Reprogramming somatic cells to pluripotent stem cells (iPSCs) is now routinely accomplished by overexpression of the four Yamanaka factors (OCT4, SOX2, KLF4, MYC, also OSKM)1. These iPSCs can be derived from patients’ somatic cells and differentiated toward diverse fates, serving as a resource for basic and translational research. However, mechanistic insights into regulators and pathways that initiate the pluripotency network remain to be resolved. In particular, naturally-occurring molecules that activate endogenous OCT4 and replace exogenous OCT4 in human iPSC reprogramming have yet to be found. Using a heterokaryon reprogramming system we identified NKX3-1 as an early and transiently expressed homeobox transcription factor. Upon knockdown of NKX3-1, iPSC reprogramming is abrogated. NKX3-1 functions downstream of the IL6-STAT3 regulatory network to activate endogenous OCT4. Importantly, NKX3-1 substitutes for exogenous OCT4 to reprogram both mouse and human fibroblasts at comparable efficiencies and generate fully pluripotent stem cells. Our findings establish an essential role for NKX3-1, a prostate-specific tumor suppressor, in iPSC reprogramming.
Activation of the endogenous pluripotency program is a barrier to reprogramming2. One strategy to elucidate the mechanism is to replace key transcription factors such as OCT4 in the reprogramming cocktail. In murine reprogramming Oct4 has been replaced in multiple ways by (i) activating OCT4 target genes, such as with NR5A23 or artificial transcription factors4; (ii) inducing epigenetic changes at the Oct4 gene locus such as demethylation with Tet15 or addition of BrdU6; (iii) overexpressing Sall4 or inducing Sall4 expression with small molecule DZNep7; (iv) enhancing the mesenchymal-epithelial transition (MET) with E-cadherin overexpression8 or TGF-β inhibition (via small molecule9 or micro-RNAs10,11); (v) through the seesaw model, replacing Oct4 with endoderm lineage specifiers that repress the ectoderm lineage12.
In contrast to murine reprogramming, the only known factors that can replace OCT4 in human reprogramming are a synthetic fusion protein of the lineage specifier GATA3-VP16, albeit at three orders of magnitude lower reprogramming efficiency than OCT4,13 and two microRNA clusters (mir302/367 and mir200c/302/369)11. The lack of evidence for a naturally occurring transcription factor capable of directly activating OCT4 suggests the existence of unidentified factors critical for human iPSC formation.
We sought to discover early transient regulators with a role in reprogramming prior to endogenous OCT4 activation. The heterogeneity of asynchronously reprogramming cell populations inherent in iPSC reprogramming impedes detection of critical early factors. Therefore, we used the efficient heterokaryon reprogramming system to generate a molecular map of early reprogramming using RNA-seq and ATAC-seq. Heterokaryons were generated through fusion of human fibroblasts with mouse embryonic stem cells (ESCs)14,15. Our results reveal a previously unrecognized role for NKX3-1 as an activator of endogenous OCT4 and replacement factor for exogenous oncogenic OCT4 in mouse and human reprogramming to pluripotency.
We postulated that heterokaryon reprogramming could reveal early regulators of reprogramming that are transiently expressed and would be missed during heterogeneous iPSC generation. Our previous heterokaryon analyses revealed that key pluripotency genes such as OCT4 and NANOG are induced by day 1, the earliest time-point we assayed15. To identify pluripotency regulators induced prior to these later markers of reprogramming, we conducted an RNA-seq time course with six time points in the first 24h (0.5h, 1h, 2h, 6h, 12h, and 24h) using human fibroblasts and fibroblasts co-cultured with mouse ESCs (co-cul) as a control for changes due to autocrine or paracrine effects (Supplementary Fig. 1a,b). The H1 ESC line from ENCODE was used as a reference human ESC transcriptome. Transcript abundance of human and mouse reads were accurately mapped using a concatenated mouse-human transcriptome, as previously described15.
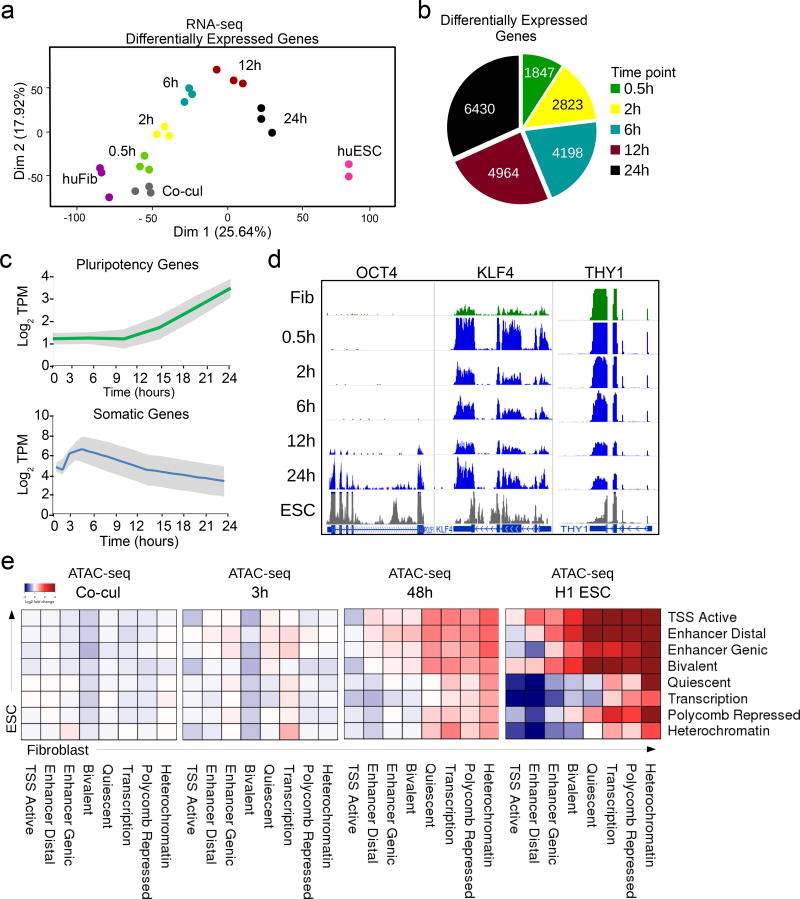
Principal component analysis of these time points revealed that heterokaryon reprogramming proceeds in a defined trajectory towards the human ESC state with high reproducibility (Fig. 1a). It was characterized by a progressive increase in the number of differentially expressed (DE) genes reaching 6430 genes by 24h (Fig. 1b, Supplementary Table 1). Specifically, upregulation of pluripotency and downregulation of fibroblast genes was detectable within the first few hours, as measured by aggregating gene expression of 331 pluripotency-associated genes and 597 fibroblast-associated genes defined by the MSigDB (Fig. 1c). For instance, KLF4 expression preceded OCT4 expression and was accompanied by a downregulation of THY1 (Fig. 1d), consistent with previous reports1. Hierarchical clustering revealed five major gene trajectories, including several transient gene expression patterns, and upregulation of a gene signature characteristic of embryonic development as fibroblast reprogramming progressed (Supplementary Fig. 1b, Supplementary Fig. 2a). Our previous RNA-seq analysis indicated that 75% of the top three categories of human ESC signature genes are upregulated in heterokaryons by day two post-fusion16, further supporting the robustness of heterokaryon reprogramming towards a human ESC fate (Supplementary Table 2).
Figure 1. Nuclear reprogramming of human fibroblasts after fusion with mouse ESCs.
(a) Principal component analysis plot of RNA-seq data shows a continuous trajectory of transcriptome changes in human fibroblasts towards human ESCs over time post-fusion in heterokaryons (n= 3 biological replicates). (b) Pie chart depicting the number of differentially expressed (DE) human genes at each time point during early reprogramming. (c) Line plot showing average RNA expression during reprogramming from 331 somatic and 597 embryonic associated genes taken from the molecular signature database (MSigDB). Gray shades represent the standard deviation from the median. (d) RNA-seq tracks for human OCT4, KLF4, and THY1 genes over the time course of heterokaryon reprogramming. (n= 3 biological replicates). (e) Chromatin state transition matrix. Using chromatin states mapped and defined by the Roadmap Epigenomics Consortium, the heatmap represents the transition from human fibroblast (columns) to embryonic stem cell (rows). The color spectrum from blue to red represents the log-fold change of the median ATAC-seq peak signal of each transition at each time point relative to time 0 (fibroblast alone).
At 12 hours, we observed that among the core pluripotency network, human OCT4 was the first to show signs of transcriptional activation (Fig. 1d). To identify the earliest regulators that functionally impact the dynamics of reprogramming at an epigenetic level, we performed ATAC-seq on heterokaryons undergoing reprogramming at 3h and 48h post-fusion as well as in human fibroblasts and co-cul as controls. ATAC-seq data from the H1 ESC line was used as a reference17. We observed that 49 out of 56 (87.5%) possible transitions from a fibroblast to an ESC chromatin state are mirrored by ATAC-seq in heterokaryons by 48h (Fig. 1e). These changes in accessibility attest to the overall efficacy of heterokaryon as a reprogramming system in this short time period. Although median peak accessibility is not different by 3h (Fig. 1e) a subset of genomic regions exhibits a much higher level of chromatin accessibility than starting fibroblasts (Fig. 2a). We hypothesized these could contain motifs of early regulators not evident in heterogeneous iPSC reprogramming. We employed motif-enrichment analysis, using fibroblast accessible regions as a control for the baseline, to identify factors contributing to early reprogramming (Fig. 2b).
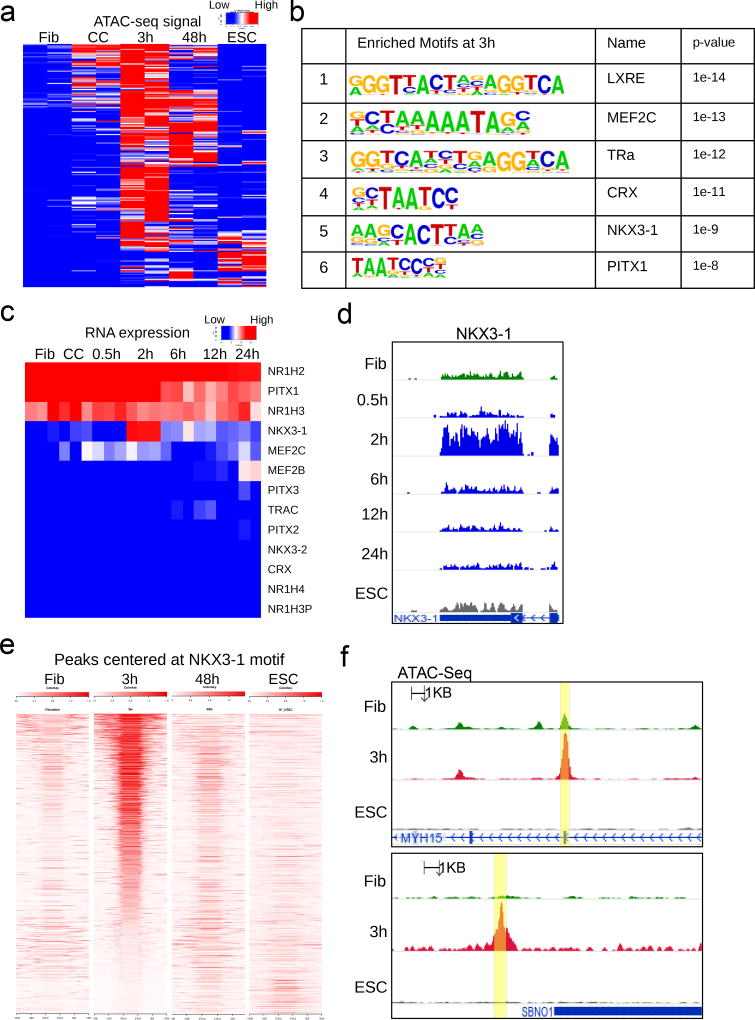
Figure 2. Motifs at early accessible chromatin and gene expression dynamics identify NKX3-1.
(a) Heatmap of ATAC-seq signal at genomic regions that become more accessible at 3h post-fusion heterokaryons (mouse ESC × human fibroblast) compared to unfused human fibroblast. Signals from those same regions are also shown for human fibroblast, human fibroblast from mouse ESC co-culture, 48h post-fusion, and human ESC as a reference. (b) Motif enrichment at 3h peaks that increased in accessibility, using fibroblast peaks as background. Motif enrichment was calculated with the cumulative binomial distribution at 3h peaks that increased in accessibility, using fibroblast as background. P-value is not adjusted for multiple testing. Benjamini-adjusted q-value calculated by Homer software is reported as 0.0000 for all motifs shown. (n = 53043 target sequences) (c) Expression by RNA-seq of candidate genes obtained from motif enrichment in 2b and their close family members. (d) RNA-seq signal track at the human NKX3-1 locus over the time course of heterokaryon reprogramming. (n= 3 biological replicates). (e) Density heatmap of ATAC-seq signal at 1705 genomic regions, centered at the NKX3-1 motif and ranked by accessibility at three hours. (f) ATAC-seq tracks of two representative peaks containing the NKX3-1 motif shown in 2e (yellow highlights). (n= 3 biological replicates).
To reduce our candidate list based on motif enrichment, we examined the expression of the top hits and their closely-related family members in our RNA-seq time course (Fig. 2c). NKX3-1 exhibited a transient expression pattern at 2h, while other transcription factors were either not expressed, expressed at very low levels, or did not change over the time course (Fig. 2c,d). We found that at 3h, differential ATAC-seq peaks centered on the NKX3-1 motif (Fig. 2e). These peaks of accessibility were greatly diminished in late stage 48h heterokaryons and end stage embryonic cells, suggesting that NKX3-1 expression and activity is transient during reprogramming (Fig. 2e,f).
The role of Nkx3-1 in iPSC reprogramming was tested with a loss of function experiment using three shRNAs, each targeting a different site on the Nkx3-1 messenger RNA, including the 3’UTR. Acute loss of Nkx3-1 after transduction of mouse embryonic fibroblasts (MEFs) with OSKM resulted in reduced colony formation, suggesting a critical role for Nkx3-1 in reprogramming (Fig. 3a). Importantly, the paucity of iPSC colonies generated by shNkx3-1 transduced cells was not due to cell death or growth inhibition, as the cells transduced with shNKX3-1 exhibited comparable proliferation rates and frequencies of apoptosis to control cells transduced with a scrambled shRNA (Fig. 3b,c, Supplementary Fig. 3a,b).
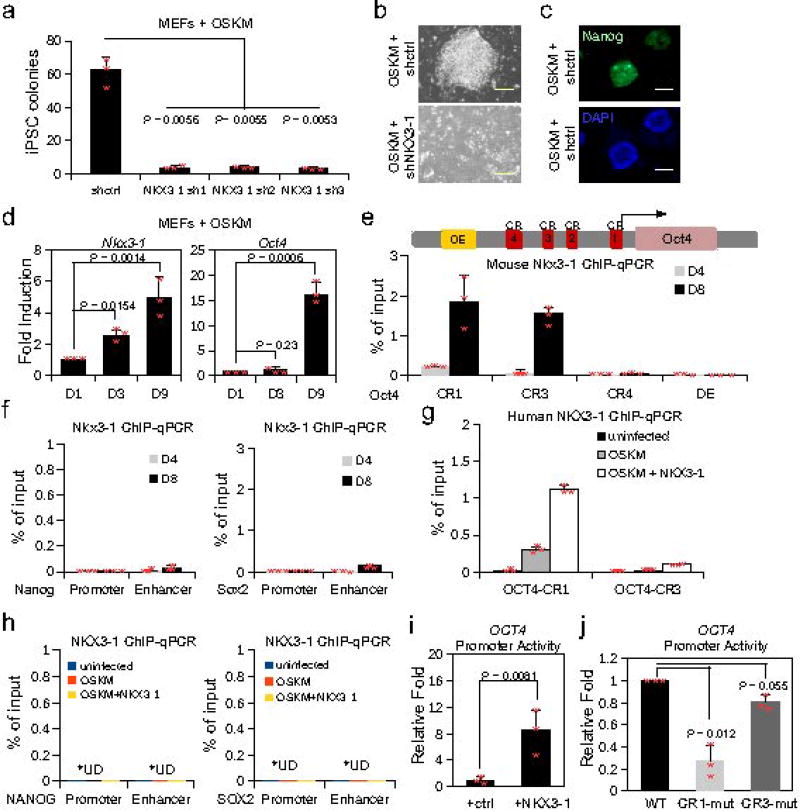
Figure 3. NKX3-1 induces Oct4 expression and is required for iPSC formation.
(a) iPSC colonies of MEFs transduced with OSKM and shcontrol (shctrl) or shNKX3-1, counted 10 days post transduction (3 biological replicates). (b) Phase contrast images of MEFs transduced with OSKM and shctrl or shNKX3-1 at day 10 post transduction (3 biological replicates). (c) Staining for NANOG+ iPSC colonies. Bar corresponds to 100 µm. (3 biological replicates). (d) Fold induction of Nkx3-1 and Oct4 expression at days 3 and 9 of iPSC reprogramming, relative to day 1 post OSKM transduction (n=3 biological replicates). (e) ChIP-qPCR of NKX3-1 in MEFs at the Oct4 conserved regulatory regions (CR1, CR3, and CR4) and distal enhancer (DE). ChIP was performed at day 4 (D4) and 8 (D8) in the presence of IL6 (n=3 biological replicates). (f) ChIP-qPCR of NKX3-1 in MEFs at the Nanog and Sox2 conserved regulatory regions. ChIP was performed on day 4 (D4) and 8 (D8) in the presence of IL6 (n=3 biological replicates). (g) ChIP-qPCR of NKX3-1 in human fibroblasts at the OCT4 conserved regulatory regions (CR1 and CR3) under indicated conditions. ChIP was performed on day 10 (n=3 biological replicates). (h) ChIP-qPCR of NKX3-1 in human fibroblasts at the NANOG and SOX2 regulatory regions under indicated conditions. ChIP was performed on day 10 (n=3 biological replicates). U.D. = undetected. (i) A plasmid containing the WT 5kb of the human OCT4 promoter driving luciferase was transfected into human fibroblasts, and was either co-transfected with a plasmid encoding NKX3-1 or a control plasmid. Luciferase activity 48h post transfection, normalized to control cells (n=3 biological replicates). (j) A plasmid containing the WT 5kb of the human OCT4 promoter, a human OCT4 promoter with a mutated NKX3-1 motif at the conserved region 1 (CR1), and a human OCT4 promoter with a mutated NKX3-1 motif at the CR3 driving luciferase was transfected into human fibroblasts. Luciferase activity 48h post transfection, normalized to control cells (n = 3 biological replicates). Unpaired Student’s t-test was used, data represent mean ± s.d. Statistical source data for a, d–j in Supplementary Table 5.
We hypothesized that NKX3-1 could function at least in part by inducing the expression of critical pluripotency genes by binding to their regulatory elements during reprogramming. We detected Nkx3-1 mRNA at day 9 of iPSC formation, but not in mature iPSC, consistent with its transient expression in heterokaryon reprogramming (Fig. 3d). Notably, Nkx3-1 expression preceded Oct4 activation suggesting Nkx3-1 is upstream of Oct4. We identified NKX3-1 motifs at the enhancers of pluripotency genes, defined by chromHMM18, including at the Oct4, Sox2, and Nanog loci. By ChIP-qPCR, we detected significant NKX3-1 binding at the Oct4 promoter conserved regions (CR1 and CR3), shared by mouse and human, but only minimal binding at the Sox2 and Nanog loci in MEFs transduced with OSKM (Fig. 3e,f)19,20. NKX3-1 occupancy at the Oct4 promoter was detectable at day 4 of OSKM reprogramming and further enriched at day 8 (Fig. 3e), correlating with endogenous Oct4 expression (Fig. 3d). In agreement with results in mouse cells, NKX3-1 occupancy was also detected at CR1 and CR3 of the human OCT4 promoter and negligible at SOX2 and NANOG in human fibroblasts undergoing iPSC reprogramming (Fig. 3g,h). Further, ectopic expression of NKX3-1 in human fibroblasts increased luciferase reporter activity driven by the human OCT4 promoter, suggesting NKX3-1 directly activates OCT4 (Fig. 3i). Moreover, when the NKX3-1 motif was mutated at CR1, there was a significant loss of OCT4 promoter activity, providing further evidence that NKX3-1 regulates OCT4 expression (Fig. 3j).
Since NKX3-1 binds to and activates the OCT4 promoter during reprogramming, we investigated whether it is sufficient to replace OCT4 in the reprogramming cocktail. We transduced MEFs and human fibroblasts with lentiviral vectors encoding either NKX3-1, SOX2 and KLF4 (NSK) or OCT4, SOX2, and KLF4 (OSK) as a positive control. We omitted MYC from the cocktail as it is dispensable for reprogramming21. We observed a comparable frequency of NANOG+ iPSC colonies in NSK and OSK transduced mouse or human fibroblasts after 12 and 28 days, respectively (Fig. 4a,b). By contrast, colony formation was greatly reduced in NK and NS conditions (Supplementary Fig. 4a), indicating NKX3-1 is only able to substitute for OCT4 in the reprogramming cocktail, consistent with our ChIP-qPCR data. These experiments were performed with constitutive NKX3-1 expression. A prediction from our heterokaryon reprogramming time course is that NKX3-1 is only required briefly at the onset of reprogramming. To test whether transient expression of NKX3-1 is sufficient to replace OCT4 in the reprogramming cocktail, transient NKX3-1 expression was induced by transfecting an NKX3-1 plasmid in lieu of lentiviral NKX3-1 into fibroblasts. As plasmids are not integrated into the genome, NKX3-1 expression is lost in the course of cell division. Although one round of transfection yielded minimal iPSC colonies, a second dose of NKX3-1 plasmid was capable of generating nearly comparable numbers of iPSC colonies as stably expressed NKX3-1 (Fig. 4c).
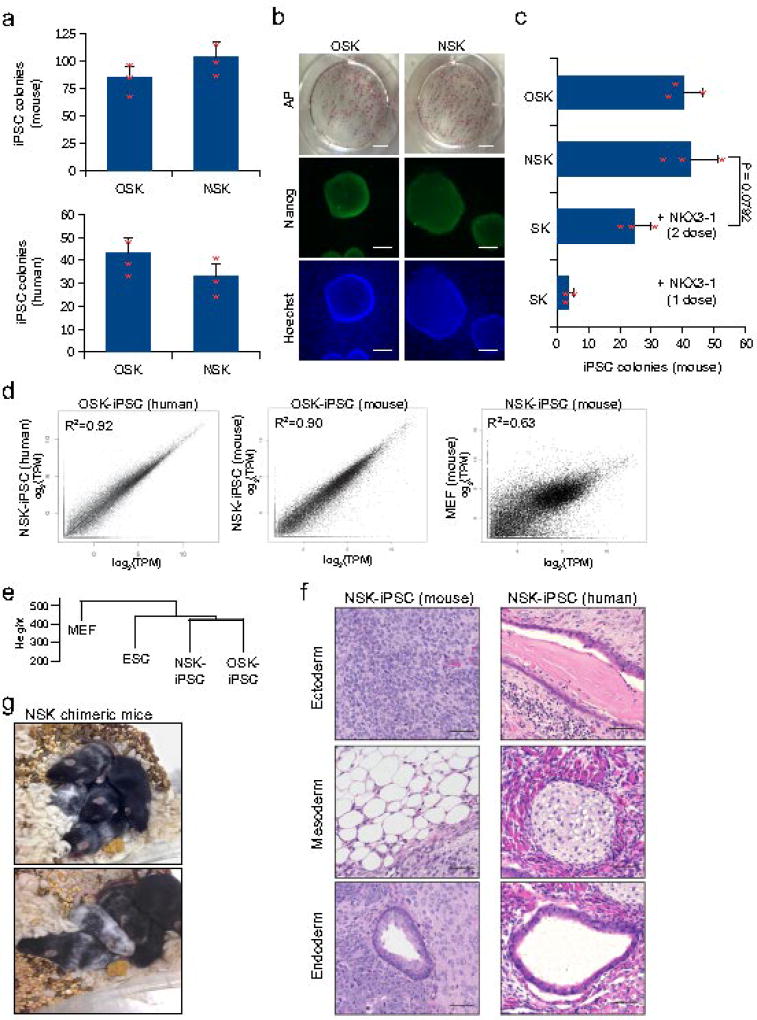
Figure 4. NKX3-1 can substitute for OCT4 to generate pluripotent mouse and human iPSCs.
(a) Histogram plot shows number of iPSC colonies formed when OCT4 is replaced with NKX3-1 in the reprogramming cocktails of mouse and human. NANOG+ iPSC colonies were counted at day 10 (mouse) and day 23 (human) after transduction with NSK or OSK cocktails (n = 3 biological replicates). Unpaired Student’s t-test was used and data represents mean ± s.d. (b) Representative immunofluorescence images of AP, NANOG, and Hoechst staining of human OSK- and NSK-derived iPSC colonies. AP image bar corresponds to 50mm. Nanog and Hoechst image bar corresponds to 100 µm. (3 biological replicates with similar results). (c) Histogram plot showing number of iPSC colonies formed when an NKX3-1 plasmid was transfected into MEFs constitutively expressing SOX2 and KLF4, MEFs constitutively expressing NKX3-1, SOX2, and KLF4, and MEFs constitutively expressing OCT4, SOX2, KLF4. iPSC colonies were counted at day 10 after transduction (n = 3 biological replicates). Unpaired Student’s t-test was used and data represents mean ± s.d. (d) Scatterplot of gene-wise comparison of the entire transcriptome of OSK- and NSK-derived iPSC in mouse and human. (n = 45798 transcripts) (e) Hierarchical clustering of the entire transcriptome of NSK-derived iPSCs, OSK-derived iPSCs, ESCs, and MEFs. (f) Histological sections of teratomas of mouse and human NSK-induced iPSCs were stained with haematoxylin and eosin (left, mouse: neuroectoderm (top), adipose tissue (middle), pseudostratified ciliated epithelium29 (bottom) and right, human: keratinized epithelium (top), cartilage tissue (middle) and pseudostratified ciliated epithelium (bottom)). Teratomas were surgically removed after 2 weeks (mouse iPSC) or 4 weeks (human iPSC). Tissues were fixed in formalin at 4 °C, embedded in paraffin wax, and sectioned at a thickness of 4 µm. Sections were stained with haematoxylin and eosin. Bar corresponds to 50 µm. (2 biological replicates with similar results). (g) Images show two litters of chimeric mice each derived from two separate C57B6 derived NSK- iPSC clones injected into C57B6 albino blastocysts. (3 biological replicates with similar results). Statistical source data and exact P values for c can be found in Supplementary Table 5.
To measure whether NSK-derived colonies are similar to more traditional OSK-derived iPSCs, we compared the transcriptional profiles between the iPSC clones by RNA-seq. Gene pair-wise comparisons between NSK-derived and OSK-derived iPSCs revealed similar transcriptome profiles in both mouse and human cells (Fig. 4d). Additionally, mouse and human NSK-derived and OSK-derived iPSCs displayed similar expression levels of OCT4, even after extensive passaging, indicating NSK-derived iPSC are stably pluripotent (Supplementary Fig. 4b,c). NKX3-1 expression in both NSK-derived and OSK-derived iPSCs was low, consistent with transgene silencing and a hallmark of successful iPSC reprogramming (Supplementary Fig. 4b). Hierarchical clustering of NSK-derived iPSCs, OSK-derived iPSCs, ESCs, and MEFs revealed NSK-derived iPSCs cluster closest to OSK-derived iPSCs (Fig. 4e). Further, the pattern of expression of pluripotency genes by NSK-derived iPSCs mirrored that of OSK-derived iPSCs in both mouse and human cells (Supplementary Fig. 4d,e). Teratoma formation assays of mouse and human NSK-derived iPSCs yielded tumors containing cells representative of all three germ layers, indicating NSK-derived iPSCs are pluripotent (Fig. 4f). Injection of NSK-derived iPSCs from C57B6 MEFs into C57B6 albino blastocysts resulted in chimeric mice, evident from their mostly black coat color, further demonstrating that NSK-derived iPSCs are fully pluripotent (Fig. 4g).
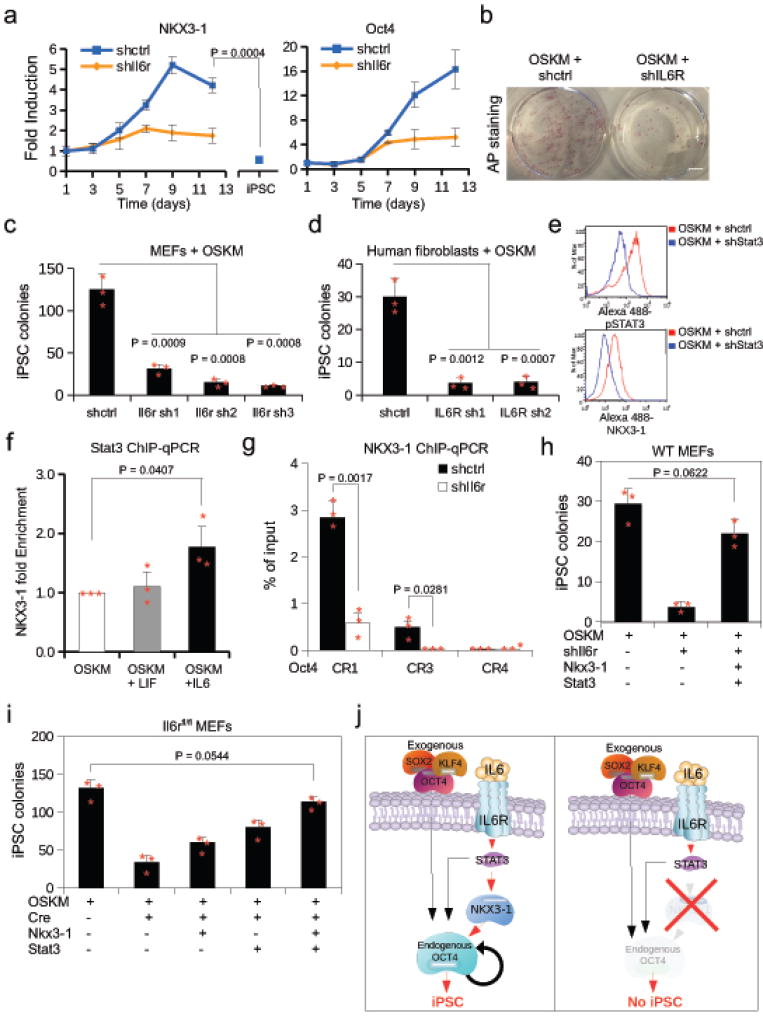
We noted that the expression kinetics of NKX3-1 mirror that of the IL6R during heterokaryon reprogramming, transiently peaking at 2h post-fusion (Supplementary Fig. 5a). Consistent with this finding, genes associated with canonical IL6-signaling including MYC and STAT3 are among the DE genes (2h and 6h, respectively) (Supplementary Table 3). To directly test the role of IL6 signaling in heterokaryon reprogramming, we knocked-down the IL6R and found NKX3-1 activation is impaired at 2h post-fusion (Supplementary Fig. 5b). To determine its role in iPSC reprogramming, shRNA-mediated knock-down of Il6r in MEFs resulted in blunted Nkx3-1 expression, suggesting that Nkx3-1 activation is downstream of Il6-signaling in iPSC reprogramming (Fig. 5a). Strikingly, knock-down of Il6r resulted in a significant reduction in iPSC colony formation in both mouse and human fibroblasts transduced with OSKM (Fig. 5b,c,d, Supplementary Fig. 5c), suggesting that IL6R signaling is essential to iPSC reprogramming.
Figure 5. NKX3-1 functions downstream of the IL6-STAT3 cascade during iPSC induction.
(a) Expression time course in MEFs transduced with OSKM of Nkx3-1 and Oct4 expression after shIl6r or shctrl by qRT-PCR relative to day 1. Values were normalized to Gapdh. (n= 3 biological replicates). Unpaired Student’s t-test was used and data represents mean ± s.d. (b) Cultures of MEFs stained for alkaline phosphatase 10 days after transduction with shctrl or shIl6r plus OSKM. (3 biological replicates). (c and d) NANOG+ iPSC colony count from reprogrammed mouse embryonic fibroblasts (MEFs) or human BJ fibroblasts transduced with shRNAs targeting Il6r/IL6R or a control shRNA (n = 3 biological replicates). Unpaired Student’s t-test was used and data represents mean ± s.d. (e) Protein expression of p-STAT3 and NKX3-1 in MEFs transduced with OSKM and shRNA to Stat3 (shStat3) or a scramble control (shctrl) by intracellular flow cytometry. (n= 2 biological replicates with similar results). (f) STAT3 ChIP-qPCR at the Nkx3-1 promoter in MEFs treated with IL6 or LIF (n = 3 biological replicates). Unpaired Student’s t-test was used and data represents mean ± s.d. (g) ChIP-qPCR of NKX3-1 at the Oct4 promoter conserved regions CR1, CR3, and CR4 in which Il6r was knocked-down at day 8 post OSKM infection (n = 3 biological replicates). Unpaired Student’s t-test was used and data represents mean ± s.d. (h) MEFs transduced with various combinations of plasmids as indicated by + symbol. Colonies were counted 10 days post OSKM transduction (n = 3 biological replicates). Unpaired Student’s t-test was used and data represents mean ± s.d. (i) Il6rfl/fl MEFs were transduced with various plasmids as indicated by + symbol. Colonies were counted 10 days post OSKM transduction. (n = 3 biological replicates). Unpaired Student’s t-test was used and data represents mean ± s.d. (j) Schematic of function of NKX3-1 in the IL6-STAT3 signaling pathway during reprogramming to iPSC. Statistical source data and exact P values for a, c–d, and f–i can be found in Supplementary Table 5.
As STAT3 is downstream of IL6R, we investigated whether Nkx3-1 induction is STAT3-dependent by performing a knock-down of Stat3 expression by shRNA in MEFs transduced with OSKM. In the presence of Stat3 shRNA, NKX3-1 protein levels were markedly decreased at the single cell level as measured by flow cytometry (Fig. 5e). To determine whether STAT3 binds to the Nkx3-1 locus, we performed ChIP-qPCR in MEFs undergoing reprogramming and found that STAT3 occupancy is significantly increased after addition of IL6 cytokine, but not its GP130 family member LIF, in MEFs transduced with OSKM (Fig. 5f). These results suggest NKX3-1 functions downstream of the IL6-Stat3 signaling pathway during iPSC induction. Moreover, knock-down of Il6r by shRNA in MEFs undergoing reprogramming resulted in impaired NKX3-1 binding at the Oct4 CR1 and CR3 regulatory regions, suggesting an IL6-STAT3-NKX3-1-OCT4 signaling cascade (Fig. 5g).
Since Nkx3-1 expression is dependent on IL6R, we reasoned that NKX3-1 might rescue iPSC reprogramming in MEFs in which IL6R was knocked down. As STAT3 binds to the Oct4 locus in ESCs22, we hypothesized that NKX3-1 and STAT3 could cooperatively activate Oct4 during reprogramming. Indeed, when NKX3-1 and STAT3 were co-expressed, colony formation upon knock-down of Il6r mRNA by shRNA was rescued (Fig. 5h). To determine more precisely whether NKX3-1 could rescue reprogramming in cells lacking IL6R signal transduction, we genetically ablated the receptor by Cre-mediated excision of the Il6r in Il6rfl/fl MEFs. In the absence of Il6r, overexpression of NKX3-1 alone rescued iPSC colony formation (Fig. 5i). In addition, STAT3 overexpression alone also rescued iPSC reprogramming in the absence of IL6 signaling, in accordance with its role in activating Nkx3-1 (Fig. 5i). In the absence of Il6r, rescue of colony formation was complete upon co-expression of STAT3 and NKX3-1, indicating that the two are additive and suggesting that STAT3 has a dual function in activating Nkx3-1 and Oct4 (Fig. 5i). The residual iPSC colonies formed after Cre-mediated excision of Il6r are likely due to incomplete floxing, as suggested by the persistence of low level Nkx3-1 expression (Supplementary Fig. 5e). These data demonstrate that signaling via IL6R is essential to iPSC reprogramming. Further, they implicate Nkx3-1 as a target gene of STAT3 that can rescue Il6r deficiency during iPSC formation. Together our data highlight a previously unrecognized role for NKX3-1 in mouse and human iPSC reprogramming, and describe an IL6-STAT3-NKX3-1-OCT4 signaling cascade critical for the generation of iPSC (Fig. 5j).
We report the unexpected discovery of NKX3-1 as a reprogramming factor. NKX3-1 motif is enriched at accessible genomic regions during early heterokaryon reprogramming and largely absent in the final ESC state, suggesting that NKX3-1 target sites are genomic elements characteristic of an early epigenetic transition. Human NKX3-1 has a striking early transient expression profile in heterokaryons, supporting the notion that the cell fusion system constitutes a potent discovery tool for transient regulators. Heterokaryon studies have elucidated different facets of reprogramming such as (1) IL6-mediated regulation of Pim 1, a serine-threonine kinase that promotes survival15, and (2) activation-induced cytidine deaminase (AID), known primarily for its role in the generation of antibody diversity, as an active DNA demethylase crucial to both heterokaryon and iPSC reprogramming14. Notably in this study, we demonstrate that heterokaryon findings are translatable not only to mouse but also human reprogramming.
Previous work established NKX3-1 as a prostate specific tumor suppressor23,24. NKX3-1 is also associated with self-renewal of luminal cells in the prostate25 and knockdown of NKX3-1 in prostate cancer stem cells impedes their self-renewal capacity26,27, suggesting that NKX3-1 may be a “stemness” gene in multiple contexts. Nkx3-1 is expressed at day 6.5 in the mouse embryo, suggesting a role in early development28. Together, these findings highlight a previously unrecognized role of NKX3-1 in reprogramming and expand avenues by which activation of Oct4 and pluripotency can be achieved.
We demonstrate that NKX3-1 acts downstream of the IL6-Stat3 signaling cascade during OSKM reprogramming, and implicate Nkx3-1 as a key target of STAT3. IL6R is essential to OSKM reprogramming in mouse and human cells, and in its absence reprogramming is abrogated. Senescence associated secretion of IL6 has been implicated during in vivo reprogramming29, consistent with our results, which demonstrate a requirement for IL6 signaling. This suggests that a fibroblast subpopulation undergoing senescence may be the source of IL6 during iPSC formation. Work on IL6R trans-signaling suggests a mechanism whereby a subpopulation of IL6R expressing cells shed the receptor to create a soluble form that complexes with IL6, providing IL6 signaling to its IL6R negative neighbors30. In our experiments, a partial knockdown of Il6r is sufficient to block iPSC colony formation (Fig. 5c,d, Supplementary Fig. 5c), suggesting that trans-IL6-signaling is not the primary mechanism, but instead IL6 signaling acts through membrane-bound IL6 receptors.
We show that constitutively expressed NKX3-1 can replace OCT4 in the OSKM cocktail, suggesting that NKX3-1 plays a role in reprogramming at least partly through activation of endogenous OCT4. Importantly, as predicted from our heterokaryon time course, transient expression of NKX3-1 is sufficient to replace OCT4 in iPSC reprogramming. We demonstrate that NSK-derived iPSCs produce all three germ layers and contribute to the generation of chimeric mice, evidence of pluripotency. However, germline transmission and/or tetraploid complementation data would be definitive. We provide evidence that mechanistic insights from heterokaryon reprogramming can be translated to iPSC reprogramming. Reprogramming efficiencies of the NSK cocktail are very similar to the OSK cocktail in both mouse and human fibroblasts, making this the first discovery of a naturally occurring protein capable of replacing OCT4 during reprogramming to pluripotency in human cells.
As a tumor suppressor, NKX3-1 replacement of the oncogene OCT4 during iPSC induction may have broader implications in reprogramming. In the absence of NKX3-1, cells exposed to UV or γ-irradiation retain γH2AX-positive puncta in the nucleus for a longer period of time31. This DNA damage response upregulates p16INK4a and p21CIP1, leading to senescence and the impairment of successful reprogramming32, whereas promoting DNA repair leads to more efficient reprogramming33. NKX3-1 may thus maintain genetic integrity during reprogramming, although direct evidence is lacking. This work provides a paradigm for identifying non-oncogenic reprogramming factors which may be preferred for clinical applications to circumvent detrimental effects such as spontaneous reactivation of OCT4. NKX3-1 also acts as a transcriptional repressor23 and down-regulates TWIST1, an epithelial-mesenchymal transition (EMT) associated gene, in the prostate tumor-derived LNCaP cell line34. Inhibition of EMT genes is critical for MET to occur35, an important event in fibroblast reprogramming. Consistent with these findings, overexpression of NKX3-1 in fibroblasts results in the activation of MET genes and down-regulation of EMT genes (Supplementary Fig. 5f,g,h). This suggests NKX3-1 may play a dual role by activating the endogenous locus of the pluripotency master regulator OCT4 and simultaneously allowing for the MET transition to occur by repressing EMT-associated genes.
Methods
Heterokaryon generation and isolation
Heterokaryons were generated and isolated as previously described15. Briefly, 500,000 human MRC5 fibroblast (ATCC® CCL171™) and 3×106 GFP mouse ESC were co-cultured overnight in stem cell medium. After fusion with PEG (Roche), heterokaryons were isolated by FACS using BD FACS ARIA and suspended in RLT buffer.
ChIP
Cells were crosslinked in 1% formadehyde for 15 min, with constant agitation, and quenched with 150mM glycine for 5 min. Micrococcal nuclease (New England Biolabs, Inc.) was used to fragment chromatin to a range of 200–400 bp. Chromatin was incubated with indicated antibody overnight at 4°C with constant agitation. Protein A Dynabeads was incubated with antibody-protein complexes for 2 hr at 4°C with constant agitation. After decrosslinking, DNA was isolated using a MinElute column (Qiagen). See Table S4 for primer sequence. For ChIP-qPCR, Sybr Green Taq 2× Master Mix (4309155, Life Technologies) was used to amplify DNA fragments.
RNA-seq library construction
Library samples were prepared as previously described15. Briefly, Poly-A+ RNA was isolated from total RNA using oligo-dT magnetic beads (Illumina TruSeq V2). Following first (Superscript III, Life Technologies) and second strand synthesis, cDNA libraries were constructed according to standard Illumina protocols. RNA and DNA integrity and quality was measured using an Agilent Bioanalyzer 2100. High throughput sequencing was performed using the Illumina HiSeq 2000. At least 25 million 50 bp reads were obtained for each sample.
RNA-seq mapping and gene expression quantification
RNA-seq reads were mapped to a concatenated genome sequence of human GRCh38/hg38 and mouse GRCm38/mm10 annotations using STAR/2.4.2a36. Following mapping, human and mouse transcript abundance were individually quantified using rsem/1.2.21. Expression levels were normalized by transcripts per million (TPM).
PCA and hierarchical clustering
The DESeq2 package in R3.1.1 was used to identify DE genes. Heatmaps were created using the heatmap.2 function in the gplots package in R3.1.1. PCA was performed across DE genes among all samples in the FactoMineR package in R3.1.1.
ATAC-seq library generation and analysis
ATAC-seq libraries were prepared as described17. Briefly from 20–25,000 sorted heterokaryons or fibroblasts (for co-culture). Reads were mapped to a concatenated human plus mouse genome with Bowtie 2.0 and reads originating from the mitochondria and those with low mapping quality scores (below 10) were removed. For motif enrichment, regions of increased accessibility at 3 hours versus fibroblast were identified with bdgdiff command in the MACS 2.0 package, and then were used as input into Homer 4.7 with human fibroblast peaks as background. For figure 2E, MACS 2.0 was used to call peaks; DESEQ2 used tag counts per peak genome-wide to identify regions of increased accessibility at 3 hours, and Homer 4.7 software was used with the findMotifsGenome.pl software to peaks containing the NKX3-1 motif (CisBP). Chromatin state transitions were mapped by using peaks were at least 30% of the peak overlapped a chromatin state as defined by Roadmap Epigenomics Consortium37. The 18-state map was condensed in the following way: States 1–4 were merged as TSS Active, states 7 and 8 were merged as Enhancer Genic, states 9 and 10 were Enhancer Distal, states 14 and 15 were Bivalent, states 16 and 17 were Polycomb Repressed. States 6, 11, and 12 were removed for simplicity. The starting cell type for the transition matrix was IMR90 and the ending was H1 ES cell. DESEQ2 was used to generate log2fold change of ATAC-seq signal at each time point relative to time 0 (unfused MRC-5 fibroblasts). For each transition (for example Enhancer Distal to Heterochromatin) at each time point (for example 3hr vs fibroblast), the median log2fold change of the overlapping peaks was plotted according to the color scale.
Virus production
For virus production, 5×106 HEK293T cells were seeded into 10 cm dishes and transfected with the vector of interest and appropriate packaging plasmids. Medium was changed 24 h later. Supernatants were collected at 48h passed through a 45 µm filter and concentrated by adding ⅓ volume of Lenti-X Concentrator solution (Clonetech). The solution was incubated at 4°C for 24h, then spun down for 45 mins and resuspended as 40X concentrate. Concentrated virus was aliquoted and stored at −80 °C until use.
iPSC generation and propagation
60,000 MEFs or 60,000 BJ fibroblast (ATCC® CRL2522™) were seeded into a 6-well plate. The next day, cells were transduced using concentrated viral supernatant (Lenti-X concentrator, Clonetech, PT4421-2). 2i was added two days post-transduction and MEF medium (DMEM, 20% FBS, sodium pyruvate, non-essential amino acids (NEAA), β-mercaptoethanol, and penstrep antibiotic) was changed daily. iPS colonies were scored on day 10 based on Nanog positivity. For human iPSCs, OSKM-transduced cells were re-seeded on mitomycin-treated feeders on day 6 post-transduction. To propagate clones, single colonies were picked and expanded into mitomycin-treated feeders and expanded for at least four passages. Mouse iPSCs were cultured in knock-out (KO) DMEM, 15 % KO serum (KSR), LIF, NEAA, L-glutamine, β-mercaptoethanol, and penstrep antibiotic. Human iPSCs were cultured in KO DMEM, 20% KSR, 10% pluriton conditioned media, NEAA, L-glutamine, FGF, β-mercaptoethanol, and penstrep antibiotic.
Immunofluorescence
Cells were fixed in 4% paraformadehyde and stained with antibodies as indicated. Images were acquired using an epifluorescent microscope (Axioplan2, Carl Zeiss MicroImaging, Inc.), Fluar ×20/0.75 or ×40/0.90 objective lens, and a digital camera (ORCA-ER C4742-95, Hamamatsu Photonics). The software used for acquisition was OpenLab 4.0.2 (Improvision).
AP staining
Alkaline phosphatase detection was performed according to protocols in the Alkaline Phosphatase Staining Kit II (Stemgent, 00-0055).
Luciferase Assay
OCT4-luciferase reporter plasmid was transfected into human MRC5 fibroblasts. After 48h, cells were lysed and luciferin was added using the Pierce Firefly Luciferase Glow Assay Kit (Thermo Fisher Scientific). Luminescence was measured using Tecan Infinite M1000 PRO plate reader. Site-directed mutagenesis was performed to mutate the NKX3-1 motifs at CR1 and CR3 of the OCT4-promoter region using the primers listed in Supplementary Table 4.
Real-time-PCR
cDNA synthesis and real-time PCR was performed as previously described (Brady et al., 2013). Briefly, cDNA was synthesized using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, #4368814). Real-time PCR was performed using an ABI 7900HT Real-time PCR system using SYBR Green Master Mix (Applied Biosystems, #4309155). Data are presented as the mean ± s.e.m. Comparisons between groups used the Student’s t-test assuming two-tailed distributions.
Statistics and Reproducibility
All in vitro iPSC reprogramming related experiments were repeated independently at least three times with similar results. All in vivo experiments were repeated independently at least two times with similar results. All graphs show mean values with error bars signifying standard deviation (s.d.) except for Fig. 1c. Centre is plotted as median. Exact P-values for each experiment are provided in Supplementary Table 5. Two-tailed Student’s t-test was performed for Figs. (3a, 3d, 3i, 3j, 4c, 5a, 5c, 5d, 5f–5i), and Supplementary Figs. (5b, 5e, 5g, and 5h). Motif enrichment was calculated with the cumulative binomial distribution using Homer package in R for Fig. 2b. The r2 coefficient was extracted after performing a linear regression model in R for Fig. 4d. DE expressed genes were determined through DESeq2 package in R through using negative binomial generalized linear models for Supplementary Fig. 2a. All in vitro iPSC reprogramming related experiments were repeated independently at least three times with similar results. All in vivo experiments were repeated independently at least two times with similar results. Replicates from failed experiments due to technical issues were excluded from analysis. Details on sample sizes and reproducibility are in the figure legends.
Teratoma formation assay and generation of chimeric mice
All experiments were performed in accordance with ethical guidelines approved by Institutional Animal Care and Use Committee of Stanford University (protocol numbers: SCRO-689, APLAC-10509, APLAC-9859, and APLAC-12002). This study is compliant with all relevant ethical regulations regarding animal research. For teratoma formation analysis, 2×106 iPSCs were washed twice with PBS and then subcutaneously injected into athymic nude 4-week-old male mice (Hsd:Athymic Nude-Foxn1nu; ENVIGO) for mice and 4-week-old male mice (NOD.Cg-Prkdcscid; The Jackson Laboratory). Teratomas were surgically removed after two weeks. Tissues were fixed in formalin at 4 °C, embedded in paraffin wax, and sectioned at a thickness of 4 µm. Sections were stained with haematoxylin and eosin for pathological examination. For generation of chimeric mice, NSK-iPSCs were injected into blastocysts of albino B6 mouse (jax, B6(Cg)Tyrc-2J/J) at 10–20 cell per blastocysts. The injected blastocysts were, then, implanted into CD1 pseudo mothers for chimeric mice.
Code Availability
Custom parameter for codes are listed in the reporting summary. Specific codes can be obtained from the authors upon request.
Data Availability
RNA–seq data and ATAC-seq that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession codes GSE103509 and GSE103535, respectively (Figures 1, 2, 4). A superseries of both datasets can be found at GSE103536. Previous published RNA-seq data that were re-analyzed here are available from GEO under accession code GSE46104. Source data from Fig. 3, 4, 5 and Supplementary Fig. 3, 4, 5 have been provided as Supplementary Table 5. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgments
We apologize to those investigators whose important work we were unable to cite or describe due to space constraints. We thank D. Burns for critical discussions of the manuscript and P. Chu (Comparative Medicine, Stanford) for technical assistance. We thank the Stanford Shared FACS Facility (SSFF) and FACS Core Facility in the Stanford Lokey Stem Cell Research Building for technical support. This work was supported by F32 GM112425-02 to T.M., Bio-X Graduate Research Fellowships to J.J.B., NSF Graduate Research Fellowship to G.M., GSK Sir James Black Program for Drug Discovery Postdoctoral Fellowship to A.P., and the Baxter Foundation, California Institute for Regenerative Medicine (CIRM) grants RB1-01292 and US National Institutes of Health (NIH) grants U01 HL100397, R01 AG009521, and R01 AG020961 to H.M.B.
Footnotes
Author Contribution: The study was designed by T.M. and H.M.B. T.M. performed the majority of the experiments. G.M., J.J.B., A.P., H.Z., and V.S. also performed experiments. T.M. and G.M. performed data analysis. The manuscript was written by T.M., G.M., and H.M.B.
References
- 1.Takahashi K, Yamanaka S. A decade of transcription factor-mediated reprogramming to pluripotency. Nature Reviews Molecular Cell Biology. 2016;17:183–193. doi: 10.1038/nrm.2016.8. [DOI] [PubMed] [Google Scholar]
- 2.Buganim Y, et al. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heng J-CD, et al. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell stem cell. 2010;6:167–174. doi: 10.1016/j.stem.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 4.Eguchi A, et al. Reprogramming cell fate with a genome-scale library of artificial transcription factors. Proc Natl Acad Sci USA. 2016;113:E8257–E8266. doi: 10.1073/pnas.1611142114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao Y, et al. Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in reprogramming. Cell stem cell. 2013;12:453–469. doi: 10.1016/j.stem.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Long Y, Wang M, Gu H, Xie X. Bromodeoxyuridine promotes full-chemical induction of mouse pluripotent stem cells. Cell research. 2015;25:1171–1174. doi: 10.1038/cr.2015.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hou P, et al. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341:651–654. doi: 10.1126/science.1239278. [DOI] [PubMed] [Google Scholar]
- 8.Redmer T, et al. E-cadherin is crucial for embryonic stem cell pluripotency and can replace OCT4 during somatic cell reprogramming. EMBO Rep. 2011;12:720–726. doi: 10.1038/embor.2011.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan F, Qian C, Tang K, Abd-Allah SM, Jing N. Inhibition of transforming growth factor β (TGF-β) signaling can substitute for Oct4 protein in reprogramming and maintain pluripotency. J Biol Chem. 2014;290:4500–4511. doi: 10.1074/jbc.M114.609016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anokye-Danso F, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell stem cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyoshi N, et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell stem cell. 2011;8:633–638. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Shu J, et al. Induction of pluripotency in mouse somatic cells with lineage specifiers. Cell. 2013;153:963–975. doi: 10.1016/j.cell.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montserrat N, et al. Reprogramming of human fibroblasts to pluripotency with lineage specifiers. Cell stem cell. 2013;13:341–350. doi: 10.1016/j.stem.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 14.Bhutani N, et al. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2009;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brady JJ, et al. Early role for IL-6 signalling during generation of induced pluripotent stem cells r evealed by heterokaryon RNA-Seq. Nat Cell Biol. 2013;15:1244–1252. doi: 10.1038/ncb2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhattacharya B, et al. Gene expression in human embryonic stem cell lines: unique molecular signature. Blood. 2003;103:2956–2964. doi: 10.1182/blood-2003-09-3314. [DOI] [PubMed] [Google Scholar]
- 17.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nordhoff V, et al. Comparative analysis of human, bovine, and murine Oct-4 upstream promoter sequences. Mamm Genome. 2001;12:309–317. doi: 10.1007/s003350010279. [DOI] [PubMed] [Google Scholar]
- 20.Jerabek S, Merino F, Schöler HR, Cojocaru V. OCT4: dynamic DNA binding pioneers stem cell pluripotency. Biochim Biophys Acta. 2013;1839:138–154. doi: 10.1016/j.bbagrm.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 21.Xu Y, et al. Transcriptional Control of Somatic Cell Reprogramming. Trends Cell Biol. 2016;26:272–288. doi: 10.1016/j.tcb.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 22.Do DV, et al. A genetic and developmental pathway from STAT3 to the OCT4-NANOG circuit is essential for maintenance of ICM lineages in vivo. Genes Dev. 2013;27:1378–1390. doi: 10.1101/gad.221176.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dutta A, et al. Identification of an NKX3.1-G9a-UTY transcriptional regulatory network that controls prostate differentiation. Science. 2016;352:1576–1580. doi: 10.1126/science.aad9512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhatia-Gaur R, et al. Roles for Nkx3.1 in prostate development and cancer. Genes Dev. 1999;13:966–977. doi: 10.1101/gad.13.8.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, et al. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461:495–500. doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin J, et al. The PSA(−/lo) prostate cancer cell population harbors self-renewing long-term tumor-propagating cells that resist castration. Cell stem cell. 2012;10:556–569. doi: 10.1016/j.stem.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He WW, et al. A novel human prostate-specific, androgen-regulated homeobox gene (NKX3.1) that maps to 8p21, a region frequently deleted in prostate cancer. Genomics. 1997;43:69–77. doi: 10.1006/geno.1997.4715. [DOI] [PubMed] [Google Scholar]
- 28.Dai H-Q, et al. TET-mediated DNA demethylation controls gastrulation by regulating Lefty-Nodal signalling. Nature. 2016;538:528–532. doi: 10.1038/nature20095. [DOI] [PubMed] [Google Scholar]
- 29.Mosteiro L, et al. Tissue damage and senescence provide critical signals for cellular reprogramming in vivo. Science. 2016;354 doi: 10.1126/science.aaf4445. [DOI] [PubMed] [Google Scholar]
- 30.Rose-John S. IL-6 trans-signaling via the soluble IL-6 receptor: importance for the pro-inflammatory activities of IL-6. Int J Biol Sci. 2012;8:1237–1247. doi: 10.7150/ijbs.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowen C, Gelmann EP. NKX3.1 activates cellular response to DNA damage. Cancer Res. 2010;70:3089–3097. doi: 10.1158/0008-5472.CAN-09-3138. [DOI] [PubMed] [Google Scholar]
- 32.Banito A, et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23:2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gong L, et al. p53 isoform Δ133p53 promotes efficiency of induced pluripotent stem cells and ensures genomic integrity during reprogramming. Sci Rep. 2016;6:37281. doi: 10.1038/srep37281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eide T, Ramberg H, Glackin C, Tindall D, Taskén KA. TWIST1, A novel androgen-regulated gene, is a target for NKX3-1 in prostate cancer cells. Cancer Cell Int. 2013;13:4. doi: 10.1186/1475-2867-13-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li R, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell stem cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 36.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kundaje A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chu J, et al. Non-invasive intravital imaging of cellular differentiation with a bright red-excitable fluorescent protein. Nat Methods. 2014;11:572–578. doi: 10.1038/nmeth.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA–seq data and ATAC-seq that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession codes GSE103509 and GSE103535, respectively (Figures 1, 2, 4). A superseries of both datasets can be found at GSE103536. Previous published RNA-seq data that were re-analyzed here are available from GEO under accession code GSE46104. Source data from Fig. 3, 4, 5 and Supplementary Fig. 3, 4, 5 have been provided as Supplementary Table 5. All other data supporting the findings of this study are available from the corresponding author on reasonable request.