Abstract
Immobilization of the tendonand ligament has beenshown to result in a rapid and significant decrease in material properties. It has been proposed that tissue degradation leading to tendon rupture or pain in humansmayalso be linked to mechanical unloading following focal tendon injury. Hence, understanding the remodeling mechanism associated with mechanical unloading has relevance for the human conditions of immobilization (e.g., casting), delayed repair of tendon ruptures, and potentially overuse injuries as well. This is the first study to investigate the time course of gene expression changes associated with tissue harvest and mechanical unloading culture in an explant model. Rat tail tendon fascicles were harvested and placed in culture unloaded for up to 48 h and then evaluated using qRT-PCR for changes in two anabolic and four catabolic genes at 12 time points. Our data demonstrates that Type I Collagen, Decorin, Cathepsin K, and MMP2 gene expression are relatively insensitive to unloaded culture conditions. However, changes in both MMP3 and MMP13 gene expression are rapid, dramatic, sustained, and changing during at least the first 48 h of unloaded culture. This data will help to further elucidate the mechanism for the loss of mechanical properties associated with mechanical unloading in tendon.
Keywords: tendon, quantitative real-time PCR, mechanobiology, matrix metalloproteinases, Collagen type I, Decorin
Tendons and ligaments are dense fibrous connective tissues primarily composed of Type I Collagen cross-linked with the proteoglycan Decorin. This structure provides high tensile strength and enables transmission of force from muscle to bone in the case of tendons, and from bone to bone for ligaments. Immobilization of tendon and ligament has been shown to upset normal homeostasis and result in tissue catabolism and matrix remodeling.1–5 A relationship between mechanical load reduction and a rapid and significant decrease in material properties has been consistently demonstrated in animal models.3,4,6–11 In vitro explant models have been utilized to further investigate changes in tissue mechanical properties associated with load reduction. Rabbit patellar tendon subunits cultured under no load for 1 week demonstrated a 43.8% decrease in tangent modulus compared to Fresh controls.12 Further, rat tail tendons exposed to no load had a reduction in elastic modulus of 23%13 to 42.9%14 after 1 week and 43.7% after 3 weeks in culture.15
Although the precise mechanisms involved in extracellular matrix remodeling associated with mechanical unloading are not known, they are likely associated with increased matrix metalloprotease (MMP) activity. Previous studies have demonstrated that the gene expression and protein levels of interstitial collagenase (MMP-13), was increased in rat tail tendon explants following 24 h of mechanical unloading in culture.14,16–18 This catabolic response can be largely abrogated by static or cyclic loading in culture unless the cytoskeleton of tendon cells is disrupted by treatment with cytochalasin-D.16,17 These results support the concept of a cytoskeletally based mechanotransduction pathway in tendon.14,16,18,19
Clinically three types of tendon injuries have been described: acute rupture,20 rupture associated with tissue degradation20–22 and chronic pain associated with tissue degradation.20,22–24 Acute rupture is generally related to a single, high-force incident. The etiology of tissue degradation leading to tendon rupture or pain, however, is less understood. It has been proposed that these conditions, often classified as “repetitive stress’’ or ‘‘overuse’’ injuries, may, in fact, be linked to mechanical unloading following focal tendon injury.14,18,20,25 Specifically, local unloading of tendon cells around the injury site is hypothesized to increase collagenase activation and/or production, leading to focal tissue degradation. This hypothesis is supported by clinical studies showing that tissue degeneration in biopsies from ruptured human tendons appears to be an active, cell-mediated process.20,21,23–32
Hence, understanding the remodeling mechanism associated with mechanical unloading has relevance for the human conditions of immobilization (e.g., casting), delayed repair of tendon ruptures and potentially overuse injuries as well. To investigate the effects of mechanical load on tendon remodeling in isolation from the influence of other physiologic variables present in vivo during tendon injury conditions, we and others have utilized the in vitro rat tail tendon fascicle model.13–18,33–37 To date, no study has investigated the time course of gene expression changes associated with tissue harvest, mechanical unloading, and culture in this explant model. Such data is important for under-standing both the kinetics and magnitude of the response of tendon cells to mechanical unloading as well as for establishing the baseline expression levels from which gene changes due to mechanical loading protocols can be compared.
Therefore, the purpose of the current study was to examine the effects of unloading on the expression of various anabolic and catabolic genes in rat tendon at several times over 48 h in culture. Specifically, we evaluated the expression of Collagen type I and Decorin, as these constituents are the most prevalent components of tendon extracellular matrix. Wealso examined several catabolic genes which have been identified as either up- or downregulated in biopsies from degenerate tendon: Cathepsin K,30 interstitial collagenase (MMP1 in human, MMP13 in rat),31,38 gelatinase (MMP2),24,31 and stromelysin (MMP3).20,24,29,31 We hypothesized that the expression of catabolic genes (Cathepsin K, MMP13, MMP2, MMP3) would be continuously upregulated in unloading conditions14,16–18 and the expression of anabolic genes (Collagen type I and Decorin) would be maintained or downregulated in a reciprocal fashion.19,37 Further, we hypothesized that the expression levels of all genes would reach new (i.e., different from Fresh) homeostatic levels by 48 h in culture.
MATERIALS AND METHODS
Tissue Harvest and Sample Preparation
Four adult male Sprague-Dawley rats were sacrificed by CO2 asphyxiation. The tail was harvested and the ventral fascicles pulled and placed into culture medium (DMEM containing L-Glutamine, HEPES, Penicillin, Streptomycin, Amphotercin B, and 10% FBS). Only fascicles with a diameter between 250 and 300 μm were used. Fascicles were cleaned of their superficial sheath and cut to 2.5-cm pieces. Five fascicle pieces were pooled to make 12 individual fascicle samples for each rat. Each sample was approximately 8 mg wet weight.
Mechanical Unloading
Fascicle samples were placed into individual culture wells containing 5 mL of culture medium [DMEM containing L-Glutamine, HEPES, Penicillin Streptomycin, Amphotercin B, and 10% fetal bovine serum (FBS)], and the plates were cultured in an incubator at 5% CO2, 37°C. The time at which the tail was harvested was considered time zero. At various time points of mechanical unloading in culture [Fresh (12 min), 30 min and 1, 2, 4, 6, 8, 12, 24, 28, 32, and 48 h], one fascicle-sample per rat (n=4 rats) was removed from culture for gene expression analysis. The relative expression of Collagen Type I, Decorin, Cathepsin K, MMP2, MMP3, and MMP13 was determined at each unloading time with respect to Fresh. 18S was used as a housekeeping gene.20
RNA Harvest, Transcription, and Quantification
Samples were incubated in Trizol® Reagent for 8 days. Subsequently, RNA was harvested according to the manufacturer’s instructions using a high salt solution (0.8 M sodium citrate, 1.2 M NaCl) in conjunction with isopropanol for the precipitation step.39 The RNA pellet was resuspended in 100 μL RNase free water. Reverse transcription was performed using TaqMan® Reverse Transcription reagents from ABI (Foster City, CA) according to the manufacturer’s instructions using 20 μL RNA in a total reaction volume of 100 μL
Real-Time Quantitative PCR
PCR was performed on an ABI 7500 system (Foster City, CA). 18S was measured using TaqMan® rRNA Control Reagents (VIC® dye) and TaqMan® Universal PCR Master Mix from ABI (Foster City, CA). Collagen Type I, Decorin, Cathepsin K, MMP2, MMP3, and MMP13 were measured using Power SYBR® Green PCR Master Mix from ABI. The primer sets for all experimental genes (Table 1) were designed using Primer Express software with one primer spanning an exon junction to prevent amplification of genomic DNA (verified by melting curve analysis). The primer concentration used for all genes was 50 nM. A five point relative standard curve (R2 =0.99) was created for each gene using cDNA made from RNA harvested from rat tail tendon fibroblasts grown in culture and stimulated with IL1-β. The relative expression for each sample was determined from the relative standard curve,40,41 eliminating the need for the efficiencies of each reaction to be equal, which is a requirement when using the ΔΔCt method for data analysis. In cases where the gene expression level was above the highest value on the standard curve, samples were diluted and both the gene of interest and the housekeeping gene were reevaluated in the diluted sample. In the event that a gene expressed below the lowest point of the standard curve (Table 2), the sample was assigned the lowest expression value of the standard curve. All assays were performed in triplicate using 2 μL cDNA in a 25 μL reaction volume. Triplicates were averaged and then normalized to the relative 18S expression for that sample, giving the normalized relative expression value.
Table 1.
Primer Sets for Gene Expression
| Forward Primer | Reverse Primer | |
|---|---|---|
| Decorin | GAGGCCTCTGGCATAATCCC | CCCAGATCAGAACACTGCACC |
| Collagen Type I | CCAGGGCTCCAACGAGATCG | CGGTGTGACTCGTGCAGCCA |
| Cathepsin K | CTATGGCTGTGGAGGCGGCTA | TCCTGCCCCACATACGGGTAA |
| MMP2 | TGATGCTTTTGCTCGGGCCT | TCTCCATGCTCCCAGCGTCC |
| MMP3 | TGGACCTCCCACAGACTCCCC | CCCCGCAGGGTGCTGACTG |
| MMP13 | GCACACGCTTTTCCTCCTGGA | AAGTTGTAGCCTTTGGAGCTGCTTGT |
Table 2.
Samples with Relative Gene Expression Off Scale Low
| Gene and Time Point |
||
|---|---|---|
| Rat | MMP2 | MMP13 |
| 1 | 12, 24 h | Fresh, 1, 2, 24, 28 h |
| 2 | Fresh, 30 min, 12 h | Fresh, 30 min, 1, 2, 12 h |
| 3 | Fresh | |
| 4 | Fresh | |
Data Analysis and Statistics
We assessed whether the expression of Decorin, Collagen Type I, Cathepsin K, MMP2, MMP3, and MMP13 were significantly different in unloaded tendon fascicles than in Fresh fascicles. For each gene, the normalized relative expression value for each rat sample at each time point was divided by the Fresh value for that animal. Accordingly, an expression ratio of one at a given time point means the gene is expressed the same as Fresh. These expression ratios at each time point were then logged to base 2 and used as the basis for a bootstrapping simulation (n=4 rats per time point). The bootstrap simulation was run 1000 times using each set of gene-time data.42
A mean was calculated directly from the bootstrap data and the 95% confidence interval was defined from the limits of the middle 95% of the bootstrap data. Means and confidence limits were subsequently unlogged and plotted. For a given gene and time point, two criteria were used to define a significant change in expression from Fresh: (1) statistical significance— the 95% confidence interval did not include an expression ratio of one and (2) biological significance—the geometric mean was at least twofold different from Fresh (mean expression ratio greater than 2 or less than 0.5).43
To determine whether the expression of each gene had stabilized or was still changing with respect to time between 24 and 48 h of culture, linear regression analysis was performed between time (independent variable) and the mean expression ratio at 24, 28, 32, and 48 h (dependent variable). If the regression coefficient was significantly different than zero ( p < 0.05), the gene was considered significantly changing with respect to time in that interval.
RESULTS
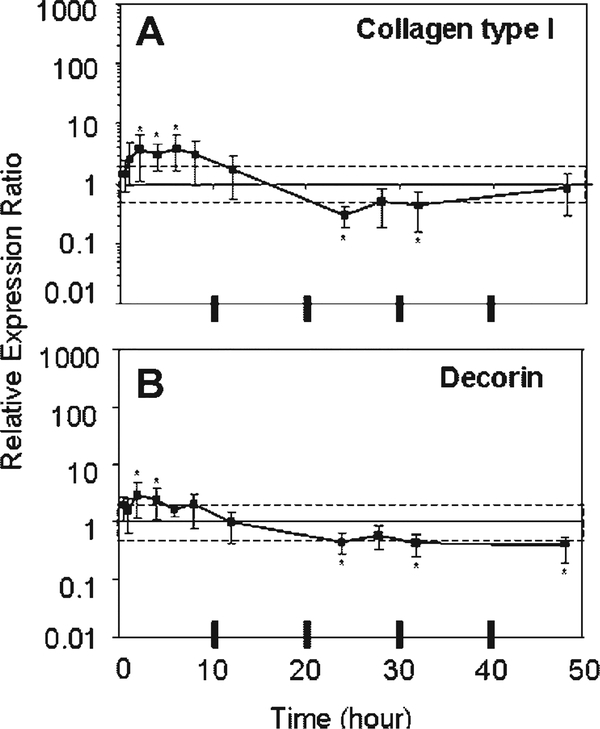
Type I Collagen and Decorin exhibited small and fluctuating changes in gene expression with mechanical unloading in culture (Fig. 1). In general, the expression of these anabolic genes was significantly increased (two-to fourfold) with respect to Fresh at 2–6 h of unloading culture and then significantly decreased to the same degree at 24 h. Subtle differences between the genes and their significance with respect to Fresh can be noted in Figure 1. Type I Collagen expression was still increasing significantly with respect to time between 24 and 48 h of unloading ( p=0.03), but had returned to levels that were not different from Fresh. Whereas Decorin expression was not changing in this time interval ( p=0.46), but remained significantly less than Fresh.
Figure 1.
Means and upper and lower limits of the 95% confidence intervals of the bootstrap data for relative expression ratio of (A) Collagen Type I and (B) Decorin are plotted. [Relative expression ratios used for bootstrapping at each time point (n=4) were derived from the normalized expression of each sample divided by normalized expression of Fresh, where 18S was the housekeeping gene.] An expression ratio of one means no change from Fresh. The dashed box indicates a two-fold change from Fresh. *Denotes significant difference from Fresh.
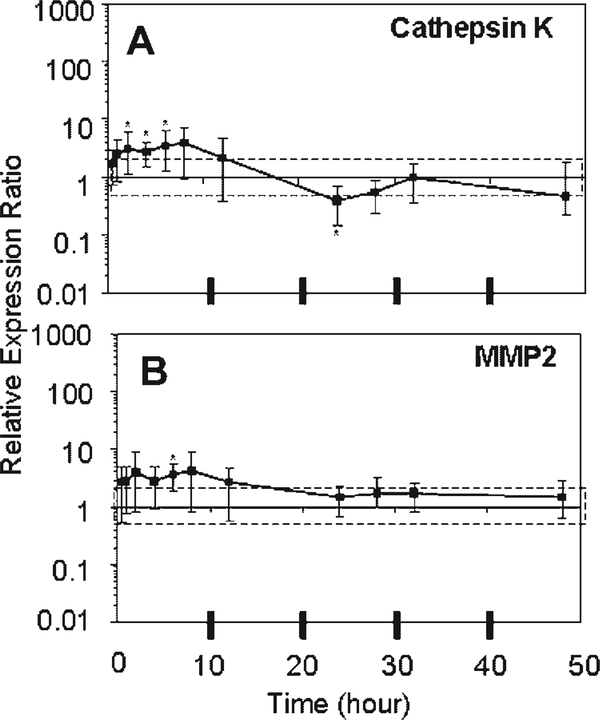
Cathepsin K demonstrated small but significant increases (two to fourfold) in expression relative to Fresh in the early period of unloading culture (2–6 h) and decreased expression with respect to Fresh (0.4-fold) at 24 h (Fig. 2). MMP2 demonstrated a slight increase in expression throughout the 48 h of unloading culture, which was significantly different from Fresh only at 6 h (Fig. 2). Neither Cathepsin K ( p=0.96) nor MMP2 ( p=0.62) expression changed over time between 24 and 48 h and had returned to levels that were not different from Fresh.
Figure 2.
Means and upper and lower limits of the 95% confidence intervals of the bootstrap data for relative expression ratio of (A) Cathepsin K and (B) MMP2 are plotted. [Relative expression ratios used for bootstrapping at each time point (n=4) were derived from the normalized expression of each sample divided by normalized expression of Fresh, where 18S was the house-keeping gene.] An expression ratio of one means no change from Fresh. The dashed box indicates a twofold change from Fresh. *Denotes significant difference from Fresh.
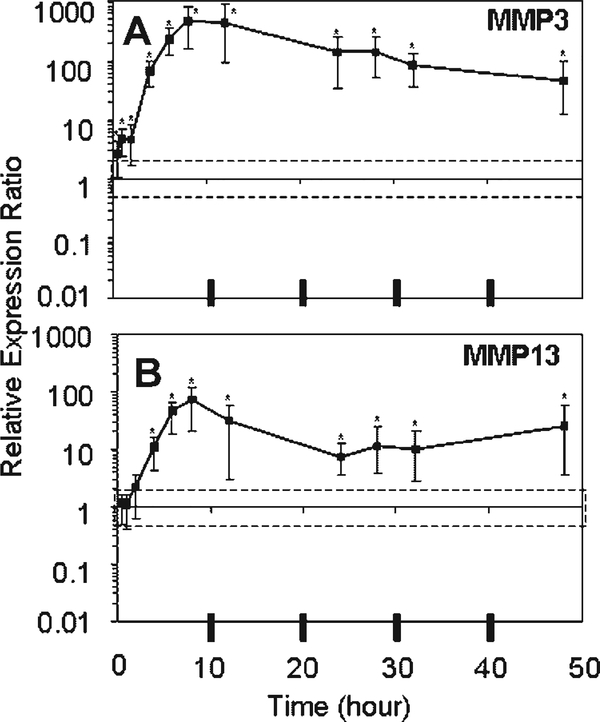
The gene expression profiles for MMP3 and MMP13 showed rapid, dramatic, and sustained responses to unloading culture (Fig. 3). MMP3 expression was significantly higher than Fresh at every time point evaluated, peaking at a 447-fold increase at 8 h of unloading and declining to a 44-fold increase at 48 h. There was a trend toward continued decreases in MMP3 expression between 24 and 48 h of unloading culture ( p=0.06). MMP13 expression became significantly increased with respect to Fresh by 4 h of unloading, peaking at a 72-fold increase at 8 h and remaining significantly increased thereafter. MMP13 expression was increasing significantly with respect to time between 24 and 48 h of unloading ( p=0.03).
Figure 3.
Means and upper and lower limits of the 95% confidence intervals of the bootstrap data for relative expression ratio of (A) MMP3 and (B) MMP13 are plotted. [Relative expression ratios used for bootstrapping at each time point (n=4) were derived from the normalized expression of each sample divided by normalized expression of Fresh, where 18S was the housekeeping gene.] An expression ratio of one means no change from Fresh. The dashed box indicates a twofold change from Fresh. *Denotes significant difference from Fresh.
DISCUSSION
This is the first study to examine the expression profile of several anabolic and catabolic extracellular matrix genes over a time course of in vitro unloading in the rat tail tendon fascicle model. In partial support of our first hypothesis, the two anabolic genes evaluated (Collagen Type I and Decorin), exhibited only small and fluctuating changes in gene expression over the time course of unloading and two of the catabolic genes evaluated (MMP3 and MMP13) demonstrated rapid, dramatic, and sustained upregulation throughout the 48 h of mechanical unloading in culture. However, contrary to our hypothesis, two of the catabolic genes (Cathepsin K and MMP2) also exhibited only small fluctuating changes in gene expression over the time course of unloading. Our second hypothesis, that gene expression levels of all genes would reach new (i.e., different from Fresh) homeostatic levels by 48 h in culture was also only partially supported. The expression of Decorin, MMP2, and Cathepsin K had stabilized between 24 and 48 h in culture ( p>0.46), but only decorin expression stabilized at a level different from Fresh. Although the expression of Type I Collagen was still changing over time between 24 and 48 h ( p=0.03), it had returned to Fresh levels. The expression of MMP3 ( p=0.06), and MMP13 ( p=0.03) was still changing over time between 24 and 48 h and MMP13 expression in particular did not appear to be changing toward Fresh levels. The expression of both MMP3 and MMP13 would need to be evaluated at longer time points in unloading culture to determine if a new homeostatic set point is reached for these genes.
Type I Collagen and Decorin are the primary constituents of the tendon extracellular matrix. Decorin is a proteoglycan that decorates the collagen fibrils in the tendon, and contains a single sulfated glycosaminogly-can (GAG) chain. Both Type I Collagen and Decorin gene expression were slightly but significantly increased (two-to fourfold) between 2–6 h of mechanical unloading and significantly decreased to the same degree by 24 h. Type I Collagen expression approached Fresh levels by 48 h, and Decorin expression remained significantly down- regulated. No other study has examined the expression of these genes in this model system, but Collagen and Decorin expression levels were reported not to change in a rabbit MCL explant model after 4 h of mechanical unloading.44 At the protein level, no change in total collagen or sulfated glycosaminoglycan (GAG) content was measured in tendon fascicles after 24 h37 or 1 week13 of unloading culture. Together these results suggest that the rate of Collagen Type I and Decorin deposition and breakdown are not significantly changed in response to mechanical unloading in this model system.
Cathepsin K and MMP2 are proteases that have been identified inhumanbiopsy samples from injured tendons and ligaments.24,30,31 Cathepsin K is a serine protease with specificity for Type I Collagen.45,46 MMP2 is one of a family of proteases known as matrix metalloproteases. The substrates of MMP2 include Collagens I, II, III, IV, V, VII, X, and XI as well as gelatin.47 In addition to cleaving other matrix proteins and proteoglycans, MMP2 is also known to activate both MMP9 and MMP13.47 Neither Cathepsin K nor MMP2 have been previously examined in mechanical unloading experiments. In this study, both Cathepsin K and MMP2 gene expression were slightly but significantly increased (two-to fourfold) between 2 and 6 h of mechanical unloading, and then stabilized at Fresh levels beyond 24 h in unloaded culture. Like type I Collagen and Decorin, Cathepsin K and MMP2 gene expression appear to be relatively insensitive to the unloaded culture conditions.
MMP3 (stromelysin-1) and MMP13 (collagenase-1) are also matrix metalloproteases. Considered primarily a proteoglycanase, MMP3 has substrate specificity for a range of matrix components and is also known to activate MMP7, MMP8, and MMP13.47 MMP13 is considered predominantly a collagenase; however, it, too, has substrate specificity for a range of matrix components and is known to activate MMP2 and MMP9.47 In the rodent, MMP13 is the predominant interstitial collagenase, whereas the predominant interstitial collagenase in humans is MMP1.48 Both MMP320,23,24,29,31 and interstitial collagenase (MMP1 in humans)20,31 have been shown to be regulated in injured tendons and ligaments. In this study, the gene expression of both MMP3 and MMP13 was dramatically increased with respect to Fresh over the entire 48 h of unloading culture. We note that the MMP13 expression level for all of the Fresh samples fell below the detection level and was assigned the lowest measurable value. Therefore, the actual fold changes in MMP13 expression are likely to be even greater than those reported here. No previous unloading study has investigated MMP3 gene expression levels in the rat tail tendon model; however, an explant model of canine cartilage showed no change in MMP3 with unloading for 6 days,49 and in vivo immobilization of the rat intervertebral disc demonstrated an upregulation of MMP3 at 72 h.50 Upregulation of MMP13 after 24 h of mechanical unloading in culture has been shown in the rat tail tendon fascicle model.14,16–18,51 Although increases in gene expression do not necessarily correlate to increases in protein level or activation of these enzymes, dramatic and sustained increases in MMP3 and MMP13 during mechanical unloading may imply downstream matrix degradation and remodeling.
Collagen Type I, Decorin, Cathepsin K, MMP2 each exhibited small and fluctuating changes in gene expression over the time course of unloading. The expression of Decorin, MMP2, and Cathepsin K stabilized between 24 and 48 h in culture, at or—in the case of Decorin— slightly lower than the levels of Fresh tendon. Although the expression of Type I Collagen was still changing significantly with respect to time at 48 h, it had rebounded to Fresh levels. These outcomes suggest that these four genes are both relatively insensitive to the unloaded culture conditions, and have reached new homeostatic expression levels at or near the levels of Fresh tendon after approximately 48 h in unloaded culture conditions.
In contrast, the gene expression of both MMP3 and MMP13 was rapidly and dramatically increased with respect to Fresh by 30 min (MMP3) or 4 h (MMP13) in culture, and remained increased over the entire 48 h of unloading. Furthermore, at least MMP13 expression was still changing significantly with respect to time at 48 h. That the upregulation of MMP13 is a result of mechanical unloading and not other conditions of the tissue culture is demonstrated by studies that show that MMP13 upregulation in unloaded tendon explants can be largely abrogated by static or cyclic loading in culture.16,17 These results suggest that these two genes are highly sensitive to mechanical unloading and have not reached new homeostatic levels after 48 h in unloaded culture conditions. We speculate that the expression of these mechanosensitive genes would reach new homeostatic levels when the tendon cells reestablish their new mechanostat set point in the unloaded condition.19
Despite evidence of increased collagenase (MMP13) expression in unloaded tendon, Lavagnino and colleagues15 found no difference in number of fibrils, mean fibril diameter, mean fibril density, and fibril size distribution between Fresh and rat tendons cultured under no load for 21 days. In our previous work, no decrease in total collagen or evidence of denatured collagen was seen in rat tail tendon cultured under no load for 7 days.13 These studies suggest that the matrix remodeling associated with decreased mechanical properties of cultured rat tail tendon may not occur on a macroscopic scale that is detectable by gross morphologic or biochemical assays, or that the remodeling does not directly involve degradation of collagen. Because MMP3 and MMP13 have a wide range of substrates, decreases in mechanical properties may be related to a more subtle degradation of ECM components involved in the cross linking and or stabilization of the tendon structure (e.g., proteoglycans, minor collagens, fibronectin). However, significantly smaller collagen fibrils have been shown in a hind limb disuse model of the rat Achilles tendon,52 alternately suggesting that the in vivo environment may allow for a more extensive response to mechanical unloading than can occur in an explant culture model.
The purported strength of an in vitro explant model is that it provides a well-controlled experimental system to manipulate variables of interest and investigate their effect on tissue structure, function, and cell biology. Further, an explant model maintains cells within their native extracellular matrix, such that mechanical load conditions, are transmitted to cells in a physiologic manner. However, the utility of an explant model could be limited for some mechanobiology experiments if the outcomes of interest are shifted dramatically and rapidly in unloaded culture conditions. For example, to use this explant model to study the effect of mechanical overloading on MMP3 and MMP13 gene expression, our data suggest that immediately upon retrieving explants from the animal, they must be placed in culture mechanical conditions that maintain these genes at or near normal levels. Because it is unlikely that placing the explants in such “homeostatic load conditions” could be achieved either within minutes or simultaneously for all explanted samples, interpretation of subsequent mechanical overloading conditions on MMP3 or MMP13 gene expression could be hampered by the dramatic and rapidly changing expression of these genes in the default unloaded conditions. Certainly, this tendon explant model would lend itself to the investigation of mechanical reloading on MMP3 or MMP13 expression. However, reloading would need to be initiated at the same time for all samples (unless sufficient culture time had elapsed such that the expression of the genes of interest had reached homeostatic levels) and interpretation of the results would be dependent on the time that reloading was introduced. These examples demonstrate the complexity of appropriately using explant models for mechanobiology research.
In summary, this is the first study to investigate the time course of gene expression changes associated with tissue harvest and mechanical unloading culture in an explant model. Such data are important for understanding both the kinetics and magnitude of the response of tendon cells to mechanical unloading as well as for establishing the baseline expression levels from which gene changes due to mechanical loading protocols can be compared. Our data demonstrates that Type I Collagen, Decorin, Cathepsin K, and MMP2 gene expression are relatively insensitive to unloaded culture conditions. However, changes in both MMP3 and MMP13 gene expression are rapid, dramatic, sustained, and changing during at least the first 48 h of unloaded culture. Future work will investigate whether changes in MMP3 and/or MMP13 gene expression coincide with changes in the activity or levels of these proteins in unloaded tendon explants over time. These studies will help to further elucidate the mechanism for the loss of mechanical properties associated with mechanical unloading in tendon.
ACKNOWLEDGMENTS
This grant was support by grants from the Whitaker Foundation (RG-02–0968) and National Institutes of Health (AR049858).
REFERENCES
- 1.Matsumoto F, Trudel G, Uhthoff HK, et al. 2003. Mechanical effects of immobilization on the Achilles’ tendon. Arch Phys Med Rehabil 84:662–667. [DOI] [PubMed] [Google Scholar]
- 2.Murrell GA, Lilly EG III, Goldner RD, et al. 1994. Effects of immobilization on Achilles tendon healing in a rat model. J Orthop Res 12:582–591. [DOI] [PubMed] [Google Scholar]
- 3.Binkley JM, Peat M. 1986. The effects of immobilization on the ultrastructure and mechanical properties of the medial collateral ligament of rats. Clin Orthop Relat Res 203:301–308. [PubMed] [Google Scholar]
- 4.Almeida-Silveira MI, Lambertz D, Perot C, et al. 2000. Changes in stiffness induced by hindlimb suspension in rat Achilles tendon. Eur J Appl Physiol 81:252–257. [DOI] [PubMed] [Google Scholar]
- 5.Uchida H, Tohyama H, Nagashima K, et al. 2005. Stress deprivation simultaneously induces over-expression of inter-leukin-1beta, tumor necrosis factor-alpha, and transforming growth factor-beta in fibroblasts and mechanical deterioration of the tissue in the patellar tendon. J Biomech 38:791–798. [DOI] [PubMed] [Google Scholar]
- 6.Loitz BJ, Zernicke RF, Vailas AC, et al. 1989. Effects of short-term immobilization versus continuous passive motion on the biomechanical and biochemical properties of the rabbit tendon. Clin Orthop Relat Res 244:265–271. [PubMed] [Google Scholar]
- 7.Majima T, Yasuda K, Fujii T, et al. 1996. Biomechanical effects of stress shielding of the rabbit patellar tendon depend on the degree of stress reduction. J Orthop Res 14:377–383. [DOI] [PubMed] [Google Scholar]
- 8.Noyes FR, Torvik PJ, Hyde WB, et al. 1974. Biomechanics of ligament failure. II. An analysis of immobilization, exercise, and reconditioning effects in primates. J Bone Joint Surg Am 56:1406–1418. [PubMed] [Google Scholar]
- 9.Woo SL, Matthews JV, Akeson WH, et al. 1975. Connective tissue response to immobility. Correlative study of biome-chanical and biochemical measurements of normal and immobilized rabbit knees. Arthritis Rheum 18:257–264. [DOI] [PubMed] [Google Scholar]
- 10.Woo SL, Gomez MA, Woo YK, et al. 1982. Mechanical properties of tendons and ligaments. II. The relationships of immobilization and exercise on tissue remodeling. Biorheology 19:397–408. [DOI] [PubMed] [Google Scholar]
- 11.Yamamoto N, Ohno K, Hayashi K, et al. 1993. Effects of stress shielding on the mechanical properties of rabbit patellar tendon. J Biomech Eng 115:23–28. [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto E, Iwanaga W, Miyazaki H, et al. 2002. Effects of static stress on the mechanical properties of cultured collagen fascicles from the rabbit patellar tendon. J Biomech Eng 124:85–93. [DOI] [PubMed] [Google Scholar]
- 13.Abreu EL, Leigh D, Derwin KA. Effect of altered mechanical load conditions on the structure and function of cultured tendon fascicles J Orthop Res (in press). [DOI] [PubMed] [Google Scholar]
- 14.Arnoczky SP, Lavagnino M, Egerbacher M, et al. 2007. Matrix metalloproteinase inhibitors prevent a decrease in the mechanical properties of stress-deprived tendons: an in vitro experimental study. Am J Sports Med 35:763–769. [DOI] [PubMed] [Google Scholar]
- 15.Lavagnino M, Arnoczky SP, Frank K, et al. 2005. Collagen fibril diameter distribution does not reflect changes in the mechanical properties of in vitro stress-deprived tendons. J Biomech 38:69–75. [DOI] [PubMed] [Google Scholar]
- 16.Arnoczky SP, Tian T, Lavagnino M, et al. 2004. Ex vivo static tensile loading inhibits MMP-1 expression in rat tail tendon cells through a cytoskeletally based mechanotransduction mechanism. J Orthop Res 22:328–333. [DOI] [PubMed] [Google Scholar]
- 17.Lavagnino M, Arnoczky SP, Tian T, et al. 2003. Effect of amplitude and frequency of cyclic tensile strain on the inhibition of MMP-1 mRNA expression in tendon cells: an in vitro study. Connect Tissue Res 44:181–187. [DOI] [PubMed] [Google Scholar]
- 18.Lavagnino M, Arnoczky SP, Egerbacher M, et al. 2006. Isolated fibrillar damage in tendons stimulates local collagenase mRNA expression and protein synthesis. J Biomech 39:2355–2362. [DOI] [PubMed] [Google Scholar]
- 19.Lavagnino M, Arnoczky SP. 2005. In vitro alterations in cytoskeletal tensional homeostasis control gene expression in tendon cells. J Orthop Res 23:1211–1218. [DOI] [PubMed] [Google Scholar]
- 20.Jones GC, Corps AN, Pennington CJ, et al. 2006. Expression profiling of metalloproteinases and tissue inhibitors of metalloproteinases in normal and degenerate human achilles tendon. Arthritis Rheum 54:832–842. [DOI] [PubMed] [Google Scholar]
- 21.Kannus P, Jozsa L. 1991. Histopathological changes preceding spontaneous rupture of a tendon. A controlled study of 891 patients. J Bone Joint Surg Am 73:1507–1525. [PubMed] [Google Scholar]
- 22.Astrom M, Rausing A. 1995. Chronic Achilles tendinopathy. A survey of surgical and histopathologic findings. Clin Orthop Relat Res 316:151–164. [PubMed] [Google Scholar]
- 23.Ireland D, Harrall R, Curry V, et al. 2001. Multiple changes in gene expression in chronic human Achilles tendinopathy. Matrix Biol 20:159–169. [DOI] [PubMed] [Google Scholar]
- 24.Alfredson H, Lorentzon M, Backman S, et al. 2003. cDNA-arrays and real-time quantitative PCR techniques in the investigation of chronic Achilles tendinosis. J Orthop Res 21: 970–975. [DOI] [PubMed] [Google Scholar]
- 25.Riley GP, Harrall RL, Constant CR, et al. 1994. Tendon degeneration and chronic shoulder pain: changes in the collagen composition of the human rotator cuff tendons in rotator cuff tendinitis. Ann Rheum Dis 53:359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dalton S, Cawston TE, Riley GP, et al. 1995. Human shoulder tendon biopsy samples in organ culture produce procollagenase and tissue inhibitor of metalloproteinases. Ann Rheum Dis 54:571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jarvinen M, Jozsa L, Kannus P, et al. 1997. Histopathological findings in chronic tendon disorders. Scand J Med Sci Sports 7:86–95. [DOI] [PubMed] [Google Scholar]
- 28.Lo IK, Marchuk LL, Hart DA, et al. 1998. Comparison of mRNA levels for matrix molecules in normal and disrupted human anterior cruciate ligaments using reverse transcription-polymerase chain reaction. J Orthop Res 16:421–428. [DOI] [PubMed] [Google Scholar]
- 29.Lo IK, Marchuk LL, Hollinshead R, et al. 2004. Matrix metalloproteinase and tissue inhibitor of matrix metalloproteinase mRNA levels are specifically altered in torn rotator cuff tendons. Am J Sports Med 32:1223–1229. [DOI] [PubMed] [Google Scholar]
- 30.Nakase T, Takeuchi E, Sugamoto K, et al. 2000. Involvement of multinucleated giant cells synthesizing cathepsin K in calcified tendinitis of the rotator cuff tendons. Rheumatology (Oxford) 39:1074–1077. [DOI] [PubMed] [Google Scholar]
- 31.Riley GP, Curry V, DeGroot J, et al. 2002. Matrix metalloproteinase activities and their relationship with collagen remodelling in tendon pathology. Matrix Biol 21:185–195. [DOI] [PubMed] [Google Scholar]
- 32.Spindler KP, Clark SW, Nanney LB, et al. 1996. Expression of collagen and matrix metalloproteinases in ruptured human anterior cruciate ligament: an in situ hybridization study. J Orthop Res 14:857–861. [DOI] [PubMed] [Google Scholar]
- 33.Arnoczky SP, Lavagnino M, Whallon JH, et al. 2002. In situ cell nucleus deformation in tendons under tensile load; a morphological analysis using confocal laser microscopy. J Orthop Res 20:29–35. [DOI] [PubMed] [Google Scholar]
- 34.Screen HR, Lee DA, Bader DL, et al. 2003. Development of a technique to determine strains in tendons using the cell nuclei. Biorheology 40:361–368. [PubMed] [Google Scholar]
- 35.Screen HR, Lee DA, Bader DL, et al. 2004. An investigation into the effects of the hierarchical structure of tendon fascicles on micromechanical properties. Proc Inst Mech Eng [H] 218: 109–119. [DOI] [PubMed] [Google Scholar]
- 36.Screen HR, Shelton JC, Chhaya VH, et al. 2005. The influence of noncollagenous matrix components on the micromechanical environment of tendon fascicles. Ann Biomed Eng 33:1090–1099. [DOI] [PubMed] [Google Scholar]
- 37.Screen HR, Shelton JC, Bader DL, et al. 2005. Cyclic tensile strain upregulates collagen synthesis in isolated tendon fascicles. Biochem Biophys Res Commun 336:424–429. [DOI] [PubMed] [Google Scholar]
- 38.Backman C, Boquist L, Friden J, et al. 1990. Chronic achilles paratenonitis with tendinosis: an experimental model in the rabbit. J Orthop Res 8:541–547. [DOI] [PubMed] [Google Scholar]
- 39.Chomczynski P, Mackey K. 1995. Short technical reports. Modification of the TRI reagent procedure for isolation of RNA from polysaccharide- and proteoglycan-rich sources. Biotechniques 19:942–945. [PubMed] [Google Scholar]
- 40.User Bulletin No 2. 1997. ABI PRISM 7700 Sequence Detection System; p 1–36.
- 41.Johnson MR, Wang K, Smith JB, et al. 2000. Quantitation of dihydropyrimidine dehydrogenase expression by real-time reverse transcription polymerase chain reaction. Anal Biochem 278:175–184. [DOI] [PubMed] [Google Scholar]
- 42.Efron B, Tibshirani RJ. 1993. An introduction to the bootstrap. New York: Chapman and Hall. [Google Scholar]
- 43.Dheda K, Huggett JF, Bustin SA, et al. 2004. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques 37:112–119. [DOI] [PubMed] [Google Scholar]
- 44.Majima T, Marchuk LL, Shrive NG, et al. 2000. In-vitro cyclic tensile loading of an immobilized and mobilized ligament autograft selectively inhibits mRNA levels for collagenase (MMP-1). J Orthop Sci 5:503–510. [DOI] [PubMed] [Google Scholar]
- 45.Dwyer KW, Provenzano PP, Muir P, et al. 2004. Blockade of the sympathetic nervous system degrades ligament in a rat MCL model. J Appl Physiol 96:711–718. [DOI] [PubMed] [Google Scholar]
- 46.Garnero P, Borel O, Byrjalsen I, et al. 1998. The collagenolytic activity of cathepsin K is unique among mammalian proteinases. J Biol Chem 273:32347–32352. [DOI] [PubMed] [Google Scholar]
- 47.Somerville RP, Oblander SA, Apte SS. 2003. Matrix metalloproteinases: old dogs with new tricks. Genome Biol 4: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jeffery JJ. 2007. Interstitial collagenases In: Parks WC, Mecham RP, editors. Matrix metalloproteinases. San Diego: Academic Press. [Google Scholar]
- 49.Kuroki K, Cook JL, Stoker AM, et al. 2005. Characterizing osteochondrosis in the dog: potential roles for matrix metalloproteinases and mechanical load in pathogenesis and disease progression. Osteoarthritis Cartilage 13:225–234. [DOI] [PubMed] [Google Scholar]
- 50.Maclean JJ, Lee CR, Grad S, et al. 2003. Effects of immobilization and dynamic compression on intervertebral disc cell gene expression in vivo. Spine 28:973–981. [DOI] [PubMed] [Google Scholar]
- 51.Virchenko O, Skoglund B, Aspenberg P. 2004. Parecoxib impairs early tendon repair but improves later remodeling. Am J Sports Med 32:1743–1747. [DOI] [PubMed] [Google Scholar]
- 52.Nakagawa Y, Totsuka M, Sato T, et al. 1989. Effect of disuse on the ultrastructure of the achilles tendon in rats. Eur J App Physiol Occup Physiol 59:239–242. [DOI] [PubMed] [Google Scholar]