Abstract
Background
In Klebsiella pneumoniae, mgrB and components of pmrHFIJKLM operon play a major role in colistin resistance.
Methods
We analyzed 23 nonduplicating colistin-resistant K. pneumoniae isolates, collected during the years 2011–2015, for the possible mechanism underlying their nonsusceptibility to colistin. Isolates were tested for their minimum inhibitory concentrations and antibiotic resistance determinants and genotyped by multilocus sequence typing (MLST). The MLST genes, antibiotic-resistant genes, and the genes of two component system (TCS), including mgrB, PhoQ/PhoP, pmrA/B, and CrrAB, were investigated by PCR amplification and Sanger sequencing.
Results
All isolates were distributed in eight sequence types (STs) and showed mutations either in mgrB or PhoP genes. ISKpn14 was found in 10, ISKpn28 in four, and IS903 in three isolates. One isolate showed deletion of a single nucleotide in mgrB open reading frame causing premature stop codon. L26Q substitution in PhoP was found in five isolates.
Conclusion
The mutations in mgrB were mostly mediated by insertion elements (IS). ISKpn14 is the major IS while ISKpn28 is reported for the first time in mediating mgrB disruption. IS903, an IS5 family member, involved in mgrB disruption in three ST-152 NDM-1-positive isolates, was previously responsible for omp-36 disruption in our carbapenem-resistant K. pneumoniae and appears to contribute to transform the isolates into a pan-drug ones. Also, the abundance of insertion sites in mgrB indicates the plasticity of this gene. In our isolates, IS-mediated colistin resistance appears to be a later phenomenon than mutation in PhoP gene.
Keywords: colistin resistance, mgrB gene mutations, insertion elements, Klebsiella pneumoniae
Introduction
Colistin has been the only hope so far for the treatment in worst scenarios of antibiotic-resistant bacterial infections, including carbapenem-resistant Enterobacteriaceae. Recent reports indicate a colistin resistance in multidrug-resistant (MDR) Klebsiella pneumoniae to approximately 45%.1 The main mechanism attributed to colistin resistance is the upregulation of PhoQ/PhoP, a two-component system (TCS), which in turn activates the pmrHFIJKLM operon. Mutations in PhoQ/PhoP as well as CrrAB signaling system are found to exert resistance against colistin.2 A significant molecular mechanism that has recently been described in colistin resistance in K pneumoniae is the mutation/inactivation of mgrB gene encoding for a 47–amino acid transmembrane protein that exerts a negativefeedback effect on PhoQ/PhoP. An inactivation of this gene is reported to upregulate the PhoQ/PhoP TCS, thus conferring resistance against colistin.3 A plasmid-mediated mobile colistin resistance gene (mcr-1,-2, and -3 variants) has recently been reported from several parts of the globe indicating increased horizontal transfer of this resistance.4
Materials and methods
Twenty-three nonrepetitive, confirmed, multidrug, colistin-resistant (ColR) K. pneumoniae isolates were studied. These isolates were collected, screened, and stored in the microbiology lab during 2011–2015. Single colonies were picked up and grown in liquid Tryptic Soy Broth (TSB) medium at 37°C overnight. Minimum inhibitory concentrations (MICs) of antibiotics for these isolates were determined using Micro Vitek 2 (bioMerieux, Durham, NC, USA). MIC breakpoints for carbapenemase were defined according to the modified 2010 CLSI guidelines.5 ColR K. pneumoniae isolates were further subjected to analysis for mgrB, PhoP/PhoQ, pmrA/B, and CrrAB genes. Polymerase chain reaction (PCR) amplification of plasmid-borne mobile colistin resistance (mcr) genes was carried out using the primers described by Liu et a1.6 Resistance genes and the genes for multilocus sequence typing, virulence factors, mgrB, PhoP/Q, PmrA/B, and CrrB were amplified using their specific primers, reported earlier.7–10 The amplicon of these PCR reactions was sequenced on an ABI 3100 DNA analyzer (Life Technologies, USA). The sequences were analyzed using SeqMan software (Lasergene, DNAstar) and blasted against the GenBank (NCBI, USA) sequence database. The type of insertion elements (IS) present in mgrB gene was determined by IS Finder.11 The evolutionary histories were inferred using the neighbor-joining method. The evolutionary distances were computed using the maximum composite likelihood method, for ISKpn14 and IS903 nucleotide sequences separately. Evolutionary analyses were conducted using Mega6 software.12
Ethics approval
The K. pneumoniae isolates studied in this report are part of the collection of pathogens in the Microbiology Laboratory of our hospital collected as routine hospital procedure. Hence, no ethical clearance/patient consent was needed.
Results
Here we present our observations on the 23 ColR K. pneumoniae isolates from Saudi Arabia; probably the first report on the mutations in mgrB and other regulatory systems involved in colistin resistance.
Demography, MIC, sequence typing, and carbapenem resistance genes
The isolates were collected from different sources including blood, wound, tracheal aspirate, urine, and so on from the patients admitted to different wards of King Abdul-Aziz Medical City, National Guards Hospital Riyadh. All isolates belonged to MDR group of K. pneumoniae (resistant to more than four groups of antibiotics, including carbapenem). OXA-48 and/or NDM-1 genes were the major carbapenemase determinants. None of the isolates was positive for KPC, VIM, or IMP gene. The distribution of resistance genes, virulence factors, and sequence types (STs) among isolates is given in Table 1.
Table 1.
Showing the details of demography, MICs, virulence factors, and determinants of carbapenem resistance
| Isolate # | ST | Index date | Sample source | IS element in mgrB | mgrB/PhoP point mutations | Virulence factors | Colist | Imipen | Merop | Carbapenemase |
|---|---|---|---|---|---|---|---|---|---|---|
| 52 | 974 | 22-Oct-11 | Wound | No insert | PhoP/L26Q | mrkD, entB | R | R | R | OXA-48 |
| 54 | 37 | 5-Sep-11 | Abdominal drain | No insert | PhoP/L26Q | mrkD, entB | R | 32R | 32R | OXA-48 |
| 62 | 709 | 1-Sep-11 | Blood | No insert | PhoP/L26Q | iut, kfu, entB | R | 4R | 4R | OXA-48 |
| 67 | 348 | 2-Jul-11 | Rectal | No insert | PhoP/L26Q | mrkD, entB | R | R | 8R | OXA-48 |
| 90 | 37 | 5-Jun-12 | Blood | No insert | PhoP/L26Q | mrkD, entB | R | R | R | OXA-48 |
| 114 | 16 | 18-Nov-12 | Trachea | ISKpn14 | No mutation | ybtS, mrkD, entB | R | 8R | 16R | OXA-48 |
| 120 | 48 | 3-Nov-12 | Rectal | ISKpn14 | No mutation | ybtS, mrkD, entB | R | NA | 16R | OXA-48 |
| 124 | 152 | 17-Sep-12 | Urine | IS903 | No mutation | ybtS, mrkD, entB | R | 4R | 4R | OXA-48, NDM-1 |
| 154 | 152 | 31-Jan-13 | Trachea | IS903 | No mutation | ybtS, mrkD, entB | R | 4R | 4R | OXA-48, NDM-1 |
| 166 | 15 | 13-Jul-13 | Wound | ISKpn14 | PhoP/L26Q | ybtS, mrkD, entB, kfu | R | 16R | 16R | OXA-48 |
| 178 | 15 | 22-Nov-13 | Other | ISKpn14 | No mutation | ybtS, mrkD, entB, kfu | R | 8R | 16R | OXA-48 |
| 197 | 14 | 15-May-14 | Urine | ISKpn14 | No mutation | ybtS, mrkD, entB | R | 4R | 4R | ND |
| 218 | 101 | 20-Oct-14 | Urine | ISKpn14 | No mutation | ybtS, mrkD, entB, kfu | R | 16R | 16R | OXA-48, NDM-1 |
| 236 | 22 | 23-Jan-15 | Wound | No insert | mgrB/▲t@22 | mrkD, entB | R | 16R | 16R | OXA-48 |
| 261 | 14 | 15-May-15 | Urine | ISKpn28 | No mutation | ybtS, mrkD, entB, K2, kfu | R | 16R | 16R | OXA-48, NDM-1 |
| 278 | 307 | 3-Jul-15 | Blood | ISKpn14 | No mutation | mrkD, entB | R | 21 | 21 | OXA-48 |
| 287 | 101 | 21-Jul-15 | Trachea | ISKpn14 | No mutation | ybtS, mrkD, entB, kfu | R | 16R | 16R | OXA-48 |
| 300 | 152 | 25-Oct-15 | Blood | IS903 | No mutation | ybtS, mrkD, entB | R | 11 | 4R | ND |
| 303 | 101 | 3-Nov-15 | Urine | ISKpn14 | No mutation | ybtS, mrkD, entB, kfu | R | 16R | 16R | OXA-48, NDM-1 |
| 305 | 14 | 27-Oct-15 | Trachea | ISKpn28 | No mutation | ybtS, mrkD, entB | R | 16R | 16R | NDM-1 |
| 309 | 14 | 8-Nov-15 | Blood | ISKpn28 | No mutation | ybtS, mrkD, entB, K2, kfu | R | 16R | 16R | NDM-1 |
| 313 | 14 | 30-Nov-15 | Blood | ISKpn28 | No mutation | ybtS, mrkD, entB | R | 16R | 16R | NDM-1 |
| 318 | 14 | 7-Dec-15 | Urine | ISKpn14 | No mutation | ybtS, mrkD, entB, K2, kfu | R | 21 | 16R | OXA-48 |
Abbreviations: IS, insertion element; MICs, minimum inhibitory concentrations; NA, not available; ND, not detected; ST, sequence type; R, resistant; I, intermediate.
mgrB and other components of colistin resistance mechanism
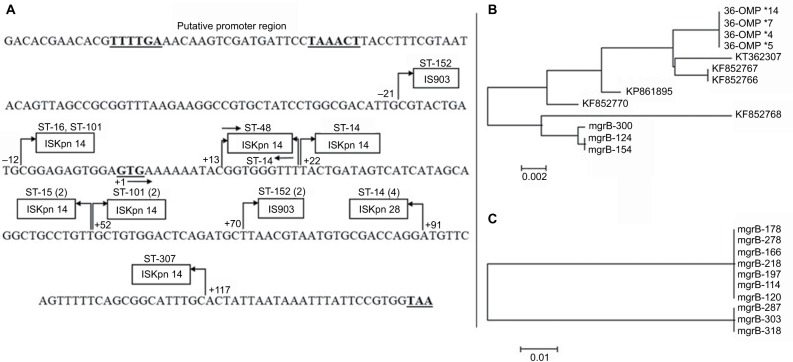
We were unable to detect the plasmid-borne mobile ColR genes mcr-1, -2, or -3 in any of our isolates. Of the 23 resistant isolates, 18 had mutations in their mgrB gene. Seventeen isolates carried IS elements while one had a nucleotide deletion in the open reading frame (ORF) of mgrB gene. Three types of elements belonging to three different families of IS were encountered in either ORF or the promoter of mgrB gene. Ten isolates were carrying ISKpn14, four isolates had ISKpn28, and three had IS903 in their mgrB gene. Out of 10 isolates with ISKpn14 insertion, four had it at position +52 of mgrB gene, two at +22, and two at −12 (in promoter region, upstream of start codon). All isolates carrying ISKpn28 belonged to ST-14. Three isolates with IS903 element, all belonging to ST-152, had it at +70 of the mgrB gene (Figure 1A). Insert sequences were clearly marked for their left and right inverted repeats (IRs). The IR sequences were as follows: ISKpn14 IRL: CCAACTTA, IRR: TAAGTTGG; for ISKpn 903 IRL: GCTTTGTTG, IRR: CAACAAAGC and for ISKpn28 IRL: ATATTGCAATT, IRR: AATTGCAATAT. The mgrB sequences with these insertions are deposited in the GenBank with accession numbers MG930931, MG930932, MG930933, MG930934, MG930935, MG930936, and MG930937. One isolate had a deletion of one nucleotide (“T”) at position +22 of the mgrB gene sequence, resulting in a premature stop codon at position 9 of the mgrB protein. Five isolates had L26Q mutation, whereas one isolate had an additional mutation S72L in the PhoP gene of TCS. No mutations were found in other compartments such as PhoQ, PmrA/B, and CrrAB of the signaling system.
Figure 1.
(A) mgrB gene sequence with the three ISs inserted at different positions in the open reading frame and the promoter region of the gene. The bold-face TTTTGA and TAAACT are −35 and −10 boxes of the promoter, whereas GTG and TAA are start and stop codons, respectively. The arrows and the numbers at their base indicate the orientation and the upstream (−) and downstream (+) positions of IS from the start codon. The numbers in parentheses indicate the multiples of isolates with the particular sequence type; (B) Phylogenetic relationship based on comparisons of IS903 insert sequence in omp-36 gene of our 2010 outbreak isolates in and the mgrB gene of the present study population and six other GenBank depositions; (C) Phylogenetic relationship based on ISKpn14 sequence among isolates of this study carrying the insert. The bar in (B) and (C) is the divergence scale denoting number of substitutions per site.
Abbreviations: IS, insertion element; KP, Klebsiella pneumoniae; ST, sequence type.
Discussion
This is probably the first report from Saudi Arabia describing the molecular mechanisms for colistin resistance in K. pneumoniae. Although certain mutations appear to be clone-specific; a broad polyclonal involvement of colistin resistance was observed in our isolates. All our 23 ColR K. pneumoniae isolates are negative for plasmid-borne mcr gene underscoring the involvement of chromosomal-mediated resistance. This is in line with the observations made recently by Sonnevend et al.13 It has been observed that greater colistin resistance resulted in K. pneumoniae by the mutation/inactivation of mgrB than the mutations in the genes of the TCSs (pmrA/pmrB or PhoP/PhoQ).3,14 We also encountered a similar situation, and 88% of our ColR isolates showed mutations in their mgrB gene alone.
Although IS5-like insertion element has been reported to be the main element for the mgrB disruption in K. pneumoniae, insertion of ISKpn14 (a member of IS1 family) is also sporadically reported.8,15 However, we found ISKpn14 to be the major IS and 59% of our isolates have ISKpn14- mediated mgrB gene disruption. The high incidence of ISKpn14 positivity in our isolates is not a mere expansion of a single clone because these isolates belonged to at least five different STs. The orientation and site of insertion are also varying (Figure 1A). Though ISKpn14 is inserted at as many as six locations in the mgrB gene, site +52 appears to have a recombination event. Four isolates from two different STs had this IS element in both orientations. As our isolates carrying ISKpn14 ranged from year 2012 to 2015, there are variations in the IS sequences. A phylogenetic tree (Figure 1C) comparing the ISKpn14 sequences from these isolates segregated the older clones from the recent ones indicating their divergence through single-nucleotide polymorphisms.
The novel finding of this study is the incidence of 1100 bp ISKpn28 (a member of IS481 family) in four isolates. To the best of our knowledge, this is the first report on ISKpn28 involvement in mgrB disruption in K. pneumoniae from any part of the globe. Interestingly, all isolates shared 100% similarities in their ISKpn28 sequence and all had the same integration site (+91) in the mgrB gene and could be the result of clonal expansion of a single isolate with this mutation. According to our records, ST-14 started circulating in our hospital only from mid-2014 and the isolates carrying ISKpn28 in this study are from year 2015, indicating the insertion to be a recent event. ST-14 with mgrB mutations has recently been reported from several parts of the world and appears to have become an endemic ColR clone.14
Another significant finding of this study is the presence of IS903 at +70 of mgrB, in three K. pneumoniae isolates belonging to ST-152, a clone found to be typically an NDM-1 gene carrier in our carbapenem-resistant (CR) K. pneumoniae isolates (data not shown). IS903, a member of IS5 family, is implicated in antibiotic resistance, either as a carrier of resistance gene or an element for modifying the targets of antibiotic action.16 Position +70 (from start codon) of the mgrB gene sequence appears to be a hotspot for insertion of IS903. GenBank DNA sequence depositions from different countries including China, Greece, and France reported the same +70 position, as the site of integration, for IS903 into the mgrB gene of their isolates.3,16 It suggests that there had been a recombination event at this location in the chromosome of the bacteria. We have previously reported a disruption of omp-36 gene in four CR K. pneumoniae isolates from ST-29 because of IS903 insertion of similar size and the same IR sequences as found in our present ColR isolates, in carbapenem-resistant but colistin-susceptible K. pneumoniae isolates from year 2010.9 However, one of the three isolates with IS903-mediated mgrB disruption also showed an insertion of IS903 in its omp-36 gene indicating its potential role not only in colistin resistance but also for carbapenem. A similar mechanism for tigecycline resistance in K. pneumoniae has been reported, where IS5 is integrated into the promoter region of a putative efflux pump operon called kpgABC.17 More studies are needed to evaluate the spacers that possibly induce the mobilization of these elements from plasmids to the chromosome to transform the MDR pathogen into a pan-drug-resistant one.18 The phylogenetic analysis comparing the IS903 sequences from omp-36 and mgrB in our isolates as well as the GenBank-deposited IS5 insert sequences for mgrB disruption from other countries showed a largely congruent distribution of the isolates (Figure 1B).
Deletions of single nucleotide causing a frame shift resulting in premature stop codon are widely reported in colistin resistance in K. pneumoniae and other bacteria.3,8,15 We found that one isolate belonging to ST-22 carried a deletion of a nucleotide (“T” at +22) in mgrB ORF causing a frame shift and premature stop codon and resulting in a shortened nonfunctional protein of only nine amino acids.
Several nonsynonymous mutations including one at L26Q in PhoP gene have been reported to cause colistin resistance;3 five of our isolates had L26Q mutation in PhoP gene. Due to the abundance of studies reporting the involvement of IS elements in disruption of mgrB as well as PhoP mutations in colistin resistance, we assume that both the insertional inactivation of mgrB and the L26Q mutation in PhoP are the cause for colistin resistance in these isolates. It is interesting to note that in all our isolates from year 2011 till mid-2012, L26Q mutation was the only probable mechanism of colistin resistance and the isolates showed no insertional element in their mgrB gene. MgrB disruption through ISs is a later phenomenon than the PhoP mutations. These data, though on a small number of isolates, support the notion that under stressful situations such as antibiotic exposure, the bacteria require a more robust structural change in the genome to bring crucial beneficial variations, and this change is impossible through simple point mutations; ISs do this job more efficiently.19,20 In conclusion, our report depicts the increased role of IS elements in colistin resistance in K. pneumoniae through mgrB disruption. Also, the abundance of insertion sites (as many as nine places) in mgrB indicates the plasticity of this gene.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Cheng YH, Lin TL, Pan YJ, Wang YP, Lin YT, Wang JT. Colistin resistance mechanisms in Klebsiella pneumoniae strains from Taiwan. Antimicrob Agents Chemother. 2015;59(5):2909–2913. doi: 10.1128/AAC.04763-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lippa AM, Goulian M. Feedback inhibition in the PhoQ/PhoP signaling system by a membrane peptide. PLoS Genet. 2009;5(12):e1000788. doi: 10.1371/journal.pgen.1000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olaitan AO, Diene SM, Kempf M, et al. Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int JAntimicrob Agents. 2014;44(6):500–507. doi: 10.1016/j.ijantimicag.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 4.Rolain JM, Kempf M, Leangapichart T, et al. Plasmid-mediated MCR-1 gene in colistin-resistant clinical isolates of Klebsiella pneumoniae in France and Laos. Antimicrob Agents Chemother. 2016;60(11):6994–6995. doi: 10.1128/AAC.00960-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CLSI Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twentieth Informational Supplement. 2010 M100-S20. [Google Scholar]
- 6.Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 7.Jayol A, Nordmann P, Brink A, Poirel L. Heteroresistance to colistin in Klebsiella pneumoniae associated with alterations in the PhoPQ regulatory system. Antimicrob Agents Chemother. 2015;59(5):2780–2784. doi: 10.1128/AAC.05055-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cannatelli A, Giani T, D’Andrea MM, et al. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob Agents Chemother. 2014;58(10):5696–5703. doi: 10.1128/AAC.03110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uz Zaman T, Aldrees M, Al Johani SM, Alrodayyan M, Aldughashem FA, Balkhy HH. Multi-drug carbapenem-resistant Klebsiella pneumoniae infection carrying the OXA-48 gene and showing variations in outer membrane protein 36 causing an outbreak in a tertiary care hospital in Riyadh, Saudi Arabia. Int J Infect Dis. 2014;28:186–192. doi: 10.1016/j.ijid.2014.05.021. [DOI] [PubMed] [Google Scholar]
- 10.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43(8):4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34(Database issue):D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sonnevend A, Ghazawi A, Alqahtani M, et al. Plasmid-mediated colistin resistance in Escherichia coli from the Arabian Peninsula. Int J Infect Dis. 2016;50:85–90. doi: 10.1016/j.ijid.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 14.Otter JA, Doumith M, Davies F, et al. Emergence and clonal spread of colistin resistance due to multiple mutational mechanisms in carbapenemase-producing Klebsiella pneumoniae in London. Sci Rep. 2017;7(1):12711. doi: 10.1038/s41598-017-12637-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poirel L, Jayol A, Bontron S, et al. The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J Antimicrob Chemother. 2015;70(1):75–80. doi: 10.1093/jac/dku323. [DOI] [PubMed] [Google Scholar]
- 16.Olaitan AO, Rolain JM. Interruption of mgrB in the mediation of colistin resistance in Klebsiella oxytoca. Int J Antimicrob Agents. 2015;46(3):354–355. doi: 10.1016/j.ijantimicag.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 17.Nielsen LE, Snesrud EC, Onmus-Leone F, et al. IS5 element integration, a novel mechanism for rapid in vivo emergence of tigecycline nonsusceptibility in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2014;58(10):6151–6156. doi: 10.1128/AAC.03053-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang W, Wang G, Sebra R, et al. Emergence and evolution of multidrug-resistant Klebsiella pneumoniae with both blaKPC and blaCTx-M integrated in the chromosome. Antimicrob Agents Chemother. 2017;61(7):e00076–e00017. doi: 10.1128/AAC.00076-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Papadopoulos D, Schneider D, Meier-Eiss J, Arber W, Lenski RE, Blot M. Genomic evolution during a 10,000-generation experiment with bacteria. Proc Natl Acad Sci U S A. 1999;96(7):3807–3812. doi: 10.1073/pnas.96.7.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider D, Lenski RE. Dynamics of insertion sequence elements during experimental evolution of bacteria. Res Microbiol. 2004;155(5):319–327. doi: 10.1016/j.resmic.2003.12.008. [DOI] [PubMed] [Google Scholar]