Abstract
Multicopy plasmids are extremely abundant in prokaryotes but their role in bacterial evolution remains poorly understood. We recently showed that the increase in gene copy number per cell provided by multicopy plasmids could accelerate the evolution of plasmid-encoded genes. In this work, we present an experimental system to test the ability of multicopy plasmids to promote gene evolution. Using simple molecular biology methods, we constructed a model system where an antibiotic resistance gene can be inserted into Escherichia coli MG1655, either in the chromosome or on a multicopy plasmid. We use an experimental evolution approach to propagate the different strains under increasing concentrations of antibiotics and we measure survival of bacterial populations over time. The choice of the antibiotic molecule and the resistance gene is so that the gene can only confer resistance through the acquisition of mutations. This "evolutionary rescue" approach provides a simple method to test the potential of multicopy plasmids to promote the acquisition of antibiotic resistance. In the next step of the experimental system, the molecular bases of antibiotic resistance are characterized. To identify mutations responsible for the acquisition of antibiotic resistance we use deep DNA sequencing of samples obtained from whole populations and clones. Finally, to confirm the role of the mutations in the gene under study, we reconstruct them in the parental background and test the resistance phenotype of the resulting strains.
Keywords: Genetics, Issue 135, Plasmid, antibiotic resistance, evolution, evolutionary rescue, multicopy plasmids, mutation, experimental evolution
Introduction
Antibiotic resistance in bacteria is a major health problem1. At a fundamental level, the spread of antibiotic resistance in pathogenic bacteria is a simple example of evolution by natural selection2,3. Put simply, the use of antibiotics generates selection for resistant strains. A key problem in evolutionary biology, therefore, is to understand the factors that influence the ability of bacterial populations to evolve resistance to antibiotics. Selection experiments have emerged as a very powerful tool to investigate the evolutionary biology of bacteria, and this field has produced incredible insights into a wide range of evolutionary problems4,5,6. In experimental evolution, bacterial populations initiated from a single parental strain are serially passaged under defined and tightly controlled conditions. Some of the mutations that occur during the growth of these cultures increase bacterial fitness, and these spread through the cultures by natural selection. During the experiment, samples of the populations are periodically cryogenically preserved to create a non-evolving frozen fossil record. A wide number of approaches can be used to characterize evolving bacterial populations, but the two most common methods are fitness assays, that measure the ability of evolved bacteria to compete against their distant ancestors, and whole genome sequencing, that is used to identify the genetic changes that drive adaptation. Following pioneering work by Richard Lenski and colleagues7,8, the standard approach in experimental evolution has been to challenge a relatively small number of replicate populations (typically <10) with adapting to a new environmental challenge, such as new carbon sources, temperature, or a predatory phage.
Infections caused by antibiotic resistant bacteria become a big problem when resistance is high enough that it is not possible to increase antibiotic concentrations to lethal levels in patient tissues. Clinicians are therefore interested in what allows bacteria to evolve resistance to high doses of antibiotic that are above this threshold antibiotic concentration, the clinical breakpoint. How to study this experimentally? If a small number of bacterial populations are challenged with a high dose of antibiotic, as in a Lenski-style experiment, then the most likely outcome is that the antibiotic will drive all of the populations to extinction. At the same time, if the dose of antibiotic that is used is low, below the minimal inhibitory concentration (MIC) of the parental strain, then it is unlikely that the bacterial populations will evolve clinically relevant levels of resistance, especially if resistance carries a large cost. One compromise between these two scenarios is to use an "evolutionary rescue" experiment9,10,11. In this approach, a very large number of cultures (typically >40) is challenged with doses of antibiotics that increase over time, typically by doubling antibiotic concentration every day12. The hallmark of this experiment is that any population that does not evolve increased resistance will be driven to extinction. Most populations that are challenged in this way will be driven extinct, but a small minority will persist by evolving high levels of resistance. In this paper, we show how this experimental design can be used to investigate multicopy plasmid contribution to the evolution of resistance.
Bacteria acquire resistance to antibiotics through two principal routes, chromosomal mutations, and acquisition of mobile genetic elements, mostly plasmids13. Plasmids play a key role in the evolution of antibiotic resistance because they are able to transfer resistance genes between bacteria by conjugation14,15. Plasmids can be divided into two groups according to their size and biology: "small", with high copy number per bacterial cell and "large", with low copy number16,17. The role of large plasmids in the evolution of antibiotic resistance has been extensively documented because they include conjugative plasmids, which are key drivers of the dissemination of resistance and multi resistance among bacteria15. Small multicopy plasmids are also extremely common in bacteria17,18, and they often code for antibiotic resistance genes19. However, the role of small multicopy plasmids in the evolution of antibiotic resistance has been studied to a lesser extent.
In a recent work, we proposed that multicopy plasmids could accelerate the evolution of the genes they carry by increasing gene mutation rates due to the higher gene copy number per cell12. Using an experimental model with E. coli strain MG1655 and the β-lactamase gene blaTEM-1 it was shown that multicopy plasmids accelerated the rate of appearance of TEM-1 mutations conferring resistance to the third-generation cephalosporin ceftazidime. These results indicated that multicopy plasmids might play an important role in the evolution of antibiotic resistance.
Here, we present a detailed description of the method we have developed to investigate the multicopy plasmid-mediated evolution of antibiotic resistance. This method has three different steps: first, insertion of the gene under study either in a multicopy plasmid or the chromosome of the host bacteria. Second, use of experimental evolution (evolutionary rescue) to assess the potential of the different strains to adapt to the selective pressure. And third, determining the molecular basis underlying plasmid-mediated evolution using DNA sequencing and reconstructing the suspected mutations individually in the parental genotype.
Finally, although the protocol described here was designed to investigate the evolution of antibiotic resistance, one can argue that this method could be generally useful to analyze the evolution of innovations acquired by mutations in any multicopy plasmid-encoded gene.
Protocol
1. Construction of the Experimental System Encoding Antibiotic Resistance Gene
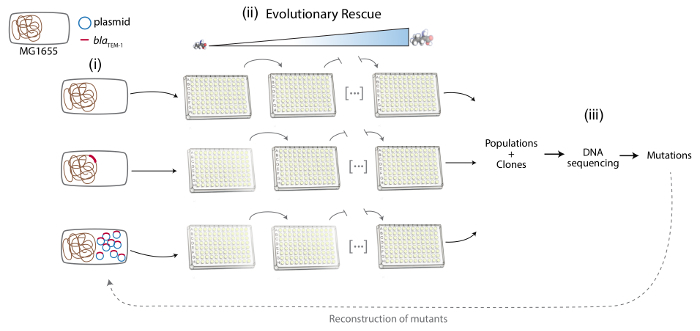
Note: Here E. coli MG1655 was used as the recipient strain of the plasmid- or chromosome-encoded antibiotic resistance gene. The antibiotic resistance gene is encoded in the chromosome or a multicopy plasmid in an otherwise isogenic strain (Figure 1).
- Insertion of the antibiotic resistance gene in the λ phage integration site (attB)20 of the chromosome of MG1655.
- Amplify 500 bp-long regions at both sides of the chromosomal attB site by PCR. Use oligonucleotides YbhC-F (5'- CCTGTACCGTACAGAGTAAT-3') and attB-R (5'- GCCCGCCACCCTCCGGGCCGGTATAAAAAAGCAGGCTTCA-3') for the left homology region and attB-F (5'- AGCGCCCTAGCGCCCGCTCCTTATACTAACTTGAGCGAAA-3') and YbhB/R (5'- TGGCGATAATATTTCACCGC-3') for the right one. Amplify the blaTEM-1 resistance gene using primers Tem1-pBAD-F (5'- TGAAGCCTGCTTTTTTATACCGGCCCGGAGGGTGGCGGGC-3') and Tem1-pBAD-R (5'- TTTCGCTCAAGTTAGTATAAGGAGCGGGCGCTAGGGCGCT-3'). Design primers with approximately 20 bp (in the 5' end) of sequence showing complementarity to the fragment that is going to be fused to.
- Fuse homology regions to the antibiotic resistance gene PCR product (including its promoter region) using isothermal assembly21, at 50 °C for 30 min.
- Electroporate MG1655 cells containing plasmid pKOBEG. Note: pKOBEG is a thermosensitive vector that contains the λ Red machinery, promoting homology-based allelic exchanges between the chromosome and PCR products22.
- Use 1 µL of the product from step 1.1.223 into 40 µL of electrocompetent cells in 2 mm cuvettes at 4 °C and 2.5 kV. Resuspend cells in 1 mL of LB broth + 0.2% arabinose to maintain the expression of the λ Red machinery.
- Transfer the total volume to a microfuge tube and incubate 2 h at 30 °C (permissive temperature for pKOBEG) with intense shaking (200 rpm in an orbital shaker) to allow for the phenotypic expression of the resistance markers inserted in the chromosome.
- Plate the cells on LB agar containing the appropriate antibiotic to select for the antibiotic resistance gene (ampicillin or carbenicillin at 100 mg/l for blaTEM-1).
- Plate 100 µL on one Petri dish, and spin down the rest of the volume, resuspend it in 100 µL of fresh LB broth and plate it on another Petri dish. Incubate overnight at 42 °C. Note: This temperature is non-permissive for the replication of pKOBEG. The colonies able to grow in these conditions will have lost pKOBEG and will present the resistance gene integrated into the attB site (for simplicity this strain will be called MG1655::resA from here on).
- Test for the loss of pKOBEG by replicating the colonies of interest in plates with chloramphenicol (the antibiotic against which pKOBEG contains a resistance marker). Note: Lack of growth in chloramphenicol plates at the permissive temperature of 30 ºC is indicative of the absence of the plasmid.
- In order to verify clones, PCR amplify the construction using external primers YbhC-Extern (5'-TTTGTGACCAGAAGACCGCA-3') and YbhB-Extern (5'-CTCATCAGTAACGATCTGCG-3'), verify through gel electrophoresis the correct size of the amplicon and sequence the purified product to ensure that the sequence of the inserted gene is correct.
- Insert the antibiotic resistance gene in a multicopy plasmid. Note: Because plasmids with very-high copy number tend to impose a large reduction in bacterial host fitness, we recommend the use of multicopy plasmids with a natural origin of replication, such as p3655 (pSU18T-pBADgfp2, ColE1-type origin of replication24). These plasmids usually present a moderate copy number of about 15-20 copies per cell.
- PCR amplify the antibiotic resistance gene of choice (including promoter region), phosphorylate the PCR product and ligate it to the PCR-amplified backbone of the plasmid:
- PCR amplify the antibiotic resistance gene; for blaTEM-1 use primers Tem1-pBAD-F (5'- TGAAGCCTGCTTTTTTATACCGGCCCGGAGGGTGGCGGGC-3') and Tem1-pBAD-R (5'- TTTCGCTCAAGTTAGTATAAGGAGCGGGCGCTAGGGCGCT-3').
- Phosphorylate the purified product using T4 polynucleotide kinase, following manufacturer's instructions.
- PCR amplify the plasmid backbone using a high-fidelity polymerase and oligonucleotides pBAD-F (5'-CGTTGATCGGCACGTAAGAG-3') and pBAD-R (5'-AAACGACGGCCAGTGCCAAG-3').
- Ligate both PCR fragments using T4-ligase following manufacturer's guidelines.
- Electroporate, as described previously, 40 µL of electrocompetent E. coli DH5-α cells with a maximum of 1 µL of the ligation product and select on the appropriate antibiotics for the antibiotic resistance gene (ampicillin or carbenicillin at 100 mg/L for blaTEM-1) and the plasmid backbone. Note: Supplementing LB with arabinose here is unnecessary.
- Extract the plasmid using the commercial mini-prep kit of the reader's choice:
- Harvest 5 mL of an overnight culture by centrifuging at 12,000 x g at room temperature. Resuspend the cells in 250 µL of resuspension solution and mix well. Add 250 µL of lysis solution and mix well. Add 350 µL of neutralization and mix well.
- Transfer the supernatant to the DNA binding column provided with the kit, centrifuge for 1 min and discard the flow through. Wash twice the column with 500 µL of washing solution, centrifuging for one minute and discarding the flowthrough at each wash. Centrifuge the empty column for 1 additional min to remove residual washing buffer.
- Place the column in a clean tube and add 50 µL of ultra-pure water to the membrane to elute the purified DNA. Centrifuge for 1-2 min and collect the flow through that contains the purified plasmid. Sanger sequence the gene of interest in the plasmid using primers from outside the inserted sequence to confirm that the sequence is correct and that no mutations are present in the resistance gene prior to the evolution experiments.
- Once the sequence is verified, electroporate the plasmid in the E. coli MG1655 strain, as described above. Note: This yields strain MG1655/pRESA.
2. Evolutionary Rescue Approach to Experimentally Evolve Antibiotic Resistance (Figure 1)
Streak out the strains under study on LB plates: MG1655::resA and MG1655/pRESA plus the susceptible parental strain, MG1655. Note: Plasmid pRESA should be stable in MG1655, so there is no need to add antibiotics to the LB plates. Mutation rates in E. coli are low enough so once the construction is verified it is not necessary to verify plasmid sequence at each step.
Prepare 96-well plates with 200 µL of LB in each well and inoculate 48 isolated colonies of each strain in independent wells (one plate per strain). Incubate the plates overnight at 37 °C and 200 rpm. Keep a frozen stock of these founder populations. Note: To prevent and control for culture cross-contamination in the 96-well plates, use a checkerboard plate design by intercalating inoculated wells with bacteria-free medium throughout the 96-well plate. Use this plate design during the entire experimental evolution.
Start the evolutionary rescue experiment by inoculating 2 µL from each well of the plates with the founder populations in new 96-well plates with 198 µL of LB in each well with a sub-inhibitory concentration of the antibiotic under study. For the evolutionary rescue approach start with ¼ or 1/8 of the minimal inhibitory concentration (MIC, determined previously25) of the antibiotic in LB for each of the three strains and double the concentration of the antibiotic daily. Use this approach to maximize the chances of populations to acquire resistance mutations26. Incubate the plates at 37 °C and 200 rpm for 20 h. Note: Measure the overnight OD of the founder populations. If there are significant differences in OD among strains correct the initial inoculum to start the experiment with the same number of cells per well.
Every day, measure the OD of each population after overnight culture and perform a transfer of the cultures, as described in 2.3, to new 96-well plates with double the concentration of the antibiotic than the day before. Incubate the plates 20 h at 37 °C and 200 rpm. Note: The 1:100 dilution factor produces approximately 6-7 generations per day.
In parallel, propagate control populations of each strain in the same conditions as described in 2.3 and 2.4, but in the absence of antibiotics. Note: Control populations will help discriminate between mutations arising due to the presence of the antibiotics and general mutations helping bacteria to adapt to the experimental conditions. Parallel mutations arising in the absence of antibiotics are likely to be helping bacteria to adapt to the experimental conditions and not related to antibiotic resistance.
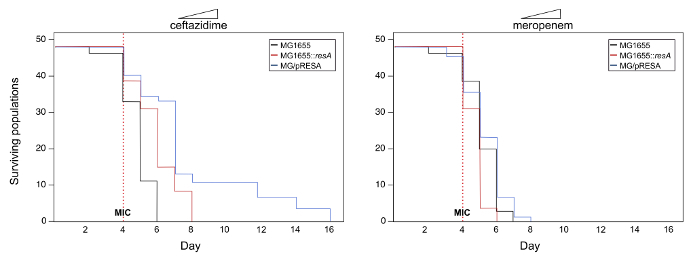
Track the number of surviving populations every day by measuring the absorbance at a wavelength of 600 nm (OD) of the cultures using a plate reader. Note: Optical density values lower than 0.1 indicate the extinction of the population. See Figure 2 for an example of survival curves.
Keep a frozen stock of all of the populations periodically (every 3-5 days).
Use log-rank tests [package "survival" in RStudio (Version 0.99.486)] to determine statistical differences in the survival of populations of the different strains over time under increasing concentration of antibiotics12. Note: This experiment will determine if multicopy plasmids potentiate the evolution of antibiotic resistance for the particular antibiotic and gene under study.
3. Molecular Basis of the Evolution of Antibiotic Resistance (Figure 1)
Perform total DNA extractions from a representative number of populations and clones across strains and treatments. Include the parental strains (MG1655, MG1655::resA and MG1655/pRESA) to be able to detect the mutations accumulating during the experiment. Note: For examples of DNA extraction kits see the table of materials. Each kit has different protocols; follow manufacturers instructions.
Quantify DNA quality and concentration. There are different methods to determine DNA quality and concentration. Determine the quality measuring the ratios of absorbance 260 nm/280 nm and 260 nm/230 nm. Use a fluorescent, DNA-binding dye to measure DNA concentration following manufacturer's instructions, and agarose gel electrophoresis to confirm that there is no DNA degradation or RNA contamination.
Use deep DNA sequencing from samples of whole populations and individual clones to investigate the genetic basis of antibiotic resistance. Note: We conducted all sequencing at the Wellcome Trust Centre for Human Genetics, Oxford, UK. For more details see San Millan et al. 201612.
Use the breseq 0.26.1 pipeline27,28 to detect the mutations, using polymorphism mode to estimate the frequency of mutations in populations. Compare the different evolved genomes to the parental genome to detect mutations that have accumulated during the experiment.
Compare the mutations accumulated in parallel in the control populations with those of the populations evolved under increasing concentration of antibiotics to differentiate between mutations helping bacteria to adapt to the general experimental conditions (those found in the control populations) and those involved in antibiotic resistance (those found exclusively in populations subjected to antibiotic pressure). Note: The number of parallel mutations is usually low, so different parallel mutations between treatments can be easily assessed. Apart from the mutations in the antibiotic resistance gene under study it is quite likely to find other mutations associated with antibiotic resistance in the chromosome, such mutations in porins or efflux systems12.
Reconstruct the mutations in the antibiotic resistance gene in the parental strain using the same approach as in section 1 of this protocol. Follow points 1.1 and 1.2 of the protocol to introduce the new mutations in the parental background (using the evolved genes as a PCR template). Analyze the antibiotic resistance phenotypes of these new constructions to confirm the role of the mutations.
Representative Results
In our previous work, the evolution the β-lactamase gene blaTEM-1 towards conferring resistance to the third generation cephalosporin ceftazidime12 was investigated. This gene was selected because, although TEM-1 does not confer resistance to ceftazidime, mutations in blaTEM-1 can expand the range of activity of TEM-1 to hydrolyze cephalosporins such as ceftazidime29. Mutations in antibiotic resistance enzymes such as β-lactamases or aminoglycoside modifying enzymes leading to changes in their range of activity are common29,30. This experimental system is ideal to explore the evolution of this type of enzymes. For a detailed report of a successful experiment following this protocol please see San Millan et al. 201612.
Here, an example of the possible outcomes of this experimental system is presented to illustrate the protocol (note that the data used for this example is not real). To investigate the potential role of multicopy plasmids in the evolution of the antibiotic resistance gene under study in this example (let's call it resA), we develop the experimental system following section 1 of the protocol described above. The experiments produce three strains: MG1655, MG1655::resA and MG1655/pRESA. The evolution of resistance to two different ß-lactam antibiotics (ceftazidime and meropenem) was tested following the steps described in section 2 of the protocol. Figure 2 shows the survival curves of the populations under study. In this example, there is a significant increase in the survival of populations belonging to MG1655/pRESA evolving in ceftazidime compared to those from MG1655 or MG1655::resA (log-rank test, P< 0.05). On the other hand, in the case of meropenem, there are no significant differences in the survival of the populations belonging to the different strains (log-rank test, P> 0.05). Therefore, these results suggest that the presence of gene resA in a multicopy plasmid potentiates the evolution of resistance to ceftazidime but not to meropenem.
In the final step of the experiment, the molecular basis of antibiotic resistance is investigated, as explained in section 3 of the protocol. First, DNA sequencing will reveal the mutations in resA that could be responsible for the resistance phenotype. And second, reconstruction of resA mutations in the parental MG1655 (both in the chromosome and plasmid) will confirm or discard their role on the antibiotic resistance phenotype.
Figure 1. Schematic representation of the different phases of the protocol. From left to right: (i) Construction of the experimental system: MG1655, MG1655::resA and MG1655/pRESA. Bacterial chromosome is represented in brown, the plasmid in blue and the resA gene in red. (ii) Evolutionary rescue approach to experimentally evolve antibiotic resistance: several populations of the different strains are propagated under increasing concentration of the antibiotic. (iii) Analysis of the molecular basis of antibiotic resistance: sequencing of DNA samples from the evolved populations and clones, detection of the antibiotic resistance mutations and reconstruction of these mutations in the parental strain. Please click here to view a larger version of this figure.
Figure 2. Survival curves with increasing concentrations of antibiotics. Representation of the number of viable populations belonging to strains MG1655, MG1655::resA, and MG1655/pRESA over time. 48 populations of each strain were propagated under increasing concentrations of antibiotics ceftazidime and meropenem, starting with 1/8 of the MIC on day 1 and doubling the antibiotic concentration every day. The red dashed vertical line represents the MIC of the antibiotics under study. Note that in the case of ceftazidime there are significant differences in the survival of the populations belonging to different strains over time (log-rank test, P< 0.05). Crucially, only populations carrying the plasmid are able to survive up to high-level concentrations of antibiotic. On the other hand, in the case of meropenem, there are no significant differences in the survival of the different populations over time (log-rank test, P> 0.05). Please click here to view a larger version of this figure.
Discussion
We present a new protocol combining molecular biology, experimental evolution and deep DNA sequencing designed to investigate the role of multicopy plasmids in the evolution of antibiotic resistance in bacteria. Although this protocol combines techniques from different fields, all the methods required to develop it are simple, and can be performed in a regular microbiology laboratory. The most critical steps in the protocol probably are the construction of the model system strains and the reconstruction of the mutations observed after the experimental evolution (which are performed using the exact same method). However, the isothermal assembly system21, simplifies significantly this protocol so any user with an intermediate level of experience in molecular biology can implement it.
Another critical step of the protocol is the experimental evolution under increasing concentrations of antibiotics. As an example, this protocol starts the experiment with ¼-1/8 of the MIC of the strains and then doubling the concentration of antibiotic every day. However, a lower rate of antibiotic change could increase the chance of evolutionary rescue from extinction26. Therefore, the rate of change of antibiotic concentrations is one of the parameters that can be modified to promote the evolution of antibiotic resistance.
DNA sequencing and analysis are also key aspects of the experimental design. Results are more straightforward when sequencing is performed on DNA samples both from whole populations and from individual clones, at different time points in the experiment. Sequencing results from populations will reveal general differences in the mutation profiles among treatments, as well as selective sweeps of beneficial mutations over time and potential events of clonal interference. When analyzing sequences from populations, it is better to filter mutations that never surpassed 10% frequency in any population. Sequences from individual clones help confirm the results obtained from populations and, most importantly, reveal the specific combinations between the different mutations observed at the population level. These specific associations may help uncover epistatic interactions between mutations, which play a critical role in bacterial adaptation31.
Using this method, we have recently shown that multicopy plasmids accelerate the evolution of antibiotic resistance, first by increasing the rate of appearance of novel mutations and then by amplifying the effect of mutations due to increased gene dosage12. Therefore, we developed the method as a tool to investigate the evolution of antibiotic resistance, but it may have a much broader range of applications. Namely, this system could be used to investigate the ability of any bacterial gene to evolve towards a new or improved function in a more general way. This system could be used, for example, to test the ability of a metabolic enzyme/pathway to use new carbon substrates32. Also, it could be used instead of hypermutators (bacteria with a defect in the cellular systems involved in DNA mismatch repair) to investigate adaptive gene evolution in bacteria, avoiding the mutational bias introduced by hypermutators.
Disclosures
The authors have nothing to disclose.
Acknowledgments
This work was supported by the Instituto de Salud Carlos III (Plan Estatal de I+D+i 2013-2016): grants CP15-00012, PI16-00860, and CIBER (CB06/02/0053), co-financed by the European Development Regional Fund ''A way to achieve Europe'' (ERDF). JAE is supported by the Atracción de talento program of the government of the region of Madrid (2016-T1/BIO-1105) and the I+D Excelencia of the Spanish Ministerio de Economía, Industria y Competitividad (BIO2017-85056-P). ASM is supported by a Miguel Servet Fellowship from the Instituto de Salud Carlos III (MS15/00012) co-financed by The European Social Fund "Investing in your future" (ESF) and ERDF.
References
- Neill J. TACKLING DRUG-RESISTANT INFECTIONS GLOBALLY: FINAL REPORT AND RECOMMENDATIONS. Review on Antimicrobal Resistance. 2016.
- Palmer AC, Kishony R. Understanding, predicting and manipulating the genotypic evolution of antibiotic resistance. Nat Rev Genet. 2013;14(4):243–248. doi: 10.1038/nrg3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean RC, Hall AR, Perron GG, Buckling A. The population genetics of antibiotic resistance: integrating molecular mechanisms and treatment contexts. Nat Rev Genet. 2010;11(6):405–414. doi: 10.1038/nrg2778. [DOI] [PubMed] [Google Scholar]
- Buckling A, Maclean RC, Brockhurst MA, Colegrave N. The Beagle in a bottle. Nature. 2009;457(7231):824–829. doi: 10.1038/nature07892. [DOI] [PubMed] [Google Scholar]
- Barrick JE, Lenski RE. Genome dynamics during experimental evolution. Nature Reviews Genetics. 2013;14(12):827–839. doi: 10.1038/nrg3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena SF, Lenski RE. Evolution experiments with microorganisms: The dynamics and genetic bases of adaptation. Nature Reviews Genetics. 2003;4(6):457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- Lenski RE, Rose MR, Simpson SC, Tadler SC. Long-Term Experimental Evolution in Escherichia coli. I. Adaptation and Divergence During 2,000 Generations. The American Naturalist. 1991;138(6):1315–1341. [Google Scholar]
- Bennett AF, Dao KM, Lenski RE. Rapid evolution in response to high-temperature selection. Nature. 1990;346(6279):79–81. doi: 10.1038/346079a0. [DOI] [PubMed] [Google Scholar]
- Bell G. Evolutionary rescue and the limits of adaptation. Philosophical Transactions of the Royal Society B-Biological Sciences. 2013;368(1610) doi: 10.1098/rstb.2012.0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G, Gonzalez A. Adaptation and Evolutionary Rescue in Metapopulations Experiencing Environmental Deterioration. Science. 2011;332(6035):1327–1330. doi: 10.1126/science.1203105. [DOI] [PubMed] [Google Scholar]
- Bell G, Gonzalez A. Evolutionary rescue can prevent extinction following environmental change. Ecology Letters. 2009;12(9):942–948. doi: 10.1111/j.1461-0248.2009.01350.x. [DOI] [PubMed] [Google Scholar]
- San Millan A, Escudero JA, Gifford DR, Mazel D, MacLean RC. Multicopy plasmids potentiate the evolution of antibiotic resistance in bacteria. Nature Ecology & Evolution. 2016;1:0010. doi: 10.1038/s41559-016-0010. [DOI] [PubMed] [Google Scholar]
- Alekshun MN, Levy SB. Molecular mechanisms of antibacterial multidrug resistance. Cell. 2007;128(6):1037–1050. doi: 10.1016/j.cell.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405(6784):299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- Carattoli A. Plasmids and the spread of resistance. Int J Med Microbiol. 2013;303(6-7):298–304. doi: 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Smillie C, Garcillán-Barcia MP, Francia MV, Rocha EP, de la Cruz F. Mobility of plasmids. Microbiol Mol Biol Rev. 2010;74(3):434–452. doi: 10.1128/MMBR.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Millan A, Heilbron K, Maclean RC. Positive epistasis between co-infecting plasmids promotes plasmid survival in bacterial populations. ISME J. 2013. [DOI] [PMC free article] [PubMed]
- Stoesser N, et al. Evolutionary History of the Global Emergence of the Escherichia coli Epidemic Clone ST131. MBio. 2016;7(2) doi: 10.1128/mBio.02162-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Millan A, et al. Multiresistance in Pasteurella multocida is mediated by coexistence of small plasmids. Antimicrob Agents Chemother. 2009;53(8):3399–3404. doi: 10.1128/AAC.01522-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero JA, et al. Unmasking the ancestral activity of integron integrases reveals a smooth evolutionary transition during functional innovation. Nat Commun. 2016;7:10937. doi: 10.1038/ncomms10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6(5):343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaveroche MK, Ghigo JM, d'Enfert C. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 2000;28(22):97. doi: 10.1093/nar/28.22.e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roux F, Binesse J, Saulnier D, Mazel D. Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector. Appl Environ Microbiol. 2007;73(3):777–784. doi: 10.1128/AEM.02147-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Performance standards for antimicrobial susceptibility testing; 19th ed. Approved standard M100-S19. Wayne, PA, USA: Clinical and Laboratory Standards Institute; 2009. [Google Scholar]
- Lindsey HA, Gallie J, Taylor S, Kerr B. Evolutionary rescue from extinction is contingent on a lower rate of environmental change. Nature. 2013;494(7438):463–467. doi: 10.1038/nature11879. [DOI] [PubMed] [Google Scholar]
- Deatherage DE, Barrick JE. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol. 2014;1151:165–188. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, et al. Identifying structural variation in haploid microbial genomes from short-read resequencing data using breseq. BMC Genomics. 2014;15:1039. doi: 10.1186/1471-2164-15-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salverda ML, De Visser JA, Barlow M. Natural evolution of TEM-1 β-lactamase: experimental reconstruction and clinical relevance. FEMS Microbiol Rev. 2010;34(6):1015–1036. doi: 10.1111/j.1574-6976.2010.00222.x. [DOI] [PubMed] [Google Scholar]
- Ramirez MS, Tolmasky ME. Aminoglycoside modifying enzymes. Drug Resist Updat. 2010;13(6):151–171. doi: 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerison ER, Desai MM. Genomic investigations of evolutionary dynamics and epistasis in microbial evolution experiments. Current Opinion in Genetics & Development. 2015;35:33–39. doi: 10.1016/j.gde.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toll-Riera M, San Millan A, Wagner A, MacLean RC. The Genomic Basis of Evolutionary Innovation in Pseudomonas aeruginosa. PLoS Genet. 2016;12(5):1006005. doi: 10.1371/journal.pgen.1006005. [DOI] [PMC free article] [PubMed] [Google Scholar]


