Abstract
Heat shock protein 70 (Hsp70) is a molecular chaperone that binds to “client” proteins and protects them from protein degradation. Hsp70 is essential for the survival of many cancer cells, but it is not yet clear which of its clients are involved. Using structurally distinct chemical inhibitors, we found that many of the well-known clients of the related chaperone, Hsp90, are not strikingly responsive to Hsp70 inhibition. Rather, Hsp70 appeared to be important for the stability of the RIP1 (RIPK1) regulators: cIAP1/2 (BIRC1 and BIRC3), XIAP, and cFLIPS/L (CFLAR). These results suggest that Hsp70 limits apoptosis and necroptosis pathways downstream of RIP1. Consistent with this model, MCF7 breast cancer cells treated with Hsp70 inhibitors underwent apoptosis, while co-treatment with z-VAD.fmk switched the cell death pathway to necroptosis. In addition, cell death in response to Hsp70 inhibitors was strongly suppressed by RIP1 knockdown or inhibitors. Thus, these data indicate that Hsp70 plays a previously unrecognized and important role in suppressing RIP1 activity.
Introduction
Elevated expression of Hsp70 correlates with poor survival and resistance to chemotherapeutics1–4. Hsp70 is generally thought to inhibit both the extrinsic and intrinsic pathways of apoptosis5 by protecting important “clients”, such as the oncoproteins Raf-1 and Akt-1, from degradation6–8, However, this model is largely based on analogy to the related chaperone, Hsp909,10. Inhibitors of Hsp90 are well-known to release clients from that chaperone, leading to protein degradation and, ultimately, apoptotic cell death11,12. It is not clear whether Hsp70’s activity is restricted to these “Hsp90-like” functions or if it plays a broader or even parallel role.
The molecular roles of Hsp70 in cancer have been elusive, in part, because of a lack of selective chemical inhibitors. A number of recent reports have created the first generation of Hsp70 inhibitors, including VER-1550088, MAL3-10113 and JG-9814. These molecules belong to distinct chemical families and have non-overlapping binding sites15. For example, JG-98 is an allosteric inhibitor that binds tightly to a deep pocket16 that is conserved in members of the Hsp70 family14. Importantly, JG-98 and its analogs have been found to be relatively selective for members of the Hsp70 family, based on results from pulldowns 17, over-expression and point mutations 18–21. The mechanism of JG-98 is to block a key allosteric transition in Hsp70 that favors degradation of some Hsp70-bound clients 19,21. Other compounds bind different locations and have distinct mechanisms22. For example, VER-155008 competes for binding of nucleotide to Hsp70 8 and MAL3-101 binds to a distinct allosteric site 23. Although JG-98 is relatively non-toxic (EC50 > 20 μM) to normal mouse embryonic fibroblasts (MEFs), it has anti-proliferative activity (EC50 ~ 400 nM) in multiple cancer cell lines14 and its analogs kill tamoxifen-resistant cells24. Similar selectivity for transformed cells is observed using Hsp70 inhibitors belonging to other chemical series8,25. The consistency of this result is important because parallel activity across chemically distinct molecules often suggests that the activity is mediated by the intended target. Based on all of these recent findings, we envisioned JG-98 and other new Hsp70 inhibitors as promising chemical tools for better understanding the chaperone’s specific molecular roles in cancer.
Using multiple, structurally distinct Hsp70 inhibitors, we found that Hsp90 clients, such as Akt or Raf1, are only weakly degraded after treatment. Rather, the stability of the RIP1 regulators, IAP1/2, XIAP, and cFLIPS/L, seemed sensitive to Hsp70 activity. Indeed, in MDA-MB-231 breast cancer cells, the kinetics of cell death correlated better with the loss of the RIP1 regulators than with degradation of Hsp90 clients. Consistent with a role in limiting RIP1 activation, treatment with Hsp70 inhibitors led to apoptotic cell death, but co-administration with z-VAD-fmk switched the cells to a necroptotic pathway. Further, cell death in response to Hsp70 inhibitors required RIP1 activity, as shown using RIP1 knockdown and selective RIP1 kinase inhibitors. Thus, although Hsp70 is likely to have multiple clients, its activity on RIP1 seems to be especially important in cell survival. These findings may help guide the selection of Hsp70-selective biomarkers and potentially accelerate the discovery of clinical candidates.
Materials and Methods
Reagents and Antibodies
Inhibitors
The following reagents were purchased from Sigma-Aldrich: Necrostatin-1, Bortezomib; Enzo: z-VAD.fmk; Millipore: Necrosulfonamide; LC Labs: 17-DMAG; StressMarq: VER-155008; and Teva Pharmaceuticals: Etoposide. JG-98 was synthesized and characterized as previously described14. All compounds were suspended in DMSO and the final solvent concentration in the assays was held to 1%.
Antibodies
The following antibodies were purchased from Enzo: Hsp72 (C92F3A-5), XIAP (ADI-AAM-050), c-IAP1 (ALX-803-335), Caspase-8 (ALX-804-429); SCBT: GAPDH (sc-32233), Hsp90 (sc-7947), Raf-1 (sc-133), Caspase-8 (sc-6136), anti-rat (sc-2006), goat IgG (sc-2028); CellSignal: Akt-1 (2967), Cleaved Caspase-3 (9664), c-IAP2 (3130); BD Pharmingen: RIP1 (610459), Cdk4 (559693), cytochrome c (556433), Bcl-xL (610746); Molecular Probes: COXIV (A21347); Alexis: FLIP (ALX-804-428); Millipore: Smac (567365).
Tissue Culture
MDA-MB-231 WT cells were maintained in DMEM (Invitrogen), supplemented with 10% Fetal Bovine Serum (FBS), 1% Penicillin-Streptomycin, and non-essential amino acids. Jurkat cells were grown in RPMI 1640 (Corning), supplemented with GlutaMax. MDA-MB-231 and Jurkat cells overexpressing Bcl-xL and RIP1 knockout Jurkat cells were all created as previously described26.
Cell Line Authentication
All cell lines were purchased from American Type Culture Collection (ATCC) and experiments performed on cells that were passaged less than 20 times.
Cell Growth Assays
Cell growth was analyzed using either the MTT or Cell Titer Glo assays as previously described14 or trypan blue exclusion, as indicated in the text. To supplement these assays, light microscopy, annexin and Hoescht staining were used to report on cell viability. EC50 values were calculated from 12-point concentration curves, using serial 2-fold dilutions. When noted, cells were pre-treated with z-VAD.fmk (40 μM), Necrostatin-1 (20 μM), Necrosulfonamide (20 μM), or a combination for 1 hour before addition of designated drug.
Flow Cytometry
MDA-MB-231 cells were detached using Accutase (BD Biosciences) and washed with PBS before staining with annexinV-APC (BD Biosciences) for 15 minutes at room temperature. Cells were washed again with PBS before addition of DAPI (100 μg/mL) immediately before analysis.
Immunoprecipitations
Ripoptosome
Cells were pre-treated with z-VAD.fmk (20 μM) to limit cleavage of RIP1. Following compound treatments, cells were incubated for 24 hours before proceeding with lysis, immunoprecipitation, and western blots, as previously described27.
Hsp70 binding to XIAP
MCF-7 cell extracts were prepared in chilled lysis buffer (50 mM Tris (pH 8), 150 mM NaCl, 1 mM ATP, 10 mM KCl, 5 mM Mg(OAc)2, 1% NP-40) supplemented with protease inhibitor cocktail (Roche Applied Science). The total protein concentration was adjusted to 5 mg of protein in 1 mL of cell extract. PureProteome Protein G magnetic beads (Millipore) were incubated with 6 μg of a specific antibody for Hsp70 (Santa Cruz Biotechnology) or XIAP (Enzo Life Sciences) or nonspecific mouse IgG (Santa Cruz Biotechnology) for 30 minutes at room temperature with mixing, followed by antibody crosslinking with bis(sulfosuccinimidyl) suberate (Thermo Scientific) for 1 hour at room temperature with mixing. The crosslinking reaction was quenched with 1 M Tris (pH 7.5) for 1 hour at room temperature with mixing. Equal 100 μL samples of cell lysate were each pre-cleared by incubation with 50 μL of protein G beads for 1 hour at room temperature with mixing. Protein complexes were immunoprecipitated by incubation of the pre-cleared lysate (1 mg total protein per IP) with 50 μL of antibody-crosslinked protein G beads for 1 hour at room temperature with mixing. The immunocomplexes were washed 3 times with 500 μL of wash buffer (PBS (pH 7.4), 0.1% Tween-20) and eluted with 0.1 M glycine (pH 2.6). Samples were run on a 10% Tris-glycine gel (Bio-Rad) and transferred to PVDF membrane (Thermo Scientific). Proteins were visualized by Western blot with antibodies against Hsp70 (1:1000, Santa Cruz Biotechnology) and XIAP (1:500, Enzo Life Sciences).
Fluorescence and Light Microscopy
Cells were visualized using an Olympus IX83 Inverted Microscope. Nuclear morphology was examined by Hoechst 33258 staining (Sigma). For each experiment, at least 10 individual frames were examined and representative panels chosen for presentation. For quantification, we manually counted 200 cells from three independent experiments.
Proteolytic Profiling (“N-terminal Degradomics”)
Eight liters of T-cell leukemia Jurkat A cells were grown in four separate 3 L spinner flasks to a density of 0.8 x 106 cells/mL (see below). The cells were then treated for 16 hours with 1) 10 μM JG98 alone, 2) 10 μM JG98 and 20 μM necrosulfamide (NSA), 3) 10 μM JG98 and 20 μM of cell permeable pan caspase inhibitor zVAD(OMe)-fmk, or 4) left untreated. For the combination treatments, the cells were pre-treated with NSA or zVAD for 1 hour prior to the addition of JG98. Cell growth was monitored using a Scepter cell counter (Millipore inc.) to estimate the amount of cell death and cell morphology changes monitored under a Zeiss Observer Z1 microscope to confirm the expected phenotype. Free N-termini were detected as previously described (see below). In short, cell lysates were collected and the cells lysed using 0.1% Trion X-100 (~0.8–1.2 x 109 cells per sample), free N-termini biotinylated using a cleavable peptide ester (TEVest4B) and subtiligase, protein fragments captured on Neutravidin beads and trypsin-digested. Each sample was then fractionated by HPLC into 12 fractions, for a total of 48 LC-MS/MS injections on an Orbitrap Velos Pro (Thermo Scientific inc.). Peptide identification was performed using Protein Prospector (v. 5.10.15 or higher) (University of California, San Francisco). All spectra were searched using the full human SwissProt database (downloaded 2013/06/27) with random sequence database for false discovery rate determination. Search parameters included: fixed modification cysteine carbamidomethylation; variable modifications N-terminal aminobutyric acid, methionine-loss (N-terminus) and methionine oxidation; up to one missed tryptic cleavages; C-terminal trypsin cleavage; non-specific cleavage N-terminus; parent mass tolerance 30 ppm; fragment mass tolerance 0.5 Da. Expectation value cut-off was adjusted to maintain <1% false-discovery rate at the peptide level in each sample. The analysis was performed using in-house scripts.
Results
Rapid kinetics of cell death in response to Hsp70 inhibitors suggests that it has distinct clients
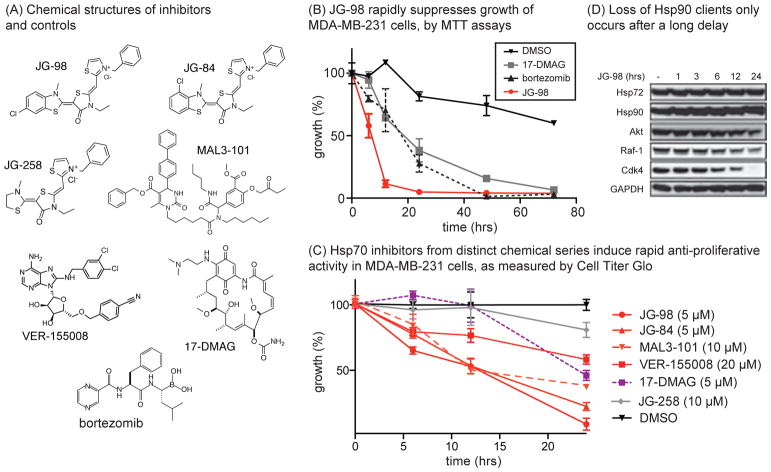
As a first step towards identifying clients important in Hsp70-mediated cell survival, we measured the kinetics of cell growth in response to the Hsp70 inhibitor, JG-98 (Fig 1a). In these experiments, we used MDA-MB-231 breast cancer cells, because they had previously been shown to be sensitive to Hsp70 inhibitors14. Using MTT assays, we found that the growth of JG-98 treated MDA-MB-231 cells was reduced within 10 to 12 hours (Fig. 1b). In contrast, the response to the Hsp90 inhibitor, 17-DMAG, or the proteasome inhibitor, bortezomib, occurred over longer times, ~18 to 24 hours (Fig 1b). To confirm this result using a distinct cell growth measure, we also performed CellTiterGlo assays on treated MDA-MB-231 cells and found that the kinetics in response to multiple, different Hsp70 inhibitors: JG-98, JG-84, VER-155008 or MAL3-101, was markedly faster than treatment with Hsp90 inhibitors (Fig 1c). As mentioned above, it was important to note that Hsp70 inhibitors from multiple chemical series showed similar responses, giving confidence in the role of that target. It is well known that treatment with Hsp90 inhibitors leads to degradation of Hsp90 clients, such as Akt, Raf-1 and Cdk4, and that the kinetics of that process parallels cell death 28,29. To see if these same clients were involved in Hsp70-mediated cell proliferation, we performed Western blots on JG-98 treated lysates. Surprisingly, we found that it took ~12 to 24 hours for the levels of Akt, Raf-1 and Cdk4 to be reduced (Fig. 1d), which is relatively late into the response. This result suggested that the mechanism(s) of Hsp70 inhibitors might not be simply explained by “Hsp90-like” effects in these cells.
Figure 1. Inhibition of Hsp70 Causes Rapid Anti-Proliferative Activity.
(A) Chemical structures of Hsp70 inhibitors (JG-98, JG-84, MAL3-101 and VER-155008), controls (JG-258) and inhibitors of Hsp90 (17-DMAG) and the proteasome (bortezomib). (B) JG-98 (5 μM) suppress growth of MDA-MB-231 cancer cells with relatively rapid kinetics compared to 17-DMAG (5 μM) or bortezomib (40 nM), as monitored by MTT assays. Results are the average of two independent experiments performed in quintuplicates. The error bars represent the standard error of the mean (SEM). (C) Hsp70 inhibitors induce rapid cell growth effects, as measured by Cell Titer Glo. Results are the average of experiments performed in triplicate. Errors are SEM. (D) Chaperone clients are degraded relatively late after treatment with JG-98 (10 μM), after onset of effects on cell growth. Results are representative of experiments performed in duplicate.
Inhibitors of Hsp70 induce apoptosis, independent of Bcl-xL status
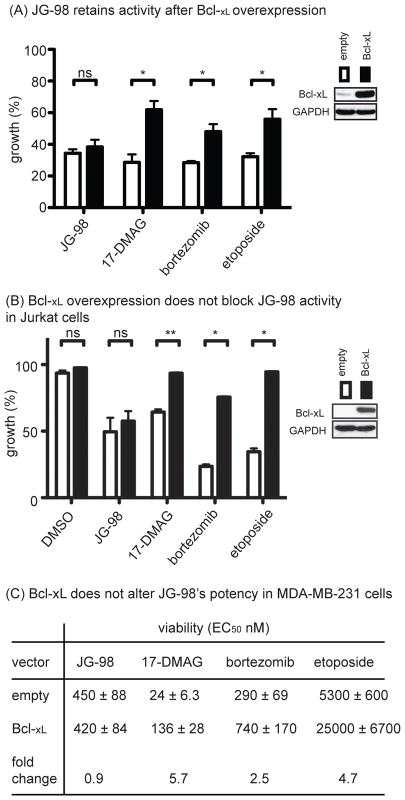
To better understand the response to Hsp70 inhibition, we then asked whether treated MDA-MB-231 cells underwent apoptosis. This is an important step because MTT and CellTiterGlo assays primarily report on proliferation and not necessarily on cell death. Indeed, we found that JG-98 induced classical apoptosis features, including morphological changes consistent with programmed cell death (Fig S1a) and positive annexin staining (Fig S1b). This result is consistent with previous reports that JG-98 activates caspase cleavage14. To further probe this idea, we overexpressed the Bcl-2 family member, Bcl-xL, a mitochondrial anti-apoptotic protein, in MDA-MB-231 cells. These cells are considered type II cells that undergo apoptosis through caspase-8 and the mitochondrial pathway30, so high Bcl-xL levels would be expected to block JG-98-mediated cell death. Importantly, we also performed parallel studies in leukemia-derived Jurkat cells, because it is known that over-expression of Bcl-xL only incompletely inhibits RIP1-depedent cell death in MDA-MB-231 cells27. Surprisingly, we found that overexpression of Bcl-xL did not impede cytotoxicity in response to JG-98 in either cell type (Figs. 2a, b). As controls, we confirmed that Bcl-xL overexpression partially suppressed cell death in response to the control molecules, 17-DMAG, bortezomib or etoposide, in either MDA-MB-231 (see Fig 2a) or Jurkat (see Fig 2b) cells. To understand the impact of Bcl-xL over-expression on the potency of the compounds, we performed dose dependence studies in MDA-MB-231 cells and calculated EC50 values. Consistent with the single concentration data, we found that over-expression of Bcl-xL did not alter the EC50 for JG-98 (Fig 2c); whereas the EC50 of 17-DMAG, bortezomib and etoposide were significantly increased (2.5- to 5.7-fold). The mechanism of cell death did not seem to be affected, because treatment of the Bcl-xL overexpressing cells with JG-98 led to morphological features that were still consistent with apoptosis (Fig. S2a). Based on the mitochondrial retention of COX IV in treated MDA-MB-231 cells, this apoptotic activity also seemed to occur without global disruption of mitochondria (Fig S2b). Thus, inhibition of Hsp70 appeared to initiate a mitochondrial death pathway that was independent of Bcl-2 family status.
Figure 2. Hsp70 Inhibitor Activity is Independent of Bcl Status.
(A) Overexpression of Bcl-xL does not block JG-98 cytotoxicity. MDA-MB-231 cells were treated for 24 hrs with JG-98 (10 μM), 17-DMAG (10 μM), bortezomib (40 nM) or etoposide (20 μM). MTT results are the mean average of triplicates and error bars represent SEM. *p value < 0.05. (B) Jurkat cells over-expressing Bcl-xL were treated with indicated compounds for 24 hours. Viability was determined by trypan blue staining. Results are the average of two experiments performed in triplicate. Error is SEM. **p value < 0.01. (C) Bcl-xL overexpression provides partial resistance to compounds, except JG-98. MDA-MB-231 cells were treated for 72 hours and cell growth determined by MTT assays. Results are the average of two independent experiments performed in triplicate. Error is SEM.
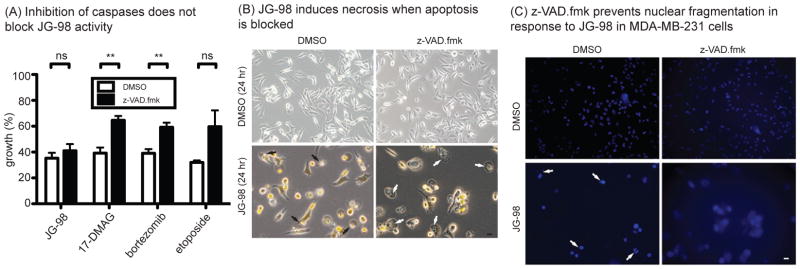
When caspase-mediated apoptosis is blocked, Hsp70 inhibitors trigger an alternative, necroptosis pathway
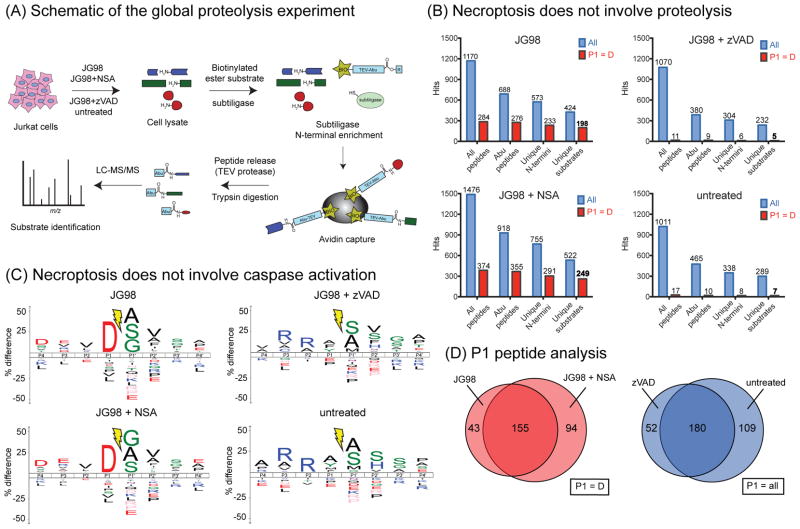
The caspase inhibitor, z-VAD.fmk, is known to partially suppress cell death in response to Hsp90 inhibitors31. However, we found that this compound did not block proliferation induced by JG-98 (Fig. 3a). Rather, we observed that 51 ± 13% cells pre-treated with z-VAD.fmk before addition of JG-98 displayed a swollen cytoplasm containing numerous granules (Fig. 3b). This morphology was not observed in the absence of v-VAD.fmk. The swollen cytoplasm feature is reminiscent of necrotic cell death, suggesting that JG-98 might be activating an alternative pathway if apoptosis was blocked. Consistent with this idea, we observed nuclear fragmentation in only 4 ± 1% of cells that were pre-treated with z-VAD.fmk prior to JG-98, compared to 40 ± 16% in the absence of z-VAD.fmk (Fig. 3c). To further explore whether JG-98 might be activating an alternative death pathway, we profiled the peptide fragments produced from proteolysis in cells treated with JG-98, using a recently described mass spectrometry-based method termed “N-terminal degradomics” (Fig 4a) 32. Briefly, this method profiles the neo peptide termini that are produced by proteolysis during cell death. Treatment of Jurkat cells with JG-98 produced a fragment profile consistent with caspase activation, as judged by the prominent aspartate in the P1 position (Fig 4b,c and Table S1). As expected, treatment with z-VAD.fmk nearly completely blocked this signature. Moreover, when z-VAD.fmk was used as a co-treatment with JG-98, it also suppressed the caspase response (Fig 4b,c), with the peptide profile resembling the untreated cells. These results support the idea that treatment with JG-98 initiates caspase-mediated apoptosis, but that the cells die by an alternative, necroptosis, pathway when caspases are inhibited. Perhaps more broadly, these findings also show that necroptosis occurs in the absence of dramatic proteolysis, a feature of this alternative cell death pathway that had not previously been described.
Figure 3. Inhibition of Caspases with z-VAD-fmk Does not Block JG-98’s Activity.
(A) Inhibition of caspases by z-VAD.fmk does not suppress the activity of JG-98. Results are the average of three independent experiments performed in quintuplicate. Error bars represent SEM. Cells were pretreated with z-VAD.fmk (40 μM) for 1 hour prior to addition of compounds (same concentrations as in panel A) and MTT assays performed after 24 hrs. **p value < 0.01. ns = not significant. Error is SEM. (B) Cells were treated as in panel A and visualized using an Olympus IX83 Inverted Microscope. Black arrows indicate apoptotic cells; white arrows indicate necrotic cells. (C) Hoechst 33258 staining of MDA-MB-231 cells pretreated with z-VAD.fmk (40 μM) for 1 hour prior to addition of JG-98 (10 μM) for 24 hours. Cells pre-treated with z-VAD.fmk show no nuclear fragmentation (white arrows), even after co-administration of JG-98. Scale bar is 10 μm.
Figure 4. Proteolytic Signature of Cells Treated with JG-98 Supports Activation of Both an Apoptotic and Alternative Cell Death Pathway.
(A) Schematic overview of the proteolytic profiling study (aka “N-terminal degradomics”). (B) Overview of the peptide fragments produced in response to compound treatments. More than 45% of the substrates identified after JG-98 treatment feature a P1 = D, suggesting the activation of caspases. In general, there was minimal proteolysis in untreated cells or cells treated with JG-98 and z-VAD. Aminobutyric acid (Abu). (C) Logo plots show that JG-98 activates apoptosis in Jurkat cells and NSA is unable to block this activity. However, z-VAD reversed the proteolytic profile to one that approximated untreated cells. (D) The caspase activation profile (P1=D peptides) was similar between cells treated with JG-98 or JG-98 plus NSA, while the pattern of peptides (P1 = all) was similar between JG98 + zVAD and untreated cells. Experiments repeated in biological duplicate and techincal triplicate.
Hsp70 limits RIP1 activation by stabilizing E3 ligases
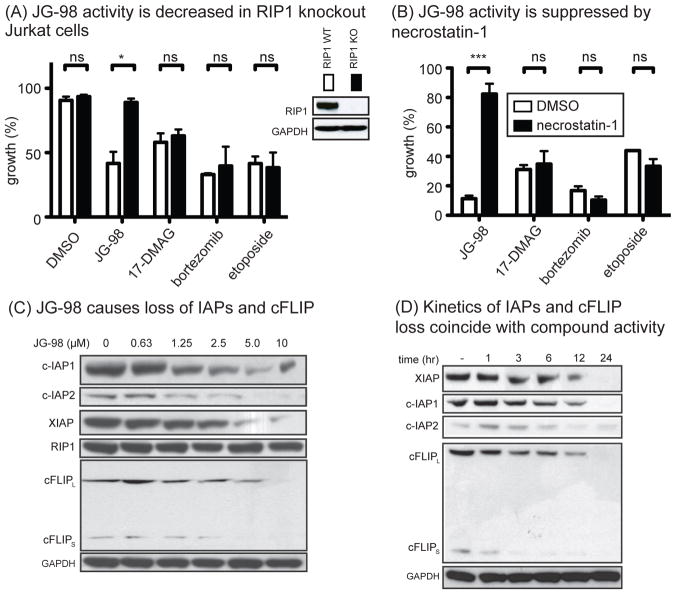
RIP1 is an important signaling regulator that directs cells into either the apoptotic or necroptotic cascades27,33. Because JG-98 appeared to initiate both pathways, we hypothesized that RIP1 might be important in Hsp70-mediated cell survival. To test this idea, we treated Jurkat cells lacking RIP1. Strikingly, JG-98 was not cytotoxic in RIP1 knockout Jurkat cells (Fig. 5a). Similarly, necrostatin-1, an inhibitor of RIP1’s kinase activity34, protected MDA-MB-231 cells from treatment with JG-98 (Figs. 5b). Conversely, treatment with 17-DMAG, bortezomib or etoposide was not affected by loss of RIP1 protein or RIP1 activity (see Figs 5a,b). Thus, cell death by JG-98 appeared to be uniquely dependent on RIP1 kinase activity. RIP1 is constitutively ubiquitinated by the E3 ubiquitin ligases XIAP, c-IAP1, c-IAP2, and cFLIPS/L35. Ubiquitination of RIP1 is linked to turnover of the kinase, but also to non-degradation pathways that modulate RIP1 activity36. Indeed, ubiquitination of RIP1 is thought to protect against necroptosis37. Hsp70 is known to protect a number of proteins from degradation, so it seems possible that it may normally stabilize RIP1 itself or the E3 ligases. To test this idea, we treated MDA-MB-231 cells with JG-98 and examined the levels of RIP1 and its E3 ligases by Western blots. Although inhibition of Hsp70 did not significantly impact RIP1 protein levels under these conditions, it induced a striking loss of XIAP, c-IAP1/2 and cFLIPS/L (Fig. 5c). To test whether this pathway might be linked to JG-98 treatment, we examined the kinetics by which the levels of c-IAP, XIAP and cFLIPS/L are reduced. Indeed, loss of the E3 ubiquitin ligases closely paralleled the kinetics of cell death, with cFLIPS levels diminished within 1 to 3 hours and the levels of the other ligases beginning to decrease between 6 and 12 hours (Fig. 5d). This result sharply contrasts with the delay in Hsp90 client destabilization that we observed earlier (see Fig. 1b), suggesting that Hsp70 regulation of RIP1 may be more closely linked to activity in these cells. To understand the mechanism of turnover, we deleted the RING domain from XIAP (XIAPΔRING) and compared its stability to full length XIAP (XIAPFL) in the presence of JG-98 in MDA-MB-231 cells (Fig S3a). XIAP is known to degraded by self-ubiquitination through this domain38,39. We found that XIAPΔRING was resistant to treatment with JG-98 (Fig S3b), consistent with activation of its normal turnover pathway through the ubiquitin proteasome system. Finally, we wanted to test whether Hsp70 inhibitors from different structural classes also trigger loss of the E3 ligases. These types of experiments are important for confirming on-target activity because, as mentioned above, the different inhibitors have distinct chemical structures and they bind different allosteric sites on the chaperone15. Indeed, we found that treatment with JG-84, VER-155008 or MAL3-101 also destabilized c-IAP1 in MDA-MB-231 cells, while a structurally-related negative control, JG-258, had no effect (Fig S3c).
Figure 5. JG-98 Activity Occurs through a RIP1-Dependent Process.
(A) RIP1 KO Jurkat cells are resistant to JG-98. Viability was determined by trypan blue exclusion. Cells were treated for 24 hrs with JG-98 (10 μM), 17-DMAG (10 μM), bortezomib (40 nM), etoposide (20 μM). Results are the average of three independent experiments performed in triplicate. ns = not significant; * p < 0.05. (B) JG-98 activity requires RIP1 kinase. MDA-MB-231 cells were pretreated with 20 μM necrostatin-1 for 1 hour prior to addition of compounds. Viability was determined by three independent MTT assays performed in quintuplicate. Error is SEM. ns = not significant; ***p < 0.0001. (C) JG-98 induces degradation of RIP1 modulators. MDA-MB-231 cells were treated for 24 hours. Results represent experiments performed in triplicate. (D) RIP1 regulators are rapidly degraded in response to JG-98. MDA-MB-231 cells were treated with JG-98 (10 μM). Results are representative of duplicates.
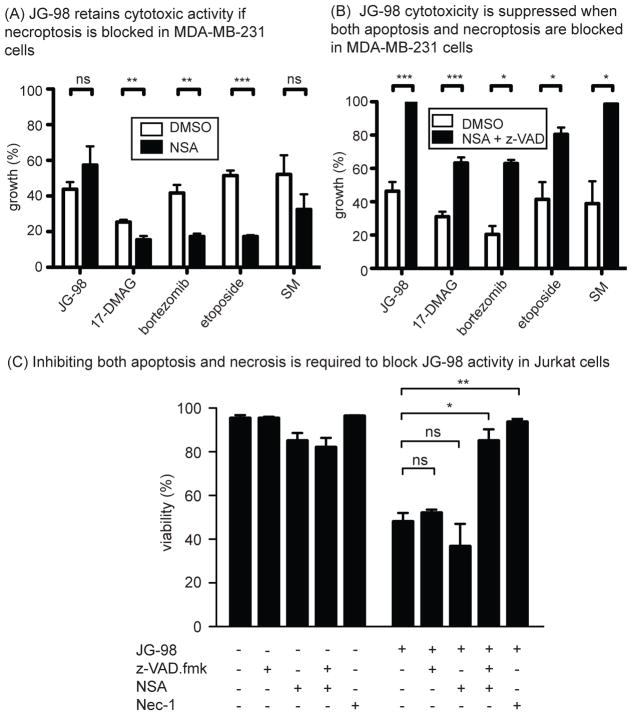
Blocking caspase-mediated apoptosis and necroptosis prevents cell death in response to Hsp70 inhibitors
Next, we wondered whether blocking necroptosis would make cells resistant to JG-98. However, we found that inhibition of this pathway by necrosulfonamide (NSA) did not block JG-98 effects on proliferation (Fig 6a). Further, the proteolytic fragment signature of these treated cells suggested that they may still die by caspase-mediated apoptosis (see Fig 4d). Then, to understand if blocking both apoptosis and necroptosis might limit JG-98 mediated activity, we pre-treated MDA-MB-231 (Fig 6b) or Jurkat cells (Fig 6c) with both z-VAD.fmk and NSA. We found that the combination partially blocked JG-98-mediated cell death. Importantly, a distinct Hsp70 inhibitor, VER-155008, also required co-treatment with both z-VAD-fmk and NSA to protect against activity in MDA-MB-231 cells (Fig S4a). To explore this phenomenon more broadly, we tested JG-98 in combination with v-VAD.fmk or z-VAD.fmk plus NSA in cancer cells derived from a variety of tissues. Consistent with the results from the MDA-MB-231 and Jurkat cells, z-VAD.fmk alone was unable to protect cells derived from breast (MCF7, SK-BR-3, T-47D), lung (A549) or colon (HT-29) (Fig S4b), while cervical-derived HeLa cells were partially protected (EC50 increased 2-fold). Next, we used the combination of z-VAD.fmk and NSA and found that both were required to protect MDA- MCF7, SK-BR-3 and Jurkat cells. Interestingly, A549, T47-D and HT-29 cells were not fully protected by this combination and the effect of JG-98 may have even been mildly exacerbated (see Fig S4b). These results suggest that Hsp70 may play addition roles in some cell types, perhaps through additional mechanisms such as lysosomal cell death40.
Figure 6. JG-98 Activity Requires Both Apoptosis and Necroptosis.
(A) Inhibition of necroptosis alone does not restore growth of cells treated with JG-98. Cells were pretreated with necrosulfonamide (NSA, 20 μM) for 1 hour prior to addition of compounds. Error is SEM. (B) Inhibition of both apoptosis and necroptosis is necessary to fully suppress JG-98 activity. Cells were pretreated with both 40 μM z-VAD.fmk and 20 μM Necrosulfonamide for 1 hour prior to addition of compounds. Cell growth was determined by three independent MTT assays performed in quintuplicate. *p value < 0.05, **p value < 0.01, ***p value < 0.001, ns = not significant. (C) Jurkat cells were pretreated with z-VAD.fmk (40 μM), NSA, both, or necrostatin-1 for 1 hour prior to addition of JG-98 (10 μM). Inactivation of both apoptosis and necroptosis was necessary to prevent activity. Cell viability was determined by trypan blue exclusion. Results are the average of three independent experiments performed in triplicate.
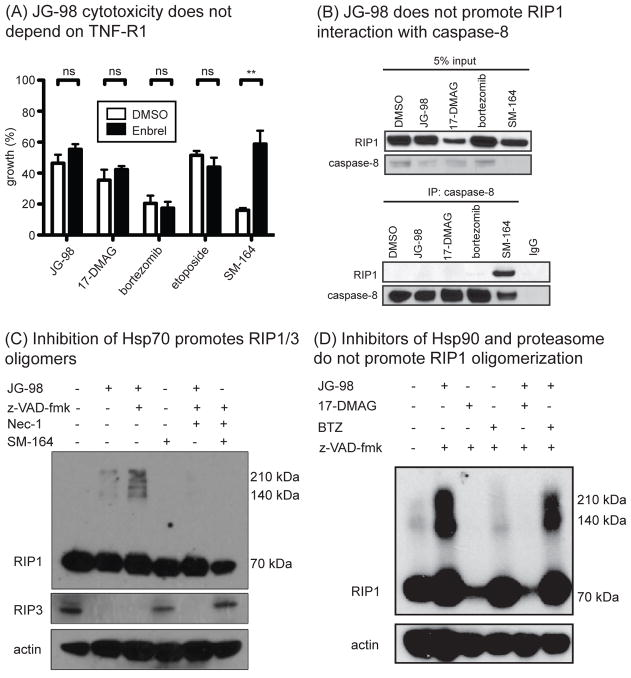
Hsp70 blocks oligomerization and activation of RIP1
Finally, we wanted to explore additional, possible mechanisms by which Hsp70 could inhibit RIP1 activation. One major role for RIP1 is as a central scaffold in the TNF-Receptor 1 (TNF-R1) complex33. It has been shown that autocrine-TNF-producing cells, such as MDA-MB-231, are sensitive to small molecule mimetics of the Smac/DIABLO protein and that this cytotoxicity proceeds through TNF-R127. We investigated this possibility for JG-98, and found that a neutralizing antibody against TNF-R1 (Enbrel) blocked cell death by a Smac mimetic (SM), but that it was unable to prevent JG-98 induced cell death (Fig. 7a). Thus, JG-98 appears to act independently of TNF-R1, but with a mechanism distinct from Smac mimetics. A cytosolic death complex containing RIP1, termed the ripoptosome, has been also shown to direct both apoptosis and necroptosis27. A key event in formation of the ripoptosome is binding of RIP1 to caspase-8. To determine if treatment with JG-98 triggered this event, we immunoprecipitated caspase-8 and performed Western blots with a RIP1 antibody. While Smac mimetic gave the expected result, JG-98 did not promote the interaction (Fig. 7b), suggesting that JG-98 does not trigger formation of the ripoptosome. Lastly, we tested whether Hsp70 might play a role in RIP1 oligomerization. It has recently been reported that RIP1 and RIP3 forms stable, oligomeric, amyloid-like structures that are required for necroptosis41, which appear as stable, high molecular mass bands on SDS-PAGE. At the same time, it is well known that Hsp70 broadly inhibits the formation of amyloids formed from diverse proteins, such as amyloid beta, huntingtin and tau42. Thus, it seemed logical that Hsp70 might counteract conversion of RIP1 into its amyloid state, such that treatment with JG-98 might relieve this suppression. To test this idea, we treated MDA-MB-231 cells with JG-98 or the combination of JG-98 and z-VAD.fmk (to induce necroptosis) and looked for stable, high molecular mass oligomers of RIP1. We found that treatment with JG-98 promoted conversion of RIP1 to the oligomeric state, concurrent with depletion of its binding partner, RIP3 (Fig 7c). Similar effects were observed with the other Hsp70 inhibitors, JG-84 and VER-155008 (Fig S5). Again, this mechanism was distinct from that used by SM-164, which had no effect on RIP1 oligomerization in this system (see Fig 7d). Further, RIP1 oligomerization did require kinase activity in this context, as Nec-1 could block the process. Importantly, treatment with 17-DMAG or bortezomib did not impact RIP1 oligomerization (Fig 7d). Together, these results suggested that Hsp70 plays a key role in suppressing RIP1 activation, possibly through effects on the stability of its E3 ligases and on its self-association.
Figure 7. Hsp70 Limits RIP1/3 Oligomerization.
(A) JG-98 toxicity does not depend on TNF signaling. MDA-MB-231 cells were pre-treated with Enbrel (5 μg/mL) for 1 hour prior to addition of compounds. Viability was determined by MTT assay. Results are the average of three independent experiments performed in quintuplicate. Error is SEM. **p value < 0.01 (B) JG-98 does not induce formation of a RIP1-caspase-8 complex. MDA-MB-231 cells were treated for 24 hours with indicated compounds. SM-164 was used at 100 nM. (C) Treatment with JG-98 (10 μM) favored formation of a high molecular mass oligomer of RIP1 and a reduction of RIP3. Co-treatment with z-VAD-fmk exacerbated this effect. Necrostatin could block the effects of JG-98. Results are representative of experiments performed in triplicate. (D) Treatment with 17-DMAG or bortezomib (BTZ) did not change RIP1 oligomerization.
Discussion
Knockdown of Hsp70 has been shown to reduce proliferation and enhance sensitivity to chemotherapy in multiple cancer models1–4. Because Hsp70 itself is not an oncogene, it was presumed that it acts through a “non-oncogene addiction” mechanism, in which the chaperone stabilizes a subset of oncogenes to protect against cell death6,43. Hsp90 is known to play a similar functional role12, yet more is known about the mechanisms by which that chaperone creates a non-oncogene addiction. Specifically, Hsp90 is known to directly bind and stabilize a subset of pro-survival clients, including Akt, Her-2, Raf-1 and cdk444. Although individual clients of Hsp70 have been suggested6–8 and some shared clients seem likely45, we wondered whether these two chaperones might also have distinct clients. This question became possible to address, thanks to the recent development of Hsp70 inhibitors by multiple research groups. By analogy, the clients of Hsp90 were revealed after the serendipitous discovery of geldanamycin and the development of its more potent analogs, such as 17-DMAG46. Thus, given recent advances in Hsp70 inhibitor discovery, it seemed timely to ask whether Hsp70 might have its own clients. Here, we used a suite of Hsp70 inhibitors with different binding sites and mechanisms to explore Hsp70’s pro-survival function. We focused on MDA-MB-231 and Jurkat cells, because of existing genetic data on the importance of Hsp70 in these cells, but it is important to stress that Hsp70 may play additional roles in other cells (see Fig S4B).
Our results suggest an unexpected model in which Hsp70 protects against cell death through suppressing RIP1 activation. In part, this mechanism appears to involve Hsp70-mediated stabilization of the E3 ubiquitin ligases of RIP1, such as c-IAPs and XIAP (see Fig 5). Importantly, we found that the kinetics of cell death correlate better with loss of these E3 ligases than with the turnover of the classical Hsp90 clients, such as Raf-1. This result could be particularly important for the future of Hsp70 inhibitor development, as these E3 ligases might be biomarkers for Hsp70 engagement. Again, an analogy to Hsp90 is illuminating because loss of the Hsp90 clients, such as Raf-1 and Akt, is often used to estimate engagement of that target. Another mechanism of Hsp70-mediated cell survival appeared to involve suppression of RIP1/3 amyloid formation. Specifically, we found that treatment with JG-98 or other Hsp70 inhibitors relieved this suppression and allowed RIP1 oligomers to form (see Fig 7). In retrospect, this finding might have been expected, based on the widespread role of Hsp70 in preventing amyloid formation47. Although this mechanism has been best described in the neurodegenerative disease literature, it seems logical that it would also play this part in cancer.
Hsp70 has been linked to a wide number of cell survival aspects. We hypothesize that some of these activities might be consolidated and understood through the functions on RIP1, a key, upstream “hub” of multiple cell death pathways. Indeed, our results suggest that, at least in MDA-MB-231 and Jurkat cells, this mechanism might be particularly important because RIP1 knockdown or co-treatment with necrostatin was sufficient to completely suppress JG-98’s activity. However, many important mysteries remain. For example, we observed that cell death in response to Hsp70 inhibitor was independent of Bcl-xL and it was coupled with an unusual relationship to cytochrome c release. Further, it has been shown that RIP1 itself is a client of Hsp9048 and that Hsp90 inhibitors can impact necroptosis in fibrosarcoma cells49. Thus, it seems compelling to envision that Hsp90 and Hsp70 may cooperate in suppressing the RIP1 pathway, in a currently unknown way. It is even possible that combinations of Hsp70 and Hsp90 inhibitors could be synergistic in this setting.
Finally, this work may accelerate the search for drug candidates that target Hsp70. Based on expression data, biochemical studies and knockdowns, members of the Hsp70 family appear to be compelling, potential drug targets50,51. However, progress has been slowed by lack of mechanistic knowledge. Our results are potentially important in that context because they suggest that IAP family members may be biomarkers of Hsp70 inhibition. If that finding is found to be robust across many cell types and by different groups, it would be expected to boost efforts to discover, optimize and deploy clinical candidates to test the central hypothesis of Hsp70 as an anti-cancer target.
Supplementary Material
Implications.
These findings clarify the role of Hsp70 in pro-survival signaling and suggest IAPs as potential new biomarkers for Hsp70 inhibition.
Acknowledgments
The authors would like to thank the members of the Gestwicki and Duckett labs for assistance. MAL3-101 was supplied by Jeff Brodsky (U. Pittsburgh).
Grant Support
Funding for this work was provided by the NIH (NS095690) and the University of Michigan’s Comprehensive Cancer Center (CA046592).
Footnotes
Disclosure of Potential Conflicts of Interest
The authors claim no competing financial interests.
Author’s Contributions
S.R.S, X.L., O.J., M.Z., L.C.C., H.S. and J.C. conceived, designed and performed experiments. I.M., J.A.W., C.D. and J.E.G. conceived, designed and supervised the project. All of the authors contributed to writing the manuscript.
Figs. S1, S2, S3, S4, S5 and Table S1 are available.
References
- 1.Nylandsted J, Brand K, Jaattela M. Heat shock protein 70 is required for the survival of cancer cells. Ann N Y Acad Sci. 2000;926:122–125. doi: 10.1111/j.1749-6632.2000.tb05605.x. [DOI] [PubMed] [Google Scholar]
- 2.Nanbu K, et al. Prognostic significance of heat shock proteins HSP70 and HSP90 in endometrial carcinomas. Cancer detection and prevention. 1998;22:549–555. doi: 10.1046/j.1525-1500.1998.00069.x. [DOI] [PubMed] [Google Scholar]
- 3.Rohde M, et al. Members of the heat-shock protein 70 family promote cancer cell growth by distinct mechanisms. Genes Dev. 2005;19:570–582. doi: 10.1101/gad.305405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciocca DR, Calderwood SK. Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones. 2005;10:86–103. doi: 10.1379/CSC-99r.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brodsky JL, Chiosis G. Hsp70 molecular chaperones: emerging roles in human disease and identification of small molecule modulators. Curr Top Med Chem. 2006;6:1215–1225. doi: 10.2174/156802606777811997. [DOI] [PubMed] [Google Scholar]
- 6.Davenport EL, et al. Targeting heat shock protein 72 enhances Hsp90 inhibitor-induced apoptosis in myeloma. Leukemia. 2010;24:1804–1807. doi: 10.1038/leu.2010.168. [DOI] [PubMed] [Google Scholar]
- 7.Powers MV, Clarke PA, Workman P. Dual targeting of HSC70 and HSP72 inhibits HSP90 function and induces tumor-specific apoptosis. Cancer cell. 2008;14:250–262. doi: 10.1016/j.ccr.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 8.Williamson DS, et al. Novel adenosine-derived inhibitors of 70 kDa heat shock protein, discovered through structure-based design. J Med Chem. 2009;52:1510–1513. doi: 10.1021/jm801627a. [DOI] [PubMed] [Google Scholar]
- 9.da Silva VC, Ramos CH. The network interaction of the human cytosolic 90 kDa heat shock protein Hsp90: A target for cancer therapeutics. Journal of proteomics. 2012;75:2790–2802. doi: 10.1016/j.jprot.2011.12.028. [DOI] [PubMed] [Google Scholar]
- 10.Jolly C, Morimoto RI. Role of the heat shock response and molecular chaperones in oncogenesis and cell death. J Natl Cancer Inst. 2000;92:1564–1572. doi: 10.1093/jnci/92.19.1564. [DOI] [PubMed] [Google Scholar]
- 11.Matts RL, et al. A systematic protocol for the characterization of Hsp90 modulators. Bioorg Med Chem. 2011;19:684–692. doi: 10.1016/j.bmc.2010.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nature reviews. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 13.Fewell SW, et al. Small molecule modulators of endogenous and co-chaperone-stimulated Hsp70 ATPase activity. J Biol Chem. 2004;279:51131–51140. doi: 10.1074/jbc.M404857200. [DOI] [PubMed] [Google Scholar]
- 14.Li X, et al. Analogs of the Allosteric Heat Shock Protein 70 (Hsp70) Inhibitor, MKT-077, as Anti-Cancer Agents. ACS medicinal chemistry letters. 2013;4 doi: 10.1021/ml400204n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Assimon VA, Gillies AT, Rauch JN, Gestwicki JE. Hsp70 protein complexes as drug targets. Curr Pharm Des. 2013;19:404–417. doi: 10.2174/138161213804143699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rousaki A, et al. Allosteric drugs: the interaction of antitumor compound MKT-077 with human Hsp70 chaperones. J Mol Biol. 2011;411:614–632. doi: 10.1016/j.jmb.2011.06.003. S0022-2836(11)00627-9 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wadhwa R, et al. Selective toxicity of MKT-077 to cancer cells is mediated by its binding to the hsp70 family protein mot-2 and reactivation of p53 function. Cancer Res. 2000;60:6818–6821. [PubMed] [Google Scholar]
- 18.Fontaine SN, et al. Isoform-selective Genetic Inhibition of Constitutive Cytosolic Hsp70 Activity Promotes Client Tau Degradation Using an Altered Co-chaperone Complement. J Biol Chem. 2015;290:13115–13127. doi: 10.1074/jbc.M115.637595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taguwa S, et al. Defining Hsp70 Subnetworks in Dengue Virus Replication Reveals Key Vulnerability in Flavivirus Infection. Cell. 2015;163:1108–1123. doi: 10.1016/j.cell.2015.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Young ZT, et al. Stabilizing the Hsp70-Tau Complex Promotes Turnover in Models of Tauopathy. Cell chemical biology. 2016;23:992–1001. doi: 10.1016/j.chembiol.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang AM, et al. Activation of Hsp70 reduces neurotoxicity by promoting polyglutamine protein degradation. Nature chemical biology. 2013;9:112–118. doi: 10.1038/nchembio.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, Shao H, Taylor IR, Gestwicki JE. Targeting Allosteric Control Mechanisms in Heat Shock Protein 70 (Hsp70) Curr Top Med Chem. 2016;16:2729–2740. doi: 10.2174/1568026616666160413140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wisen S, et al. Binding of a small molecule at a protein-protein interface regulates the chaperone activity of hsp70-hsp40. ACS chemical biology. 2010;5:611–622. doi: 10.1021/cb1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koren J, 3rd, et al. Rhodacyanine derivative selectively targets cancer cells and overcomes tamoxifen resistance. PLoS ONE. 7:e35566. doi: 10.1371/journal.pone.0035566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taldone T, et al. Heat shock protein 70 inhibitors2.2,5'-thiodipyrimidines, 5-(phenylthio)pyrimidines, 2-(pyridin-3-ylthio)pyrimidines, and 3-(phenylthio)pyridines as reversible binders to an allosteric site on heat shock protein 70. J Med Chem. 2014;57:1208–1224. doi: 10.1021/jm401552y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galban S, et al. Cytoprotective effects of IAPs revealed by a small molecule antagonist. Biochem J. 2009;417:765–771. doi: 10.1042/BJ20081677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tenev T, et al. The Ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol Cell. 2011;43:432–448. doi: 10.1016/j.molcel.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 28.Chandarlapaty S, et al. SNX2112, a synthetic heat shock protein 90 inhibitor, has potent antitumor activity against HER kinase-dependent cancers. Clin Cancer Res. 2008;14:240–248. doi: 10.1158/1078-0432.CCR-07-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caldas-Lopes E, et al. Hsp90 inhibitor PU-H71, a multimodal inhibitor of malignancy, induces complete responses in triple-negative breast cancer models. Proc Natl Acad Sci U S A. 2009;106:8368–8373. doi: 10.1073/pnas.0903392106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scaffidi C, et al. Two CD95 (APO-1/Fas) signaling pathways. EMBO J. 1998;17:1675–1687. doi: 10.1093/emboj/17.6.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Georgakis GV, et al. Inhibition of heat shock protein 90 function by 17-allylamino-17-demethoxy-geldanamycin in Hodgkin's lymphoma cells down-regulates Akt kinase, dephosphorylates extracellular signal-regulated kinase, and induces cell cycle arrest and cell death. Clin Cancer Res. 2006;12:584–590. doi: 10.1158/1078-0432.CCR-05-1194. [DOI] [PubMed] [Google Scholar]
- 32.Agard NJ, et al. Global kinetic analysis of proteolysis via quantitative targeted proteomics. Proc Natl Acad Sci U S A. 2012;109:1913–1918. doi: 10.1073/pnas.1117158109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christofferson DE, Li Y, Yuan J. Control of life-or-death decisions by RIP1 kinase. Annu Rev Physiol. 2014;76:129–150. doi: 10.1146/annurev-physiol-021113-170259. [DOI] [PubMed] [Google Scholar]
- 34.Degterev A, et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol. 2008;4:313–321. doi: 10.1038/nchembio.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feoktistova M, et al. cIAPs block Ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol Cell. 2011;43:449–463. doi: 10.1016/j.molcel.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wertz IE, Dixit VM. Signaling to NF-kappaB: regulation by ubiquitination. Cold Spring Harbor perspectives in biology. 2010;2:a003350. doi: 10.1101/cshperspect.a003350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moquin DM, McQuade T, Chan FK. CYLD deubiquitinates RIP1 in the TNFalpha-induced necrosome to facilitate kinase activation and programmed necrosis. PLoS One. 2013;8:e76841. doi: 10.1371/journal.pone.0076841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galban S, Duckett CS. XIAP as a ubiquitin ligase in cellular signaling. Cell death and differentiation. 2010;17:54–60. doi: 10.1038/cdd.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Y, Fang S, Jensen JP, Weissman AM, Ashwell JD. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science. 2000;288:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]
- 40.Kirkegaard T, et al. Hsp70 stabilizes lysosomes and reverts Niemann-Pick disease-associated lysosomal pathology. Nature. 2010;463:549–553. doi: 10.1038/nature08710. [DOI] [PubMed] [Google Scholar]
- 41.Li J, et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 2012;150:339–350. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci. 2005;6:11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- 43.Sabnis AJ, et al. Combined chemical-genetic approach identifies cytosolic HSP70 dependence in rhabdomyosarcoma. Proc Natl Acad Sci U S A. 2016;113:9015–9020. doi: 10.1073/pnas.1603883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taipale M, et al. Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell. 2012;150:987–1001. doi: 10.1016/j.cell.2012.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pratt WB, Morishima Y, Murphy M, Harrell M. Chaperoning of glucocorticoid receptors. Handb Exp Pharmacol. 2006:111–138. doi: 10.1007/3-540-29717-0_5. [DOI] [PubMed] [Google Scholar]
- 46.Schulte TW, Neckers LM. The benzoquinone ansamycin 17-allylamino-17-demethoxygeldanamycin binds to HSP90 and shares important biologic activities with geldanamycin. Cancer chemotherapy and pharmacology. 1998;42:273–279. doi: 10.1007/s002800050817. [DOI] [PubMed] [Google Scholar]
- 47.Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 48.Lewis J, et al. Disruption of hsp90 function results in degradation of the death domain kinase, receptor-interacting protein (RIP), and blockage of tumor necrosis factor-induced nuclear factor-kappaB activation. J Biol Chem. 2000;275:10519–10526. doi: 10.1074/jbc.275.14.10519. [DOI] [PubMed] [Google Scholar]
- 49.Vanden Berghe T, Kalai M, van Loo G, Declercq W, Vandenabeele P. Disruption of HSP90 function reverts tumor necrosis factor-induced necrosis to apoptosis. J Biol Chem. 2003;278:5622–5629. doi: 10.1074/jbc.M208925200. [DOI] [PubMed] [Google Scholar]
- 50.Powers MV, et al. Targeting HSP70: the second potentially druggable heat shock protein and molecular chaperone? Cell Cycle. 2010;9:1542–1550. doi: 10.4161/cc.9.8.11204. [DOI] [PubMed] [Google Scholar]
- 51.Rodina A, et al. Affinity purification probes of potential use to investigate the endogenous Hsp70 interactome in cancer. ACS Chem Biol. 2014;9:1698–1705. doi: 10.1021/cb500256u. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.