLINE abstracts where it appears, SD represents ‘Standard Deviation’ and appears together with many PGx genes. More robust networkbased ranking algorithms may be able to detect common ambiguous genes if we assume that common ambiguous genes such as SD and MS are randomly distributed and associated with all other human genes. Standard network ranking algorithms such as PageRank algorithm (Page et al., 1999) can be used to detect the random distribution of common ambiguous genes in the whole gene-gene network and the non-random enrichment of PGx genes in PGx-specific sub-networks.
In this study, we conducted comprehensive precision and recall study for only one seed gene, CYP2C9, due to the time-consuming manual evaluation process for precision calculation. For precision evaluation, we manually examined a randomly selected 20% of all 13,581 genes extracted, starting with seed gene CYP2C9 for HGnC genes and 2093 genes for PharmGKB genes. For each gene, we examined its name and the MEDLINE sentences in which it appeared to determine if it was a PGx gene. If a different seed, for example VKORC1, is used; the extracted gene list will be different from the 13,581 genes extracted with seed CYP2C9. Manual examination of a portion of the gene list with a different seed will be needed for precision calculation. However, we expect that the precision and recall will not differ significantly as long as a good seed is used. In our study, we have demonstrated that there are significant overlaps in extracted and ranked gene lists when three different PGx genes CYP2C9, CYP2D6 and VKORC1 are used as seeds (Tables 3 and 4). In addition, the top-ranked genes for each seed are also typical PGx genes, further strengthening our belief that as long as a good PGx gene is used as seed, the actual PGx genes in the final gene lists will be similar. One of the goals of this study is to create a comprehensive and accurate PGx-specific gene lexicon to facilitate drug-gene relationship extraction from free text; it will be important to conduct a quantitative study using state-of-art PGx drug-gene extraction algorithms to demonstrate the degree of improvements of a precise and comprehensive PGx gene lexicon on subsequent drug-gene extraction. It is possible that the effects of the gene lexicon on certain types of algorithms (e.g. statistical or machine learning approaches) will be greater than on others (e.g. NLP-based approaches).
6. Conclusion
A high-quality PGx-specific gene lexicon is important for automatic drug-gene relationship extraction tasks. In this study, we have developed a bootstrapping learning technique to rank 33,310 human genes and 2388 PharmGKB genes on their relevance to drug response. The algorithm uses one seed PGx gene to iteratively extract and rank genes that co-occur using 20 million MEDLINE abstracts. Compared to randomly ranked genes (precision: 0.032, recall: 0.013, F1: 0.018), the algorithm has achieved significantly better performance (precision: 0.861, recall: 0.548, F1: 0.662) for the top 2.5% of ranked genes. In addition, our algorithm has achieved similar performance in ranking PharmGKB genes (precision: 1.000, recall: 0.500, F1: 0.667). This algorithm is domain independent and can be used to rank other types of genes.
Acknowledgements
Both Xu and Wang have conceived the idea, designed and implemented the algorithms, and prepared the manuscript. Xu is funded by Case Western Reserve University faculty starting grant and Wang is funded by ThinTek.
Biography
Rong Xu is an Assistant Professor in the Medical Informatics Division, Case Western Reserve University, Cleveland, USA. She received her PhD in Biomedical Informatics in 2010 from Stanford University, USA. Her research interests include natural language processing, text mining, systems biology and imaging informatics.
QuanQiu Wang is the founder of ThinTek LLC at Palo Alto, CA, USA. He received his MS in Computer Science from Stanford University, USA. His research interests include biomedical knowledge acquisition, semantic search, natural language processing, text mining, knowledge integration, ontology and machine learning.
Contributor Information
Rong Xu, Medical Informatics Division, Case Western Reserve University, Cleveland, OH 44106, USA.
QuanQiu Wang, Thin Tek LLC, Palo Alto, CA 94306, USA, quanqiu@thintek.com.
References
- Ahlers CB, Fiszman M, Demner-fushman D, Lang F and Rindesch TC (2007) ‘Extracting semantic predications from MEDLINE citations for pharmacogenomics’, Pacific Symposium on Biocomputing, pp.209–220. [PubMed] [Google Scholar]
- Bosch TM, Doodeman VD, Smits PH, Meijerman I, Schellens JH and Beijnen JH (2006) Pharmacogenetic screening for polymorphisms in drug-metabolizing enzymes and drug transporters in a Dutch population’, Molecular Diagnnosis & Therapy, Vol. 10, No. 3, pp.175–185. [DOI] [PubMed] [Google Scholar]
- Chang JT and Altman RB (2004) ‘Extracting and characterizing gene-drug relationships from the literature’, Pharmacogenetics, Vol. 14, pp.577–586. [DOI] [PubMed] [Google Scholar]
- Coulet AM, Shah N, Garten Y, Musen M and Altman RB (2010) ‘Using text to build semantic networks for pharmacogenomics’, Journal of Biomedical Informatics, Vol. 43, pp.1009–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JC, Furstenthal L, Desai AA, Norris T, Sutaria S, Fleming E and Ma P (2009) ‘The microeconomics of personalized medicine: today’s challenge and tomorrow’s promise’, Nature reviews Drug discovery, Vol. 8, pp.279–286. [DOI] [PubMed] [Google Scholar]
- Dong C, Wong ML and Licinio J (2009) Sequence variations of ABCB1, SLC6A2, SLC6A3, SLC6A4, CREB1, CRHR1 and NTRK2: association with major depression and antidepressant response in Mexican-Americans’, Molecular Psychiatry, Vol. 14, pp.1105–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans WE and McLeod HL (2003) ‘Pharmacogenomics: drug disposition, drug targets, and side effects’, The New England Journal of Medicine, Vol. 348, pp.538–549. [DOI] [PubMed] [Google Scholar]
- Garten Y and Altman RB (2009) ‘Pharmspresso: a text-mining tool for extraction of pharmacogenomic concepts and relationships from full text’, BMC Bioinformatics, Vol. 10, p.S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garten Y, Coulet A and Altman RB (2010) ‘Recent progress in automatically extracting information from the pharmacogenomic literature’, Pharmacogenomics, Vol. 11, pp1467–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen NI, Brunak S and Altman RB (2009) ‘Generating genome-scale candidate gene lists for pharmacogenomics’, Clinical Pharmacology and Therapeutics, Vol. 86, pp.183–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haveliwala T (2003) ‘Topic-sensitive PageRank: a context-sensitive ranking algorithm for web search’, IEEE Transactions on Knowledge and Data Engineering, Vol. 15, No. 9, pp.784–796. [Google Scholar]
- Herget M and Tampé R (2007) ‘Intracellular peptide transporters in human- compartmentalization of the “peptidome”‘, Pflugers Arch, Vol. 453, No. 5, pp.591–600. [DOI] [PubMed] [Google Scholar]
- Jenssen TK, Laegreid A, Komorowski J and Hovig E (2001) ‘A literature network of human genes for high-throughput analysis of gene expression’, Nature Genetics, Vol. 1, pp.21–28. [DOI] [PubMed] [Google Scholar]
- Junnila S, Kokkola A, Karjalainen-Lindsberg ML, Puolakkainen P and Monni O (2010) ‘Genome-wide gene copy number and expression analysis of primary gastric tumors and gastric cancer cell lines’, BMC Cancer, doi: 10.1186/1471-2407-10-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapucuoglu N, Coban T, Raunio H, Pelkonen O, Edwards RJ, Boobis AR and Iscan M (2003) ‘Expression of CYP3A4 in human breast tumour and non-tumour tissues’, Cancer Letter, Vol. 202, No. 1, pp.17–23. [DOI] [PubMed] [Google Scholar]
- Klein TE, Chang JT, Cho MK, Easton KL, Fergerson R, Hewett M, Lin Z, Liu Y, Liu S, Oliver DE, Rubin DL, Shafa F, Stuart JM and Altman RB (2001) ‘Integrating genotype and phenotype information: an overview of the PharmGKB project - pharmacogenetics research network and knowledge base’, The Pharmacogenomics Journal, Vol. 1, No. 3, pp.167–170. [DOI] [PubMed] [Google Scholar]
- Morrison JL, Breitling R, Higham DJ and Gilbert DR (2005) ‘GeneRank: using search engine technology for the analysis of microarray experiments’, BMC Bioinformatics, Vol. 6, p.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page L, Brin S, Motwani R and Winograd T (1999) The PageRank Citation Ranking: Bringing Order to the Web, Technical Report, Stanford InfoLab [Google Scholar]
- Piacentini S, Polimanti R, Moscatelli B, Re MA, Fuciarelli R, Manfellotto D and Fuciarelli M (2010) ‘Glutathione S-transferase gene polymorphisms and air pollution as interactive risk factors for asthma in a multicentre Italian field study: a preliminary study’, Annals of Human Biology, Vol. 37, No. 3, pp.427–439. [DOI] [PubMed] [Google Scholar]
- Swen JJ, Huizinga TW, Gelderblom H, de Vries EGE, Assendelft WJJ, Kirchheiner J and Guchelaar H (2007) ‘Translating pharmacogenomics: challenges on the road to the clinic’, Plos Medicine, Vol. 4, p.e209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theobald M, Shah N and Shrager J (2009) ‘Extraction of conditional probabilities of the relationships between drugs, diseases, and genes from PubMed guided by relationships in PharmGKB’, AMIA Summit on Translational Bioinformatics, pp.124–128. [PMC free article] [PubMed] [Google Scholar]
- Vecsler M, Loebstein R, Almog S, Kurnik D, Goldman B, Halkin H and Gak E (2006) ‘Combined genetic profiles of components and regulators of the vitamin K-dependent gammacarboxylation system affect individual sensitivity to warfarin’, Thrombosis Haemostasis, Vol. 95, No. 2, pp.205–211. [DOI] [PubMed] [Google Scholar]
- Weiss ST (2008) ‘Creating and evaluating genetic tests predictive of drug response’, Nature Review Drug Discovery, Vol. 7, pp.568–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Liu M, Zheng W, Zhao Z and Xu H (2012) ‘Ranking gene-drug relationships in biomedical literature using latent dirichlet allocation’, Pacific Symposium on Biocomputing, pp.422–433. [PMC free article] [PubMed] [Google Scholar]
- Xu R, Das AM and Garber A (2009a) ‘Investigation of unsupervised pattern learning techniques for bootstrap construction of a medical treatment lexicon’, NAACL BioNLP, pp.63–70. [Google Scholar]
- Xu R, Morgan A, Das AM and Garber A (2009b) ‘Unsupervised method for extracting machine understandable medical knowledge from a large free text collection’, Proceedings of AMIA Symposium, pp.709–713. [PMC free article] [PubMed] [Google Scholar]
- Xu R, Supekar K, Morgan A, Das AM and Garber A (2008) ‘Unsupervised method for automatic construction of a disease dictionary from a large free text collection’, Proceedings of AMIA Symposium, pp.820–824. [PMC free article] [PubMed] [Google Scholar]
- Xu R and Wang Q-Q (2012) ‘A knowledge-driven conditional approach to extract pharmacogenomics specific drug-gene relationships from free text’, Journal of Biomedical Informatics, Vol. 5, pp.827–834. [DOI] [PMC free article] [PubMed] [Google Scholar]