Abstract
Purpose
Long noncoding RNA PVT1 is dysregulated in some human tumors and has been found to increase the risk of tumor progression and poor prognosis. This study aimed to reanalyze the effect of PVT1 on tumorous prognosis.
Materials and methods
The effect of PVT1 on metastasis and survival were analyzed by univariate logistic regression and Cox proportional hazards model for 32 types of cancer in the Cancer Genome Atlas database (TCGA), and the relationship between PVT1 level and expression of relative genes was assessed by Pearson correlation analysis. RevMan5.3 and STATA14.0 were used to estimate pooled effects of PVT1 on cancer prognosis with data from TCGA and published studies.
Results
In TCGA data, high PVT1 expression tended to increase the risk of TNM progression and decreased the overall survival (OS) time in most of cancers. The pooled effect of PVT1 on TNM (pooled-OR=1.46, 95% CI: 1.29–1.65) and OS (pooled HR=1.32, 95% CI: 1.22–1.43), calculated from 37 and 48 cohorts, identified that high PVT1 expression promoted the metastasis and poor prognosis of cancer. Furthermore, the pooled ORs of 2.77 (95% CI: 1.65–4.66), 4.32 (95% CI: 1.99–9.36), 1.35 (95% CI: 1.01–1.80), 1.62 (95% CI: 1.21–2.18) and 1.48 (95% CI: 1.02–2.15) provided evidence that PVT1 played a role in lymph node metastasis, depth of invasion, distant metastasis, differentiation and lymphatic invasion; while the expression of 24 identified target genes was significantly associated with PVT1 level, and high PVT1 expression dependently decreased the OS time under the influence of co-expression genes (OR=1.29, 95% CI: 1.25–1.32) in high-throughput RNA sequencing merging data. In addition, the expression of PVT1 could be upregulated by smoking, with the pooled OR being 1.09 (95% CI 1.01–1.16).
Conclusion
PVT1 is a dependent biomarker for tumorous prognosis surveillance. However, the reference value of PVT1 needs further study.
Keywords: PVT1, Cancer, biomarker, survive, metastasis
Introduction
Cancer is a major public health problem worldwide.1 Although a decrease of overall cancer mortality has been observed in the last two decades,2 it still causes 22% of noncommunicable disease deaths,3 due to the complex pathogenesis and few target treatments. Numerous experimental studies have attempted to reveal the mechanisms of tumorigenesis to provide clues for prevention, early diagnosis and target treatment. Developments in biotechnology have gradually unveiled some of the mysteries of cancer, with the identification of abundant DNA biomarkers, transcripts, protein and epigenetics. Noncoding RNAs (ncRNAs), which are the largest component of human genome transcripts, especially long noncoding RNAs (lncRNAs), have been confirmed as participating in diverse cellular processes from normal development to cancer.4 Increasing evidence suggests that lncRNAs could be the key regulators interacting with other components such as proteins, RNAs and DNAs;5,6 while regulating lncRNAs have been shown to have aberrant expression in tumor tissues.7–9 To date, overwhelming numbers of cancer-related lncRNAs have been reported in the database of the Cancer Genome Atlas (TCGA) project10,11 as well as the noncode database (http://www.noncode.org) and LNCipedia (http://www.lncipedia.org). Because of being involved in the onset and development of cancer, some key lncRNAs are expected to play a crucial role in cancer detection, diagnosis and therapy.
lncRNA PVT1, homologous to the mouse plasmacytoma variant translocation gene (Pvt1), lies in human chromosome 8q24.21 and has attracted widespread attention.12 Recent studies showed that PVT1 was dysregulated in some human tumors, such as gastric cancer, nonsmall-cell lung cancer, colorectal cancer, esophageal cancer, pancreatic cancer, and hepatocellular carcinoma.10,13,14 High PVT1 expression was further found to increase the risk of tumor progression and poor prognosis.13,15–17 Furthermore, studies of its mechanism suggested that aberrant level of PVT1 was linked to proliferation, angiogenesis and metastasis in human malignancies.18,19 PVT1 might act as an effective biomarker for tumorous prognosis surveillance. Nevertheless, the association between PVT1 expression and cancer prognosis is still not clear. Moreover, no meta-analysis has investigated the common effect of PVT1 on prognosis of most cancers based on multisource data.
The present study aimed to analyze the pooled effect of PVT1 on cancer prognosis by meta-analysis and further explore possibly common target genes of PVT1 in most cancers.
Materials and methods
TCGA sequencing data
High-throughput RNA sequencing (RNA-Seq) and clinical data of different cancers were downloaded from https://portal.gdc.cancer.gov/projects/(TCGA database). Because it did not show in RNA-Seq data of most cancers, PVT1 expression level was redownloaded from https://xenabrowser.net/heatmap/(TCGA database). All the expression data were transformed into the format of log2 (Illumina Hiseq Pancan normalized numbers+1) after deleting the samples with no PVT1 data or where PVT1 was detected in corresponding normal tissues. Only the samples with clinical and PVT1 expression data were further enrolled for prognosis analysis.
Literature search strategy
English or Chinese studies on the role of PVT1 in human cancer were searched in PubMed, EMBASE, Cochrane Library, China National Knowledge Infrastructure, and Wanfang databases with the keywords PVT1 and cancer. The references of retrieved papers and conference reports were also searched to identify relevant studies. The last search date was May 8, 2017.
Selection criteria of reported research
The titles and abstracts of searched articles were checked by 4 authors (YC, CW, ZS, and DW) after duplicates were removed. Then, the full text of eligible articles was retrieved. Eligible articles should have the following criteria: 1) the expression of PVT1 was analyzed by prognosis of human cancer; 2) the expression of PVT1 was detected in cancer tissue or circulating blood by reverse transcription polymerase chain reaction (RT-PCR), fluorescence in-situ hybridization or RNA-Seq; 3) the high and low groups of PVT1 expression were divided by the mean/median or ROC curve; and 4) HRs for survival (overall survival [OS], recurrence-free survival [RFS], disease-free survival [DFS], and progression-free survival [PFS]) or ORs for metastasis (tumor-node-metastasis [TNM], lymphatic invasion [LI], lymph node metastasis [LNM], depth of invasion [DOI], distant metastasis [DM], and differentiation [DIF]) were provided or could be calculated from the available data. Furthermore, if more than 1 report from the same cohort was published, only the most recent publication was included. Consensus in searching and exclusion was resolved by discussion of 3 other investigators (XC, YZ, and DH) if needed.
Data extraction and quality assessment
Four authors (YY, SW, HJ, and GZ) extracted the following data by an extraction form: first author’s name, published year, region of cohort, cancer type, sample size, and HRs/ORs (95% CI). The quality of studies was assessed by New-castle–Ottawa Scale (NOS) and the score ≥ 6 was considered as high quality.
Statistical methods
Univariate logistic regression was applied to analyze the risk of high PVT1 expression on metastasis as well as the effect of smoking on PVT1 expression, whereas the PVT1 effects on OS of 32 types of cancer were assessed by the Cox proportional hazards model with TCGA data. In the meta-analysis, the heterogeneity among studies was tested by inconsistency (I2) and Q tests (chi-square test). The fixed effects model was used to estimate the pooled effect with no statistical heterogeneity found (I2<50%, PQ>0.05); otherwise, a random effects model was used. Publication bias was assessed by Begg’s and Egger’s tests, as trim and fill analysis was used to adjust the pooled effects, if necessary.20 In addition, Engauge Digitizer 4.1 was used to analyze HRs and 95% CIs, when they were not provided directly in some studies. The correlations between PVT1 and other genes were estimated by Pearson correlation analysis. All tests, being considered statistically significant with P<0.05, were two sided and performed by STATA 14.0 and Review Manager 5.3 (Cochrane network).
Results
The effect of PVT1 on prognosis of TCGA cancers
The 9,451 patients with 32 types of cancer were divided into high and low groups with the cutoff point being the median of the PVT1 level which was stably detected in tissues. As shown in Table 1, high PVT1 expression tended to deteriorate the prognosis of most cancers. Furthermore, it significantly increased the risk of TNM progression of breast cancer, head and neck cancer, kidney clear cell carcinoma (KIRC), and kidney papillary cell carcinoma (KIRP), with ORs being 1.168 (P=0.022), 1.947 (P<0.001), 1.593 (P<0.001) and 1.841 (P<0.001), and significantly decreased the OS time of adrenocortical cancer, KIRC, lower grade glioma, pancreatic cancer, prostate cancer, sarcoma, thyroid cancer, endometrioid cancer, and ocular melanomas (UVM) with the HRs being 1.953 (P=0.001), 1.508 (P<0.001), 1.519 (P<0.001), 1.292 (P=0.002), 2.010 (P=0.041), 1.235 (P=0.043), 1.741 (P=0.006), 2.329 (P=0.005), 1.471 (P<0.001), and 3.718 (P<0.001). Conversely, the OS time of stomach cancer was significantly increased with high PVT1 expression (HR=0.848, P=0.046). In addition, smoking significantly increased the expression of PVT1 in KIRP (OR=1.319, P=0.007) and lung adenocarcinoma (OR=1.205, P=0.034).
Table 1.
The effect of PVT1 on TNM and survival as well as smoking on PVT1 expression
| Cancer types | Cases (high/low) | TNM ([III+IV]/[I+ II])
|
Overall survival (high/low)
|
Smoking (yes/no)
|
|||
|---|---|---|---|---|---|---|---|
| P value | OR (95% CI) | P value | HR (95% CI) | P value | OR (95% CI) | ||
| ACC | 79 (39/40) | 0.050 | 1.611 (1.000–2.595) | 0.001 | 1.953 (1.293–2.949) | NR | NR |
| BLCA | 407 (203/204) | 0.282 | 1.103 (0.923–1.317) | 0.258 | 1.079 (0.946–1.232) | 0.736 | 0.972 (0.822–1.148) |
| BRCA | 1,095 (547/548) | 0.022 | 1.168 (1.023–1.333) | 0.927 | 0.993 (0.850–1.160) | NR | NR |
| CESC | 302 (151/151) | NR | NR | 0.395 | 1.106 (0.877–1.395) | 0.372 | 1.112 (0.881–1.404) |
| CHOL | 36 (18/18) | 0.050 | 2.450 (0.999–6.012) | 0.750 | 0.946 (0.672–1.331) | NR | NR |
| COAD | 283 (141/142) | NR | NR | 0.343 | 1.157 (0.856–1.562) | NR | NR |
| DLBC | 48 (24/24) | NR | NR | 0.869 | 1.057 (0.454–2.548) | NR | NR |
| ESCA | 184 (92/92) | 0.670 | 1.076 (0.770–1.504) | 0.627 | 1.060 (0.839–1.338) | 0.265 | 0.839 (0.617–1.142) |
| GBM | 153 (76/77) | NR | NR | 0.274 | 1.118 (0.915–1.365) | NR | NR |
| HNSC | 520 (260/260) | <0.001 | 1.947 (1.464–2.590) | 0.102 | 1.126 (0.976–1.299) | 0.256 | 1.119 (0.922–1.359) |
| KICH | 66 (33/33) | 0.058 | 1.383 (0.989–1.934) | 0.174 | 1.365 (0.871–2.139) | 0.308 | 1.222 (0.831–1.798) |
| KIRC | 533 (266/267) | <0.001 | 1.593 (1.309–1.938) | <0.001 | 1.508 (1.279–1.777) | 0.806 | 0.952 (0.645–1.406) |
| KIRP | 290 (145/145) | <0.001 | 1.841 (1.450–2.338) | 0.056 | 1.245 (0.994–1.560) | 0.007 | 1.319 (1.078–1.614) |
| LGG | 515 (257/258) | NR | NR | <0.001 | 1.519 (1.333–1.731) | NR | NR |
| LIHC | 371 (185/186) | 0.790 | 0.981 (0.854–1.128) | 0.346 | 1.049 (0.949–1.160) | NR | NR |
| LUAD | 515 (257/258) | 0.552 | 1.060 (0.875–1.285) | 0.346 | 0.941 (0.829–1.068) | 0.034 | 1.205 (1.015–1.430) |
| LUSC | 501 (250/251) | 0.975 | 1.004 (0.795–1.267) | 0.434 | 0.947 (0.826–1.086) | 0.827 | 0.972 (0.757–1.249) |
| MESO | 87 (43/44) | 0.696 | 0.883 (0.471–1.652) | 0.467 | 1.129 (0.814–1.568) | NR | NR |
| OV | 304 (152/152) | NR | NR | 0.923 | 0.994 (0.884–1.118) | NR | NR |
| PAAD | 178 (89/89) | 0.551 | 0.839 (0.471–1.494) | 0.002 | 1.292 (1.101–1.515) | 0.582 | 1.071 (0.839–1.367) |
| PCPG | 179 (90/89) | NR | NR | 0.960 | 0.981 (0.463–2.078) | NR | NR |
| PRAD | 497 (248/249) | NR | NR | 0.041 | 2.010 (1.028–3.929) | NR | NR |
| READ | 93 (46/47) | 0.206 | 1.461 (0.812–2.628) | 0.345 | 1.344 (0.728–2.480) | NR | NR |
| SARC | 259 (129/130) | NR | NR | 0.043 | 1.235 (1.007–1.516) | NR | NR |
| SKCM | 469 (234/235) | 0.562 | 1.063 (0.866–1.304) | 0.334 | 1.076 (0.927–1.250) | NR | NR |
| STAD | 415 (207/208) | 0.123 | 1.171 (0.958–1.433) | 0.046 | 0.848 (0.721–0.997) | NR | NR |
| TGCT | 134 (67/67) | 0.423 | 1.190 (0.777–1.822) | 0.597 | 1.237 (0.562–2.179) | NR | NR |
| THCA | 505 (252/253) | 0.904 | 1.012 (0.828–1.237) | 0.006 | 1.741 (1.171–2.590) | NR | NR |
| THYM | 120 (60/60) | NR | NR | 0.005 | 2.329 (1.286–4.217) | NR | NR |
| UCEC | 176 (88/88) | NR | NR | <0.001 | 1.471 (1.310–1.652) | NR | NR |
| UCS | 57 (28/29) | NR | NR | 0.077 | 1.276 (0.974–1.673) | NR | NR |
| UVM | 80 (40/40) | 0.086 | 1.604 (0.935–2.753) | <0.001 | 3.718 (1.956–7.067) | NR | NR |
Abbreviations: ACC, adrenocortical cancer; BLCA, bladder cancer; BRCA, breast cancer; CESC, cervical cancer; CHOL, bile duct cancer; COAD, colon cancer; DLBC, large B-cell lymphoma; ESCA, esophageal cancer; GBM, glioblastoma; HNSC, head and neck cancer; KICH, kidney chromophobe; KIRC, kidney clear cell carcinoma; KIRP, kidney papillary cell carcinoma; LGG, lower grade glioma; LIHC, liver cancer; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian cancer; NR, not reported; PAAD, pancreatic cancer; PCPG, pheochromocytoma & paraganglioma; PRAD, prostate cancer; READ, rectal cancer; SARC, sarcoma; SKCM, melanoma; STAD, stomach cancer; TGCT, testicular cancer; THCA, thyroid cancer; THYM, thymoma; UCEC, endometrioid cancer; UCS, uterine carcinosarcoma; UVM, ocular melanoma.
Published studies of PVT1 on tumorous prognosis
The literature search resulted in 22 published studies eligible for the meta-analysis (Figure S1), 19 from China,13,15,16,18,19,21–34 and 3 from Japan,35 USA,17 and Italy.36 These studies involved 2,376 patients and 11 types of cancers, whose PVT1 level was detected in tumor tissue and circulating blood by RT-PCR. The main characteristics of each study are summarized in Table S1. In addition, the included studies had high quality, with NOS scores of more than 6 for each study (data not shown).
Pooled effect of PVT1 on tumorous prognosis
Pooled effect of PVT1 on progression
Seventeen cohorts from published studies and 20 from TCGA provided the ORs of PVT1 on TNM progression in 8,128 patients. Under the random effects model, the pooled effect (pooled OR=1.46, 95% CI: 1.29–1.65) showed that high PVT1 significantly increased the risk of TNM progression (Table 2, Figure S2). Moreover, high PVT1 expression also significantly promoted the development of LNM, DOI and DM with pooled ORs of 2.77 (95% CI: 1.65–4.66), 4.32 (95% CI: 1.99–9.36) and 1.35 (95% CI: 1.01–1.80) under the random effects model (Table 2, Figure S3A–C). Furthermore, the risks of poor differentiation (pooled OR=1.62, 95% CI: 1.21–2.18) and lymphatic invasion (pooled OR=1.48, 95% CI: 1.02–2.15) were significantly increased in patients with high PVT1 expression under the fixed effects model (Table 2, Figure S3D–E).
Table 2.
The effect of high PVT1 expression on metastasis
| Type of metastasis | Cohorts | Cases | P value of heterogeneity | Pooled OR | 95% CI |
|---|---|---|---|---|---|
| TNM | 37 | 8,128 | <0.0001 | 1.46 | 1.29–1.65 |
| LNM | 11 | 1,130 | 0.001 | 2.77 | 1.65–4.66 |
| DOI | 6 | 498 | 0.007 | 4.32 | 1.99–9.36 |
| DM | 8 | 884 | 0.008 | 1.35 | 1.01–1.80 |
| DIF | 8 | 1,116 | 0.500 | 1.62 | 1.21–2.18 |
| LI | 4 | 543 | 0.130 | 1.48 | 1.02–2.15 |
Abbreviations: LNM, lymph node metastasis; DOI, depth of invasive; DM, distant metastasis; DIF, differentiation; LI, lymphatic invasion.
Pooled effect of PVT1 on survival
Forty-eight cohorts, 16 from published studies and 32 from TCGA, showed data for OS by PVT1 level in 11,022 patients. The pooled effect (pooled HR=1.32, 95% CI: 1.22–1.43) indicated that high PVT1 expression significantly decreased the OS time under the random effects model (Table 3, Figure S4). Similar effects have been shown on DFS (pooled HR=1.77, 95% CI: 1.46–2.13) and PFS (pooled HR=1.71, 95% CI: 1.45–2.00) which were reported in more than 2 cohorts (Table 3, Figure S5).
Table 3.
The effect of high PVT1 expression on prognosis
| Type of prognosis | Cohorts | Cases | P value of heterogeneity | Pooled OR | 95% CI |
|---|---|---|---|---|---|
| OS | 48 | 11,022 | <0.0001 | 1.32 | 1.22–1.43 |
| DFS | 8 | 975 | 0.750 | 1.77 | 1.46–2.13 |
| PFS | 3 | 304 | 0.110 | 1.71 | 1.45–2.00 |
Abbreviations: OS, overall survival; DFS, disease-free survival; PFS, progression-free survival.
Pooled effect of smoking on PVT1 expression
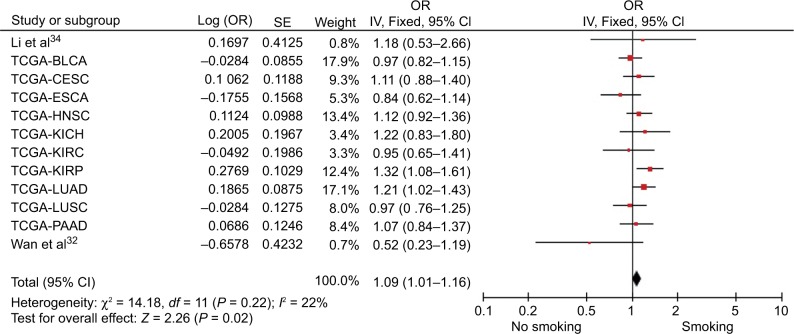
In addition, the effects of smoking on PVT1 expression were also reported in 12 cohorts within 3,600 patients. Figure 1 depicts that smoking significantly increased the level of PVT1 expression under the fixed effect model (pooled OR=1.09, 95% CI: 1.01–1.16).
Figure 1.
Pooled OR of smoking on PVT1 expression.
Abbreviations: SE, standard error; IV, inverse variance methods; BLCA, bladder cancer; CESC, cervical cancer; ESCA, esophageal cancer; HNSC, head and neck cancer; KICH, kidney chromophobe; KIRC, kidney clear cell carcinoma; KIRP, kidney papillary cell carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; PAAD, pancreatic cancer; TCGA, the Cancer Genome Atlas database.
Correlation of PVT1 and relative genes in TCGA cancers
Thirty-four relative genes of PVT1 in cancers were summarized by systematic review. Besides 6 co-expression genes in 8q24, 28 target genes of transcriptional regulation of PVT1 were identified by functional experiments in a variety of human cell lines.30–33,35,37–50 The correlations between them and PVT1 were assessed with the merging RNA-Seq data of 32 types of cancers in TCGA. The PVT1 level was positive relative to the expression of 30 genes except for miR-30a (r=0.0196, P=0.06384), whereas miR-1206, miR-146a and miR-195 could not be detected by RNA-Seq technique (Table 4). However, the relative coefficient of miR-152 (r=0.0382, P=0.00031) and TSHR (r=0.0897, P<0.00001) was <0.1, which was considered as no correlation. Because of significant correlations between PVT1 and co-expression genes, the effects of PVT1 on OS time were further assessed by Cox proportional hazards model by controlling the expression of MYC, miR-1207, miR-1208, miR-1205, miR-1205 and miR-1204 in 9451 patients of 32 types of cancer; as shown in Table 5, high PVT1 expression still decreased the OS time with the HR being 1.29 (95% CI: 1.25–1.32).
Table 4.
The characteristic of identified genes and correlation with PVT1 in 8,927 patients of 32 cancers
| Gene name | Genes identified in systematic review
|
Correlation with PVT1
|
|||
|---|---|---|---|---|---|
| Gene type | Relationship | Cancers identified | Coefficient | P value | |
| MIR1204 | miRNA | Co-expression | NR | 0.2234 | 0.00000 |
| MIR1205 | miRNA | Co-expression | NR | 0.2664 | 0.00000 |
| MIR1206 | miRNA | Co-expression | NR | – | – |
| MIR1208 | miRNA | Co-expression | NR | 0.1430 | 0.00000 |
| MYC | mRNA | Co-expression | NR | 0.1918 | 0.00000 |
| MIR1207(-5p) | miRNA | Co-expression and target | BRCA | 0.1982 | 0.00000 |
| BCL2 | mRNA | Target | OS* | 0.1666 | 0.00000 |
| CASP3 | mRNA | Target | CRC | 0.1722 | 0.00000 |
| CCND1 | mRNA | Target | OS* | 0.1774 | 0.00000 |
| CD151 | mRNA | Target | GC | 0.1761 | 0.00000 |
| CDKN1A(p21) | mRNA | Target | PAAD, NSCLC | 0.1735 | 0.00000 |
| CDKN2A(p16) | mRNA | Target | GC | 0.1766 | 0.00000 |
| CDKN2B(p15) | mRNA | Target | NSCLC, GC | 0.1661 | 0.00000 |
| DDB2 | mRNA | Target | NSCLC | 0.1755 | 0.00000 |
| EZH2 | mRNA | Target | CESC, THCA, NSCLC | 0.1799 | 0.00000 |
| FASN | mRNA | Target | OS* | 0.1769 | 0.00000 |
| FGF2 | mRNA | Target | GC | 0.1384 | 0.00000 |
| HIF1A(HIF1a) | mRNA | Target | GC | 0.1667 | 0.00000 |
| LASP1 | mRNA | Target | ESCC | 0.1714 | 0.00000 |
| LATS2 | mRNA | Target | NSCLC | 0.1700 | 0.00000 |
| MIR146A | miRNA | Target | PRAD | – | – |
| MIR152 | miRNA | Target | GC | 0.0382 | 0.00031 |
| MIR186 | miRNA | Target | GC | 0.1146 | 0.00000 |
| MIR195 | miRNA | Target | CESC, OS* | – | – |
| MIR200B | miRNA | Target | CESC | 0.1965 | 0.00000 |
| MIR203A | miRNA | Target | ESCC | 0.1381 | 0.00000 |
| MIR30A | miRNA | Target | GC | 0.0196 | 0.06384 |
| MIR424 | miRNA | Target | CESC | 0.1641 | 0.00000 |
| SMAD3 | mRNA | Target | CESC | 0.1713 | 0.00000 |
| SMAD4 | mRNA | Target | CRC | 0.1673 | 0.00000 |
| SNAI1 | mRNA | Target | GC | 0.1752 | 0.00000 |
| TIMP1 | mRNA | Target | OV | 0.1800 | 0.00000 |
| TP53(p53) | mRNA | Target | OV | 0.1759 | 0.00000 |
| TSHR | mRNA | Target | THCA | 0.0897 | 0.00000 |
Abbreviations: NR, not reported; BRCA, breast cancer; OS*, osteosarcoma; CRC, colorectal cancer; GC, gastric cancer; PAAD, pancreatic cancer; NSCLC, nonsmall-cell lung cancer; CESC, cervical cancer; THCA, thyroid cancer; ESCC, esophageal squamous cell carcinoma; PRAD, prostate cancer; OV, ovarian cancer; CRC, colorectal cancer.
Table 5.
The effect of PVT1 and coexpression genes on OS time under Cox proportional hazards model
| Gene | Wald | P value | HR (95% CI) |
|---|---|---|---|
| MYC | 71.07 | <0.0001 | 1.077 (1.058–1.095) |
| miR-1207 | 5.933 | 0.015 | 1.011 (1.002–1.019) |
| miR-1208 | 3.985 | 0.046 | 1.011 (1.001–1.021) |
| miR-1205 | 19.593 | <0.0001 | 1.017 (1.009–1.025) |
| miR-1204 | 313.171 | <0.0001 | 0.847 (0.831–0.862) |
| PVT1 | 263.402 | <0.0001 | 1.285 (1.247–1.324) |
Sensitivity analysis
Sensitivity analysis was conducted for the association between PVT1 expression and TNM as well as OS. Each diagnosis test was deleted in turn to examine the influence of the removed data on the overall OR/HR. High PVT1 expression still significantly increased the risk of TNM and OS throughout (data not shown).
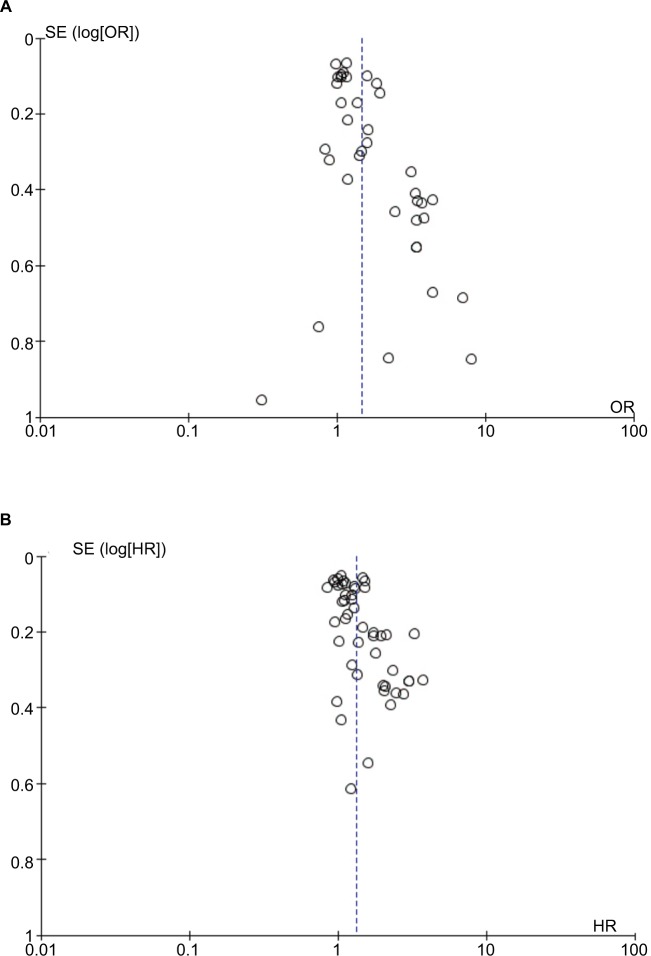
Publication bias
Publication bias was checked for the effects of PVT1 expression on TNM and OS (Figure 2). Begg’s test showed significant rank correlation in studies of PVT1 effect on TNM (Z=2.45, Pr> |z|=0.015) and OS (Z=3.01, Pr>|z|=0.003). Given this result, we performed Egger’s test where evidence of significance publication bias was found for TNM (r=1.81, 95% CI: 0.98–2.64, P<0.0001) and OS (r=1.85, 95% CI: 0.95–2.74, P<0.0001). Consequently, we performed trim and fill analysis; the adjusted pooled-OR of TNM and pooled-HR of OS were 1.31 (95% CI: 1.16–1.49) and 1.15 (95% CI: 1.11–1.18) with P<0.0001 for heterogeneity of both.
Figure 2.
Funnel plots to evaluate the publication bias of PVT1 expression on (A) TNM and (B) OS.
Abbreviations: OS, overall survival; SE, standard error.
Discussion
This study aimed to assess the effect of PVT1 expression on cancer prognosis. The pooled effect showed that high PVT1 expression significantly increased the risk of poor differentiation and cancer metastasis, and significantly decreased the survival time of patients. Furthermore, the expression of PVT1 was significantly correlated to that of genes playing important roles in tumorigenesis. PVT1 could act as an effective biomarker for tumorous prognosis surveillance.
lncRNAs have been shown to act as master regulators of gene expression and thus could play a critical role in various biological functions and disease processes including cancer. With the advances in the RNA-Seq technique and improvement of bioinformatics, a large number of tumor associated lncRNAs have recently been discovered through genomics studies. However, only a few lncRNAs have been fully explored to understand the role in cancers such as regulation of transcription, translation, protein modification and the formation of RNA–protein or protein–protein complexes. PVT1 has been identified as an oncogene and highly correlated with Myc which participates in oncogene activation through Akt/c-Myc signaling pathway.12,13,51,52 Further research into the mechanism found PVT1 could target genes such as LASP1, p15, p16, EZH2, TSHR, and FOXM1 to promote tumor cell proliferation, migration and invasive capability in some cancers.25,28,34,48 Moreover, PVT1 was confirmed to promote protein stability of MYC, RSPO1, NOP2 and increase the level of them.52–54 High PVT1 expression was also linked to poor prognosis of some cancers with most patients from China (Table S1). The relationship between PVT1 expression and cancer prognosis is still ambiguous and should be identified by more samples from other groups. For stable detection in 32 types of TCGA cancer, it was found that PVT1 tended to increase the risk of TNM progression and decrease the OS time in most of them.
To further explore the unbiased effect of PVT1 on cancer prognosis, we performed a meta-analysis with the cohorts from TCGA and other published studies. The adjusted pooled OR of 1.31 (95% CI: 1.16–1.49) and pooled HR of 1.15 (95% CI: 1.11–1.18) indicated that high PVT1 increased the risk of TNM progression and decreased the overall survival time. The effect of high PVT1 on depth of invasive, lymph node metastasis, distant metastasis as well as poor differentiation and lymphatic invasion also provided direct and epidemiological evidence of PVT1 participating in cancer metastasis. Furthermore, five co-expression genes and 24 identified target genes were significantly associated with PVT1 in all TCGA cancers; although the relative coefficients of them were too small to declare a high degree of correlation, which might be caused by heterogeneity of huge samples of data. Moreover, high PVT1 expression dependently decreased the OS time of patients by controlling the influence of co-expressed genes. In addition, it was shown that the expression of PVT1 could be upregulated by smoking to deteriorate the cancer prognosis.55–57 This suggested that PVT1 expression was an effectively common biomarker for human tumorous prognosis surveillance.
Some meta-analyses focused on the association between lncRNAs such as BANCR,58 HOTTIP,59 CCAT2,60 and metastasis as well as prognosis of cancer; all based on lncRNAs detected by RT-PCR from the cohorts of published studies. To search for an applicably common biomarker for prognosis surveillance, we focused on the effect of PVT1 expression, detected by RT-PCR and RNA-Seq on metastasis and survival as well as the correlation of PVT1 and possible target genes in all common cancer. In the present study, a new way was provided to improve the traditional meta-analysis, by which useful information not published in the literature could be found through published databases to provide more comprehensive evidence. More importantly, the possible mechanism and the common effect of PVT1 on prognosis of all cancers was proved. Furthermore, we provide coefficient references for correlation analysis of gene expression with huge amounts of RNA-Seq data. To our best knowledge, this is the first improved meta-analysis of PVT1 effect on cancer prognosis with the data from the cohorts of TCGA and published studies.
Our study also contains some limitations. Since PVT1 was identified as a common biomarker for cancer prognosis surveillance, there was no reference value to distinguish poor prognosis patients with PVT1 expression detected by RT-PCR or RNA-Seq. Second, although 24 possible target genes were proved to be significantly related to PVT1 expression in most cancers by pooled analysis, the mechanism of their involvement in PVT1 deteriorating cancer prognosis was still clear. Therefore, studies are needed to explore the reference value of PVT1 expression detected by RT-PCR/RNA-Seq to distinguish poor prognosis as well as the role of PVT1 target genes in cancer prognosis.
Conclusion
This improved meta-analysis is the first to demonstrate the effect and possible mechanism of PVT1 on cancer prognosis. The expression of PVT1 could be a biomarker for tumorous prognosis surveillance.
Supplementary materials
The flow chart of meta-analysis.
The pooled effect value of PVT1 on TNM.
Abbreviations: IV, inverse variance methods; SE, standard error; UVM, ocular melanoma; SKCM, melanoma; STAD, stomach cancer; TGCT, testicular cancer; THCA, thyroid cancer; READ, rectal cancer; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; LIHC, liver cancer; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney clear cell carcinoma; KIRP, kidney papillary cell carcinoma; HNSC, head and neck cancer; ESCA, esophageal cancer; ACC, adrenocortical cancer; BLCA, bladder cancer; BRCA, breast cancer; CHOL, bile duct cancer; TCGA, the Cancer Genome Atlas database.
The pooled effect value of PVT1 on (A) LNM, (B) DOI, (C) DM, (D) DIF, and (E) LI.
Abbreviations: DIF, differentiation; DM, distant metastasis; DOI, depth of invasive; IV, inverse variance methods; LI, lymphatic invasion; LNM, lymph node metastasis; SE, standard error.
The pooled effect value of PVT1 on OS.
Abbreviations: IV, inverse variance methods; OS, overall survival; SE, standard error; ACC, adrenocortical cancer; BLCA, bladder cancer; BRCA, breast cancer; CESC, cervical cancer; CHOL, bile duct cancer; COAD, colon cancer; DLBC, large B-cell lymphoma; ESCA, esophageal cancer; GBM, glioblastoma; HNSC, head and neck cancer; KICH, kidney chromophobe; KIRC, kidney clear cell carcinoma; KIRP, kidney papillary cell carcinoma; LGG, lower grade glioma; LIHC, liver cancer; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian cancer; PAAD, pancreatic cancer; PCPG, pheochromocytoma & paraganglioma; PRAD, prostate cancer; READ, rectal cancer; SARC, sarcoma; SKCM, melanoma; STAD, stomach cancer; TGCT, testicular cancer; THCA, thyroid cancer; THYM, thymoma; UCEC, endometrioid cancer; UCS, uterine carcinosarcoma; UVM, ocular melanoma.
The pooled effect value of PVT1 on (A) DFS and (B) PFS.
Abbreviations: DFS, disease-free survival; IV, inverse variance methods; PFS, progression-free survival; SE, standard error.
Table S1.
Basic data for all included studies in the meta-analysis
| Author | Year | Country | Cancer type | Case (high/low) | Metastasis | Prognosis | Smoking on PVT1 OR (95% CI) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||
| TNM
|
LI
|
LNM
|
DOI
|
DM
|
DIF
|
Outcomes | HR (95% CI) | ||||||
| OR (95% CI) | OR (95% CI) | OR (95% CI) | OR (95% CI) | OR (95% CI) | OR (95% CI) | ||||||||
| Yuan et al17 | 2015 | China | GC (tissue) | 112 (55/56) | 3.346 (1.498–7.475) | 2.337 (1.086–5.030) | 2.337 (1.086–5.030) | 11.846 (4.858–28.884) | 2.075 (0.183–23.573) | NR | OS DFS |
2.280 (1.054–4.930) 2.210 (1.111–4.395) |
NR |
| Cui et al19 | 2015 | China | NSCLC (tissue) | 108 (53/55) | 3.168 (1.588–8.243) | NR | 2.289 (1.054–4.972) | 11.026 (4.505–26.983) | 2.118 (0.186–24.073) | 1.054 (0.461–2.408) | OS DFS |
1.72 (1.14–3.25) 1.98 (1.16–4.35) |
Irrelevant data |
| Xu et al23 | 2016 | China | GC (tissue) | 190 (96/94) | NR | NR | NR | NR | NR | NR | DFS DSS |
1.75 (1.25–2.56) 1.64 (1.15–2.39) |
NR |
| Takahashi et al4 | 2014 | Japan | CRC (tissue) | 164 (133/31) | 3.462 (1.494–8.022) | 2.119 (0.915–4.908) | 2.968 (1.282–6.870) | NR | 0.750 (0.075–7.451) | 1.281 (0.267–6.149) | DFS | 2.532 (1.152–10.747) | NR |
| Li et al18 | 2016 | China | CRC (tissue) | 30 (15/15) | 0.750 (0.169–3.327) | NR | NR | NR | NR | NR | DFS | 5.84 (0.42–81.19) | NR |
| Zheng et al16 | 2015 | China | ESCA (tissue) | 77 (39/38) | 3.450 (1.346–8.841) | NR | NR | NR | NR | NR | NR | NR | NR |
| Huang et al10 | 2015 | China | PAAD (tissue) | 85 (67/18) | 3.442 (1.170–10.127) | NR | 0.276 (0.088–0.863) | 1.829 (0.614–5.444) | NR | 1.550 (0.275–8.738) | OS | 3.013 (1.574–6.673) | NR |
| Yang et al1 | 2014 | China | NSCLC (tissue) | 82 (65/17) | 7.000 (1.829–26.788) | NR | 9.911 (2.093–46.925) | NR | NR | NR | OS | 3.273 (2.184–6.937) | NR |
| Huang et al12 | 2016 | China | SCLC (tissue) | 120 (60/60) | NR | NR | 9.000 (3.939–20.566) | NR | 11.455 (3.683–35.628) | NR | OS | 1.782 (1.078–2.945) | NR |
| Martini et al21 | 2016 | Italy | EOC (tissue) | Training 73 (NR) Testing 126 (NR) |
NR NR |
NR NR |
NR NR |
NR NR |
NR NR |
NR NR |
OS PFS OS PFS |
2.10 (1.40–3.30) 1.70 (1.30–2.20) 1.30 (1.10–1.60) 1.60 (1.30–1.90) |
NR |
| Kong et al8 | 2015 | China | GC (tissue) | 80 (40/40) | 3.857 (1.526–9.750) | 1.833 (0.755–4.455) | NR | 6.152 (2.334–16.211) | 1.111 (1.002–1.232) | 2.333 (0.932–5.839) | OS DFS |
2.092 (1.068–4.096) 2.216 (1.130–4.345) |
NR |
| Ding et al11 | 2015 | China | HCC (tissue) | 58 (49/9) 214 (157/57) |
0.311 (0.048–2.2028) 1.417 (0.772–2.603) |
NR NR |
NR NR |
NR NR |
NR NR |
1.534 (0.367–6.412) 1.739 (0.937–3.228) |
NR RFS OS |
NR 1.653 (1.019–2.681) 1.01 (0.65–1.56) |
NR NR |
| Zhang et al15 | 2016 | China | CESC (tissue) | 90 (45/45) | NR | NR | NR | NR | NR | NR | OS | 1.60 (0.55–4.66) | NR |
| Wu et al2 | 2017 | China | PAAD (tissue) | 30 (15/15) | 8.000 (1.5224–2.042) | NR | Unuseful data | NR | NR | Unuseful data | NR | NR | NR |
| Ren et al5 | 2016 | China | GC (serum) | 28 (13/15) | 2.222 (0.426–11.603) | NR | 8.250 (1.328–51.263) | 1.212 (0.216–6.800) | 1.083 (0.926–1.267) | 0.938 (0.205–4.294) | NR | NR | NR |
| Wan et al3 | 2016 | China | NSCLC (tissue) | 105 (56/49) | 3.710 (1.584–8.694) | NR | 2.842 (1.286–6.282) | NR | NR | NR | OS PFS |
2.464 (1.214–4.999) 3.39 (1.74–6.63) |
0.518 (0.226–1.186) |
| Zhou et al22 | 2016 | China | OS* (tissue) | 53 (29/24) | NR | NR | NR | NR | NR | NR | OS | 1.23 (0.37–4.05) | NR |
| Iden et al13 | 2016 | USA | CESC (tissue) | 121 (63/58) | NR | NR | NR | NR | NR | NR | OS | 2.03 (1.01–4.08) | NR |
| Li et al7 | 2017 | China | ESCC (tissue) | 104 (52/52) | 4.429 (1.917–10.229) | NR | NR | NR | NR | 3.167 (1.235–8.117) | OS DFS |
2.75 (1.35–5.59) 1.42 (0.71–2.87) |
1.185 (0.528–2.662) |
| Liu et al6 | 2016 | China | CESC (tissue) | 85 (67/18) | 3.442 (1.170–10.127) | NR | 3.621 (1.158–11.323) | 1.829 (0.614–5.444) | 1.455 (0.402–5.260) | 1.550 (0.275–8.738) | OS | 3.013 (1.574–6.673) | NR |
| Huang et al9 | 2015 | China | RC (tissue) | 54 (39/15) | 4.400 (1.183–16.367) | NR | 2.783 (0.675–11.477) | NR | NR | NR | NR | NR | NR |
| Chen et al20 | 2016 | China | GC (serum) | 187 (112/75) | 1.175 (0.567–2.434) | 0.838 (0.458–1.532) | 1.587 (0.672–3.751) | NR | 2.112 (0.845–5.278) | 0.982 (0.501–1.927) | OS DFS |
1.455 (1.005–2.105) 1.435 (0.998–2.061) |
NR |
Abbreviations: LI, lymphatic invasion; LNM, lymph node metastasis; DOI, depth of invasive; DM, distant metastasis; DIF, differentiation; OS, overall survival; DFS, disease-free survival; PFS, progression-free survival; DSS, disease specific survival; RFS, recurrence free survival; NR, no report; GC, gastric cancer; NSCLC, non-small cell lung cancer; CRC, colorectal cancer; ESCA, esophageal carcinoma; PAAD, pancreatic cancer; SCLC, small-cell lung cancer; EOC, epithelial ovarian cancer; HCC, hepatocellular carcinoma; CESC, cervical cancer; OS*, osteosarcoma; ESCC, esophageal squamous cell carcinoma; RC, renal carcinoma.
References
- 1.Yang YR, Zang SZ, Zhong CL, Li YX, Zhao SS, Feng XJ. Increased expression of the lncRNA PVT1 promotes tumorigenesis in non-small cell lung cancer. Int J Clin Exp Pathol. 2014;7(10):6929–6935. [PMC free article] [PubMed] [Google Scholar]
- 2.Wu BQ, Jiang Y, Zhu F, Sun DL, He XZ. Long noncoding RNA PVT1 promotes EMT and cell proliferation and migration through downregulating p21 in pancreatic cancer cells. Technol Cancer Res Treat. 2017 doi: 10.1177/1533034617700559. 1533034617700559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wan L, Sun M, Liu GJ, et al. Long noncoding RNA PVT1 promotes non-small cell lung cancer cell proliferation through epigenetically regulating LATS2 expression. Mol Cancer Ther. 2016;15(5):1082–1094. doi: 10.1158/1535-7163.MCT-15-0707. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi Y, Sawada G, Kurashige J, et al. Amplification of PVT-1 is involved in poor prognosis via apoptosis inhibition in colorectal cancers. Br J Cancer. 2014;110(1):164–171. doi: 10.1038/bjc.2013.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ren X, Xiao Y, Zhang L, et al. Expression and clinical significance of PVT1 gene in blood of the patients with gastric cancer. Chin J Cancer Biother. 2016;23(05):688–691. [Google Scholar]
- 6.Liu Y, Quan X, Qin X, Y L. Effect of neoadjuvant chemotherapy on difference of expression of IncRNA in ovarian cancer patients. Pract J Cancer. 2016;31(03):384–386+389. [Google Scholar]
- 7.Li PD, Hu JL, Ma C, et al. Upregulation of the long non-coding RNA PVT1 promotes esophageal squamous cell carcinoma progression by acting as a molecular sponge of miR-203 and LASP1. Oncotarget. 2017;8(21):34164–34176. doi: 10.18632/oncotarget.15878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kong R, Zhang EB, Yin DD, et al. Long noncoding RNA PVT1 indicates a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically regulating p15 and p16. Mol Cancer. 2015;14:82. doi: 10.1186/s12943-015-0355-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang Y, Mu Z, Lin L, et al. Expression and clinical significance of lncRNA PVT1 in renal carcinoma tissues. Shandong Medical. 2015;55(36):11–13. [Google Scholar]
- 10.Huang C, Yu W, Wang Q, et al. Increased expression of the lncRNA PVT1 is associated with poor prognosis in pancreatic cancer patients. Minerva Med. 2015;106(3):143–149. [PubMed] [Google Scholar]
- 11.Ding C, Yang Z, Lv Z, et al. Long non-coding RNA PVT1 is associated with tumor progression and predicts recurrence in hepatocellular carcinoma patients. Oncol Lett. 2015;9(2):955–963. doi: 10.3892/ol.2014.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang C, Liu S, Wang H, Zhang Z, Yang Q, Gao F. LncRNA PVT1 overexpression is a poor prognostic biomarker and regulates migration and invasion in small cell lung cancer. Am J Transl Res. 2016;8(11):5025–5034. [PMC free article] [PubMed] [Google Scholar]
- 13.Iden M, Fye S, Li K, Chowdhury T, Ramchandran R, Rader JS. The lncRNA PVT1 contributes to the cervical cancer phenotype and associates with poor patient prognosis. PLoS One. 2016;11(5):e0156274. doi: 10.1371/journal.pone.0156274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Quan X, Qin X, Y L. Effect of neoadjuvant chemotherapy on difference of expression of IncRNA in ovarian cancer patients. Pract J Cancer. 2016;31(03):384–386+389. [Google Scholar]
- 15.Zhang S, Zhang G, Liu J. Long noncoding RNA PVT1 promotes cervical cancer progression through epigenetically silencing miR-200b. APMIS. 2016;124(8):649–658. doi: 10.1111/apm.12555. [DOI] [PubMed] [Google Scholar]
- 16.Zheng X, Hu H, Li S. High expression of lncRNA PVT1 promotes invasion by inducing epithelial-to-mesenchymal transition in esophageal cancer. Oncol Lett. 2016;12(4):2357–2362. doi: 10.3892/ol.2016.5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan CL, Li H, Zhu L, Liu Z, Zhou J, Shu Y. Aberrant expression of long noncoding RNA PVT1 and its diagnostic and prognostic significance in patients with gastric cancer. Neoplasma. 2016;63(3):442–449. doi: 10.4149/314_150825N45. [DOI] [PubMed] [Google Scholar]
- 18.Li Q, Dai Y, Wang F, et al. Differentially expressed long non-coding RNAs and the prognostic potential in colorectal cancer. Neoplasma. 2016;63(6):977–983. doi: 10.4149/neo_2016_617. [DOI] [PubMed] [Google Scholar]
- 19.Cui D, Yu CH, Liu M, Xia QQ, Zhang YF, Jiang WL. Long non-coding RNA PVT1 as a novel biomarker for diagnosis and prognosis of non-small cell lung cancer. Tumour Biol. 2016;37(3):4127–4134. doi: 10.1007/s13277-015-4261-x. [DOI] [PubMed] [Google Scholar]
- 20.Chen J, Li Y, Zheng Q, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–219. doi: 10.1016/j.canlet.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 21.Martini P, Paracchini L, Caratti G, et al. lncRNAs as novel indicators of patients’ prognosis in stage I epithelial ovarian cancer: a retrospective and multicentric study. Clin Cancer Res. 2017;23(9):2356–2366. doi: 10.1158/1078-0432.CCR-16-1402. [DOI] [PubMed] [Google Scholar]
- 22.Zhou Q, Chen F, Zhao J, et al. Long non-coding RNA PVT1 promotes osteosarcoma development by acting as a molecular sponge to regulate miR-195. Oncotarget. 2016;7(50):82620–82633. doi: 10.18632/oncotarget.13012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu MD, Wang Y, Weng W, et al. A positive feedback loop of lncRNAPVT1 and FOXM1 facilitates gastric cancer growth and invasion. Clin Cancer Res. 2017;23(8):2071–2080. doi: 10.1158/1078-0432.CCR-16-0742. [DOI] [PubMed] [Google Scholar]
Acknowledgments
The authors are grateful for the contribution of TCGA. This study was supported by Research Project of Shenzhen Health and Family Planning System (201604130086, SZBC2017003).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.GBD 2015 Mortality and Causes of Death Collaborators Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388(10053):1459–1544. doi: 10.1016/S0140-6736(16)31012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hashim D, Boffetta P, La Vecchia C, et al. The global decrease in cancer mortality: trends and disparities. Ann Oncol. 2016;27(5):926–933. doi: 10.1093/annonc/mdw027. [DOI] [PubMed] [Google Scholar]
- 3.Pineros M, Znaor A, Mery L, Bray F. A global cancer surveillance framework within noncommunicable disease surveillance: making the case for population-based cancer registries. Epidemiol Rev. 2017;39(1):161–169. doi: 10.1093/epirev/mxx003. [DOI] [PubMed] [Google Scholar]
- 4.Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661–5667. doi: 10.1038/onc.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73(13):2491–2509. doi: 10.1007/s00018-016-2174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhamija S, Diederichs S. From junk to master regulators of invasion: lncRNA functions in migration, EMT and metastasis. Int J Cancer. 2016;139(2):269–280. doi: 10.1002/ijc.30039. [DOI] [PubMed] [Google Scholar]
- 8.Yu X, Li Z. Long non-coding RNA HOTAIR: a novel oncogene (review) Mol Med Rep. 2015;12(4):5611–5618. doi: 10.3892/mmr.2015.4161. [DOI] [PubMed] [Google Scholar]
- 9.Raveh E, Matouk IJ, Gilon M, Hochberg A. The H19 Long non-coding RNA in cancer initiation, progression and metastasis – a proposed unifying theory. Mol Cancer. 2015;14:184. doi: 10.1186/s12943-015-0458-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weidle UH, Birzele F, Kollmorgen G, Rüger R. Long non-coding RNAs and their role in metastasis. Cancer Genomics Proteomics. 2017;14(3):143–160. doi: 10.21873/cgp.20027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JS. Exploring cancer genomic data from the cancer genome atlas project. BMB Rep. 2016;49(11):607–611. doi: 10.5483/BMBRep.2016.49.11.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colombo T, Farina L, Macino G. Paci PVT1: a rising star among oncogenic long noncoding RNAs. Biomed Res Int. 2015;2015:304208. doi: 10.1155/2015/304208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui D, Yu CH, Liu M, Xia QQ, Zhang YF, Jiang WL. Long non-coding RNA PVT1 as a novel biomarker for diagnosis and prognosis of non-small cell lung cancer. Tumour Biol. 2016;37(3):4127–4134. doi: 10.1007/s13277-015-4261-x. [DOI] [PubMed] [Google Scholar]
- 14.Cumbo F, Fiscon G, Ceri S, Masseroli M, Weitschek E. TCGA2BED: extracting, extending, integrating, and querying The Cancer Genome Atlas. BMC Bioinformatics. 2017;18(1):6. doi: 10.1186/s12859-016-1419-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan CL, Li H, Zhu L, Liu Z, Zhou J, Shu Y. Aberrant expression of long noncoding RNA PVT1 and its diagnostic and prognostic significance in patients with gastric cancer. Neoplasma. 2016;63(3):442–449. doi: 10.4149/314_150825N45. [DOI] [PubMed] [Google Scholar]
- 16.Yang YR, Zang SZ, Zhong CL, Li YX, Zhao SS, Feng XJ. Increased expression of the lncRNA PVT1 promotes tumorigenesis in non-small cell lung cancer. Int J Clin Exp Pathol. 2014;7(10):6929–6935. [PMC free article] [PubMed] [Google Scholar]
- 17.Iden M, Fye S, Li K, Chowdhury T, Ramchandran R, Rader JS. The lncRNA PVT1 contributes to the cervical cancer phenotype and associates with poor patient prognosis. PLoS One. 2016;11(5):e0156274. doi: 10.1371/journal.pone.0156274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng X, Hu H, Li S. High expression of lncRNA PVT1 promotes invasion by inducing epithelial-to-mesenchymal transition in esophageal cancer. Oncol Lett. 2016;12(4):2357–2362. doi: 10.3892/ol.2016.5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang C, Liu S, Wang H, Zhang Z, Yang Q, Gao F. LncRNA PVT1 overexpression is a poor prognostic biomarker and regulates migration and invasion in small cell lung cancer. Am J Transl Res. 2016;8(11):5025–5034. [PMC free article] [PubMed] [Google Scholar]
- 20.Soeken KL, Sripusanapan A. Assessing publication bias in meta-analysis. Nurs Res. 2003;52(1):57–60. doi: 10.1097/00006199-200301000-00009. [DOI] [PubMed] [Google Scholar]
- 21.Chen J, Li Y, Zheng Q, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–219. doi: 10.1016/j.canlet.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 22.Huang Y, Mu Z, Lin L, et al. Expression and clinical significance of lncRNA PVT1 in renal carcinoma tissues. [Accessed July 05, 2018];Shandong Medical. 2015 55(36):11–13. Available from: http://www.sdyy.cbpt.cnki.net/WKD/WebPublication/paperDigest.aspx?paperID=aedb6d35-739c-40ed-85bd-d23caab689ef. [Google Scholar]
- 23.Liu Y, Quan X, Qin X, Y L. Effect of neoadjuvant chemotherapy on difference of expression of IncRNA in ovarian cancer patients. Pract J Cancer. 2016;31(03):384–386+389. [Google Scholar]
- 24.Ren X, Xiao Y, Zhang L, et al. Expression and clinical significance of PVT1 gene in blood of the patients with gastric cancer. [Accessed July 05, 2018];Chin J Cancer Biother. 2016 23(05):688–691. Available from: http://www.biother.org/zgzlswzlzz/ch/reader/view_abstract.aspx?file_no=20160517&flag=1. [Google Scholar]
- 25.Xu MD, Wang Y, Weng W, et al. A positive feedback loop of lncRNA-PVT1 and FOXM1 facilitates gastric cancer growth and invasion. Clin Cancer Res. 2017;23(8):2071–2080. doi: 10.1158/1078-0432.CCR-16-0742. [DOI] [PubMed] [Google Scholar]
- 26.Li Q, Dai Y, Wang F, et al. Differentially expressed long non-coding RNAs and the prognostic potential in colorectal cancer. Neoplasma. 2016;63(6):977–983. doi: 10.4149/neo_2016_617. [DOI] [PubMed] [Google Scholar]
- 27.Huang C, Yu W, Wang Q, et al. Increased expression of the lncRNA PVT1 is associated with poor prognosis in pancreatic cancer patients. Minerva Med. 2015;106(3):143–149. [PubMed] [Google Scholar]
- 28.Kong R, Zhang EB, Yin DD, et al. Long noncoding RNA PVT1 indicates a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically regulating p15 and p16. Mol Cancer. 2015;14:82. doi: 10.1186/s12943-015-0355-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding C, Yang Z, Lv Z, et al. Long non-coding RNA PVT1 is associated with tumor progression and predicts recurrence in hepatocellular carcinoma patients. Oncol Lett. 2015;9(2):955–963. doi: 10.3892/ol.2014.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang S, Zhang G, Liu J. Long noncoding RNA PVT1 promotes cervical cancer progression through epigenetically silencing miR-200b. APMIS. 2016;124(8):649–658. doi: 10.1111/apm.12555. [DOI] [PubMed] [Google Scholar]
- 31.Wu BQ, Jiang Y, Zhu F, Sun DL, He XZ. Long noncoding RNA PVT1 promotes EMT and cell proliferation and migration through downregulating p21 in pancreatic cancer cells. Technol Cancer Res Treat. 2017 doi: 10.1177/1533034617700559. 1533034617700559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wan L, Sun M, Liu GJ, et al. Long noncoding RNA PVT1 promotes non-small cell lung cancer cell proliferation through epigenetically regulating LATS2 expression. Mol Cancer Ther. 2016;15(5):1082–1094. doi: 10.1158/1535-7163.MCT-15-0707. [DOI] [PubMed] [Google Scholar]
- 33.Zhou Q, Chen F, Zhao J, et al. Long non-coding RNA PVT1 promotes osteosarcoma development by acting as a molecular sponge to regulate miR-195. Oncotarget. 2016;7(50):82620–82633. doi: 10.18632/oncotarget.13012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li PD, Hu JL, Ma C, et al. Upregulation of the long non-coding RNA PVT1 promotes esophageal squamous cell carcinoma progression by acting as a molecular sponge of miR-203 and LASP1. Oncotarget. 2017;8(21):34164–34176. doi: 10.18632/oncotarget.15878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahashi Y, Sawada G, Kurashige J, et al. Amplification of PVT-1 is involved in poor prognosis via apoptosis inhibition in colorectal cancers. Br J Cancer. 2014;110(1):164–171. doi: 10.1038/bjc.2013.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martini P, Paracchini L, Caratti G, et al. lncRNAs as novel indicators of patients’ prognosis in stage I epithelial ovarian cancer: a retrospective and multicentric study. Clin Cancer Res. 2017;23(9):2356–2366. doi: 10.1158/1078-0432.CCR-16-1402. [DOI] [PubMed] [Google Scholar]
- 37.Beck-Engeser GB, Lum AM, Huppi K, Caplen NJ, Wang BB, Wabl M. Pvt1-encoded microRNAs in oncogenesis. Retrovirology. 2008;5:4. doi: 10.1186/1742-4690-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siwarski D, Müller U, Andersson J, et al. Structure and expression of the c-Myc/Pvt1 megagene locus. Curr Top Microbiol Immunol. 1997;224:67–72. doi: 10.1007/978-3-642-60801-8_6. [DOI] [PubMed] [Google Scholar]
- 39.Carramusa L, Contino F, Ferro A, et al. The PVT-1 oncogene is a Myc protein target that is overexpressed in transformed cells. J Cell Physiol. 2007;213(2):511–518. doi: 10.1002/jcp.21133. [DOI] [PubMed] [Google Scholar]
- 40.Liu E, Liu Z, Zhou Y. Carboplatin-docetaxel-induced activity against ovarian cancer is dependent on up-regulated lncRNA PVT1. Int J Clin Exp Pathol. 2015;8(4):3803–3810. [PMC free article] [PubMed] [Google Scholar]
- 41.Kong W. Regulation of miR-1207-5p by lncRNA PVT1 and its effect on breast cancer development. Tianjing Medical University [M]; 2016. [Accessed July 05, 2018]. Available from: https://max.book118.com/html/2017/0730/125183433.shtm. [Google Scholar]
- 42.Chen X, Gao G, Liu S, et al. Long noncoding RNA PVT1 as a novel diagnostic biomarker and therapeutic target for melanoma. Biomed Res Int. 2017;2017:7038579. doi: 10.1155/2017/7038579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang H, Xie L, Zhu Y, et al. Possible mechanism of lnc RNA-PVT1 on cisplatin-induced DNA damage and repair. J Guangdong Medical Collge. 2016;34(6):572–577. [Google Scholar]
- 44.Shen CJ, Cheng YM, Wang CL. LncRNA PVT1 epigenetically silences miR-195 and modulates EMT and chemoresistance in cervical cancer cells. J Drug Target. 2017;25(7):637–644. doi: 10.1080/1061186X.2017.1307379. [DOI] [PubMed] [Google Scholar]
- 45.Liu HT, Fang L, Cheng YX, Sun Q. LncRNA PVT1 regulates prostate cancer cell growth by inducing the methylation of miR-146a. Cancer Med. 2016;5(12):3512–3519. doi: 10.1002/cam4.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li T, Meng XL, Yang WQ. Long noncoding RNA PVT1 acts as a “sponge” to inhibit microRNA-152 in gastric cancer cells. Dig Dis Sci. 2017;62(11):3021–3028. doi: 10.1007/s10620-017-4508-z. [DOI] [PubMed] [Google Scholar]
- 47.Huang T, Liu HW, Chen JQ, et al. The long noncoding RNA PVT1 functions as a competing endogenous RNA by sponging miR-186 in gastric cancer. Biomed Pharmacother. 2017;88:302–308. doi: 10.1016/j.biopha.2017.01.049. [DOI] [PubMed] [Google Scholar]
- 48.Zhou Q, Chen J, Feng J, Wang J. Long noncoding RNA PVT1 modulates thyroid cancer cell proliferation by recruiting EZH2 and regulating thyroid-stimulating hormone receptor (TSHR) Tumour Biol. 2016;37(3):3105–3113. doi: 10.1007/s13277-015-4149-9. [DOI] [PubMed] [Google Scholar]
- 49.Gao YL, Zhao ZS, Zhang MY, Han LJ, Dong YJ, Xu B. Long non-coding RNA PVT1 facilitates cervical cancer progression via negative regulating of miR-424. Oncol Res. 2017;25(8):1391–1398. doi: 10.3727/096504017X14881559833562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zuo Q. The characteristics and roles of non-coding RNAs miR-141/miR-22 and PVT1 on tumour growth and metastasis in gastric cancer. Third Military Medical University [D]; 2016. [Google Scholar]
- 51.Tseng YY, Bagchi A. The PVT1-MYC duet in cancer. Mol Cell Oncol. 2015;2(2):e974467. doi: 10.4161/23723556.2014.974467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Riquelme E, Suraokar MB, Rodriguez J, et al. Frequent coamplification and cooperation between C-MYC and PVT1 oncogenes promote malignant pleural mesothelioma. J Thorac Oncol. 2014;9(7):998–1007. doi: 10.1097/JTO.0000000000000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sarver AL, Murray CD, Temiz NA, Tseng YY, Bagchi A. MYC and PVT1 synergize to regulate RSPO1 levels in breast cancer. Cell Cycle. 2016;15(7):881–885. doi: 10.1080/15384101.2016.1149660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang F, Yuan JH, Wang SB, et al. Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology. 2014;60(4):1278–1290. doi: 10.1002/hep.27239. [DOI] [PubMed] [Google Scholar]
- 55.Wang J, Qiu M, Xu Y, et al. Long noncoding RNA CCAT2 correlates with smoking in esophageal squamous cell carcinoma. Tumour Biol. 2015;36(7):5523–5528. doi: 10.1007/s13277-015-3220-x. [DOI] [PubMed] [Google Scholar]
- 56.Liu Y, Luo F, Xu Y, et al. Epithelial-mesenchymal transition and cancer stem cells, mediated by a long non-coding RNA, HOTAIR, are involved in cell malignant transformation induced by cigarette smoke extract. Toxicol Appl Pharmacol. 2015;282(1):9–19. doi: 10.1016/j.taap.2014.10.022. [DOI] [PubMed] [Google Scholar]
- 57.Thai P, Statt S, Chen CH, Liang E, Campbell C, Wu R. Characterization of a novel long noncoding RNA, SCAL1, induced by cigarette smoke and elevated in lung cancer cell lines. Am J Respir Cell Mol Biol. 2013;49(2):204–211. doi: 10.1165/rcmb.2013-0159RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu L, Li M, Pu L, Ding Y, Liu J, Xiong S. The different prognostic value of long non-coding RNA BANCR in human cancers. Minerva Med. 2017;108(1):97–100. doi: 10.23736/S0026-4806.16.04670-X. [DOI] [PubMed] [Google Scholar]
- 59.Hu L, Li M, Ding Y, Pu L, Liu J, Xiong S. Long non-coding RNA HOTTIP, a novel potential prognostic marker in cancers. Minerva Med. 2017;108(1):95–97. doi: 10.23736/S0026-4806.16.04668-1. [DOI] [PubMed] [Google Scholar]
- 60.Fan YH, Fang H, Ji CX, Xie H, Xiao B, Zhu XG. Long noncoding RNA CCAT2 can predict metastasis and poor prognosis: a meta-analysis. Clin Chim Acta. 2017;466:120–126. doi: 10.1016/j.cca.2017.01.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The flow chart of meta-analysis.
The pooled effect value of PVT1 on TNM.
Abbreviations: IV, inverse variance methods; SE, standard error; UVM, ocular melanoma; SKCM, melanoma; STAD, stomach cancer; TGCT, testicular cancer; THCA, thyroid cancer; READ, rectal cancer; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; LIHC, liver cancer; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney clear cell carcinoma; KIRP, kidney papillary cell carcinoma; HNSC, head and neck cancer; ESCA, esophageal cancer; ACC, adrenocortical cancer; BLCA, bladder cancer; BRCA, breast cancer; CHOL, bile duct cancer; TCGA, the Cancer Genome Atlas database.
The pooled effect value of PVT1 on (A) LNM, (B) DOI, (C) DM, (D) DIF, and (E) LI.
Abbreviations: DIF, differentiation; DM, distant metastasis; DOI, depth of invasive; IV, inverse variance methods; LI, lymphatic invasion; LNM, lymph node metastasis; SE, standard error.
The pooled effect value of PVT1 on OS.
Abbreviations: IV, inverse variance methods; OS, overall survival; SE, standard error; ACC, adrenocortical cancer; BLCA, bladder cancer; BRCA, breast cancer; CESC, cervical cancer; CHOL, bile duct cancer; COAD, colon cancer; DLBC, large B-cell lymphoma; ESCA, esophageal cancer; GBM, glioblastoma; HNSC, head and neck cancer; KICH, kidney chromophobe; KIRC, kidney clear cell carcinoma; KIRP, kidney papillary cell carcinoma; LGG, lower grade glioma; LIHC, liver cancer; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian cancer; PAAD, pancreatic cancer; PCPG, pheochromocytoma & paraganglioma; PRAD, prostate cancer; READ, rectal cancer; SARC, sarcoma; SKCM, melanoma; STAD, stomach cancer; TGCT, testicular cancer; THCA, thyroid cancer; THYM, thymoma; UCEC, endometrioid cancer; UCS, uterine carcinosarcoma; UVM, ocular melanoma.
The pooled effect value of PVT1 on (A) DFS and (B) PFS.
Abbreviations: DFS, disease-free survival; IV, inverse variance methods; PFS, progression-free survival; SE, standard error.
Table S1.
Basic data for all included studies in the meta-analysis
| Author | Year | Country | Cancer type | Case (high/low) | Metastasis | Prognosis | Smoking on PVT1 OR (95% CI) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||
| TNM
|
LI
|
LNM
|
DOI
|
DM
|
DIF
|
Outcomes | HR (95% CI) | ||||||
| OR (95% CI) | OR (95% CI) | OR (95% CI) | OR (95% CI) | OR (95% CI) | OR (95% CI) | ||||||||
| Yuan et al17 | 2015 | China | GC (tissue) | 112 (55/56) | 3.346 (1.498–7.475) | 2.337 (1.086–5.030) | 2.337 (1.086–5.030) | 11.846 (4.858–28.884) | 2.075 (0.183–23.573) | NR | OS DFS |
2.280 (1.054–4.930) 2.210 (1.111–4.395) |
NR |
| Cui et al19 | 2015 | China | NSCLC (tissue) | 108 (53/55) | 3.168 (1.588–8.243) | NR | 2.289 (1.054–4.972) | 11.026 (4.505–26.983) | 2.118 (0.186–24.073) | 1.054 (0.461–2.408) | OS DFS |
1.72 (1.14–3.25) 1.98 (1.16–4.35) |
Irrelevant data |
| Xu et al23 | 2016 | China | GC (tissue) | 190 (96/94) | NR | NR | NR | NR | NR | NR | DFS DSS |
1.75 (1.25–2.56) 1.64 (1.15–2.39) |
NR |
| Takahashi et al4 | 2014 | Japan | CRC (tissue) | 164 (133/31) | 3.462 (1.494–8.022) | 2.119 (0.915–4.908) | 2.968 (1.282–6.870) | NR | 0.750 (0.075–7.451) | 1.281 (0.267–6.149) | DFS | 2.532 (1.152–10.747) | NR |
| Li et al18 | 2016 | China | CRC (tissue) | 30 (15/15) | 0.750 (0.169–3.327) | NR | NR | NR | NR | NR | DFS | 5.84 (0.42–81.19) | NR |
| Zheng et al16 | 2015 | China | ESCA (tissue) | 77 (39/38) | 3.450 (1.346–8.841) | NR | NR | NR | NR | NR | NR | NR | NR |
| Huang et al10 | 2015 | China | PAAD (tissue) | 85 (67/18) | 3.442 (1.170–10.127) | NR | 0.276 (0.088–0.863) | 1.829 (0.614–5.444) | NR | 1.550 (0.275–8.738) | OS | 3.013 (1.574–6.673) | NR |
| Yang et al1 | 2014 | China | NSCLC (tissue) | 82 (65/17) | 7.000 (1.829–26.788) | NR | 9.911 (2.093–46.925) | NR | NR | NR | OS | 3.273 (2.184–6.937) | NR |
| Huang et al12 | 2016 | China | SCLC (tissue) | 120 (60/60) | NR | NR | 9.000 (3.939–20.566) | NR | 11.455 (3.683–35.628) | NR | OS | 1.782 (1.078–2.945) | NR |
| Martini et al21 | 2016 | Italy | EOC (tissue) | Training 73 (NR) Testing 126 (NR) |
NR NR |
NR NR |
NR NR |
NR NR |
NR NR |
NR NR |
OS PFS OS PFS |
2.10 (1.40–3.30) 1.70 (1.30–2.20) 1.30 (1.10–1.60) 1.60 (1.30–1.90) |
NR |
| Kong et al8 | 2015 | China | GC (tissue) | 80 (40/40) | 3.857 (1.526–9.750) | 1.833 (0.755–4.455) | NR | 6.152 (2.334–16.211) | 1.111 (1.002–1.232) | 2.333 (0.932–5.839) | OS DFS |
2.092 (1.068–4.096) 2.216 (1.130–4.345) |
NR |
| Ding et al11 | 2015 | China | HCC (tissue) | 58 (49/9) 214 (157/57) |
0.311 (0.048–2.2028) 1.417 (0.772–2.603) |
NR NR |
NR NR |
NR NR |
NR NR |
1.534 (0.367–6.412) 1.739 (0.937–3.228) |
NR RFS OS |
NR 1.653 (1.019–2.681) 1.01 (0.65–1.56) |
NR NR |
| Zhang et al15 | 2016 | China | CESC (tissue) | 90 (45/45) | NR | NR | NR | NR | NR | NR | OS | 1.60 (0.55–4.66) | NR |
| Wu et al2 | 2017 | China | PAAD (tissue) | 30 (15/15) | 8.000 (1.5224–2.042) | NR | Unuseful data | NR | NR | Unuseful data | NR | NR | NR |
| Ren et al5 | 2016 | China | GC (serum) | 28 (13/15) | 2.222 (0.426–11.603) | NR | 8.250 (1.328–51.263) | 1.212 (0.216–6.800) | 1.083 (0.926–1.267) | 0.938 (0.205–4.294) | NR | NR | NR |
| Wan et al3 | 2016 | China | NSCLC (tissue) | 105 (56/49) | 3.710 (1.584–8.694) | NR | 2.842 (1.286–6.282) | NR | NR | NR | OS PFS |
2.464 (1.214–4.999) 3.39 (1.74–6.63) |
0.518 (0.226–1.186) |
| Zhou et al22 | 2016 | China | OS* (tissue) | 53 (29/24) | NR | NR | NR | NR | NR | NR | OS | 1.23 (0.37–4.05) | NR |
| Iden et al13 | 2016 | USA | CESC (tissue) | 121 (63/58) | NR | NR | NR | NR | NR | NR | OS | 2.03 (1.01–4.08) | NR |
| Li et al7 | 2017 | China | ESCC (tissue) | 104 (52/52) | 4.429 (1.917–10.229) | NR | NR | NR | NR | 3.167 (1.235–8.117) | OS DFS |
2.75 (1.35–5.59) 1.42 (0.71–2.87) |
1.185 (0.528–2.662) |
| Liu et al6 | 2016 | China | CESC (tissue) | 85 (67/18) | 3.442 (1.170–10.127) | NR | 3.621 (1.158–11.323) | 1.829 (0.614–5.444) | 1.455 (0.402–5.260) | 1.550 (0.275–8.738) | OS | 3.013 (1.574–6.673) | NR |
| Huang et al9 | 2015 | China | RC (tissue) | 54 (39/15) | 4.400 (1.183–16.367) | NR | 2.783 (0.675–11.477) | NR | NR | NR | NR | NR | NR |
| Chen et al20 | 2016 | China | GC (serum) | 187 (112/75) | 1.175 (0.567–2.434) | 0.838 (0.458–1.532) | 1.587 (0.672–3.751) | NR | 2.112 (0.845–5.278) | 0.982 (0.501–1.927) | OS DFS |
1.455 (1.005–2.105) 1.435 (0.998–2.061) |
NR |
Abbreviations: LI, lymphatic invasion; LNM, lymph node metastasis; DOI, depth of invasive; DM, distant metastasis; DIF, differentiation; OS, overall survival; DFS, disease-free survival; PFS, progression-free survival; DSS, disease specific survival; RFS, recurrence free survival; NR, no report; GC, gastric cancer; NSCLC, non-small cell lung cancer; CRC, colorectal cancer; ESCA, esophageal carcinoma; PAAD, pancreatic cancer; SCLC, small-cell lung cancer; EOC, epithelial ovarian cancer; HCC, hepatocellular carcinoma; CESC, cervical cancer; OS*, osteosarcoma; ESCC, esophageal squamous cell carcinoma; RC, renal carcinoma.