Abstract
Biofilm formation is a dynamic, multistage process that occurs in bacteria under harsh environmental conditions or times of stress. For enteric pathogens, a significant stress response is induced during gastrointestinal transit and upon bile exposure, a normal component of human digestion. To overcome the bactericidal effects of bile, many enteric pathogens form a biofilm hypothesized to permit survival when transiting through the small intestine. Here we present methodologies to define biofilm formation through solid-phase adherence assays as well as extracellular polymeric substance (EPS) matrix detection and visualization. Furthermore, biofilm dispersion assessment is presented to mimic the analysis of events triggering release of bacteria during the infection process. Crystal violet staining is used to detect adherent bacteria in a high-throughput 96-well plate adherence assay. EPS production assessment is determined by two assays, namely microscopy staining of the EPS matrix and semi-quantitative analysis with a fluorescently-conjugated polysaccharide binding lectin. Finally, biofilm dispersion is measured through colony counts and plating. Positive data from multiple assays support the characterization of biofilms and can be utilized to identify bile salt-induced biofilm formation in other bacterial strains.
Keywords: Immunology and Infection, Issue 135, Biofilm formation, enteric pathogens, Shigella, bile, bile salts, EPS matrix, dispersion
Introduction
Biofilm formation is an important bacterial survival strategy induced during harsh environmental conditions. Exposure to bactericidal compounds like antibiotics or changes in nutrient or oxygen availability induces a stressed state in bacteria that can be alleviated through biofilm formation. A biofilm is characterized by bacterial attachment to a surface or other bacteria and is accompanied by the secretion of an EPS matrix primarily composed of polysaccharides1,2,3. Biofilm formation is a dynamic process in which a cascade of events culminates in formation of a mature adherent bacterial community1,2,3. Bacteria produce adhesins to facilitate early attachment while shifting adhesin gene expression profiles to strengthen attachment during biofilm maturation. Simultaneously, EPS production occurs to coat the bacterial community in a matrix to protect the cells from the initial stressor. Bacteria contained within the biofilm are slow growing; and as such, renders most antibiotics ineffective. Furthermore, the slow growth conserves energy until conditions change to favor bacterial growth1,2,3. After the harsh conditions have passed, bacteria disperse the biofilm and resume a planktonic lifestyle1,2,3. Traditionally, biofilms are observed on surfaces and represent a persistent clinical challenge due to infection reservoirs present on catheters and in-dwelling devices1,2,3.
Biofilm formation was recently described for several enteric pathogens; bacteria that infect the small intestine or colon4. For Shigella species, infection occurs in the human colon after a transit through the majority of the gastrointestinal tract. During passage through the small intestine, Shigella is exposed to bile; a lipid-degrading detergent secreted into the intestine to facilitate digestion of lipids while simultaneously killing most bacteria5. Enteric pathogens have a unique ability to resist the bactericidal effects of bile6. Our recent analysis utilized in vivo-like combinations of glucose and bile salts to demonstrate robust biofilm formation in S. flexneri as well as other species of Shigella, pathogenic Escherichia coli, and Salmonella4. Previously, Salmonella enterica serovar Typhi was shown to form a bile-induced biofilm due to unique colonization of the gallbladder during chronic infection7,8,9,10. Additionally, prior research with Vibrio11 and Campylobacter12 demonstrated biofilm formation in response to bile. Therefore, the analyses extended the bile-induced biofilm formation observations to other pathogens and help to establish demonstration of a conserved enteric pathogen response to bile. Unlike chronic biofilms in which bacterial gene transcription is limited and cell senescence can occur1,2,3, we propose that the enteric bile-induced biofilm is more transient in nature. This transient, virulent biofilm is hallmarked by a rapid disassembly (as seen in the dispersion assay) and enhanced virulence gene expression observed in the biofilm population4,6.
As biofilm formation is a multifaceted, dynamic process and the use of bile salts as an initiating factor has only been recently described for most enteric pathogens, the tools and techniques used are unique and creative applications of traditional methods. Thus, presented here are three complimentary strategies to quantify several important characteristics of bile salt-induced biofilm formation, including bacterial adherence, production of the EPS matrix, and dispersion of viable bacteria from the biofilm. These techniques have been utilized primarily for research with Shigella; and therefore, evaluation of other enteric pathogens may require optimization. Nevertheless, positive data from all three assays support identification of biofilms and establish reproducible protocols for bile salt-induced biofilm formation.
Protocol
1. Preparation of Reagents
Bile salts medium: To prepare tryptic soy broth (TSB) containing 0.4% bile salts (weight/volume), resuspend 200 mg of bile salts in 50 mL autoclaved TSB. Filter sterilize using a 0.22 µm filter. Make fresh medium weekly. Notes: The bile salts routinely used is a 1:1 mixture of sodium cholate and sodium deoxycholate isolated from ovine and bovine gallbladders. As demonstrated previously4, the presence of glucose was required for bile salt-induced biofilm formation. TSB has added glucose relative to Luria-Bertani (LB) broth; and therefore, was sufficient to induce biofilm formation in Shigella and the other enteric pathogens analyzed. Depending on the bacteria to be analyzed, different glucose concentrations or different sugar requirements might be needed.
0.5% w/v crystal violet in water: Dissolve 2.5 g of crystal violet in 500 mL distilled water. Filter sterilize using a 0.22 µm filter.
Concanavalin A (ConA) conjugated to fluorescein isothiocyanate (FITC): Reconstitute the stock in 1x PBS. Dilute the 10 mg concentrated stock with 400 μL of 1x PBS to a final concentration of 25 µg/mL, and protect from light.
PBS + Glucose: Dissolve 0.2 g glucose in 10 mL 1x PBS (2% w/v glucose final). Filter sterilize using a 0.22 µm filter. Make fresh on the day of use.
PBS + Bile Salts: Dissolve 40 mg in 10 mL 1x PBS (0.4% w/v bile salts final). Filter sterilize using a 0.22 µm filter. Make fresh on the day of use.
PBS + glucose and bile salts: Dissolve 40 mg bile salts and 0.2 g glucose in 10 mL 1x PBS (0.4% w/v bile salts and 2% w/v glucose final). Filter sterilize using a 0.22 µm filter. Make fresh on the day of use.
Prepare LB agar plates.
Formaldehyde/glutaraldehyde fix: Add 810 µL formaldehyde (37% stock solution, 3% final concentration) and 125 µL glutaraldehyde (25% stock solution, 0.25% final concentration) to 14 mL 1x PBS. Mix thoroughly and store at 4 °C. The fix should be cold for proper use. Caution: The fix is toxic and requires hazardous waste disposal.
Antifade mountant solution: Use antifade mountant solution containing 4,6-diamidino-2-phenylindole (DAPI) stain to inhibit photobleaching of immunofluorescent microscopy samples while fluorescently staining the DNA of the bacteria.
2. Preparation of Bacteria
Grow overnight cultures of the bacterial strains to be tested by inoculating 3 mL of TSB with a single, well-isolated colony in a sterile culture tube. Incubate at 37 °C with shaking at 225 rpm for overnight incubation (16 - 24 h). NOTE: Strains should be restreaked from freezer stocks every 2 to 4 weeks, and maintained on plates no more than 2 weeks old.
3. Solid-phase Adherence Assay
NOTE: This assay quantifies adherent bacteria using a 96-well plate method. Bacteria are grown statically in flat bottom plates. Washing is performed to remove non-adherent bacteria and adherent bacteria are stained with crystal violet. The crystal violet stain binds peptidoglycan in the bacterial cell wall and can be solubilized using ethanol. The number of adherent bacteria is determined based on crystal violet retention.
Set up two 1.5 mL tubes. Label with TSB or TSB + bile salts (BS).
Add 1 mL of TSB or TSB + BS to the respective tubes.
Inoculate tubes with 20 µL of overnight culture (at a 1:50 dilution).
In a sterile, clear, flat-bottomed, tissue culture-treated 96-well plate, add 130 µL/well of uninoculated control media to three wells to serve as the blank control. Set up three control wells for each media type (TSB and TSB + BS) to be tested.
Add 130 µL/well of inoculated culture into three wells, and repeat until all experimental conditions are plated in triplicate.
Incubate for 4 - 24 h at 37 °C statically.
Using a plate reader, record the OD600. Set the control wells as 'blank.' Confirm the control medium is clear with no evidence of turbidity. If any turbidity is detected, discard the experiment. The OD600 values can be used to normalize the data if there are significant differences in growth rate between bacterial strains.
Remove the culture medium using a vacuum line by gently tilting the plate and slowly aspirating the medium at the lower edge of the well. Be sure to collect all the culture medium without disrupting the adherent bacterial population located on the plastic surface. If EPS matrix was produced during the incubation time, the matrix will be visualized as a white precipitate. Do not disturb the EPS matrix.
Gently wash the wells once with 200 µL sterile PBS. Remove the PBS wash using the vacuum line.
Invert the plate to dry. Allow a minimum of 20 min to dry. NOTE: Since the biofilm must be thoroughly dried before staining, the protocol can be paused at this step for a few hours or even overnight. The added drying time will not alter the staining procedure while incomplete drying will affect quantification of the results and reproducibility.
Add 150 µL of 0.5% crystal violet to each experimental and control well.
Incubate for 5 min at room temperature (RT).
Wash the wells once with 400 µL of distilled water. The added volume helps to remove residual crystal violet stain from the sides of the wells. Remove the wash with the vacuum line.
Wash the wells five times with 200 µL of distilled water. Remove the wash with the vacuum line. NOTE: Thorough washing is important for quantification. When the blank wells are clear from the distilled water and do not contain any residual crystal violet stain, proceed to the next step.
Invert the plate to dry, protected from light. Ensure complete drying of the plate as noted above.
Destain the wells with 200 µL of 95% ethanol. Incubate the plate on the shaker for 30 min. To avoid evaporation, particularly at higher temperatures, perform this step at 4 °C.
Using a plate reader, record the OD540. NOTE: The wavelength for maximum absorbance of crystal violet is near 590 nm. The literature reports a range from OD540 - OD5954,13,14,15,16 for crystal violet absorbance; thus choose a wavelength available based on the plate reader.
4. EPS Matrix Detection
NOTE: These complimentary assays quantify and visualize the EPS. In both, EPS is detected using a lectin to bind polysaccharides. The fluorescently-conjugated protein allows quantification (step 4.1) or visualization (step 4.2).
- Semi-quantitative detection of EPS
- Set up two 1.5 mL tubes. Label with TSB or TSB + BS.
- Add 1 mL of TSB or TSB + BS to the respective tubes.
- Inoculate tubes with 20 µL of overnight culture (a 1:50 dilution).
- In a sterile, black, flat-bottomed, tissue culture-treated 96-well plate, add 130 µL/well of uninoculated control media to three wells to serve as the blank control. Set up three control wells for each medium type to be tested. Add 130 µL/well of inoculated culture to the wells, and repeat until all experimental conditions are plated in triplicate.
- Incubate for 4 - 24 h at 37 °C statically.
- Transfer the culture medium to a clear 96-well plate with a pipette, ideally a multichannel pipette. Be cautious to collect all the culture medium without disrupting the adherent population and/or the EPS located on the plastic surface. Set aside.
- Fix the black plate for 15 min at RT using 200 µL/well of the formaldehyde/glutaraldehyde in 1x PBS.
- While the adherent population is fixing, evaluate the supernatant fraction from step 4.1.6. Using a plate reader, record the OD600. Set the control wells as 'blank.' Confirm the control medium is clear with no evidence of turbidity. If any turbidity is detected, discard the experiment.
- Alternatively, the entire assay can be performed in a black clear bottom plate. In those circumstances, the OD600 values can be recorded and the culture medium can be subsequently discarded prior to proceeding to step 4.1.7. The OD600 values can be used to normalize the data in the case there are significant differences in growth rate between bacterial strains.
- Remove the fix and dispose in the hazardous waste.
- Gently wash the wells twice with 200 µL/well of sterile PBS. Remove the PBS wash using the vacuum line by gently tilting the plate and slowly aspirating the wash at the lower edge of the well.
- Add 150 µL/well of 25 µg/mL ConA-FITC, and incubate for 15 min at RT.
- Gently wash twice with 200 µL of PBS.
- Add 150 µL of PBS to each well. Record the fluorescence at 488 nm.
- Confocal microscopy visualization of EPS production and calculation of biofilm thickness
- Set up two 1.5 mL tubes. Label with TSB or TSB + BS.
- Add 1 mL of TSB or TSB + BS to the respective tubes.
- Inoculate tubes with 20 µL of overnight culture (at a 1:50 dilution).
- Set up a sterile 24-well plate with 12 mm sterile round glass coverslips. Add 400 µL/well of uninoculated control media to three wells to serve as the blank control. Set up three control wells for each medium type to be tested.
- Add 400 µL of inoculated culture to wells in duplicate, and repeat until all experimental conditions are plated.
- Incubate for 4 - 24 h at 37 °C statically.
- Visually confirm growth in the TSB condition, growth and EPS precipitate (white) in the TSB + BS condition, and no growth and/or white precipitate in sterile control wells. Remove the supernatants.
- Fix for 15 min at RT using 200 µL/well of formaldehyde/glutaraldehyde solution in 1x PBS.
- Remove the fix and dispose in hazardous waste.
- Gently wash the wells twice with 200 µL/well of sterile PBS. Remove the PBS wash using the vacuum line.
- Add 150 µL/well of 25 µg/mL ConA-FITC and incubate for 15 min at RT.
- Gently wash twice with 200 µL/well of PBS.
- Mount with antifade mountant solution with DAPI. NOTE: The solution is a ready-made, glycerol-based mount solution containing DAPI for preserving the fluorescent label while simultaneously staining the DNA in bacterial cells for visualization as a counterstain. Follow the kit's directions and recommendations for incubation times prior to imaging samples. The DAPI staining procedure can be performed prior to applying antifade mountant solution that does not contain DAPI.
- Evaluate by confocal microscopy. On a confocal microscope, set the laser to 495 nm/519 nm excitation/emission for FITC visualization. Set a second laser to 360 nm/460 nm excitation/emission to visualize DAPI. Locate the biofilm by focusing on the DAPI channel. Use both the DAPI and FITC channels to determine the full thickness and set the upper and lower imaging borders. Record full thickness Z-stack images by capturing images every 0.25 µm after setting perimeters to the top and bottom of the z-stack. For more information on confocal microscopy, please refer to publications by Paddock et al.17,18,19,20
- Reconstruct the 3D images in ImageJ (https://imagej.nih.gov/ij/) to visualize the full biofilm formation and calculate the biofilm thickness. The average thickness obtained for S. flexneri bile-salt induced biofilms was 14 µm4.
5. Dispersion Assay
NOTE: In this assay, the disassembly of biofilm through bacterial dispersion is detected. Here, mature biofilms are established and subsequently (usually the next day), the media are replaced with PBS or supplemented PBS. The supernatant component is then assessed to quantitate the number of bacteria that have dissociated from the biofilm.
Set up two 1.5 mL tubes. Label with TSB or TSB + BS.
Add 1 mL of TSB or TSB + BS to the respective tubes.
Inoculate tubes with 20 µL of overnight culture (at a 1:50 dilution).
In a sterile, clear, flat-bottomed, tissue culture-treated 96-well plate, add 130 µL/well of uninoculated control media to three wells to serve as the blank control. Set up three control wells for each media type to be tested.
Add 130 µL/well of inoculated culture to three wells, and repeat until all experimental conditions are plated in triplicate.
Incubate the plate for 4 - 24 h at 37 °C statically.
Before continuing, warm the PBS, PBS + glucose, PBS + bile salts, and PBS + bile salts + glucose to 37 °C. Prepare these reagents fresh daily.
Using a plate reader, record the OD600. Set the control wells as 'blank.' Confirm the medium is clear with no evidence of turbidity. If any turbidity is detected, discard the experiment. The OD600 values can be used to normalize the data if there are significant differences in growth rate between bacterial strains.
Remove the culture medium using a vacuum line. Be sure to collect all the culture medium without disrupting the adherent population located on the plastic surface.
Gently wash the wells twice with 200 µL/well of sterile PBS. Remove the PBS wash using the vacuum line.
Replace the wash with 130 µL/well of the following into three wells each: PBS, PBS + glucose, PBS + bile salts, and PBS + bile salts + glucose. These reagents must be pre-warmed to 37 °C.
Incubate the plate for 30 min at 37 °C.
Carefully remove the plate from the 37 °C incubator. Transfer the supernatants to a fresh, sterile 96-well plate.
Using a 96-well plate or dilution block, prepare 10-fold (1:10) serial dilutions of the supernatant into sterile PBS. NOTE: The dilution series should range from undiluted to 10-6 depending on the bacterial strain to be tested. Pilot experiments can be performed to determine the optimal dilution range.
Spot plate 5 µL of each dilution onto LB agar using a multichannel pipette. Incubate at 37 °C overnight. Count colonies on the next day and account for the dilution factor when determining the recovery colony forming units (CFU). To calculate the percent dispersion relative to the 1x PBS control (set at 100%), divide the recovery CFU from each treatment condition by the recovery CFU from the 1x PBS control sample. NOTE: Routine media plates for the bacterial strain of interest can also be used. For example, Shigella can be plated on Congo red plates. Ensure the plates are dry for appropriate spot-plating techniques.
Representative Results
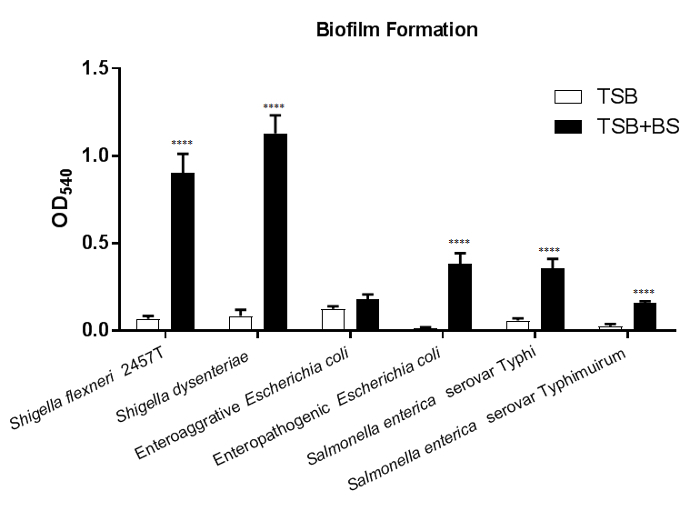
In Figure 1, biofilm formation is induced in most of the six enteric pathogens tested following growth in media containing bile salts. A significant increase in adherent bacteria after bile salts exposure is observed in nearly all strains tested. The exception is enteroaggregative E. coli (EAEC);however, note the induced observation of the Δaaf mutant4. The results indicate that additional adherence mechanisms are induced by bile salts exposure in EAEC in the absence of aggregative adherence fimbriae I (AAF/I)21. To draw conclusions from this data set, plot the OD540 values for each strain and media type tested. Comparing the values from each strain in the media control (-BS) relative to media containing bile salts will determine if bile salts significantly induce biofilm formation. Comparisons across bacterial strains and/or mutants can also be performed for further characterization analyses.
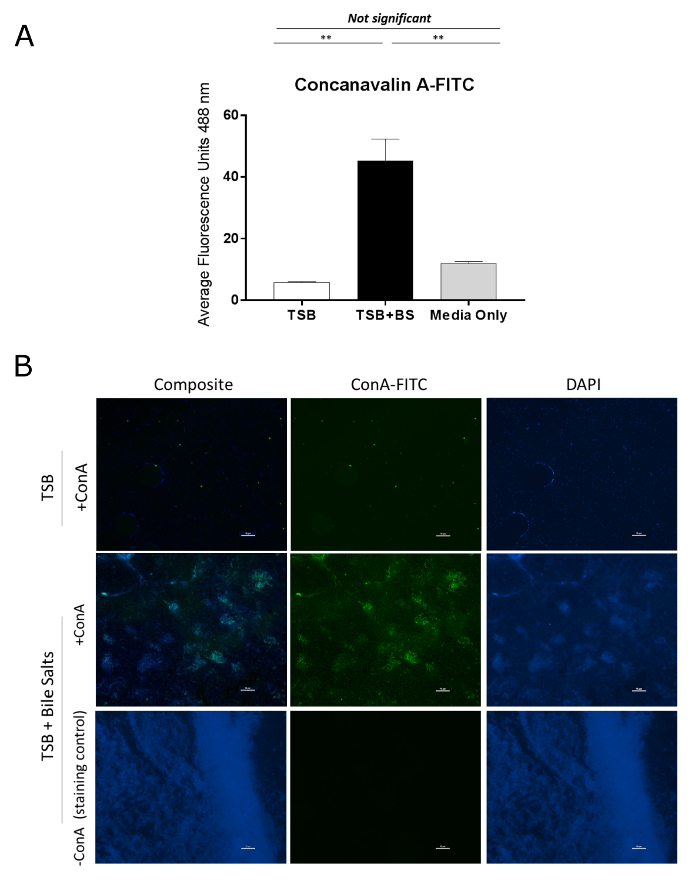
The ConA-FITC assessment of EPS is presented in Figure 2. Figure 2A represents the semi-quantitative assessment of EPS production in which ConA-FITC is used to stain the biofilms. The ConA binds to the polysaccharides in the EPS matrix and the amount retained is detected by a fluorescent plate reader. To present the data, plot the average fluorescence units for each condition and compare the biofilm-inducing conditions to the negative control. Additionally, comparisons across bacterial strains or mutants can be performed. It is critical to analyze a media-only, negative control to determine the amount of background ConA-FITC fluorescence that occurs for the plate. The TSB condition was not significantly different from the media control, while the fluorescence readings significantly increased following bile salts exposure. The data confirm that EPS production is indicative of biofilm formation and does not occur in the absence of bile salts for S. flexneri. The images of bile salt-induced biofilms in S. flexneri is represented in Figure 2B. ConA-FITC is used to stain polysaccharides in the EPS matrix while a DAPI counterstain is used to visualize the bacteria by staining the DNA. For bacteria not exposed to bile salts (negative control), only DAPI is detected. Notably, the density of bacteria is much lower than in the bile salts condition. A clear background on the FITC channel is obtained to indicate the lack of EPS production. As expected, the data demonstrate that EPS production is correlated with biofilm formation1,2,3 following bile salts exposure. The -ConA staining control in the bile salt-treated samples is provided to demonstrate the specificity of the FITC fluorescence. For these images, 1x PBS was used in place of ConA-FITC and the DAPI counterstain was maintained. Imaging can be performed with a confocal microscope to assess the thickness of biofilms with the EPS-positive samples4.
Bacterial dispersion from biofilms in Figure 3 is detected by incubating established biofilms in PBS or supplemented PBS. Physiologically, bile is reabsorbed into circulation in the small intestine and only 5% of bile enters the colon5. As Shigella invades the colonic epithelium, we hypothesize that reabsorption of bile is an important signal for biofilm dispersion. In this figure, dispersion occurs in biofilms exposed to PBS or PBS + glucose; however, dispersion is inhibited in PBS + bile salts regardless of the presence of glucose. The data demonstrate that the removal of bile salts is necessary and sufficient to induce dispersion of S. flexneri from bile salt-induced biofilms4. To plot the data, either the recovered CFU of the bacteria or the relative percent to the control (as shown here) can be plotted.
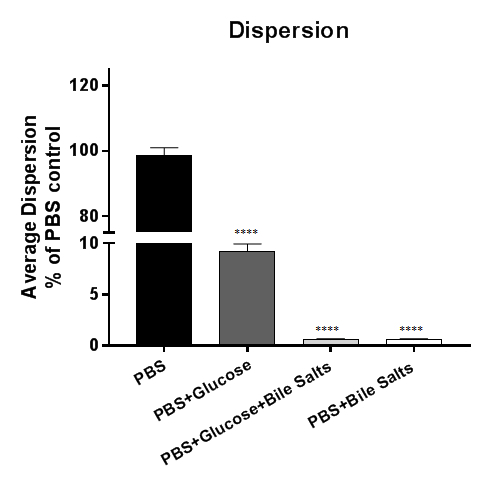
Figure 1: Bile salts induce biofilm formation in enteric pathogens. 18 h biofilms were grown on tissue-culture treated 96-well plates in either TSB or TSB + 0.4% bile salts media and stained with 0.5% crystal violet. Quantitative values from ethanol solubilization of the crystal violet stain were determined by spectrophotometry at OD540. Bile salts exposure significantly increased biofilm formation in Shigella flexneri, S. dysenteriae, enteropathogenic Escherichia coli, Salmonella enterica serovar Typhi, and S. enterica serovar Typhimurium. Statistical significance was determined by the student's T-test for each strain comparing the bile salts treatment to the media control. The standard error of the mean (SEM) is represented by the error bars. All p-values are <0.0001. Data shown are from one (n = 3) representative experiment, with a full set of data provided in a previous publication4. Please click here to view a larger version of this figure.
Figure 2: Bile salt-induced biofilm formation is characterized by production of a EPS matrix that is detected with FITC-labeled Concanavalin A. (A) Sterile black, 96-well plate were seeded with S. flexneri at a 1:50 dilution into TSB or TSB + bile salts and grown for 18 h. Biofilms were fixed, gently washed, and stained with ConA-FITC. The amount of retained Concanavalin-A FITC was determined by a fluorescent plate reader. The values plotted demonstrate EPS quantification following growth of S. flexneri grown in TSB or TSB-containing bile salts. Significance was determined by one-way ANOVA and p-values are <0.01. The SEM is represented by the error bars. Data shown are from one (n = 3) representative experiment, with a full set of data provided in a previous publication4. (B) Microscopy images from a bile salt-induced Shigella biofilm. Sterile coverslips were seeded with S. flexneri at a 1:50 dilution in TSB or TSB + bile salts and grown for 18 h. The coverslips were subsequently fixed and stained with ConA-FITC and/or counterstained with DAPI. ConA-FITC was only detected in the bile salts condition due to the presence of the EPS matrix. A coverslip stained only with DAPI in the TSB + bile salts treatment was used to demonstrate minimal background fluorescence for the FITC setting on the confocal microscope. Images for each wavelength channel and a composite overlay are provided for all treatment conditions. Scale bars indicate 10 µm. Data shown are from one (n = 3) representative experiment, with a full set of data provided in a previous publication4. Please click here to view a larger version of this figure.
Figure 3: Analysis of bacterial dispersion from bile salt-induced biofilms. 18 h bile salt-induced Shigella flexneri biofilms were exposed to PBS or PBS supplemented with glucose, bile salts, or a combination of glucose and bile salts. The supernatant was subsequently serially diluted and plated on agar plates to enumerate the colony forming units (CFUs) that dispersed the biofilm. The number of bacteria recovered was plotted relative to the PBS control. Bacteria dispersed from the biofilm in the PBS or PBS + glucose conditions, but the presence of bile salts was sufficient to inhibit bacterial dispersion from the biofilm. Data shown are from one (n = 3) representative experiment, with a full set of data provided in a previous publication4. For reference, 100-fold more bacteria were routinely recovered from the PBS control treatment (approximately 7 x 107 CFU) compared to the PBS + bile salts treatment (approximately 7 x 105 CFU). Significance was determined by one-way ANOVA and p-values are <0.0001. The SEM is represented by the error bars.
Discussion
Analysis of biofilm formation is challenging due to the dynamic nature of biofilms and the variability between strains, materials, laboratories, and assays. Here, several strategies are presented to determine biofilm formation in enteric pathogens following bile salts exposure with experimental insight provided to promote reproducibility. There are additional considerations to ensure reproducibility. First and foremost, we recommend performing at least three independent experiments each with technical triplicates to confirm observations and statistical significance due to the variation that can occur. Second, the use of positive and negative controls is vital to ensuring reproducibility of the assay. It is highly recommended to use the wild type strain as a positive control when evaluating mutants for biofilm formation. Furthermore, as genes that are important for biofilm formation are identified (for example, see Vibrio22, Salmonella7, and E. coli23,24,25), the use of biofilm mutants as negative controls will validate the reproducibility of the assay. Third, the above methods are based on focused research with Shigella and S. flexneri, in particular. Adjustments to the protocols may be required depending on the growth requirements of each enteric pathogen evaluated. Fourth, the assays measure a bacterial stress response; and therefore, it is critical to work with appropriately maintained bacteria from frozen stocks (-80 °C) or storage plates kept under refrigeration (maximum 4 °C) for no more than two weeks. It is important to note that some bacteria including the enteric pathogens in the Yersinia genus are psychrotrophs that can grow at temperatures close to 0 °C26. Since the storage at refrigerated temperatures can alter the bacterial physiology and possibly result in genotypic or phenotypic shifts27,28,29, it is highly recommended to prepare aliquots of the stock culture at -80 °C for routine inoculation into starter cultures. Furthermore, working with clinical isolates requires more frequent restreaks from -80 °C stocks as genetic shifts are rapidly observed when transitioning a clinically-derived bacterial strain into the laboratory setting30,31. Especially working with Shigella isolates, strains rapidly adapt and phenotype shifts occur often32,33. It is recommended to restreak bacteria from freezer stocks once a month, and restreak the stock plate every two weeks.
Another important factor to ensure reproducibility is fresh preparation of the bile salts media to limit variability of the assay. Bile salts media older than 1 week negatively impacted the robust phenotype observed with fresh media preparation. Moreover, depending on the pathogen and the type of exposure to be analyzed (small intestine versus colon), different sources and mixtures of bile salts may be required. For example, individual bile salts, conjugated and de-conjugated sources, or crude bile extracts that include additional bile components such as cholesterol and bilirubin may be utilized5,6. Shigella analyses have focused on bacterial adaptation to the host during transit of the small intestine prior to infection in the colon. Therefore, the use of a cholate and deoxycholate mixture mimics conditions of the small intestine5,6,34. Also with regards to the dispersion assay (section 5), it is specified that the PBS-supplemented reagents are to be made fresh daily and pre-warmed to 37 °C prior to assay. When reagents older than a few days were used, reproducibility of the assay was severely compromised. It was also critical to pre-warm these reagents to 37 °C as dispersion did not occur readily with room-temperature reagents. Finally, dispersion methods for assessment of complete titer enumeration of the biofilm can be used, which include sonication35,36 or enzymatic digestion37. The approach to the dispersion assay was directed toward assessing the effect of bile salt reabsorption as Shigella transits from the terminal ileum and into the colon. The analysis was based on the hypotheses that the loss of bile salts would enable dispersion of the transient biofilm and that Shigella must disperse the biofilm prior to causing infection in the colonic epithelium4,5. Conditions such as sonication and enzymatic digestion represent total release of microbial mass from the biofilm while the strategy presented here emulates the ileum to colon transition to capture a physiological phenotype for Shigella. Depending on the desired experimental conditions, additional methods for dispersion analysis or biofilm enumeration may be required.
The assays are designed to be moderately high-throughput with the use of 96-well plates to enable testing of multiple bacterial strains and/or conditions. To further ensure reproducibility of biofilm formation, flat-bottomed, 96-well plates that were tissue culture treated were used. For technical reasons, plate readers cannot accurately measure absorbance or fluorescence in round bottom plates. Additionally, uncoated plates offered variable adherence profiles in Shigella, which is an avenue of current investigation. The assays can be scaled to larger well formats proportionally, but will require the alterations to the protocol that can reduce experimental efficiency. For example, the culture media will need to be transferred to cuvettes to record the OD600 values with a spectrophotometer. Additionally, the crystal violet-stained biofilm can be scraped following a 30 - 60 min incubation in the 95% ethanol destain step and the contents can be transferred to a cuvette for spectrophotometer readings at OD540. Care must be taken when scrapping the contents of the biofilm in each well to avoid splashing, and we have found that scrapping is easier following a 60 min incubation.
Analysis with Shigella, Salmonella, and pathogenic E. coli4 demonstrated a repeated observation that biofilm formation occurs in an accelerated time scale (within 24 h) for enteric pathogens exposed to bile salts. Contrary to other bacteria that require 48 to 72 h to observe biofilm formation under normal conditions, bile salt-induced enteric biofilm begins as early as 4 h with the initial adherence phase. Subsequently, EPS matrix formation is evident by 6 h, and maturation occurs by 24 h. As such, the adherence phenotype at 24 h may be too robust to decipher the role of different adherence factors in biofilm formation. Performing early analyses, for example as early as 4 h, allows for identification of factors important in early biofilm formation that would otherwise be missed at a later time point. Finally, and as mentioned above in the protocol steps, bile salt-induced biofilm formation for Shigella was not detected in the absence of glucose4. Since the EPS matrix is comprised primarily of polysaccharides and is a critical step in biofilm formation for most bacteria38, the presence of glucose or other sugar in the media is most likely important to observe biofilm formation. Depending on the bacterial pathogen analyzed, different base media formulations may be required.
Biofilm formation is a multilayered process that often occurs in an open system where nutrient flux and fluid motion readily occur1,2,3. The methods are limited by the closed culture and static growth requirements; however, the methods enable assessment of biofilm formation in the laboratory setting. Upon identification of the biofilm phenotype, further experiments using fluid cells or continuous culture strategies39,40,41 may provide additional mechanistic insight into physiological or clinical significance of biofilm formation. For identification of the EPS matrix, the protocols use the polysaccharide-binding lectin ConA. ConA binds α-D-glucosyl or sterically-closely related residues42. Different lectins may be required depending on type of EPS produced for different genera or species. An alternative strategy to identify EPS production is mass spectrometry, which will provide both verification that EPS is produced and determine the composition of the EPS matrix43,44. Additionally, the use of the DAPI counterstain can result in significant brightness given the number of bacteria in the biofilm or the occurrence of extracellular DNA that is often present in the EPS matrix45,46.
There are alternatives to the above methods described in the literature. For example, the solid-phase binding assay has been reported for use in other pathogens (for example, references4,25,47,48,49). There are modifications to the protocol between publications; and therefore, the strategies presented here are an easy-to-replicate format. It is important to note that the protocols specify air-drying biofilms prior to staining (step 3.10) while some groups prefer to fix biofilms. The two different approaches may represent varying pathogen-specific strategies. Additionally, by preparing the crystal violet stain in water, a consistent biofilm formation phenotype was detected. Other groups use ethanol to solubilize crystal violet; which for Shigella, led to destabilization of biofilms and compromised reproducibility. We are currently investigating the factors that may be ethanol soluble in the Shigella biofilm. Another example is the quantification of the EPS production by ConA staining. A 96-well plate fluorescent detection method (step 4.1) is presented to determine semi-quantitative values reflecting the amount of FITC-conjugated ConA binding to the biofilm. To our knowledge, the method is the first time this strategy has been reported. EPS detection via microscopic examination is accepted in the literature and allows for a different, but complimentary, strategy to visualize EPS. The semi-quantitative fluorescence detection combined with the confocal microscopic detection of EPS certainly strengthened the data, especially given the limitations of the DAPI counterstain procedure in the microscopy analysis described above. In all, a significant strength in the set of assays described here is that each technique complements another approach to give rise to a dynamic assessment of bile salt-induced biofilm formation in enteric pathogens.
As bile salts and virulent biofilms are incorporated into the paradigm of enteric bacterial pathogenesis, it is anticipated that these methods will be used to validate models, identify roles of specific genes, identify genes of unknown function, and provide general characterization of clinical strains. This multifaceted approach allows the identification of genes important in one aspect of biofilm formation that may be lost once the mature biofilm is established, which therefore enables identification of specialized functions of each bacterial component of the biofilm. Furthermore, as evident by the fact that pathogen adaptation within the human intestine is a rapidly occurring phenomenon, the role of bile salt-induced biofilm formation is an example of a newly appreciated, conserved, and critical phenomenon that occurs in Shigella infection as well as additional enteric pathogens.
Disclosures
The authors have no disclosures.
Acknowledgments
We thank Rachael B. Chanin and Alejandro Llanos-Chea for technical assistance. We thank Anthony T. Maurelli, Bryan P. Hurley, Alessio Fasano, Brett E. Swierczewski, and Bobby Cherayil for the strains used in this study. This work was supported by the National Institute of Allergy and Infectious Diseases Grant K22AI104755 (C.S.F.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Joo H-SS, Otto M. Molecular basis of in vivo biofilm formation by bacterial pathogens. Chem Biol. 2012;19(12):1503–1513. doi: 10.1016/j.chembiol.2012.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole G, Kaplan HB, Kolter R. Biofilm Formation as Microbial Development. Annu Rev Microbiol. 2000;54(1):49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- Donlan RM. Biofilm Formation: A Clinically Relevant Microbiological Process. Clin Infect Dis. 2001;33(8):1387–1392. doi: 10.1086/322972. [DOI] [PubMed] [Google Scholar]
- Nickerson KP, et al. Analysis of Shigella flexneri resistance, biofilm formation, and transcriptional profile in response to bile salts. Infect Immun. 2017;85(6) doi: 10.1128/IAI.01067-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon JM, Kang D-J, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47(2):241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- Sistrunk JR, Nickerson KP, Chanin RB, Rasko DA, Faherty CS. Survival of the fittest: How bacterial pathogens utilize bile to enhance infection. Clin Microbiol Rev. 2016;29(4) doi: 10.1128/CMR.00031-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prouty AM, Schwesinger WH, Gunn JS. Biofilm formation and interaction with the surfaces of gallstones by Salmonella spp. Infect Immun. 2002;70(5):2640–2649. doi: 10.1128/IAI.70.5.2640-2649.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford RW, Gibson DL, Kay WW, Gunn JS. Identification of a bile-induced exopolysaccharide required for Salmonella biofilm formation on gallstone surfaces. Infect Immun. 2008;76(11):5341–5349. doi: 10.1128/IAI.00786-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford RW, Reeve KE, Gunn JS. Flagellated but not hyperfimbriated Salmonella enterica serovar Typhimurium attaches to and forms biofilms on cholesterol-coated surfaces. J Bacteriol. 2010;192(12):2981–2990. doi: 10.1128/JB.01620-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford RW, Rosales-Reyes R, Ramírez-Aguilar MdelaL, Chapa-Azuela O, Alpuche-Aranda C, Gunn JS. Gallstones play a significant role in Salmonella spp. gallbladder colonization and carriage. Proc Natl Acad Sci U S A. 2010;107(9):4353–4358. doi: 10.1073/pnas.1000862107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koestler BJ, Waters CM. Bile acids and bicarbonate inversely regulate intracellular cyclic di-GMP in Vibrio cholerae. Infect Immun. 2014;82(7):3002–3014. doi: 10.1128/IAI.01664-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson SL, Pryjma M, Gaynor EC. Flagella-mediated adhesion and extracellular DNA release contribute to biofilm formation and stress tolerance of Campylobacter jejuni. PLoS One. 2014;9(8):e106063. doi: 10.1371/journal.pone.0106063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Medina M, et al. Biofilm formation as a novel phenotypic feature of adherent-invasive Escherichia coli (AIEC) BMC Microbiol. 2009;9(1):202. doi: 10.1186/1471-2180-9-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naves P, et al. Measurement of biofilm formation by clinical isolates of Escherichia coli is method-dependent. J Appl Microbiol. 2008;105(2):585–590. doi: 10.1111/j.1365-2672.2008.03791.x. [DOI] [PubMed] [Google Scholar]
- Danese PN, Pratt LA, Dove SL, Kolter R. The outer membrane protein, Antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol Microbiol. 2000;37(2):424–432. doi: 10.1046/j.1365-2958.2000.02008.x. [DOI] [PubMed] [Google Scholar]
- Nickerson KP, McDonald C. Crohn's disease-associated adherent-invasive Escherichia coli adhesion is enhanced by exposure to the ubiquitous dietary polysaccharide maltodextrin. PLoS One. 2012;7(12):e52132. doi: 10.1371/journal.pone.0052132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddock SW. Confocal laser scanning microscopy. Biotechniques. 1999;27(5) doi: 10.2144/99275ov01. 992-6, 998-1002, 1004. [DOI] [PubMed] [Google Scholar]
- Paddock SW. Principles and practices of laser scanning confocal microscopy. Mol Biotechnol. 2000;16(2):127–149. doi: 10.1385/MB:16:2:127. [DOI] [PubMed] [Google Scholar]
- Paddock S. Over the rainbow: 25 years of confocal imaging. Biotechniques. 2008;44(5) doi: 10.2144/000112798. 643-4, 646, 648. [DOI] [PubMed] [Google Scholar]
- Paddock SW, Eliceiri KW. Laser scanning confocal microscopy: history, applications, and related optical sectioning techniques. Methods Mol Biol. 2014;1075:9–47. doi: 10.1007/978-1-60761-847-8_2. [DOI] [PubMed] [Google Scholar]
- Nataro JP, Steiner T, Guerrant RL. Enteroaggregative Escherichia coli. Emerg Infect Dis. 1998;4(2):251–261. doi: 10.3201/eid0402.980212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesper J, Lauriano CM, Klose KE, Kapfhammer D, Kraiss A, Reidl J. Characterization of Vibrio cholerae O1 El tor galU and galE mutants: influence on lipopolysaccharide structure, colonization, and biofilm formation. Infect Immun. 2001;69(1):435–445. doi: 10.1128/IAI.69.1.435-445.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjifrangiskou M, et al. Transposon mutagenesis identifies uropathogenic Escherichia coli biofilm factors. J Bacteriol. 2012;194(22):6195–6205. doi: 10.1128/JB.01012-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimpour M, et al. GlgS, described previously as a glycogen synthesis control protein, negatively regulates motility and biofilm formation in Escherichia coli. Biochem J. 2013;452(3):559–573. doi: 10.1042/BJ20130154. [DOI] [PubMed] [Google Scholar]
- Sharma VK, Kudva IT, Bearson BL, Stasko JA. Contributions of EspA Filaments and Curli Fimbriae in Cellular Adherence and Biofilm Formation of Enterohemorrhagic Escherichia coli O157:H7. PLoS One. 2016;11(2):e0149745. doi: 10.1371/journal.pone.0149745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keto-Timonen R, Hietala N, Palonen E, Hakakorpi A, Lindström M, Korkeala H. Cold Shock Proteins: A Minireview with Special Emphasis on Csp-family of Enteropathogenic Yersinia. Front Microbiol. 2016;7:1151. doi: 10.3389/fmicb.2016.01151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöntinen A, Markkula A, Lindström M, Korkeala H. Two-Component-System Histidine Kinases Involved in Growth of Listeria monocytogenes EGD-e at Low Temperatures. Appl Environ Microbiol. 2015;81(12):3994–4004. doi: 10.1128/AEM.00626-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regeard C, Mérieau A, Guespin-Michel JF. A bioluminescence assay for screening thermoregulated genes in a psychrotrophic bacterium Pseudomonas fluorescens. J Appl Microbiol. 2000;88(1):183–189. doi: 10.1046/j.1365-2672.2000.00952.x. [DOI] [PubMed] [Google Scholar]
- Markkula A, Mattila M, Lindström M, Korkeala H. Genes encoding putative DEAD-box RNA helicases in Listeria monocytogenes EGD-e are needed for growth and motility at 3°C. Environ Microbiol. 2012;14(8):2223–2232. doi: 10.1111/j.1462-2920.2012.02761.x. [DOI] [PubMed] [Google Scholar]
- Fux CA, Shirtliff M, Stoodley P, Costerton JW. Can laboratory reference strains mirror "real-world" pathogenesis? Trends Microbiol. 2005;13(2):58–63. doi: 10.1016/j.tim.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Takai S, Sekizaki T, Ozawa T, Sugawara T, Watanabe Y, Tsubaki S. Association between a large plasmid and 15- to 17-kilodalton antigens in virulent Rhodococcus equi. Infect Immun. 1991;59(11):4056–4060. doi: 10.1128/iai.59.11.4056-4060.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurelli AT, Blackmon B, Curtiss R. Loss of pigmentation in Shigella flexneri 2a is correlated with loss of virulence and virulence-associated plasmid. Infect Immun. 1984;43(1):397–401. doi: 10.1128/iai.43.1.397-401.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecko DJ, Washington O, Formal SB. Genetic and physical evidence for plasmid control of Shigella sonnei form I cell surface antigen. Infect Immun. 1980;29(1):207–214. doi: 10.1128/iai.29.1.207-214.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faherty CS, Redman JC, Rasko DA, Barry EM, Nataro JP. Shigella flexneri effectors OspE1 and OspE2 mediate induced adherence to the colonic epithelium following bile salts exposure. Mol Microbiol. 2012;85(1):107–121. doi: 10.1111/j.1365-2958.2012.08092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Oethinger M, Tuohy MJ, Procop GW, Bauer TW. Improved detection of biofilm-formative bacteria by vortexing and sonication: a pilot study. Clin Orthop Relat Res. 2009;467(5):1360–1364. doi: 10.1007/s11999-008-0609-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Ferreira T, et al. Microbial investigation of biofilms recovered from endotracheal tubes using sonication in intensive care unit pediatric patients. Braz J Infect Dis. 2016;20(5):468–475. doi: 10.1016/j.bjid.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruzzi B, Briggs RE, Swords WE, De Castro C, Molinaro A, Inzana TJ. Capsular Polysaccharide Interferes with Biofilm Formation by Pasteurella multocida Serogroup A. MBio. 2017;8(6):e01843–e01817. doi: 10.1128/mBio.01843-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne DE, Boles BR. Emerging interactions between matrix components during biofilm development. Curr Genet. 2016;62(1):137–141. doi: 10.1007/s00294-015-0527-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Li M, Gregory RL. Bacterial interactions in dental biofilm. Virulence. 2011;2(5):435–444. doi: 10.4161/viru.2.5.16140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buswell CM, Nicholl HS, Walker JT. Use of continuous culture bioreactors for the study of pathogens such as Campylobacter jejuni and Escherichia coli O157 in biofilms. Methods Enzymol. 2001;337:70–78. doi: 10.1016/s0076-6879(01)37007-6. [DOI] [PubMed] [Google Scholar]
- McBain AJ. Chapter 4 In Vitro Biofilm Models. Adv Appl Microbiol. 2009;69:99–132. doi: 10.1016/S0065-2164(09)69004-3. [DOI] [PubMed] [Google Scholar]
- Schiefer HG, Krauss H, Brunner H, Gerhardt U. Ultrastructural visualization of surface carbohydrate structures on mycoplasma membranes by concanavalin A. J Bacteriol. 1975;124(3):1598–1600. doi: 10.1128/jb.124.3.1598-1600.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liener I. The Lectins: Properties, Functions and Applications in Biology and Medicine. Elsevier Science; 1986. [Google Scholar]
- Wittmann V, Pieters RJ. Bridging lectin binding sites by multivalent carbohydrates. Chem Soc Rev. 2013;42(10):4492–4503. doi: 10.1039/c3cs60089k. [DOI] [PubMed] [Google Scholar]
- Wang S, et al. The exopolysaccharide Psl-eDNA interaction enables the formation of a biofilm skeleton in Pseudomonas aeruginosa. Environ Microbiol Rep. 2015;7(2):330–340. doi: 10.1111/1758-2229.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okshevsky M, Meyer RL. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit Rev Microbiol. 2015;41(3):341–352. doi: 10.3109/1040841X.2013.841639. [DOI] [PubMed] [Google Scholar]
- Xu D, Zhang W, Zhang B, Liao C, Shao Y. Characterization of a biofilm-forming Shigella flexneri phenotype due to deficiency in Hep biosynthesis. PeerJ. 2016;4:e2178. doi: 10.7717/peerj.2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole GA. Microtiter Dish Biofilm Formation Assay. J Vis Exp. 2011. [DOI] [PMC free article] [PubMed]
- Nickerson KP, McDonald C. Crohn's Disease-Associated Adherent-Invasive Escherichia coli Adhesion Is Enhanced by Exposure to the Ubiquitous Dietary Polysaccharide Maltodextrin. PLoS One. 2012;7(12) doi: 10.1371/journal.pone.0052132. [DOI] [PMC free article] [PubMed] [Google Scholar]


