Abstract
Objectives
The information of antimicrobial susceptibility, toxin gene, and ribotype distribution of toxigenic Clostridium difficile isolates in Taiwan remain limited.
Patients and methods
The study was conducted from January 2015 to December 2016 in 5 hospitals in Taiwan. Adults aged ≥20 years with a hospital stay for >5 days were included, and those with colectomy or intestinal infection due to other enteropathogens were excluded. Multiplex PCR was used to detect tcdA, tcdB, cdtA, cdtB, and tcdC deletions, and antimicrobial susceptibility for metronidazole, vancomycin, doxycycline, and tigecycline was investigated. Ribotypes of those isolates with tcdC deletion and tcdA+/tcdB+ were determined.
Results
Of 1112 C. difficile isolates collected from adults at 5 hospitals, 842 were toxigenic, including 749 (89.0%) tcdA+/tcdB+ isolates and 93 (11.0%) tcdA−/tcdB+. Of the toxigenic isolates, 76 (9.0%) had a tcdC deletion and were cdtA+/cdtB+, indicative of hypervirulence, and RT078 lineage, including RT126, RT127, and RT078, predominated (n=53, 76.3%). Similar to the susceptibility data in Asia countries, metronidazole or vancomycin resistance was rare, noted in 1.2% or 2.1%, respectively. Reduced doxycycline susceptibility (minimum inhibitory concentration [MIC] of ≥8 mg/L) was more common among RT078 lineage than non-RT078 lineage (75.9%, 44/58 vs 6.0%, 47/784; P<0.001). Also reduced tigecycline susceptibility (MIC ≥0.125 mg/L) was more common among RT078 lineage (20.7%, 12/58 vs 6.5%, 51/784; P<0.001).
Conclusion
In Taiwan, toxigenic C. difficile isolates remain susceptible to metronidazole and vancomycin. RT078 lineage predominated among toxigenic isolates with cdtA, cdtB, and tcdC deletion, and more often had reduced doxycycline and tigecycline susceptibility than the isolates other than RT078 lineage.
Keywords: MIC, metronidazole, vancomycin, RT126, RT127
Introduction
Clostridium difficile is the primary cause of nosocomial antibiotic-associated diarrhea, with clinical symptoms ranging from mild diarrhea to pseudomembranous colitis or toxic megacolon, with a mortality rate of up to 25%–40%.1 An epidemic C. difficile strain circulating in western countries had been assigned to the North American pulse-field type 1 (NAP1), restriction endonuclease analysis group BI, and polymerase chain reaction (PCR) ribotype 027 (sometimes referred to as BI/NAP1/027) has 3 important microbiological characters: increased production of toxin A and B (in conjunction with tcdC deletion), fluoroquinolone resistance, and production of binary toxin (encoded by cdtA and cdtB).2 Toxigenic C. difficile isolates with tcdC deletion, cdtA, and cdtB have been found in Taiwan.3–7 Three cluster cases presenting infectious diarrhea with multiple recurrences due to C. difficile RT126, belonging to RT078 lineage, were reported in 2014.3 In the following year, 3 cases infected by RT027 were reported in Taiwan, with clinical manifestations of pseudomembranous colitis, toxic megacolon or bowel perforation.4–6 In addition to the RT027 and RT078 lineage isolates, toxigenic C. difficile isolates harboring tcdB and truncated tcdA were predominant, accounting for 43.3% of infections in a district hospital in Taiwan.7 C. difficile RT027 and RT078 isolates were reported sporadically in Mainland China.8 Information about the toxin genes or ribotype distribution of toxigenic or hypervirulent C. difficile isolates in Taiwan has not been reported.
Some metronidazole- or vancomycin-resistant C. difficile isolates were observed in Taiwan. Among 403 non-duplicate isolates of C. difficile from 3 teaching hospitals in Taiwan between 2005 and 2010, all isolates were susceptible to metronidazole, but 2 had reduced susceptibility to vancomycin (minimum inhibitory concentration [MIC], 4 mg/L).9 In a retrospective study in a medical center in northern Taiwan, 2 (1.8%) isolates in 2003 showed resistance to metronidazole (MIC >32 mg/L) and 5 (4.5%) obtained in 2003 (n=1), 2006 (n=1), and 2007 (n=3) showed resistance to vancomycin (MIC >2 mg/L).10 Doxycycline and tigecycline had been suggested as potential alternatives for C. difficile infection (CDI).9,11 Tigecycline was in vitro highly active against an earlier collection, between 2001 and 2009, of clinical C. difficile isolates in Taiwan.12 In the present study, we conducted a multicenter and prospective study to investigate the distribution of toxin genes, ribotypes, and antimicrobial susceptibilities among clinical C. difficile isolates from 2015 to 2016 in Taiwan.
Patients and methods
Hospital settings
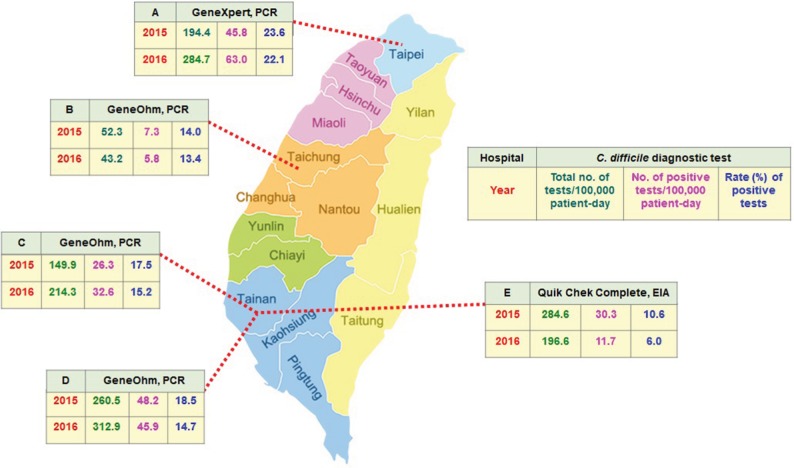
This study was conducted in the National Taiwan University Hospital (Hospital A: a medical center in northern Taiwan), Chung Shan Medical University Hospital (B: a medical center in central Taiwan), Chi-Mei Hospital (C: a medical center in southern Taiwan), National Cheng Kung University Hospital (D: a medical center in southern Taiwan), and Tainan Hospital, Ministry of Health and Welfare (E: a district hospital in southern Taiwan), as noted in Figure 1.
Figure 1.
Geographic distribution and laboratory burden of Clostridium difficile tests, expressed as total test numbers per 100,000 patient-days, positive test numbers per 100,000 patient-days, and positive rates of the 5 participating hospitals in 2015 and 2016.
Abbreviations: PCR, polymerase chain reaction; EIA, enzyme immunoassay.
C. difficile isolates collected from participating hospitals
This study was conducted from January 2015 to December 2016, and only patients aged at least 20 years were included. In routine clinical care, stool samples from those with suspected CDI were tested by GeneXpert C. difficile PCR assay (Cepheid, CA, USA) in the Hospital A, BD GeneOhm™ Cdiff PCR assay (BD Diagnostics, San Diego, CA, USA) in the Hospital B, C, and D, or C. Diff Quik Chek Complete (Alere, Waltham, MA, USA) in the Hospital E. If positive, stools were plated on cycloserine-cefoxitin-fructose agar and incubated anaerobically for 24–48 h. During the study period, fecal samples or C. difficile isolates in the Hospital A, B, C, and E, were submitted to the Hospital D for cultures and further tests. For surveillance purposes, the isolate collected >60 days after the initial one was considered a new and distinct one and those collected between 14 and 60 days after the initial isolate were regarded as recurrent ones.13 The isolates obtained within 14 days after the initial isolate were excluded.
Multiplex PCRs for toxin genes and PCR ribotyping
C. difficile isolates were sub-cultured on cycloserine-cefoxitin fructose selective plates and incubated anaerobically for 24–48 h. The colony with suspected morphology of C. difficile from the culture plate was selected for further identification and studies. A multiplex PCR detecting 16S ribosomal RNA, tpi (encoding triose phosphate isomerase, used as a C. difficile-specific marker), tcdA (encoding toxin A), tcdB (encoding toxin B), cdtA and cdtB (encoding binary toxin), and tcdC deletion, and PCR ribotyping were performed at the Hospital D, as previously described.14 Briefly, after PCR amplification, the samples were concentrated using the Gel/PCR DNA Fragments Extraction Kit (Geneaid Biotech, New Taipei, Taiwan) and separated with the QIAxcel capillary electrophoresis system (Qiagen, Hilden, Germany) using the “OM500” method and QX Alignment Marker 15 bp/3 kb (Qiagen, Hilden, Germany). The PCR ribotypes were confirmed by the WEBRIBO database (http://webribo.ages.at).
Antimicrobial susceptibility testing
MICs were determined by the agar dilution method according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (M11-A8).15 Supplemented Brucella agar deeps were obtained from Anaerobe Systems (Morgan Hill, CA, USA). Frozen defibrinated sheep blood (Hema Resources Inc., Aurora, OR, USA) was thawed to produce laked blood. On the day of testing, laked blood and antimicrobial agents were added to tubes of molten agar before pouring into agar dilution plates. The tested isolates were applied to the plates using a Steers multipronged inoculator for a final concentration of ~105 colony forming unit/spot. After 44 h of incubation at 36°C in an anaerobic chamber incubator, the plates were examined for bacterial growth and the MICs interpreted. Metronidazole and vancomycin were tested at concentrations of 0.03 to 512 mg/L, and MIC breakpoints for resistance are defined as ≥32 and ≥16 mg/L, respectively, according to CLSI 2012.15 Quality control strains included Bacteroides fragilis ATCC 25285, C. difficile ATCC 700057, Staphylococcus aureus ATCC 29213, and Enterococcus faecalis ATCC 29212. The MIC was defined as the lowest concentration that yielded no visible growth or a marked change/reduction in growth compared to the growth controls. MIC breakpoints for resistance to doxycycline and tigecycline are defined as ≥8 and ≥0.5 mg/L, respectively, according to a previous report11 and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) version 6.0.16
Statistical analysis
Statistical analysis was performed using the statistical software, SPSS, version 13.0. The χ2-test or Fisher’s test was used for categorical variables, and a 2-tailed P-value of <0.05 was considered statistically significant.
Ethical approval
The study was approved by the Institutional Review Board in each hospital: A (National Taiwan University Hospital Research Ethics Committee: 201412190RIND), B (Institutional Review Board, Chung Shan Medical University Hospital: CS14179), C (Institutional Review Board, Chi Mei Medical Center: 10402-006), D and E (Institutional Review Board, National Cheng Kung University Hospital: B-ER-103-098). The written informed consents were waived because of the retrospective studies of clinical isolates without interventions on the patients.
Results
Laboratory burden of C. difficile tests in study hospitals
In the 2-year period, the incidence of C. difficile tests performed in 5 hospitals varied greatly, ranging from 53.3 to 284.6 tests per 100,000 patient-days in 2015 and 43.2–312.9 tests per 100,000 patients-days in 2016. With a positivity rate of 10.6%–23.6% in 2015 and 6.0%–22.1% in 2016, the incidence of positive C. difficile tests ranged from 7.3 to 48.3 tests per 100,000 patient-days in 2015 and 5.8 to 63.0 tests per 100,000 patient-days in 2016 (Figure 1).
Ribotypes of C. difficile isolates
A total of 1112 C. difficile isolates were obtained from 1262 fecal samples, and 842 (75.7%) were toxigenic. By the surveillance definition, the recurrent rate of CDI was 4.6% (39/842). The numbers of toxigenic C. difficile isolates collected in Hospitals A–E were 214, 87, 147, 289, and 105, respectively (Table 1). Of 842 toxigenic isolates, the vast majority (749, 89.0%) had tcdA and tcdB (i.e., tcdA+/tcdB+), and 93 (11.0%) were tcdA−/tcdB+. Of the tcdA+/tcdB+ isolates, 10.9% (82/842) possessed cdtA and cdtB, and 76 (9.0%) also had tcdC deletion. Among participating hospitals, tcdC deletion was most often found in 18 (12.2%) isolates of 147 toxigenic isolates from Hospital C. By contrast, 3.8%–10.0% of toxigenic isolates in other 4 hospitals had a tcdC deletion. Two major lineages of 76 isolates with tcdC deletion were RT078 (58, 76.3%) and RT027 lineages (13, 17.1%) (Table 1). Major ribotypes of 58 RT078 lineage isolates included RT126 (39.7%, 23), RT127 (37.9%, 22), and RT078 (19.0%, 11). Of 13 RT027 lineage isolates, 8 belonged to RT027, 4 RT034, and 1 RT075 (Figure 2). Among 749 toxigenic isolates with tcdA and tcdB, the percentages of RT027 or RT078 lineage isolates were not significantly different, ranging from 0% to 4.1% (P=0.22) or 4.7%–10.7% (P=0.29), respectively, in five hospitals. As for 93 tcdA−/tcdB+ isolates which accounted for 10.0% of tcdB+ toxigenic isolates, 89.2% were RT017.
Table 1.
RTs of tcdB-positive Clostridium difficile isolates from 5 hospitals in Taiwan
| Toxin genes and RTs | No. (%) of isolates with indicated toxin genotypes and RTs
|
|||||
|---|---|---|---|---|---|---|
| Total (n=842) |
Hospital A (n=214) |
Hospital B (n=87) |
Hospital C (n=147) |
Hospital D (n=289) |
Hospital E (n=105) |
|
| tcdA+/tcdB+ | 749 | 204 | 85 | 121 | 265 | 74 |
| cdtA+/cdtB+ | 82 (10.9) | 21 (10.3) | 5 (5.9) | 19 (15.7) | 32 (12.1) | 5 (6.8) |
| tcdC deletion | 76 (10.1) | 20 (9.8) | 5 (5.9) | 18 (14.9) | 29 (10.9) | 4 (5.4) |
| RT027 lineage | 13 (1.7) | 2 (1.0) | 1 (1.2) | 5 (4.1) | 5 (1.9) | 0 |
| RT027 | 8 (1.1) | 1 (0.5) | 0 | 4 (3.3) | 3 (1.1) | 0 |
| RT034 | 4 (0.5) | 1 (0.5) | 1 (1.2) | 1 (0.8) | 1 (0.4) | 0 |
| RT075 | 1 (0.1) | 0 | 0 | 0 | 1 (0.4) | 0 |
| RT078 lineage | 58 (7.7) | 16 (7.8) | 4 (4.7) | 13 (10.7) | 21 (7.9) | 4 (5.4) |
| RT033 | 2 (0.3) | 2 (1.0) | 0 | 0 | 0 | 0 |
| RT078 | 11 (1.5) | 1 (0.5) | 1 (1.2) | 8 (6.6) | 1 (0.4) | 0 |
| RT126 | 23 (3.1) | 11 (5.4) | 3 (3.5) | 2 (1.7) | 7 (2.6) | 0 |
| RT127 | 22 (2.9) | 2 (1.0) | 0 | 3 (2.5) | 13 (4.9) | 4 (5.4) |
| tcdA-/tcdB+ | 93 | 10 | 2 | 26 | 24 | 31 |
| RT017 | 84 (90.3) | 8 (80.0) | 2 (100) | 25 (96.2) | 22 (91.7) | 27 (87.1) |
Abbreviation: RT, ribotype.
Figure 2.
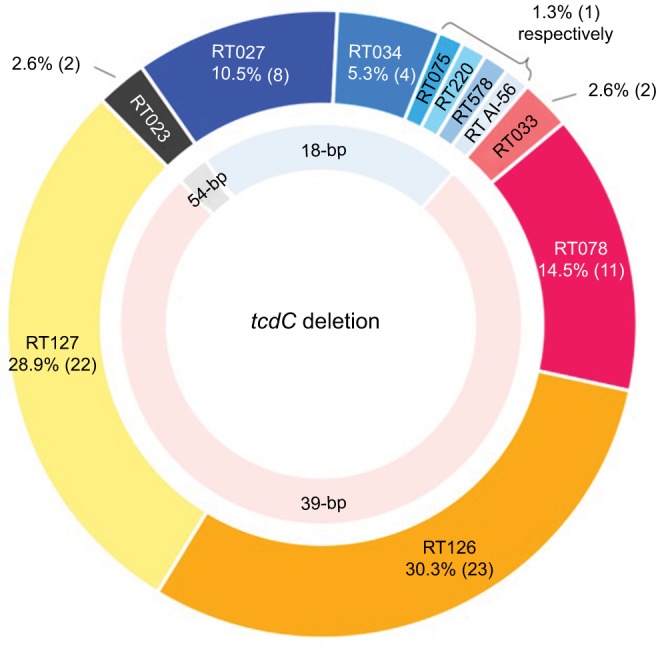
Ribotype distribution of 76 Clostridium difficile isolates with cdtA and cdtB, in which there were 3 types of tcdC deletion, 18-bp, 39-bp, and 54-bp deletion.
Abbreviation: bp, base pair.
Susceptibility data of C. difficile isolates in Taiwan
Metronidazole MICs of the 1112 C. difficile isolates ranged from <0.125 to >32 mg/L, and metronidazole resistance rate varied among hospitals, ranging from 0.6% (Hospital E) to 3.3% (Hospital C). Vancomycin MICs ranged from <0.0625 to >8 mg/L, and vancomycin resistance rate ranged from 1.1% (Hospital B) to 4.6% (Hospital C) (Table 2). As for tigecycline, MIC values ranged from ≤0.0625 to >32 mg/L, and resistance to tigecycline (arbitrarily defined as MIC ≤0.5 mg/L) was noted in 0.6%–3.2% of the isolates in 5 hospitals. In contrast, the MIC range of doxycycline was between ≤0.0625 and >32 mg/L, and doxycycline resistance (defined as doxycycline MIC ≥8 mg/L) was present in 7.7%–17.3% of the isolates from different hospitals (Table 3). However, doxycycline and tigecycline MICs of <0.0625 mg/L were noted in 51.4% and 92.3%, respectively, of all C. difficile isolates.
Table 2.
Antimicrobial susceptibilities of metronidazole and vancomycin in 1112 Clostridium difficile isolates from 5 hospitals*
| Hospital (number of isolates tested) | Metronidazole MICs (mg/L)
|
Resistance, % | Vancomycin MICs (mg/L)
|
Resistance, % | ||||
|---|---|---|---|---|---|---|---|---|
| Range | MIC50 | MIC90 | Range | MIC50 | MIC90 | |||
| A (310) | ≤0.125 to >32 | 0.5 | 1 | 1.0 | ≤0.0625 to >8 | 0.5 | 1 | 1.6 |
| B (181) | ≤0.125 to >32 | 0.25 | 1 | 1.1 | ≤0.0625 to 4 | 0.5 | 1 | 1.1 |
| C (151) | ≤0.125 to >32 | 0.5 | 1 | 3.3 | ≤0.0625 to >8 | 0.5 | 1 | 4.6 |
| D (308) | ≤0.125 to >32 | 0.5 | 1 | 1.0 | ≤0.0625 to >8 | 0.5 | 2 | 1.3 |
| E (162) | ≤0.125 to >32 | 0.25 | 1 | 0.6 | ≤0.0625 to >8 | 0.5 | 1 | 1.9 |
Note:
Resistance is defined as the breakpoints in Clinical and Laboratory Standards Institute Guidelines.15
Abbreviations: MIC50, minimum inhibitory concentration required to inhibit 50% of bacteria; MIC90, minimum inhibitory concentration required to inhibit 90% of bacteria.
Table 3.
Antimicrobial susceptibilities of doxycycline and tigecycline in 1112 Clostridium difficile isolates from 5 hospitals*
| Hospital (number of isolates tested) | Doxycycline MICs (mg/L)
|
Resistance, % | Tigecycline MICs (mg/L)
|
Resistance, % | ||||
|---|---|---|---|---|---|---|---|---|
| Range | MIC50 | MIC90 | Range | MIC50 | MIC90 | |||
| A (310) | ≤0.0625 to >32 | ≤0.0625 | 4 | 7.7 | ≤0.0625 to 1 | ≤0.0625 | ≤0.0625 | 3.2 |
| B (181) | ≤0.0625 to 32 | 0.125 | 8 | 11.0 | ≤0.0625 to 0.5 | ≤0.0625 | 0.125 | 1.7 |
| C (151) | ≤0.0625 to >32 | ≤0.0625 | 4 | 11.9 | ≤0.0625 to >32 | ≤0.0625 | ≤0.0625 | 1.3 |
| D (308) | ≤0.0625 to >32 | ≤0.0625 | 4 | 9.4 | ≤0.0625 to >32 | ≤0.0625 | ≤0.0625 | 1.9 |
| E (162) | ≤0.0625 to 32 | 0.25 | 16 | 17.3 | ≤0.0625 to 1 | ≤0.0625 | ≤0.0625 | 0.6 |
Overall, 1.2%, 2.1%, and 1.9% of toxigenic C. difficile isolates were resistant to metronidazole, vancomycin, and tigecycline, respectively, and the corresponding figure for non-toxigenic isolates was 1.5%, 1.1%, and 2.2%, respectively (Table 4). However, doxycycline resistance was observed in 10.7% and 10.7% of toxigenic and non-toxigenic isolates, respectively.
Table 4.
Antimicrobial susceptibilities of metronidazole, vancomycin, doxycycline, and tigecycline among toxigenic and non-toxigenic Clostridium difficile isolates collected from 5 hospitals
| Antimicrobial agents | Toxigenic isolates (n=842)
|
Non-toxigenic isolates (n=270)
|
||||||
|---|---|---|---|---|---|---|---|---|
| MIC range, (mg/L) | MIC50 (mg/L) | MIC90 (mg/L) | Resistance, % | MIC range, (mg/L) | MIC50 (mg/L) | MIC90 (mg/L) | Resistance, % | |
| Metronidazole* | ≤0.125 to >32 | 0.5 | 1 | 1.2 | ≤0.125 to >32 | 0.25 | 1 | 1.5 |
| Vancomycin* | ≤0.0625 to >8 | 0.5 | 2 | 2.1 | ≤0.0625 to >8 | 0.5 | 1 | 1.1 |
| Doxycycline** | ≤ 0.0625 to >32 | ≤0.0625 | 8 | 10.8 | ≤0.0625 to 32 | 0.25 | 8 | 10.7 |
| Tigecycline** | ≤0.0625 to >32 | ≤0.0625 | ≤0.0625 | 1.9 | ≤0.0625 to 1 | ≤0.0625 | ≤0.0625 | 2.2 |
Notes:
Resistance is defined as the breakpoints in Clinical and Laboratory Standards Institute Guidelines.15
MIC breakpoints for resistance to doxycycline and tigecycline are defined as ≥8 and ≥0.5 mg/L, respectively.11,16
Abbreviations: MIC50, minimum inhibitory concentration required to inhibit 50% of bacteria; MIC90, minimum inhibitory concentration required to inhibit 90% of bacteria.
Compared with toxigenic isolates of non-RT078 lineage, more toxigenic RT078 lineage isolates exhibited reduced susceptibility to doxycycline (MIC >8 mg/L: 75.9%, 44/58% vs 6.0%, 47/784; P<0.001) and tigecycline (MIC >0.125 mg/L: 20.7%, 12/58 vs 6.5%, 51/784; P<0.001).
Discussion
The incidence of CDI was reported as 45 and 42.6/100,000 patient-days in 2 retrospective studies in Hospitals A17 and D,18 respectively, and 42.4/100,000 patient-day in a prospective study in Hospital E.19 However, the present study reported the laboratory burden of C. difficile tests, either PCR or enzyme immunoassay, ranging from 43.2 to 312.8 tests/100,000 patient-days, which may be related to the real burden of CDIs or the clinical awareness of CDIs among attending physicians. The incidence of positive C. difficile tests ranged from 5.8 to 63.0 tests/100,000 patient-day and the positive test rate ranged from 6.0% to 23.6% among study hospitals. These data suggest the substantial, although geographically diverse, disease burden of CDIs in Taiwan.
In our study, although the distribution of the toxin gene pattern, ribotype or antimicrobial susceptibility varied in each hospital, the dominant ribotypes among C. difficile isolates with tcdC deletion, cdtA, and cdtB from 5 hospitals in 3 major cities in Taiwan were RT078 and RT027 lineages. C. difficile RT078 lineage isolates (such as RT126 and 127) have been linked to animal reserves or contact20 and zoonotic potential of RT078 lineage in Taiwan warrants more investigations. The emergence of severe colitis due to C. difficile RT027 and RT078 in Taiwan and Asia indicates the need for comprehensive surveillance of CDI and molecular typing of clinical C. difficile isolates in the continent with a significant burden of multidrug-resistant organisms.
Metronidazole and vancomycin are regarded as the primary therapy options for CDI, depending on the severity of disease.2 Among a collection of >800 clinical toxigenic C. difficile isolates in our study, 1.2% or 2.1% were resistant to metronidazole or vancomycin, respectively. The published literature also indicates that antimicrobial resistance to 2 commonly prescribed antimicrobial agents was rare among clinical C. difficile isolates from several southeastern and northeastern Asian countries.
Lower MICs were noted for doxycycline and tigecycline, which were suggested as alternative choices for CDI.9,11 In a study for each day of doxycycline receipt, the CDI rate was 27% lower than that of no doxycycline therapy (hazard ratio, 0.73; 95% CI: 0.56–0.96).21 Successful treatment with CDI using tigecycline-based combination regimens was reported.22 A previous study reported that in clinical isolates in Taiwan, the in vitro susceptibility rate of C. difficile to tigecycline was high.12 Both doxycycline and tigecycline revealed relatively low MICs among C. difficile isolates, except in some RT078 lineage isolates in the study. Resistance to one or more of drugs tetracycline, moxifloxacin, clindamycin, or erythromycin has been reported in 26.9% (46) of 171 C. difficile isolates in Australia, predominantly RT126 and 078.23 The clinical utility of doxycycline or tigecycline alone or in combination as therapeutic alternatives for CDI warrants further investigations.
Limitations
There were some limitations in our study. First, because the protocols or clinical algorithms to harvest C. difficile isolates from stool samples varied, we cannot compare the CDI incidence among study hospitals. Second, clinical information was not available to be correlated with the distribution of toxin genes, ribotypes, and antimicrobial susceptibilities. The clinical implications of predominant ribotypes among C. difficile isolates with tcdC deletion, cdtA, and cdtB or antimicrobial resistance in Taiwan warrant further studies. Third, the ribotypes of more than 700 tcdA+/tcdB+/cdtA−/cdtB− C. difficile isolates were not investigated, and the likelihood of the existence of other predominant ribotypes in Taiwan cannot be excluded. Finally, no local information of antibiotic consumption in pig feed was available, so the potential linkage between higher prevalence of reduced susceptibility to doxycycline and tigecycline in RT078 isolates and agricultural antibiotic use remains obscure.
Conclusion
The dominant ribotypes among C. difficile isolates with tcdC deletion, cdtA, and cdtB were RT078 and RT027 lineages. Metronidazole or vancomycin resistance remained rare in toxigenic C. difficile isolates, but reduced susceptibility to doxycycline and tigecycline was more common among RT078 than non-RT078 lineage isolates.
Acknowledgments
This study was supported by research grants from the Ministry of Health and Welfare (MOHW 107-TDU-B-211-123003) and the Ministry of Science and Technology (MOST 105-2314-B-006-078-MY3, MOST 106-2321-B-006-006, and MOST 107-2321-B-006-010).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bartlett JG. Narrative review: the new epidemic of Clostridium difficile-associated enteric disease. Ann Intern Med. 2006;145:758–764. doi: 10.7326/0003-4819-145-10-200611210-00008. [DOI] [PubMed] [Google Scholar]
- 2.Kelly CP, LaMont JT. Clostridium difficile--more difficult than ever. N Engl J Med. 2008;359:1932–1940. doi: 10.1056/NEJMra0707500. [DOI] [PubMed] [Google Scholar]
- 3.Hung YP, Lin HJ, Tsai BY, et al. Clostridium difficile ribotype 126 in southern Taiwan: a cluster of three symptomatic cases. Anaerobe. 2014;30:188–192. doi: 10.1016/j.anaerobe.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 4.Hung YP, Cia CT, Tsai BY, et al. The first case of severe Clostridium difficile ribotype 027 infection in Taiwan. J Infect. 2015;70:98–101. doi: 10.1016/j.jinf.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Lai MJ, Chiueh TS, Huang ZY, Lin JC. The first Clostridium difficile ribotype 027 strain isolated in Taiwan. J Formos Med Assoc. 2016;115:210–212. doi: 10.1016/j.jfma.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Liao TL, Lin CF, Chiou CS, Shen GH, Wang J. Clostridium difficile PCR ribotype 027 emerges in Taiwan. Jpn J Infect Dis. 2015;68:338–340. doi: 10.7883/yoken.JJID.2014.271. [DOI] [PubMed] [Google Scholar]
- 7.Hung YP, Huang IH, Lin HJ, et al. Predominance of Clostridium difficile Ribotypes 017 and 078 among toxigenic clinical isolates in southern Taiwan. PLoS One. 2016;11:e0166159. doi: 10.1371/journal.pone.0166159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang C, Cui L, Xu Y, et al. The incidence and drug resistance of Clostridium difficile infection in Mainland China: a systematic review and meta-analysis. Sci Rep. 2016;6:37865. doi: 10.1038/srep37865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liao CH, Ko WC, Lu JJ, Hsueh PR. Characterizations of clinical isolates of Clostridium difficile by toxin genotypes and by susceptibility to 12 antimicrobial agents, including fidaxomicin (OPT-80) and rifaximin: a multicenter study in Taiwan. AntimicrobAgents Chemother. 2012;56:3943–3949. doi: 10.1128/AAC.00191-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chia JH, Lai HC, Su LH, Kuo AJ, Wu TL. Molecular epidemiology of Clostridium difficile at a medical center in Taiwan: persistence of genetically clustering of A(−)B(+) isolates and increase of A(+)B(+) isolates. PLoS One. 2013;8:e75471. doi: 10.1371/journal.pone.0075471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt C, Loffler B, Ackermann G. Antimicrobial phenotypes and molecular basis in clinical strains of Clostridium difficile. Diagn Microbiol Infect Dis. 2007;59:1–5. doi: 10.1016/j.diagmicrobio.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 12.Lin YC, Huang YT, Tsai PJ, et al. Antimicrobial susceptibilities and molecular epidemiology of clinical isolates of Clostridium difficile in taiwan. Antimicrob Agents Chemother. 2011;55:1701–1705. doi: 10.1128/AAC.01440-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheitoyan-Pesant C, Abou Chakra CN, Pepin J, Marcil-Heguy A, Nault V, Valiquette L. Clinical and healthcare burden of multiple recurrences of Clostridium difficile infection. Clin Infect Dis. 2016;62:574–580. doi: 10.1093/cid/civ958. [DOI] [PubMed] [Google Scholar]
- 14.Persson S, Jensen JN, Olsen KE. Multiplex PCR method for detection of Clostridium difficile tcdA, tcdB, cdtA, and cdtB and internal in-frame deletion of tcdC. J Clin Microbiol. 2011;49:4299–4300. doi: 10.1128/JCM.05161-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing; 25th Informational Supplement. Wayne, PA: Clinical and Laboratory Standards Institute; 2015. [Google Scholar]
- 16.European Committee on Antimicrobial Susceptibility Testing Breakpoint tables for interpretation of MICs and zone diameters, version 6.0. 2016. [Accessed June 15, 2018]. Available from: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_6.0_Breakpoint_table.pdf.
- 17.Lee YC, Wang JT, Chen AC, Sheng WH, Chang SC, Chen YC. Changing incidence and clinical manifestations of Clostridium difficile-associated diarrhea detected by combination of glutamate dehydrogenase and toxin assay in Northern Taiwan. J Microbiol Immunol Infect. 2012;45:287–295. doi: 10.1016/j.jmii.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Chung CH, Wu CJ, Lee HC, et al. Clostridium difficile infection at a medical center in southern Taiwan: incidence, clinical features and prognosis. J Microbiol Immunol Infect. 2010;43:119–125. doi: 10.1016/S1684-1182(10)60019-9. [DOI] [PubMed] [Google Scholar]
- 19.Hung YP, Lin HJ, Wu TC, et al. Risk factors of fecal toxigenic or nontoxigenic Clostridium difficile colonization: impact of Toll-like receptor polymorphisms and prior antibiotic exposure. PLoS One. 2013;8:e69577. doi: 10.1371/journal.pone.0069577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keessen EC, Hensgens MP, Spigaglia P, et al. Antimicrobial susceptibility profiles of human and piglet Clostridium difficile PCR-ribotype 078. Antimicrob Resist Infect Control. 2013;2:14. doi: 10.1186/2047-2994-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doernberg SB, Winston LG, Deck DH, Chambers HF. Does doxycycline protect against development of Clostridium difficile infection? Clin Infect Dis. 2012;55:615–620. doi: 10.1093/cid/cis457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El-Herte RI, Baban TA, Kanj SS. Recurrent refractory Clostridium difficile colitis treated successfully with rifaximin and tigecycline: a case report and review of the literature. Scand J Infect Dis. 2012;44:228–230. doi: 10.3109/00365548.2011.616224. [DOI] [PubMed] [Google Scholar]
- 23.Knight DR, Riley TV. Clostridium difficile clade 5 in Australia: antimicrobial susceptibility profiling of PCR ribotypes of human and animal origin. J Antimicrob Chemother. 2016;71:2213–2217. doi: 10.1093/jac/dkw124. [DOI] [PubMed] [Google Scholar]