Abstract
The regrowth capacity of damaged neurons governs neuroregeneration and functional recovery after nervous system trauma. Over the past few decades, various intrinsic and extrinsic inhibitory factors involved in the restriction of axon regeneration have been identified. However, simply removing these inhibitory cues is insufficient for successful regeneration, indicating the existence of additional regulatory machinery. Drosophila melanogaster, the fruit fly, shares evolutionarily conserved genes and signaling pathways with vertebrates, including humans. Combining the powerful genetic toolbox of flies with two-photon laser axotomy/dendriotomy, we describe here the Drosophila sensory neuron – dendritic arborization (da) neuron injury model as a platform for systematically screening for novel regeneration regulators. Briefly, this paradigm includes a) the preparation of larvae, b) lesion induction to dendrite(s) or axon(s) using a two-photon laser, c) live confocal imaging post-injury and d) data analysis. Our model enables highly reproducible injury of single labeled neurons, axons, and dendrites of well-defined neuronal subtypes, in both the peripheral and central nervous system.
Keywords: Medicine, Issue 135, Drosophila, sensory neuron, dendritic arborization neuron, neural injury, neuroregeneration, axotomy, dendriotomy, PNS, CNS
Introduction
The inability of axons to regenerate after an injury to the central nervous system (CNS), may lead to permanent disabilities in patients, and also plays a role in the irreversible neurological deficits in neurodegenerative diseases1,2,3,4,5. The CNS environment, as well as the intrinsic growth ability of neurons, determines whether axons are able to regenerate after trauma. Extracellular factors from oligodendrocyte, astroglial, and fibroblastic sources have been shown to impede neuronal growth4,6,7,8, but the elimination of these molecules only allows for limited sprouting5. Intrinsic regeneration signals can influence regenerative success5,9 and represent potential therapeutic targets, but these processes are still not well-defined at the molecular level. Increases in trophic factor signaling or elimination of endogenous brakes, such as the Pten phosphatase10, can result in axonal regeneration in certain circumstances. Combinations of different methods found to be individually effective also only provide limited overall recovery to date11,12,13,14. Therefore, there is a desperate need to identify additional pathways for targeted therapy. In addition to the initiation of axon regrowth, whether and how axons re-wire to the correct target, reform synapse specificity, and achieve functional recovery are important unanswered questions.
In summary, current understanding of the machinery dictating axon regeneration is still very fragmentary. Part of the problem is the technical difficulty of studying axon regeneration in mammals in real-time, an approach that is costly, time-consuming, and challenging for conducting large-scale genetic screens. Drosophila melanogaster, on the other hand, has proven to be an exceptionally powerful system for the study of complex biological questions. The fruit fly has been instrumental in defining genes and signaling pathways that are strikingly conserved in humans and has been a successful model for the study of human conditions, such as neurodegenerative diseases, through the vast molecular genetics tools available to manipulate gene function15. In particular, fruit flies are considered to be an ideal tool for the discovery of genes involved in neural injury and regrowth15,16. Several fly neural injury models have been developed, including adult-head or larval ventral nerve cord (VNC) stabbing with needles, larval VNC or nerve crush with forceps, larval neuron laser axotomy, olfactory receptor neuron removal, brain explants injury, and peripheral nerve lesion by wing severance15,17,18,19,20,21,22,23. Excitingly, recent work utilizing Drosophila injury models have advanced our understanding of the cellular and genetic pathways used by the nervous system to respond to neural injuries, some of which have been shown to be conserved in mammals24,25. Again, this emphasizes the utility of this model organism for identifying novel mechanisms of neural repair.
Described here is a two-photon laser-based Drosophila larval sensory neuron injury model. A two-photon laser was first used to cut axons in zebrafish in vivo in 200326. In the same year, the first laser dendriotomy was performed in Drosophila using a pulsed nitrogen laser27. Shortly afterward, several C. elegans labs used femtosecond lasers to establish models of axon regeneration28. In 2007, Wu and colleagues compared and reported the differences between laser injuries in C. elegans induced by various types of lasers29. In 2010, axon regeneration after laser axotomy was first shown to occur in Drosophila30. Building on this extensive laser injury literature, we have developed a fly neural injury model using the two-photon laser, which allows precise induction of injury to targeted sites with minimal perturbation of neighboring tissues, providing a relatively clean system to study both the intrinsic and extrinsic properties of neuroregeneration with single-cell resolution. Specifically, we have established a set of injury methods for dendritic arborization (da) sensory neurons in both the peripheral nervous system (PNS) and CNS. Da neurons can be grouped into four distinct classes distinguished primarily by their dendrite branching complexities: class I to IV31. Our published work shows that da neuron regeneration resembles mammalian injury models at the phenotypic and molecular level: da neurons display class specific regeneration properties, with class IV but not class I or III da neurons exhibiting regeneration in the PNS; class IV da neuron axons regenerate robustly in the periphery, but their regenerative potential is dramatically reduced in the CNS, thus resembling dorsal root ganglion (DRG) neurons in mammals; enhancing mTOR activity via Pten deletion or Akt overexpression enhances axon regeneration in the fly CNS19. Utilizing this injury model, we have been performing genetic screens and have identified the RNA processing enzyme Rtca as an evolutionarily conserved inhibitory factor for axon regeneration, linking axon injury to cellular stress and RNA modification20.
In the presented paradigm, the injury is induced via laser axotomy/dendriotomy of larval class IV or III da neurons, labeled by ppk-CD4-tdGFP or 19-12-Gal4, UAS-CD4-tdGFP, repo-Gal80, respectively. The injury is performed on 2nd to 3rd instar larvae at around 48 - 72 h after egg laying (h AEL). For PNS axotomy the lesion is targeted to the section of axon ~20 - 50 µm away from the cell body, for CNS axotomy to an area of ~20 µm in diameter at the commissure junction in the VNC, and for dendriotomy to the primary dendritic branch points. The same neuron is imaged at 8 - 24 h after injury (AI) to confirm complete transection, and at 48 - 72 h AI to assess regeneration. Through time-lapse confocal imaging, the degeneration and regeneration of individual axons/dendrites that have been injured in vivo can be monitored over time.
Protocol
1. Preparation of Culture Plates and Bottles
- Preparation of grape juice agar plates
- Add 10 g of agar powder, 200 mL grape juice, and 192 mL ddH2O into a beaker and microwave for about 4-5 min, stirring intermittently until the agar is completely dissolved.
- In a fume hood, cool down the solution to approx. 60 °C. Add 4.2 mL 95% ethanol and 4.0 mL glacial acetic acid. Adjust the total volume of the solution to 400 mL with ddH2O. Mix well.
- For each 35-mm plate, add about 2-3 mL of the solution. Make approximately 120-150 plates for a total of 400 mL grape juice agar solution.
- Wait for 10 min to cool down the plates and solidify the agar solution. Pack the grape juice agar plates in self-sealed ziplock bags and store at 4°C for future use.
- Preparation of Drosophila culture bottles
- Use a blade to punch a 1.5 cm side length triangular hole on one wall of the Drosophila culture bottle and fill the hole with a 2-2.5 cm diameter ball of cotton for ventilation.
- Plug the bottle with a grape juice agar plate supplemented with about 0.5 cm3 of yeast paste.
2. Collection of Drosophila Larvae
- Set up crosses of adult flies
- Place 10 virgin females and 5 male adult flies together in a culture bottle plugged with a grape juice agar plate.
- Place the bottle bottoms up at 25°C, so that flies will lay eggs on the grape juice agar plate. Change the plate with yeast paste at least once a day. For a 2-h collection period, which permits the harvesting of larvae of a homogenous developmental stage, start with more than 20 virgin females and 10 males in Step 2.1.1.
- Culture the plate at 25°C in a 60-mm Petri dish with a wet tissue, for example, soaked in 0.5% propionic acid solution. Arrange the tissue so that it does not block oxygen flow. NOTE: The propionic acid solution is used to maintain humidity in the dish and avoid the growth of mold.
- Harvest larvae of a specific age
- Use a pair of forceps to transfer larvae of the desired stage, for example, 2nd to 3rd instar larvae at 48-72 h after egg laying (AEL), to a new grape juice agar plate without yeast paste.
- Remove the yeast from the larvae's skin by letting them crawl around on the new plate, to prevent from potential interfering of yeast with laser axotomy and imaging. Alternatively, thoroughly clean the larvae by washing them in a dish of PBS and drying briefly on a piece of tissue paper.
3. Two-photon Injury and Confocal Imaging
- Microscope setup NOTE: A confocal laser scanning microscope with a two-photon laser was used for this experiment, but other systems with an equivalent setup will also suffice. The two-photon laser (930 nm) was used for delivering injury and an Argon laser (488 nm) was used for confocal imaging of GFP.
- At the beginning of each session, turn on the two-photon laser and/or the confocal laser(s), and the microscope. Open the imaging software.
- For two-photon injury, set up the following parameters for imaging GFP with the two-photon laser at 930 nm (1,950 mW).
- Select line scan mode. Open up the pinhole all the way. Increase laser intensity to ~20% (390 mW).
- Select 512 x 512 as the frame scan. Use maximal scanning speed (typically with the pixel dwell time at 0.77 µs). Ensure that the average number is 1 and bit depth is 8 bits.
- Set gain to ~750, and the offset to 0.
- Save this pre-set experimental protocol as 2P GFP 930 Ablation, allowing for easy reuse in future experiments.
- For confocal imaging, set up the following parameters for imaging GFP with the Argon laser at 488 nm:
- Select the Acquisition tab and then Z-stack.
- Under "Laser", turn on the power for the 488 nm Argon laser.
- Go to Channels, select the 488 mm laser, and increase the laser power to 5-10%. For the pinhole, use the 1-2 airy unit (AU). Adjust the gain to 650.
- In Acquisition Mode, select 1024 x 1024 as the frame scan, use the maximum scan speed, an average number of 2, and bit depth of 8 Bit.
- Save this pre-set experimental protocol as GFP Imaging.
- Larvae anesthesia with diethyl ether and mounting
- In a fume hood, place a 60-mm glass dish in a 15-cm plastic Petri dish. Fold and lay a piece of tissue paper at the bottom of the glass dish, then place a grape juice agar plate on the tissue. Add diethyl ether into the glass dish, to the point where the tissue paper is soaked and there is a layer of liquid ether remaining in the dish. Keep the lid on at all times.
- Prepare a glass slide with one drop of halocarbon 27 oil in the center. Add 4 spots of vacuum grease onto the four corners of the slide, to later support the coverslip.
- Use forceps to pick up a cleaned larva and place it on the agar plate in the 60-mm glass dish. Cover the glass dish with its lid and wait until the larva stops moving. For PNS injury/imaging, take out the larva as soon as its tail stops twitching. For the CNS, wait until the entire larva becomes motionless, especially the head segments. NOTE: The timing of ether exposure is critical. See Discussion.
- Carefully pick up the anesthetized larva and place it head-upright into the drop of halocarbon oil on the slide. Add a coverslip on top of the slide. Use gentle pressure to push down on the coverslip, until it touches the larva (Figure 1A).
- Adjust the larva's position by gently pushing the coverslip towards the left or right to roll the larva, so that the neuron/axon/dendrite of interest is on the top and closest to the microscope lens.
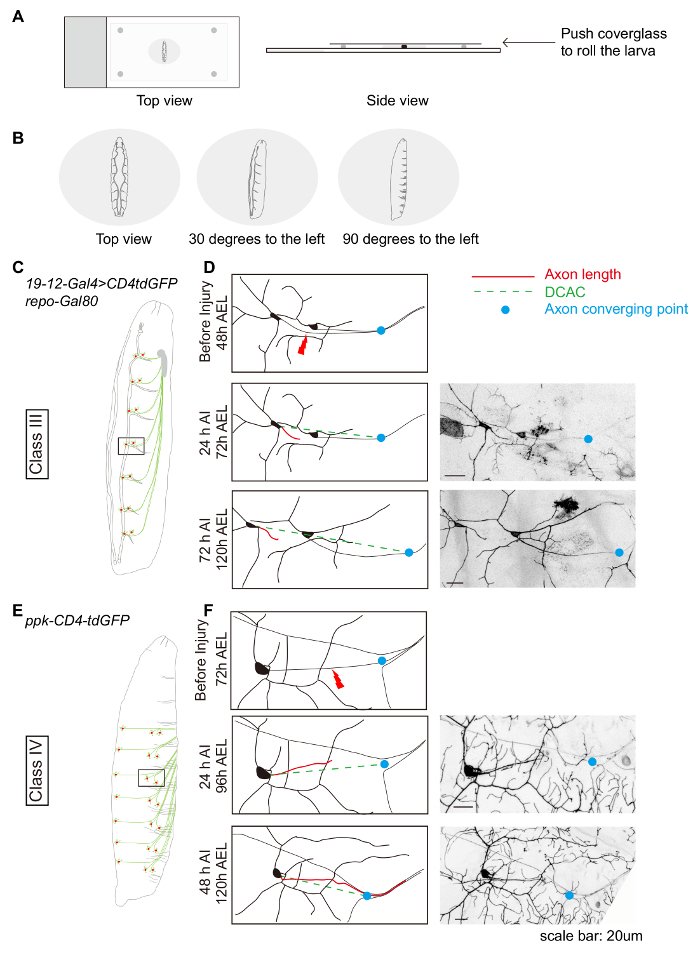
- For PNS injury, mount the larva dorsal side up, so that both the tracheas are visible. Then roll the larva ~30 degrees to the left for injuring class III da neuron axons (Figure 1B and 1C), 90 degrees for injuring class IV da neuron axons (Figure 1B and 1E), or ~30 degrees for injuring class IV da neuron dendrites (Figure 2A).
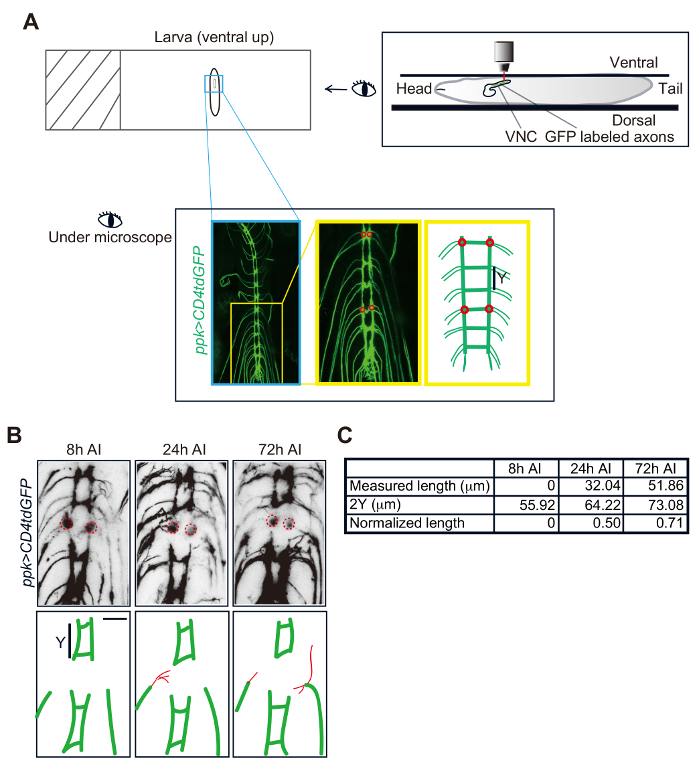
- For CNS injury, position the larva to be perfectly ventral side up (Figure 3A), so that the region of interest is closest to the microscope lens in the z-plane.
- Injury by two-photon laser
- Place the slide with the larva under the microscope and secure it in place with the slide holder on the stage. Use the 10X (0.3 NA) objective to find the larva.
- Add 1 drop of objective oil onto the coverslip, switch to the 40X (1.3 NA) objective and adjust the focus.
- Switch to the scanning mode and reuse the experimental protocol 2P GFP 930 Ablation. Make sure the pinhole is opened all the way. NOTE: The configuration needs to be optimized based on individual systems.
- Start the Live mode to locate the region of interest (ROI), and fine-tune the settings to achieve good image quality with the appropriate zoom. NOTE: The purpose of this step is to find the neuron/axon/dendrite to injure, rather than taking the best quality image. Therefore, use the minimal settings sufficient to visualize the target area, in order to avoid overexposure or photobleaching.
- Stop Live scan, so that the Crop button will become available. Let the still image serve as the roadmap. Select the Crop function and adjust the scan window to focus on the target to be injured.
- Reduce the ROI to be the size of the prospective site of injury. For example, just cover the width of an axon or a dendrite, to ensure the precision of the injury and reduce damage to neighboring tissues. If desired, zoom in on the ROI before cropping, allowing for more precise injury.
- Open a new imaging window. Reduce the scan speed and increase laser intensity. Determine the increase in laser intensity based on the tissue fluorescence signal scanned in Live mode.
- Typically, set the two-photon laser intensity starting from 25% for PNS injury and 50-100% for VNC injury. For PNS axon injury, ensure that the laser intensity is ~480 mW and pixel dwell time is 8.19 μs. For the VNC axon injury, ensure that the laser intensity and pixel dwell time are usually 965-1930 mW and 8.19-32.77 μs, respectively.
- Start Continuous scan. Leave the cursor hovering over the Continuous button. Keep a close eye on the image and stop the scan as soon as a drastic increase in fluorescence is observed. NOTE: The appearance of the fluorescence spike is due to auto-fluorescence at the injury site.
- Switch back to the Live mode by reusing the settings. Find the region of interest that was just targeted by adjusting the focus. NOTE: A good indication of successful injury is the appearance of a small crater, ring-like structure, or localized debris right at the injury site.
- Move to the next neuron and repeat from Step 3.3.5, to injure multiple neurons in a single animal. Or repeat Step 3.3.5 while gradually increasing the power and/or reducing scan speed if the initial injury was insufficient. NOTE: In the case that the laser power is too high, a large damaged area will be visible in the post-injury live scan image. Too much injury may cause the death of the larva.
- Recover the larva by carefully removing the coverslip and transferring the injured larva onto a new plate with yeast paste. Ditch several caves on the agar plate with forceps; alternatively, make an island of agar in the plate instead of using the whole plate, to reduce the possibility of the larva crawling out of the plate.
- Put the plate in a 60-mm Petri dish with wet tissue (soaked with 0.5% propionic acid solution) and culture at room temperature or 25°C. NOTE: The larva will remain in the larval stage for approximately an extra day at room temperature (22°C) compared to at 25°C.
- Post-injury confocal imaging
- Image the injured larva at desired time points by preparing the larva using the same procedure of anesthesia and mounting as in Step 3.2, then imaging with the confocal laser. NOTE: Image the larva at 24 h after injury (AI) to confirm axonal injury and at 48 h AI (class IV da neurons) or 72 h AI (class III da neurons) to assess regeneration.
- Locate the larva using the 10X objective, then switch to a 25X (0.8 NA) objective. Reuse the experimental protocol GFP Imaging.
- Click the Live button and find the same neuron injured previously.
- Set the first and last Z positions in the live scan window. Press stop and click Start experiment to acquire a Z-stack image. NOTE: Make sure that a normalization point (the axon converging point) is included when capturing images so that quantification of regeneration is possible (Figure 1D, 1F) – this is discussed further in the data analysis section.
- Switch to Image Processing, select the image just taken, and generate a maximum intensity projection. Save both the z-stack and maximum intensity projection images.
4. Data Analysis
Process and quantify images using either the imaging software or ImageJ.
- Quantification of axon regeneration in the PNS
- Calculate Regeneration percentage, which refers to the percent of regenerating axons among all the axons that were lesioned. Score an axon as regenerating as long as it regrows beyond the injury site.
- Measure Regeneration length, which is the increase in axon length. If quantifying with ImageJ, use the Segmented Line tool to trace the regenerated axon, and use Measure in the Analyze drop-down menu to obtain the length of the line representing the regenerated axon.
- Calculate Regeneration Index, which is the increase in normalized axon length. NOTE: Axon length is normalized by the distance between the cell body and the axon converging point (DCAC) – axon length/DCAC (Figure 1D and 1F). This value helps account for any axonal growth that is due to larval scaling. A positive value represents regeneration, a value of 0 means no regeneration, and a negative value means retraction.
- Quantification of dendrite regeneration
- Calculate Regeneration percentage, which is the percent of da neurons among all those severed that show obvious dendrite regrowth. NOTE: Dendrite regrowth is scored as positive if new dendrites regrow out from the retracted dendritic stem and beyond the injury site. The injury site is determined by landmarks, and in some cases, is readily visible due to the residual injury-induced autofluorescence.
- Calculate Increase of branch points, which counts the addition of new dendritic branch points after injury.
- Calculate Increase of total dendrite length, which is the cumulative length of all the new dendrites added after injury.
- Quantification of axon regeneration in the CNS NOTE: If a lesion site shows degeneration at the first time point imaged (Figure 3B), it will be included in the analysis of regeneration length and rate.
- Measure Regeneration length, which is the length of the regrowing axon. NOTE: The regrowing axon of a single lesion site is identified as it originates off the original axon route before the injury.
- Calculate Normalized regeneration length, which normalizes the regeneration length to the length of the commissure segment - the longitudinal distance between commissures ("Y" in Figure 3A and 3B). NOTE: This value helps correct for the effect of larval size differences.
- Calculate Regeneration rate, which is the percent of regenerating segments among all the segments that were lesioned, to reflect the regeneration capacity of a particular genotype.
Representative Results
Da neurons show differential regeneration potential between the peripheral and central nervous system, as well as class specificity. This provides unique opportunities to screen for novel factors that are required for axon regeneration (using class IV PNS injury), as well as those that are inhibitory for regeneration (with class IV CNS injury and class III PNS injury).
Axon regeneration in the PNS
As an example, the characterization of regeneration of class III and class IV da neurons is described. These neurons are located bilaterally in each body segment. Multiple neurons can be injured in the same larva; typically, 3-4 neurons in the right side of abdominal segments A7-A2. Class III and class IV da neurons can be visualized by 19-12-Gal4, UAS-CD4tdGFP, repo-Gal80 and ppk-CD4tdGFP, respectively. Anesthetize and mount 48-72 h AEL larvae as described, adjusting the position of the larvae so that the da neurons of interest are facing up (Figure 1A and 1B). We usually injure the class III ddaF and class IV v'ada neurons (Figure 1C and 1E). Perform axotomy and recover larvae as described. Shortly after injury (AI), the larvae will have recovered from surgery and exhibit normal locomotion. The survival rate here is typically over 80%. Discard larvae that are dead or sick. Remount the remainder as described and assess degeneration. At 24 h AI, the distal axons should have completed degeneration at this time point19, and the axon stem will be readily visible (Figure 1D and 1F). Reimage the same larvae at 48 h AI for class IV da neurons or 72 h AI for class III da neurons to assess regeneration. We usually aim to assess at least 20 injured neurons per experimental condition. In (wild type) WT animals, whereas typically ~70% of severed class IV da neurons will have regenerated beyond the injury site (Figure 1F), class III da neurons fail to regrow, evidenced by growth cone stalling (Figure 1D).
Dendrite regeneration
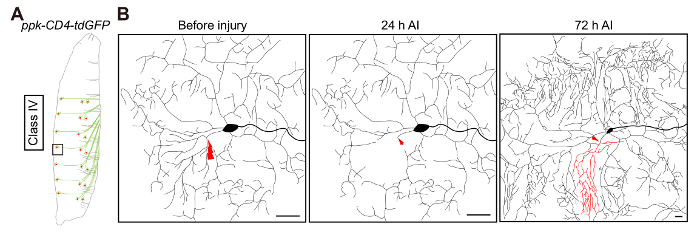
We usually perform dendriotomy on class IV da neurons ddaC (Figure 2A). As shown in the schematic diagram, the injury is targeted to the primary dendritic branch point. Based on experience, when injured at 48 h AEL, ~50% of ddaC neurons regenerate their dendrites (Figure 2B). In the remaining 50%, neighboring dendrites invade and cover the vacant space. Additionally, the regeneration potential of these neurons is reduced if injured at a later developmental stage.
Axon regeneration in the CNS
For VNC axon injury, the survival rate of larvae varies substantially and depends upon the age at which injury is induced. Based on experience, larvae of 48-72 h AEL typically have the highest survival rate (>60%) among the different stages tested. Larvae younger than 48 h AEL survive poorly after injury, while in those older than 72 h AEL it is difficult to introduce injury in the VNC. Moreover, it is easier to induce injury at the posterior commissure segments than the anterior, as these posterior segments of the VNC are closer to the ventral surface and thus more accessible by laser (Figure 3A).
To injure class IV da neuron axons in the CNS, mount the larvae as previously described (Figure 3A). Under the microscope, locate the ladder-like structure of axon bundles that form part of the VNC (Figure 3A), and perform axotomy as outlined. Degeneration is confirmed at 8 h AI and regeneration are assessed at 24 and 72 h AI. As shown in Figure 3B, axons at 8 h AI have already started to degenerate, and at 24 h AI, axon regeneration is observed while axon debris can still be found around the injury sites. WT axons show limited regrowth in the VNC and fail to reconnect the gaps generated by the injury (Figure 3B). To quantify the regeneration ability of injured axons, the regrowth length after injury is measured and the length of the commissure segment (Y in Figure 3A) is used for normalization (Figure 3C).
Figure 1: Da neuron axon regeneration in the periphery displays class specificity. (A and B) Schematic drawing showing the position of the larvae. (C) Schematic drawing of class III da neurons. (D) Axons of the class III da neurons ddaF, labeled with 19-12-Gal4, UAS-CD4-tdGFP, repo-Gal80/+, fail to regrow. (E) Schematic drawing of class IV da neurons. (F) Axons of the class IV da neurons v'ada, labeled with ppk-CD4-tdGFP/+, regrow beyond the lesion site. (D and F) Red line indicates axon length while green dashed line marks the distance between the cell body and the axon converging point (DCAC). The blue dot marks the axon converging point. Scale bar = 20 µm. Please click here to view a larger version of this figure.
Figure 2: Da neuron dendrite regeneration. (A) Illustrative representation of class IV da neurons. (B) Illustrative representation of dendrite regeneration in class IV da neuron ddaC, labeled by ppk-CD4-tdGFP/+. Laser ablation is targeted to the primary branch point and is conducted at 48 h AEL. At 24 h AI injury transection of the neurite is confirmed, and at 72 h AI regeneration is quantified. Dendrites of ddaC neurons demonstrate substantial regrowth, with new dendritic branches sprouting from the severed stem to tile the vacant space. It is worth noting that new terminal branches are continuously added to the uninjured dendrites at this developmental stage. Scale bar = 20 µm. Please click here to view a larger version of this figure.
Figure 3: Da neuron axon regeneration in the VNC. (A) Schematic drawing of a Drosophila larva mounted on a slide and imaged under the microscope. Class IV da neuron axons in the VNC visualized in a ppk-CD4tdGFP/+ larva. Two candidate commissure segments are shown in the zoomed-in image and the schematic drawing. Each of them has two injury sites (red circles). (B) Confocal images of one injured segment imaged at 8, 24 and 72 h after injury (AI). Red lines depict the regrowing axons. (C) Measurement and normalization of regrowing axons. Scale bar = 20 µm. Please click here to view a larger version of this figure.
Discussion
When setting up fly crosses, the number of females and males used can vary depending on the genotypes and the number of larvae needed for specific experiments. For WT flies, typical cross uses 10 females and 5 males. The collection window may be narrowed, depending on the accuracy of the larvae age required. For example, a 2-h collection period will yield larvae of a more homogenous population. In this case, using 20 or more virgin females to set up the crosses will help yield sufficient eggs. The yield from the first day is usually scarce, so collect the plate with eggs two or more days after setting up the chamber. When further culturing the plate with eggs, it is recommended to soak the tissue in the 60-mm Petri dish in 0.5% propionic acid solution instead of water, which will not only help maintain humidity in the dish but also avoid the growth of mold.
When setting up the devices for larvae anesthesia and mounting, make sure to use a glass dish because ether will melt through plastics. The glass dish is housed in a 15-cm plastic Petri dish in case of a leak. The tissue paper helps preserve ether in the dish so that the larva can be effectively knocked out by the ether vapor. Using amber dropping bottles to store aliquots of ether and adding ether in droplets with the glass dropper is recommended. Replenish the ether and always keep a layer of liquid ether at the bottom of the dish for optimal effect.
The timing of ether exposure is critical: under-anesthesia will lead to larva recovery in the middle of the imaging session and shaky images; an anesthesia overdose, or direct contact with the ether liquid, will cause lethality. To maximize survival and success rate, our rule of thumb is as follows: for PNS injury/imaging, take out the larva as soon as its tail stops twitching; for the CNS, wait until the entire larva becomes motionless, especially the head segments. Even a slight movement will interfere with the injury and imaging of VNC axons. Older larvae tend to take longer to knock out. It usually takes less than 2 min for larvae younger than 72 h AEL and 2-5 min for larvae older than 72 h AEL.
When setting up the intensity of the two-photon laser for injury, the value is determined by the tissue fluorescence signal obtained from the "Live" scan. The injury is introduced by "Continuous" scan as in step 3.3. The injury-induced increase in fluorescence is due to autofluorescence at the injury site and serves as a good indicator of the severance of the neurite. Typically, it takes 1-10 s to see the fluorescence spike. It is critical to control the scan time once fluorescence is elevated. For PNS injury, it is essential to stop scanning immediately. Prolonged exposure will enlarge the injury site and damage neighboring tissues. However, this does provide the opportunity to adjust scan time and manipulate the severity of the injury. A successful injury should have a lesion size diameter smaller than 3-4 μm. For VNC axon injury, the VNC axons are embedded much deeper compared to the da neuron PNS axons/dendrites, which are right underneath the skin, and thus require higher laser intensity. We usually leave the scan on for a few more seconds. This is to ensure that the entire axon bundle is severed. If the radius of the lesion sites is more than half the width of the commissure bundle, such injury sites are counted as unsuccessful. Such larvae have a low survival rate and will not be included in the analysis.
For axon regeneration, the regeneration ability is similar across larval stages. But for dendrite regeneration after a single dendrite cut, the regeneration potential is reduced after 72 h AEL19. Thus, axon injury is typically performed at 48 h-72 h AEL and dendrite injury at 48h AEL, with the assessment of regeneration at 120h AEL. Larvae of 24h AEL can also be used, but they require more careful handling, given their smaller size. Larvae pupate after 120 h AEL, making imaging more challenging. Therefore, our endpoint is usually 120 h AEL.
What is the interrelationship between degeneration and regeneration? For PNS injury, at 24 h AI, normally the distal axon/dendrite in WT has completed Wallerian degeneration while regeneration has not started at this time point. Therefore, we believe that in WT larvae, degeneration of severed neurites only has a very limited impact on regeneration, if at all. For VNC injury, axons at 8 h AI have already started to degenerate, and at 24 h AI, axon regeneration is observed while axon debris could still be found around the injury sites. The debris does not seem to block regeneration. However, it is possible that under certain circumstances, there may be an overlap or even a crosstalk between degeneration and regeneration. In fact, it has been reported that in aged mice, debris clearance after peripheral nerve damage is slower than that in young animals. Concomitantly, slower reinnervation of the neuromuscular junction was observed, which may be attributed to the greater number of obstructions regenerating axons encounter in the old animals. Surprisingly, however, axons from aged animals regenerate quickly and reinnervate neuromuscular junction sites efficiently when not confronted with debris32. This suggests that facilitating debris clearance might be a potential strategy to promote regeneration.
Compared to other neuroregeneration models, the fly sensory neuron injury model has unique advantages. Mouse models usually take weeks to months to perform and are not suitable for conducting large-scale genetic screens; C. elegans only has a primitive central nervous system which may not closely recapitulate the regeneration barriers in the mammalian CNS; different from mammals, zebrafish CNS axons regenerate robustly. The rapid life cycle of flies, the versatility of fly genetics, accessibility and stereotypical patterning of axons/dendrites of fly sensory neurons, and the characteristic regeneration properties of fly sensory neurons – subtype specific regeneration in the PNS and limited regeneration in the CNS – make Drosophila da neurons an attractive model for studying neuroregeneration. Moreover, recent studies from crushed mouse retinal ganglion cells (RGCs) also suggest that neuronal subtypes bear distinct regeneration competence; some RGC subtypes can regenerate, whereas others in the seemingly homogenous nerve bundle fail to regrow33. This important finding suggests that neuronal type-specific strategies should be exploited to promote regeneration and functional recovery and argues strongly that the induction of axon injury and subsequent regeneration analysis should be performed in a neuronal subtype-specific manner. Furthermore, the cellular and molecular determinants for this regeneration type-specificity remain largely unknown19,33. Therefore, the da neuron injury model offers the ideal paradigm to tackle these issues.
Compared to axon regeneration, studies focusing on dendrite regeneration are much scarcer. Dendrite injury does occur, such as in traumatic brain injury, stroke, and many forms of neurodegeneration, yet almost nothing is known about dendrites' ability to repair and reform neural connections. The da neuron injury model again provides a highly accessible system that displays stereotypical patterning, class specificity, and temporal regulation19, to explore this direction.
It is also worth mentioning that while it is possible to injure a labeled single axon in the PNS, the way injury is performed in the CNS results in the lesion of a bundle of axons. If desired, MARCM34 or the FLP-out clone35 approach may be used to label single axons in the CNS. Additionally, when glial cells are simultaneously labeled with mRFP (Repo-Gal4, UAS-mRFP), the accumulation of glia processes is observed specifically at the lesion site. Furthermore, the expression of Ptp99A, the fly homolog of chondroitin sulfate proteoglycan (CSPG) phosphacan/ PTPRZ1, is up-regulated at the lesion site. Ptp99A co-localizes with glial cells and surrounds the injury site, forming a ring-like structure similar to what has been reported for astroglial scars in mammals19,36,37. In conclusion, the da neuron injury model, when combined with markers of other cell types such as glial cells or immune cells, will allow in vivo real-time surveillance of the multicellular interactions between an injured neuron and its surrounding environment.
While this fly larvae sensory neuron injury model provides us opportunities to find potential neuroregeneration regulators both in the PNS and CNS, it still has several limitations. First, it is not of high throughput at the present stage. Typically, 5-6 genotypes could be screened by one person in one week. It needs to be optimized in order to perform an unbiased screen. Second, as the ether anesthesia on larvae can last from several min to no more than twenty min, it is not optimal for long-term imaging. Thus, specific time points that are representative of axon degeneration and regeneration respectively are chosen for imaging. Third, even though this protocol is introducing a way to injure axons precisely, the possibility of damage to surrounding tissues cannot be completely ruled out. In the VNC, these tissues can be glial cells, axons, and dendrites of other neurons. To minimize this potential caveat, we minimize the injury sites, apply the lowest laser power possible, and perform the same procedures in parallel to both control and experiment groups. Fourth, in the process of setting up the CNS injury paradigm, different injury sites were tested, including the sites close to the axon termini that we chose to use and the entry point of axons into the VNC. The entry points were found to be more difficult to injure because they are deeper in the tissue and more likely to move. Thus, for these practical reasons, we opt for the current method, which is more consistent, better controlled, and good for larger-scale experiments. One potential concern, as stated above, is the possibility of injuring neighboring tissues, such as the postsynaptic components and the glial cells. On the other hand, it is an outstanding question how the damaged surrounding tissues influence the degeneration and regeneration of axons. For example, how the glial scar affects axon regeneration. This presents another reason why our injury model may closely resemble injury models in mammals, in which axons and tissues around the lesion sites are usually injured simultaneously.
Laser injury followed by time-lapse microscopy is a sensitive assay for studying axon/dendrite regeneration. However, one main concern of this assay is the perceived cost, which ranges from <$10K for low-end solid-state pulse lasers, $25-100K for femtosecond lasers to >$100K for two-photon lasers. There are several good discussions about different laser systems used for axotomy29,38,39,40,41,42,43,44,45. To summarize, conventional lasers are optimal for cutting axons within about 30-50 µm from the surface. There will be more collateral damage with the nano and pico second lasers compared with the femtosecond laser, especially as the depth of the target area increases45. For injuring axons in the VNC, the depth of which is typically about 50-100 µm, it is essential to minimize tissue damage. In this case, the two-photon laser is ideal, which focuses the laser power to the focal plane, reducing collateral tissue damage without compromising tissue penetration. In conclusion, the two-photon system is costly but offers the best precision and tissue preservation. However, if only the PNS axons are the target for axotomy, conventional pulse lasers may be a more affordable alternative.
Disclosures
The authors have nothing to disclose.
Acknowledgments
We thank Jessica Goldshteyn for technical support. Work in the Song lab is funded by the NIH grant R00NS088211, and the Intellectual and Developmental Disabilities Research Center (IDDRC) New Program Development Award.
References
- Yakura JS. Recovery following spinal cord injury. 1996. [DOI] [PubMed]
- Harel NY, Strittmatter SM. Can regenerating axons recapitulate developmental guidance during recovery from spinal cord injury? Nature reviews. Neuroscience. 2006;7:603–616. doi: 10.1038/nrn1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurewicz A, Matysiak M, Raine CS, Selmaj K. Soluble Nogo-A, an inhibitor of axonal regeneration, as a biomarker for multiple sclerosis. Neurology. 2007;68:283–287. doi: 10.1212/01.wnl.0000252357.30287.1d. [DOI] [PubMed] [Google Scholar]
- Yiu G, He Z. Glial inhibition of CNS axon regeneration. Nat Rev Neurosci. 2006;7:617–627. doi: 10.1038/nrn1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, He Z. Neuronal intrinsic barriers for axon regeneration in the adult CNS. Curr Opin Neurobiol. 2010. [DOI] [PMC free article] [PubMed]
- Liu BP, Cafferty WB, Budel SO, Strittmatter SM. Extracellular regulators of axonal growth in the adult central nervous system. Philos Trans R Soc Lond B Biol Sci. 2006;361:1593–1610. doi: 10.1098/rstb.2006.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Tedeschi A, Park KK, He Z. Neuronal intrinsic mechanisms of axon regeneration. Annu Rev Neurosci. 2011;34:131–152. doi: 10.1146/annurev-neuro-061010-113723. [DOI] [PubMed] [Google Scholar]
- Schwab ME, Strittmatter SM. Nogo limits neural plasticity and recovery from injury. Curr Opin Neurobiol. 2014;27:53–60. doi: 10.1016/j.conb.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Jin Y. Intrinsic Control of Axon Regeneration. Neuron. 2016;90:437–451. doi: 10.1016/j.neuron.2016.04.022. [DOI] [PubMed] [Google Scholar]
- Park KK, et al. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science. 2008;322:963–966. doi: 10.1126/science.1161566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoffroy CG, Hilton BJ, Tetzlaff W, Zheng B. Evidence for an Age-Dependent Decline in Axon Regeneration in the Adult Mammalian Central Nervous System. Cell Rep. 2016;15:238–246. doi: 10.1016/j.celrep.2016.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoffroy CG, et al. Effects of PTEN and Nogo Codeletion on Corticospinal Axon Sprouting and Regeneration in Mice. J Neurosci. 2015;35:6413–6428. doi: 10.1523/JNEUROSCI.4013-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D, et al. Restoration of skilled locomotion by sprouting corticospinal axons induced by co-deletion of PTEN and SOCS3. Nat Commun. 2015;6:8074. doi: 10.1038/ncomms9074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, et al. Axonal regeneration induced by blockade of glial inhibitors coupled with activation of intrinsic neuronal growth pathways. Exp Neurol. 2012;237:55–69. doi: 10.1016/j.expneurol.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Bonini NM. Axon degeneration and regeneration: insights from Drosophila models of nerve injury. Annual review of cell and developmental biology. 2012;28:575–597. doi: 10.1146/annurev-cellbio-101011-155836. [DOI] [PubMed] [Google Scholar]
- Venken KJ, Simpson JH, Bellen HJ. Genetic manipulation of genes and cells in the nervous system of the fruit fly. Neuron. 2011;72:202–230. doi: 10.1016/j.neuron.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyssen M, et al. Amyloid precursor protein promotes post-developmental neurite arborization in the Drosophila brain. The EMBO journal. 2005;24:2944–2955. doi: 10.1038/sj.emboj.7600757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald JM, et al. The Drosophila cell corpse engulfment receptor Draper mediates glial clearance of severed axons. Neuron. 2006;50:869–881. doi: 10.1016/j.neuron.2006.04.028. [DOI] [PubMed] [Google Scholar]
- Song Y, et al. Regeneration of Drosophila sensory neuron axons and dendrites is regulated by the Akt pathway involving Pten and microRNA bantam. Genes Dev. 2012;26:1612–1625. doi: 10.1101/gad.193243.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, et al. Regulation of axon regeneration by the RNA repair and splicing pathway. Nat Neurosci. 2015;18:817–825. doi: 10.1038/nn.4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Forero MG, Fenton JC, Hidalgo A. The glial regenerative response to central nervous system injury is enabled by pros-notch and pros-NFkappaB feedback. PLoS Biol. 2011;9:e1001133. doi: 10.1371/journal.pbio.1001133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Soares L, Teng X, Geary M, Bonini NM. A novel Drosophila model of nerve injury reveals an essential role of Nmnat in maintaining axonal integrity. Curr Biol. 2012;22:590–595. doi: 10.1016/j.cub.2012.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, et al. Protein turnover of the Wallenda/DLK kinase regulates a retrograde response to axonal injury. J Cell Biol. 2010;191:211–223. doi: 10.1083/jcb.201006039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brace EJ, DiAntonio A. Models of axon regeneration in Drosophila. Exp Neurol. 2017;287:310–317. doi: 10.1016/j.expneurol.2016.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Collins C. Intrinsic mechanisms for axon regeneration: insights from injured axons in Drosophila. Curr Opin Genet Dev. 2017;44:84–91. doi: 10.1016/j.gde.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith JA, Terasaki M. Controlled damage in thick specimens by multiphoton excitation. Mol Biol Cell. 2003;14:1808–1817. doi: 10.1091/mbc.E02-03-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura K, et al. Distinct developmental modes and lesion-induced reactions of dendrites of two classes of Drosophila sensory neurons. J Neurosci. 2003;23:3752–3760. doi: 10.1523/JNEUROSCI.23-09-03752.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanik MF, et al. Neurosurgery: functional regeneration after laser axotomy. Nature. 2004;432:822. doi: 10.1038/432822a. [DOI] [PubMed] [Google Scholar]
- Wu Z, et al. Caenorhabditis elegans neuronal regeneration is influenced by life stage, ephrin signaling, and synaptic branching. Proc Natl Acad Sci U S A. 2007;104:15132–15137. doi: 10.1073/pnas.0707001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone MC, Nguyen MM, Tao J, Allender DL, Rolls MM. Global up-regulation of microtubule dynamics and polarity reversal during regeneration of an axon from a dendrite. Mol Biol Cell. 2010;21:767–777. doi: 10.1091/mbc.E09-11-0967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber WB, Jan LY, Jan YN. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development. 2002;129:2867–2878. doi: 10.1242/dev.129.12.2867. [DOI] [PubMed] [Google Scholar]
- Kang H, Lichtman JW. Motor axon regeneration and muscle reinnervation in young adult and aged animals. J Neurosci. 2013;33:19480–19491. doi: 10.1523/JNEUROSCI.4067-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, et al. Subtype-specific regeneration of retinal ganglion cells following axotomy: effects of osteopontin and mTOR signaling. Neuron. 2015;85:1244–1256. doi: 10.1016/j.neuron.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Grueber WB, et al. Projections of Drosophila multidendritic neurons in the central nervous system: links with peripheral dendrite morphology. Development. 2007;134:55–64. doi: 10.1242/dev.02666. [DOI] [PubMed] [Google Scholar]
- Buss A, et al. NG2 and phosphacan are present in the astroglial scar after human traumatic spinal cord injury. BMC Neurol. 2009;9:32. doi: 10.1186/1471-2377-9-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeon RJ, Jurynec MJ, Buck CR. The chondroitin sulfate proteoglycans neurocan and phosphacan are expressed by reactive astrocytes in the chronic CNS glial scar. J Neurosci. 1999;19:10778–10788. doi: 10.1523/JNEUROSCI.19-24-10778.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raabe I, Vogel SK, Peychl J, Tolic-Norrelykke IM. Intracellular nanosurgery and cell enucleation using a picosecond laser. J Microsc. 2009;234:1–8. doi: 10.1111/j.1365-2818.2009.03142.x. [DOI] [PubMed] [Google Scholar]
- Hutson MS, Ma X. Plasma and cavitation dynamics during pulsed laser microsurgery in vivo. Phys Rev Lett. 2007;99:158104. doi: 10.1103/PhysRevLett.99.158104. [DOI] [PubMed] [Google Scholar]
- Venugopalan V, Guerra A, 3rd, Nahen K, Vogel A. Role of laser-induced plasma formation in pulsed cellular microsurgery and micromanipulation. Phys Rev Lett. 2002;88:078103. doi: 10.1103/PhysRevLett.88.078103. [DOI] [PubMed] [Google Scholar]
- Bourgeois F, Ben-Yakar A. Femtosecond laser nanoaxotomy properties and their effect on axonal recovery in C. elegans. Opt Express. 2008;16:5963. doi: 10.1364/oe.16.005963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien GS, et al. Two-photon axotomy and time-lapse confocal imaging in live zebrafish embryos. J Vis Exp. 2009. [DOI] [PMC free article] [PubMed]
- Tsai PS, et al. Plasma-mediated ablation: an optical tool for submicrometer surgery on neuronal and vascular systems. Curr Opin Biotechnol. 2009;20:90–99. doi: 10.1016/j.copbio.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SH, Clark DA, Gabel CV, Mazur E, Samuel AD. The role of the AFD neuron in C. elegans thermotaxis analyzed using femtosecond laser ablation. BMC Neurosci. 2006;7:30. doi: 10.1186/1471-2202-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams W, Nix P, Bastiani M. Constructing a low-budget laser axotomy system to study axon regeneration in C. elegans. J Vis Exp. 2011. [DOI] [PMC free article] [PubMed]


