Abstract
RNA interference (RNAi) has been widely applied for uncovering the biological functions of numerous genes, and has been envisaged as a pest control tool operating by disruption of essential gene expression. Although different methods, such as injection, feeding, and soaking, have been reported for successful delivery of double-stranded RNA (dsRNA), the efficiency of RNAi through oral delivery of dsRNA is highly variable among different insect groups. The German cockroach, Blattella germanica, is highly sensitive to the injection of dsRNA, as shown by many studies published previously. The present study describes a method to demonstrate that the dsRNA encapsulated with liposome carriers is sufficient to retard the degradation of dsRNA by midgut juice. Notably, the continuous feeding of dsRNA encapsulated by liposomes significantly reduces the tubulin expression in the midgut, and led to the death of cockroaches. In conclusion, the formulation and utilization of dsRNA lipoplexes, which protect dsRNA against nucleases, could be a practical use of RNAi for insect pest control in the future.
Keywords: Biology, Issue 135, RNA Interference, Blattella Germanica, Liposome, Oral Delivery, dsRNA, Feeding
Introduction
RNAi has been demonstrated as an effective method to knockdown gene expression through a mechanism of a post-transcriptional silencing pathway triggered by dsRNA molecules in many eukaryotes1. Over the past decade of study, RNAi has become a useful tool to study the functions of genes from development to behavior by depleting the expression of specific genes via injection and/or feeding of dsRNA in various taxa of insects2,3. Due to the specificity and robustness of the depleting effect, the application of RNAi is currently being considered as a potential strategy for pest control management4,5. However, the efficiency of RNAi varies widely between insect species, depending on the different genes being targeted and the delivery methods. A growing body of evidence suggests that the instability of dsRNA, which is degraded by ribonucleases, is a critical factor in the limited efficacy of RNAi5,6. For instance, the low RNAi sensitivity in Manduca sexta has been explained by the fact that the dsRNA mixed with hemolymph was quickly degraded within 1 hour7. Similarly, the presence of alkaline nucleases in the midgut, which efficiently degrade ingested dsRNA, is strongly correlated with low RNAi efficiency in different insect orders8,9,10.
The oral delivery of dsRNA is particularly interesting for the application of RNAi in a pest control strategy, but a method to retard the degradation of dsRNA by the nucleases in the midgut has not yet been developed, which would have the potential to ensure effective RNAi through feeding. However, the unresponsiveness of RNAi to oral delivery of dsRNA has been reported by feeding large amount of dsRNA, e.g. 50 µg in Bombyx mori, or continuously feeding for 8 days (8 µg dsRNA in total) in the locust species. The German cockroach, Blattella germinica, is highly sensitive to RNAi by the injection of dsRNA11,12,13,14, but is not responsive to dsRNA through feeding. Recently, Lin et al. (2017) have demonstrated that the dsRNA encapsulated with liposome carriers results in successful RNAi to knockdown the α-tubulin gene expression in the midgut and trigger significant mortality of the German cockroach15. As the degradation of dsRNA in the midgut is the limiting factor for oral RNAi, the liposome carriers serve as a vehicle to protect dsRNA from degradation, which is readily applicable in other insects with strong nuclease activities in the gut. Of note, the reason for choosing the particular transfection reagent (see Table of Materials) we used as liposome carrier in the current protocol is that it has been tested for insect cell line transfection with less toxicity, according to the manufacturer's instructions. According to the comparison of different liposome transfection systems in Gharavi et al. (2013)16, the efficiency of transfecting small interfering RNA (siRNA) is approximately the same between this and other commercially available systems that have been used for dsRNA delivery systems in other insects17,18.Furthermore, our feeding method is careful enough to ensure the proper amount of dsRNA is ingested by each cockroach, and that the results are robust and confirmed. In summary, the present protocol and results demonstrate that using dsRNA lipoplexes improves dsRNA stability and opens the door to the design of the strategy oral delivery of RNAi, which is a promising approach for pest control in the future.
Protocol
1. Synthesis and Preparation of dsRNA
Identify the dsRNA target sites in the 3' untranslated region of the target genes. The dsTub is used for targeting the α-tubulin (tub) gene (GenBank accession number: KX228233), and dsEGFP as a negative dsRNA control is designed from the sequence of enhanced green fluorescence protein (EGFP; GenBank accession number: LC311024).
Perform standard PCR amplification to synthesize the dsRNA templates with gene-specific primers containing the T7 promoter sequence (5'-TAATACGACTCACTATAGGG-3'). The PCR amplification conditions and primer information used are those reported by Lin et al. (2017)12 (see Table of Materials).
Proceed with dsRNA preparation using a T7 Transcription Kit (see Table of Materials), following the manufacturer's instructions. Note: To yield high quantity dsRNA, mix two reactions in one tube (in total 40 µL), and incubate at 37 °C overnight.
Add 2 µL of DNase (see Table of Materials) into dsRNA samples for 15 min at 37 °C to digest DNA templates.
Add 200 µL extraction reagent (see Table of Materials) and 40 µL chloroform, then vortex.
Centrifuge for 10 min at 13,000 x g at 4 °C, then collect supernatant (150 µL) in a new microcentrifuge tube.
Precipitate dsRNA by adding 150 µL isopropanol and incubating for 15 min on ice.
Centrifuge for 15 min at 13,000 x g at 4 °C, then remove supernatant.
Add 200 µL of the 70% ethanol to wash dsRNA pellets. Centrifuge for 10 min at 13,000 x g at 4 °C.
Remove the ethanol and repeat the ethanol wash steps.
Remove the ethanol and dry dsRNA pellet by centrifugal vacuum concentrators for 3 min, then resuspend dsRNA with RNase-free water.
Determine the quantity of the purified dsRNA using a micro-volume UV-Vis spectrophotometer (see Table of Materials) according to the manufacturer's instructions and adjust to the desired concentration (e.g., 0.25 µg/µL for preparation of dsRNA lipoplexes).
Store the dsRNA samples at -20 °C for up to 3 months.
2. Preparation of dsRNA Lipoplexes
Dilute 4 µL of the transfection reagent (see Table of Materials) by adding 4 µL of 5% glucose solution. Dilute 4 µL of the dsRNA (1 µg) by adding 4 µL of 5% glucose solution.
Add the diluted liposome reagent solution immediately into the diluted dsRNA solution all at once. Vortex briefly to mix them, then incubate for 15 min at 25 °C. Note: Do not mix the solutions in the reverse order.
The dsRNA lipoplexes are ready for use. Use within 1 hour of preparation, and discard after 1 hour. Note: According to the manufacturer's instructions, the lipoplexes should be used as soon as possible.
3. Collection of Extracellular Enzymes from Hemolymph and Midgut Juice
- Insect saline buffer (10x stock)
- Mix 900 mL double distilled water with 109.3 g NaCl, 15.7 g KCl, 6.3 g CaCl2, and 8.3 g MgCl2·6H2O.
- Adjust pH to 6.8, and fill up to 1,000 mL.
- Hemolymph collection
- Anesthetize the cockroaches on ice until they do not move for 3 min.
- Hold the cockroach with two fingers (thumb and index finger), and turn the ventral side of the cockroach up.
- Use the dissecting scissor to cut off the tip of a coxa of a hind leg off. Note: Cut the connected intersection between the coxa and trochanter. The excision of the other part of coxa was less efficient to collect hemolymph.
- Squeeze the abdomen gently with middle finger, and collect the hemolymph bleeding from the incision with a 10 µL micropipette.
- Pool the hemolymph from several cockroaches (5 individuals) in a microcentrifuge tube. Note: Keep the microcentrifuge tubes on ice to prevent melanization. The average volume of hemolymph obtained from a male cockroach is 2 - 3 µL.
- Spin down the hemolymph briefly with a benchtop centrifuge for 10 s.
- Transfer desired volume of hemolymph (i.e., 10 µL), and mix it with 50 µL of 1x insect saline buffer.
- Centrifuge for 10 min at 1,000 x g at 4 °C to spin down hemocytes. Transfer the supernatant to a clean microcentrifuge tube.
- Quantify the total protein concentration using a micro-volume UV-Vis spectrophotometer by measuring A28015.
- Adjust each sample to have the same protein concentration (6 mg/µL of total protein).
- Midgut juice collection
- Anesthetize the cockroaches on ice. Fix the cockroaches ventral side up on the dissection plate with cold 1x insect saline buffer using insect pins.
- Dissect the abdomen with fine tweezers. Remove the entire gut (including the front, mid, and hind gut) to a fresh dish with 1x insect saline buffer.
- Remove the front and hind gut, and quickly transfer the midgut to a microcentrifuge tube that contains 100 µL insect saline buffer.
- Pool the midguts from six cockroaches in the same tube, and vortex for 10 s.
- Centrifuge for 10 min at 1,000 x g at 4 °C to spin down the hemocytes and gut tissues. Transfer the supernatant to a clean microcentrifuge tube.
- Quantify the total protein concentration using a micro-volume UV-Vis spectrophotometer by measuring A28015.
- Adjust each sample to have the same protein concentration (6 mg/µL of total protein).
4. dsRNA Degradation Assay
Freshly prepare 4 µL of the naked dsRNA or dsRNA lipoplexes (1 µg).
Mix dsRNA solutions with 10 µL of insect saline buffer (control), extracted enzymes from hemolymph, or midgut juice.
Add 2 µL of either EGTA (20 mM) or RNase-free water to separate samples as controls for inhibition of enzyme activities.
Incubate for 1 hour or longer (6, 12, or 24 hours) at 25 °C.
Add 200 µL extraction reagent and 40 µL chloroform, then vortex.
Centrifuge for 10 min at 13,000 x g at 4 °C, then collect supernatant (150 µL) in a new microcentrifuge tube.
Precipitate dsRNA by adding 150 µL isopropanol and incubating for 15 min on ice.
Centrifuge for 15 min at 13,000 x g at 4 °C, then remove supernatant.
Add 200 µL of the 70% ethanol to wash dsRNA pellets. Centrifuge for 10 min at 13,000 x g at 4 °C.
Remove the ethanol and repeat the ethanol wash steps.
Remove the ethanol and dry dsRNA pellet by centrifugal vacuum concentrators for 3 min. Resuspend dsRNA with 10 µL RNase-free water.
Check the integrity of treated dsRNA via gel electrophoresis on a 1.5% agarose gel.
5. Oral Administration of dsRNA
Maintain cockroaches on an ad libitum diet of dry food (dog chow). Deprive water supply from the cockroaches one day before dsRNA ingestion experiments.
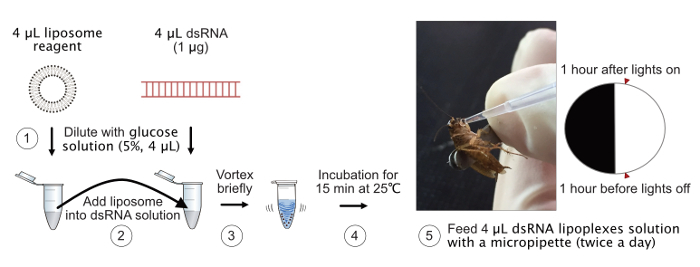
Hold a cockroach by grabbing its wings with the flexible forceps (Figure 1, step 5). Note: Do not grab the body parts of cockroaches.
Prepare the dsRNA lipoplexes, as well as naked dsRNA, for feeding, as described in section 2. Note: The dsRNA lipoplexes should be prepared freshly before feeding.
Feed 4 µL of dsRNA lipoplexes or naked dsRNA (250 ng) with a micropipette.
Slowly pipet the droplet of the dsRNA solution close to the mouthparts of cockroaches, and let the cockroaches ingest the droplet completely.
Perform the ingestion experiments twice a day (1 h after lights on and 1 h before lights off) for 8 days or 16 days continuously. Note: All cockroaches acquired water only through manual feeding with dsRNA solution or control reagents (e.g., nuclease free water, glucose solution, and liposome reagent) during ingestion experiments.
Provide water bottles to the cockroaches after 16 days of ingestion of dsRNA experiments.
6. Assess Knockdown
At chosen time points (2, 9, and 17 days after dsRNA ingestion assay), collect the dsEGFP- and dsTub-treated insects and dissect the chosen tissues (e.g., midgut).
Perform quantitative real-time PCR (qRT-PCR). The qRT-PCR amplification conditions and primer information as reported by Lin et al. (2017)12.
Assess the control and dsRNA-treated insects daily to check mortality and remove the cockroaches that are no longer moving.
Representative Results
A simplified scheme of the protocol for the oral delivery of dsRNA is presented in Figure 1, where the key steps for preparation of dsRNA lipoplexes are shown.
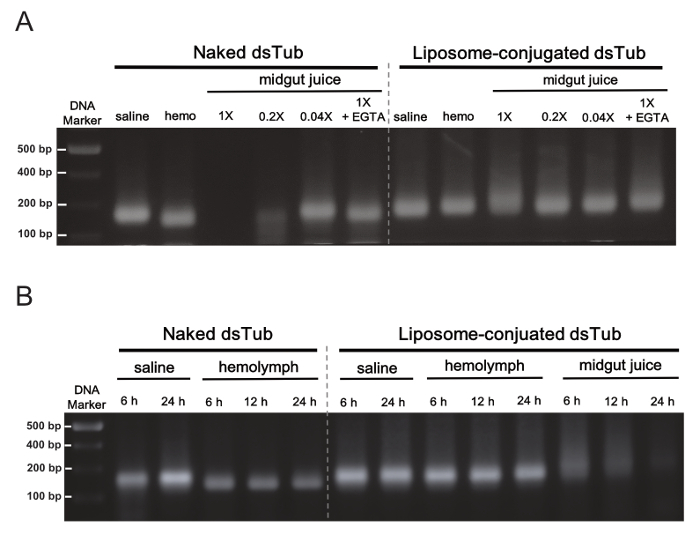
In order to investigate the protection given by liposome carriers upon dsRNA degradation in the midgut juice of B. germanica, an ex vivo assay where dsTub lipoplexes were incubated with midgut juice was conducted, and the integrity of the dsRNA was subsequently analyzed on a 1.5% agarose gel. Figure 2 shows that strong RNA nuclease activity was present in the midgut juice of B. germanica (Figure 2A), whereas the liposome carriers were able to protect dsRNA against degradation at least for 1 hour in the ex vivo incubation (Figure 2A, B). As a result, the oral ingestion of dsRNA lipoplexes was applied twice a day to increase the susceptibility of midgut tissues to RNAi in the following experiments.
Validation of RNAi response through oral delivery of dsRNA was assessed by measuring the depletion effect of tub mRNA expression in the midgut at different time points after continuous ingestion of dsRNA (Figure 3A). The tub expression in the midgut was significantly depleted after continuous ingestion of dsTub lipoplexes for 8 days. While the continuous feeding of dsRNA lasted 16 days, the tub expression in the midgut was significantly depleted in the naked dsTub and dsTub lipoplexes groups (approximately by 24 and 60%, respectively) in contrast to the controls. Figure 3B shows a consistent, significant increase of mortality after the oral administration of dsTub lipoplexes for 16 days.
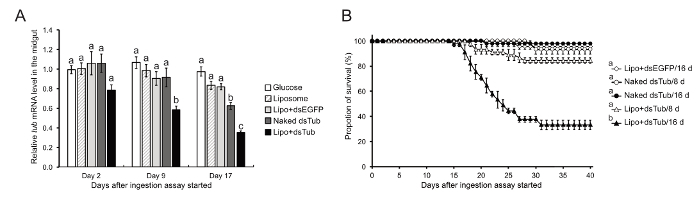
Figure 1: Simplified scheme of oral delivery of dsRNA lipoplexes for cockroaches. The key steps for preparation of dsRNA lipoplexes are represented for steps 1 to 4. The feeding technique (step 5) is shown in the photo. Please click here to view a larger version of this figure.
Figure 2: Ex vivo degradation of dsRNA by B. germanica hemolymph (Hemo) or midgut juice. (A) 1 µg of the naked or liposome-cojugated dsTub was incubated for 1 h with saline buffer and extracted enzymes from hemolymph (hemo) or midgut juice, respectively. The naked dsTub, but not liposome-conjugated dsTub, was degraded completely when incubated with 1x midgut juice. The different dilution factors of the original midgut juice (1x) are indicated. The degradation was inhibited by ion chelation due to EGTA treatment as a control. (B) Time-dependent protection of liposome-encapsulated dsTub (1 µg) against ex vivo degradation in hemolymph or midgut juice. Of note, the liposome-conjugated dsTub was degraded completely when incubated with midgut juice after 24 h. The hemolymph and midgut juice were used in the original concentration for different time periods of incubation. The results are adapted from Lin et al. (2017)12. Please click here to view a larger version of this figure.
Figure 3: Effects of RNAi response by ingestion of dsRNA on B. germanica. (A) Quantitative real-time PCR (qRT-PCR) for determining the relative tub expression in the midgut of the German cockroach after different feeding treatments. The expression levels of different treatments were normalized to the Glucose group at Day 2. Values are the mean ± SE from three independent experiments (n = 3), each with 3 - 5 biological replicates. Different letters on the bars indicate significant differences at p <0.05 (ANOVA following Tukey's HSD Post Hoc test) among different treatment groups. (B) Survivorship of the cockroaches that ingested naked dsTub or dsTub lipoplexes for 8 days (8 d) or 16 days (16 d) (each cohort of 12 - 15 individuals; n = 3 independent experiments). Values are the mean ± SE. Different letters on the treatment group indicate significant differences at p <0.05 (Kruskal-Wallis test) for the survivorship. The results are adapted from Lin et al. (2017)12. Please click here to view a larger version of this figure.
Discussion
This protocol presents a method for effective RNAi through oral delivery of dsRNA lipoplexes, involving protection against ribonuclease digestion in the midgut juice of the German cockroach. As shown in other studies in various insect species, the poor RNAi effect through oral delivery of dsRNA is mostly accounted for by the degradation of dsRNA8,9,10. This protocol produces liposomes that serve as protective vehicles in oral delivery of dsRNA against degradation in the gut. In addition, the preparation of dsRNA lipoplexes is simple and readily applicable as oral RNAi, in particular for insects with strong ribonuclease activity in the gut.
Compared to other studies that feed relatively large amounts of naked dsRNA in different insects10,18,19, this protocol uses oral administration of a relatively low amount of dsRNA (0.5 µg per day), which causes a significant depletion of target gene expression in the midgut of B. germanica. Crucial aspects of the protocol include the precise feeding method and accumulated ingestion of small amounts of dsRNA. Firstly, the feeding technique is suitable for a continuous ingestion assay and minimizes the variation of the ingested dsRNA quantity per individual insect. Second, the depletion effect of tub expression in the midgut was shown to increase from 40% at day 9 to 60% at day 17 with continuous oral administration of dsTub lipoplexes (Figure 3A). Although the naked dsRNA was rapidly degraded with ex vivo incubation of midgut juice within 1 h (Figure 2A), the continuous feeding of naked dsRNA for 16 days also resulted in a significant depletion of tub expression in the midgut (Figure 3A). Moreover, the longer period of continuous feeding of dsRNA lipoplexes for 16 days is a requirement to cause a conspicuous lethal effect by RNAi.
Although the liposome carriers (see Table of Materials) used here were originally for in vitro transfection of DNA into insect cells, there was no problem applying this liposome reagent as a vehicle of dsRNA for oral delivery in our insect model. Many studies have also successfully demonstrated the improvement of RNAi silencing in other insect species using different liposome reagents, which are designed for either DNA or RNA molecules17,20,21. Nevertheless, the mechanism of dsRNA uptake in the insect gut has not yet been completely studied, and the liposome delivery system might enhance the uptake of dsRNA into the cells, owing to its biocompatibility with phospholipid structure22. Therefore, the oral delivery of dsRNA encapsulated in the liposomes could be an appropriate strategy to achieve efficient RNAi due to the retardation of degradation by the RNases (Figure 2).
One disadvantage of this protocol might be the small amount of dsRNA in the liposome carriers, and thus may require longer periods of continuous feeding. The restriction of such a particular ratio between dsRNA to liposome (0.25 µg:1 µL) is due to the design of the commercial liposome reagent, as per the manufacturer's suggestion. The formulations in preparation of liposome nanoparticles with larger diameter size, and the different charge on the lipoplexes surface, are reported to increase the encapsulation of siRNA and RNAi silencing efficiency23,24. Therefore, the different liposome nanoparticles can be also used to improve feeding procedures.
Disclosures
The authors have nothing to disclose.
Acknowledgments
This study was supported by grants from Taiwan (Ministry of Science and Technology, MOST 100-2923-B-002-002-MY3 and 106-2313-B-002-011-MY3 to H.J.L.), the Czech Republic (Grant agency of South Bohemia University, GAJU grant 065/2017/P to Y.H.L), and Spain (Spanish Ministry of Economy and Competitiveness, grants CGL2012-36251 and CGL2015-64727-P to X.B., and the Catalan Government, grant 2014 SGR 619 to X.B.); it also received financial support from the European Fund for Economic and Regional Development (FEDER funds to X.B.).
References
- Hammond SM. Dicing and slicing: The core machinery of the RNA interference pathway. FEBS Lett. 2005;579:5822–5829. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- Bellés X. Beyond drosophila: RNAi in vivo and functional genomics in insects. Annu. Rev. Entomol. 2010;55:111–128. doi: 10.1146/annurev-ento-112408-085301. [DOI] [PubMed] [Google Scholar]
- Wynant N, Santos D, Vanden Broeck J. Chapter Five - Biological Mechanisms Determining the Success of RNA Interference in Insects. In: Jeon KW, editor. International Review of Cell and Molecular Biology. Vol. 312. Academic Press; 2014. pp. 139–167. [DOI] [PubMed] [Google Scholar]
- San Miguel K, Scott JG. The next generation of insecticides: dsRNA is stable as a foliar-applied insecticide. Pest Manag. Sci. 2016;72:801–809. doi: 10.1002/ps.4056. [DOI] [PubMed] [Google Scholar]
- Scott JG, et al. Towards the elements of successful insect RNAi. J. Insect Physiol. 2013;59:1212–1221. doi: 10.1016/j.jinsphys.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joga MR, Zotti MJ, Smagghe G, Christiaens O. RNAi efficiency, systemic properties, and novel delivery methods for pest insect control: What we know so far. Front. Physiol. 2016;7 doi: 10.3389/fphys.2016.00553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbutt JS, Bellés X, Richards EH, Reynolds SE. Persistence of double-stranded RNA in insect hemolymph as a potential determiner of RNA interference success: Evidence from Manduca sexta and Blattella germanica. J. Insect Physiol. 2013;59:171–178. doi: 10.1016/j.jinsphys.2012.05.013. [DOI] [PubMed] [Google Scholar]
- Arimatsu Y, Kotani E, Sugimura Y, Furusawa T. Molecular characterization of a cDNA encoding extracellular dsRNase and its expression in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2007;37:176–183. doi: 10.1016/j.ibmb.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Wang K, et al. Variation in RNAi efficacy among insect species is attributable to dsRNA degradation in vivo. Insect Biochem. Mol. Biol. 2016;77:1–9. doi: 10.1016/j.ibmb.2016.07.007. [DOI] [PubMed] [Google Scholar]
- Wynant N, et al. Identification, functional characterization and phylogenetic analysis of double stranded RNA degrading enzymes present in the gut of the desert locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 2014;46:1–8. doi: 10.1016/j.ibmb.2013.12.008. [DOI] [PubMed] [Google Scholar]
- Huang J-H, Belles X, Lee H-J. Functional characterization of hypertrehalosemic hormone receptor in relation to hemolymph trehalose and to oxidative stress in the cockroach Blattella germanica. Exp. Endocrinol. 2012;2:114. doi: 10.3389/fendo.2011.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-H, Lee C-M, Huang J-H, Lee H-J. Circadian regulation of permethrin susceptibility by glutathione S-transferase (BgGSTD1) in the German cockroach (Blattella germanica) J. Insect Physiol. 2014;65:45–50. doi: 10.1016/j.jinsphys.2014.05.001. [DOI] [PubMed] [Google Scholar]
- Lozano J, Kayukawa T, Shinoda T, Belles X. A role for taiman in insect metamorphosis. PLOS Genet. 2014;10:1004769. doi: 10.1371/journal.pgen.1004769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano J, Montañez R, Belles X. MiR-2 family regulates insect metamorphosis by controlling the juvenile hormone signaling pathway. Proc. Natl. Acad. Sci. 2015;112:3740–3745. doi: 10.1073/pnas.1418522112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-H, Huang J-H, Liu Y, Belles X, Lee H-J. Oral delivery of dsRNA lipoplexes to German cockroach protects dsRNA from degradation and induces RNAi response. Pest Manag. Sci. 2017;73:960–966. doi: 10.1002/ps.4407. [DOI] [PubMed] [Google Scholar]
- Gharavi J, et al. Chiral cationic polyamines for chiral microcapsules and siRNA delivery. Bioorg. Med. Chem. Lett. 2013;23:5919–5922. doi: 10.1016/j.bmcl.2013.08.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyard S, Singh AD, Wong S. Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem. Mol. Biol. 2009;39:824–832. doi: 10.1016/j.ibmb.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Luo Y, et al. Differential responses of migratory locusts to systemic RNA interference via double-stranded RNA injection and feeding. Insect Mol. Biol. 2013;22:574–583. doi: 10.1111/imb.12046. [DOI] [PubMed] [Google Scholar]
- Liu J, Smagghe G, Swevers L. Transcriptional response of BmToll9-1 and RNAi machinery genes to exogenous dsRNA in the midgut of Bombyx mori. J. Insect Physiol. 2013;59:646–654. doi: 10.1016/j.jinsphys.2013.03.013. [DOI] [PubMed] [Google Scholar]
- Airs PM, Bartholomay LC. RNA interference for mosquito and mosquito-borne disease control. Insects. 2017;8:4. doi: 10.3390/insects8010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taning CNT, et al. Oral RNAi to control Drosophila suzukii: Laboratory testing against larval and adult stages. J. Pest Sci. 2016;89:803–814. [Google Scholar]
- Wu SY, McMillan NAJ. Lipidic systems for in vivo siRNA delivery. AAPS J. 2009;11:639–652. doi: 10.1208/s12248-009-9140-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam YYC, Chen S, Cullis PR. Advances in lipid nanoparticles for siRNA delivery. Pharmaceutics. 2013;5:498–507. doi: 10.3390/pharmaceutics5030498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Tian J, Chen X. Effect of surface properties on liposomal siRNA delivery. Biomaterials. 2016;79:56–68. doi: 10.1016/j.biomaterials.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]


