Extended Data Figure 7. Cytosolic Ca2+ triggers ISC proliferation and EP differentiation into EEs.
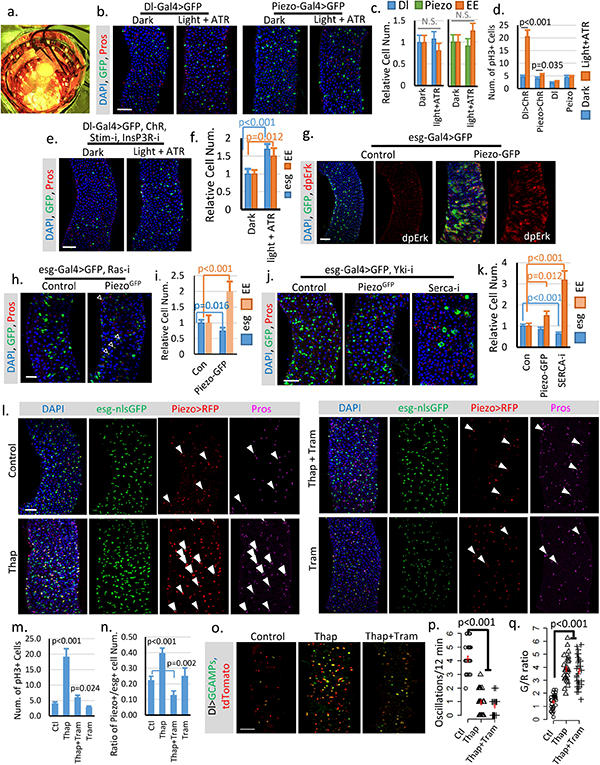
a. Image of chamber used for optogenetic activation of ChR. b,c. Flies expressing GFP only in Dl+ stem cells or Piezo+ EP (EE precursor) cells were treated under either dark or light + ATR condition for two weeks like the flies expressing ChR. No significant phenotype was induced by the treatment alone. Number of midgut areas quantified: n=29 (Dl, Dark), n=33 (Dl, light+ATR), n=31 (Piezo, Dark), n=34 (Piezo, light+ATR). Representative results from two independent replicates are shown. d. Mitosis quantification of midgut from indicated genotype/condition. Activating ChR in Dl+ cells significantly promotes stem cell proliferation. Only a mild increase of mitosis was detected in ChR active Piezo+ EP cells, suggesting that the primary effect of Ca2+ in EP cells is to promote differentiation. Data are collected from 30 guts (Dl>ChR); 30 guts (Piezo>ChR); 29 guts (Dl); guts (Piezo) from two independent replicates. pH3+ cell number is quantified from the whole midgut. e,f. Activation of CsChrimson in Dl+ stem cells with both Stim and InsP3R knocked-down shows reduced increase of stem cells and EEs compared to WT stem cells. Flies were raised at 18°C and shifted to 25°C during the experiment. Cell numbers are quantified within 10,000 μm2 area from 29 regions (dark) and 31 regions (light + ATR) from two independent replicates. g. Overexpression of Piezo in esg+ cells increases MAPK pathway activity. Phosphorylation of extracellular signal-regulated kinase (dpErk) is significantly increased in Piezo-overexpressing cells. Representative images from two independent experiments are shown. h,i. Knocking down Ras significantly reduces stem cell proliferation caused by Piezo overexpression, but does not block Piezo triggered EE differentiation. Flies were kept at 32°C for 4–5 days before analysis. esg+ and EE cell number were quantified from n=29 (control) and n=30 (PiezoGFP) midguts areas from two independent experiments. “Newborn” EEs, that are positive for both esg and Pros, are indicated by arrowheads. j,k. Knocking-down Yorkie using YkiRNAi completely blocks stem cell proliferation but not the increase of EE cells induced by either Piezo overexpression or Serca knock-down. In addition, knocking-down Serca together with Yorkie also significantly reduced stem cell number, suggesting a depletion of stem cells caused by constant EE differentiation. Cell numbers were quantified within 30 midgut areas for each genotype. l. Midguts from flies fed on control (5% sucrose), Thap (5% sucrose + 0.5μM Thapsigargin), Thap+Tram (5% sucrose + 0.5μM Thapsigargin + 10μM Trametinib), and Tram (5% sucrose + 5μM Trametinib) for 4 days. Representative images from 3 independent experiments are shown. The increase of cytosolic Ca2+ by Thap promotes stem cell proliferation, EP (enteroendocrine precursor/Piezo+ cell) production, and EE differentiation. Newborn EEs, which are positive for esg, Piezo and Pros, are indicated by white arrowheads. m. Data are collected from Quantification of mitotic cells from n=15 (control), n=16 (Thap), n=17 (Thap + Tram), and n=16 (Tram) midguts. Thap treatment triggers a significant increase in mitosis, which is largely reduced by the mitogen-activated protein kinase (MAP kinase) inhibitor Tram. n. Percentage of Piezo+ cells within esg+ cell population. Number of areas quantified: n=29 (Ctl), n=31 (Thap), n=32 (Thap+Tram), n=29 (Tram). o. Representative Ca2+ images of live midgut from control, Thap, and Thap+Tram treated flies. Similar results are collected from 4 independent guts for each condition. p,q. Thap treatment caused a reduction of oscillation frequency but an increase of average GCaMP/RFP ratio (G/R ratio). The increase of cytosolic Ca2+ by Thap is not affected by MAPK inhibition. Data are collected from 29 Cells from 3 independent guts for each condition. All data are expressed as mean + s.e.m. values (shown in red). P-values are calculated from two-tailed Student t-test with unequal variance. Scale bar: 50 μm.
