Abstract
Precise control of transgene expression is desirable in biological and clinical studies. However, because the binary feature of currently employed gene switches requires the transfer of two therapeutic expression units concurrently into a single cell, the practical application of the system for gene therapy is limited. To simplify the transgene expression system, we generated a gene switch designated as pEUI(+) encompassing a complete set of transgene expression modules in a single vector. Comprising of the GAL4 DNA-binding domain and modified EcR (GvEcR), a minimal VP16 activation domain fused with a GAL4 DNA-binding domain, as well as a modified Drosophila ecdysone receptor (EcR), the newly developed singular gene switch is highly responsive to the administration of a chemical inducer in a time- and dosage-dependent manner. The pEUI(+) vector is a potentially powerful tool for improving the control of transgene expression in both biological research and pre-clinical studies. Here, we present a detailed protocol for modulation of a transient and stable transgene expression using pEUI(+) vector by the treatment of tebufenozide (Teb). Additionally, we share important guidelines for the use of Teb as a chemical inducer.
Keywords: Genetics, Issue 135, Gene switch, ecdysone receptor, EcR, GvEcR, tebufenozide, Teb, pEUI(+), stable cell line, HEK293
Introduction
Several different gene switches have been explored for their ability to precisely regulate transgene expression in cell culture and animal models, with varying degrees of success1,2,3. Potential gene switch systems should meet several stringent criteria, including: precise directional expression of the transgene, dosage- and time-dependent responsiveness to inducers, negligible leakiness of the promoter, reversibility of transgene activation through inducer removal, and inducer toxicity levels tolerable to cell lines and laboratory animals1,2,3. Several small molecule inducers have been exploited, including tetracycline4,5, rapamycin6,7,8, mifepristone9,10, and ecdysone11,12,13,14. In general, these systems require at least two plasmids containing two or three discrete expression units, one or two of which compose a regulatory element (inducer plasmid) that binds a small molecule inducer to gain transcriptional activity; and another plasmid (effector plasmid) containing the transgene under the control of a regulatory DNA region that allows the access of the inducer-bound regulatory element. The main drawback of the binary system is that it requires the concomitant introduction of two vectors (inducer and effector) into the target cell. In transient transgene expression experiments, the simultaneous transfection of two plasmids inevitably produces a population of cells that is singly transfected with either the inducer or the effector, or co-transfected unequally. In addition, the binary system requires at least two rounds of antibiotic selection to establish stable cell lines, which is time-consuming and laborious. Thus, combining all of the components required for transgene expression and regulation into a single vector would be ideal to guarantee the concomitant expression of all the required elements in a single cell and allow for the regulation of transgene expression through treatment with small molecule inducers.
To overcome the shortcomings of binary gene switches, we recently developed a singular gene switch, using a Drosophila melanogaster EcR-based gene inducible system15. Ecdysone mediates gene expression when it binds to the EcR/ultraspiracle (USP) heterodimer, which in turn induces binding of the EcR to DNA regulatory elements3,11. The vertebrate retinoid X receptor (RXR), an ortholog of insect USP, recruits EcR to form an active transcriptional activator16. Thus, for the targeted expression of a transgene in vertebrate cells or tissue, the EcR and RXR must be co-expressed simultaneously before being stimulated by ecdysone or its agonists. Since the heterodimeric feature of the EcR-based gene switch can be influenced by the endogenous RXR level, Padidam et al. replaced the EcR DNA-binding and activation domains with heterologous GAL4 DNA-binding, herpes simplex virus protein vmw65 (VP16) activation domains fused to the EcR ligand-binding domain, which then became unresponsive to endogenous RXR and formed a homodimer to gain transcriptional activity17. The EcR-based gene switch was further improved by constructing a chimeric protein comprised of minimal VP16 activation domain combined with GvEcR, which formed a homodimer and localized in the cytosol in the absence of the ecdysone agonist, Teb16,18. By binding to Teb, the GvEcR changed its subcellular localization from the cytosol to the nucleus to recognize a hybrid promoter comprised of zebrafish E1b minimal promoter combined with a ten tandem repeated upstream activation sequences (UASs) to initiate the transcription of the target gene16.
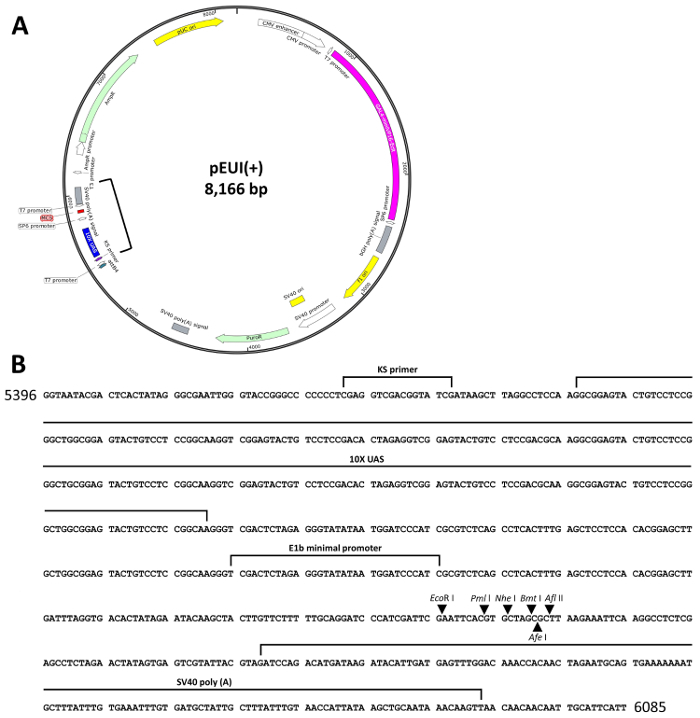
To simplify the previously reported EcR-based gene switches16,18, we combined the binary features of the system into a single vector equipped with all the required elements for stimulating transgene expression, and then designated the newly generated vector, pEUI(+) (GenBank accession number: KP123436, Figure 1A)15. The effector responding to the GvEcR driver (hereafter driver) consists of ten tandem UAS repeats next to an E1b minimal promoter followed by a multiple cloning site (MCS) with EcoRI, PmlI, NheI, BmtI, AfeI, and AflII recognition sequences (Figure 1B). Additionally, to facilitate the selection of transfected cells, an SV40 promoter-driven puromycin N-acetyl-transferase (PAC) gene was placed between the driver and effector regions to maintain stable cell lines (Figure 1).
We describe a detailed protocol for the transient and stable modulation of transgene expression using pEUI(+). In addition, we provide detailed instructions for the successful use of Teb as an ecdysone agonist.
Protocol
1. Transgene Subcloning
Choose an appropriate restriction enzyme recognition site (or sites) in the MCS of pEUI(+), and subclone the gene of interest in the desired direction (Figure 1). NOTE: The EGFP or FLAG epitope-tagged ankyrin repeat domain 13A (ANKRD13A) transgene was subcloned in EcoRI site of the pEUI(+) vector using T4 DNA polymerase in a sequence- and ligation-independent cloning method19.
Linearize the pEUI(+) with EcoRI for 1 h at 37 °C. Mix 50 ng/µL of linearized pEUI(+) with 40 ng/µL of PCR-amplified EGFP or ANKRD13A, and react with T4 DNA polymerase (1 U) for 1 min at room temperature (RT) before incubation on ice for 10 min. Add 10 µL of the reaction mixture directly to 100 µL of DH10B competent cells for the following transformation experiment. NOTE: Although several restriction enzyme recognition sites were added to the MCS of pEUI(+) (Figure 1B), we recommend subcloning the target gene into the EcoRI site, using ligation-independent cloning technology19,20.
Verify the insert by sequencing and then purify the DNA using a DNA purification kit that is suitable for transfection.
2. Transient Transfection and Teb Treatment
Prepare four cell culture dishes (60 mm diameter) containing freshly passaged HEK293 cells in 5 mL of Dulbecco's modified Eagle's medium (DMEM) supplemented with 5 - 10% fetal bovine serum (FBS) and antibiotics (100 U/mL penicillin, 100 µg/mL streptomycin). Hereafter, "cell culture medium" or simply "culture medium" represents the DMEM supplemented with FBS and antibiotics. Maintain the HEK293 cells at 37 °C.
Transfect 1 µg of purified DNA into the HEK293 cells at 50 - 60% density using 3 µL of transfection reagent. Add the transfection reagent to 200 µL of diluent (DMEM without FBS and antibiotics) containing the DNA. After the incubation of the reaction mixture for 30 min at RT, add the mixture to the HEK-293 cells on the culture dish. Use two dishes for transfection, and leave the other two untreated as controls.
After 12 - 24 h of transfection, add Teb (final concentrations, 0.2 - 10 µmol) to one of the transfected samples and to one of the non-transfected controls. Leave the other two samples untreated as experimental controls. NOTE: The Teb stock solution (50 mM dissolved in dimethyl sulfoxide) can be diluted in cell culture medium, DMEM, or phosphate-buffered saline (PBS) at an appropriate concentration before adding to the cultured cells depending on the experimental parameters. However, due to the low solubility of Teb in aqueous solution, avoid preparing a master mix with a high concentration of Teb in medium; instead, replace the complete cell culture medium with fresh medium containing Teb at the desired concentration.
Cultivate the Teb-treated and untreated cells for 12 - 48 h at 37 °C.
- If using the EGFP insert, analyze EGFP expression under a fluorescent microscope ( Figure 2). Otherwise, harvest the cells for an immunoblotting assay.
- For the immunoblotting assay, rinse the cells twice with ice-cold PBS and then scrape them out using a cell scraper.
- For piling the cells, spin down the harvests in a 15 mL conical tube at 21,000 x g for 2 min at 18-25 °C (RT).
- After discarding the PBS completely, add 0.5 mL of ice-cold mammalian protein extraction reagent (M-PER), together with protease inhibitor (5 µL of 100x protease inhibitor cocktail), to the pellet if the cells are cultured in a 60 mm dish.
- Re-suspend the cells with several rounds of pipetting, and then incubate the suspended cells for 15 min at 18 - 25 °C (RT).
- Spin the cell lysate at 21,000 x g for 10 min at 4 °C.
- Transfer the aqueous supernatant into a fresh 1.5 mL microcentrifuge tube.
- Analyze the supernatant by immunoblotting, following standard protocols with appropriate antibodies.
3. Generation of Stable HEK293 Cell Lines with Genomic Integration of the pEUI(+) Gene Switch
Transfect 5 - 10 µg of the purified pEUI(+) with the transgene into HEK293 cells that are 50 - 60% confluent in a 90 mm diameter cell culture dish, using 15 - 30 µL of the transfection reagent. Add the transfection reagent to 500 µL of diluent (DMEM without FBS and antibiotics) containing the DNA. After the incubation of the reaction mixture for 30 min at RT, add the mixture to HEK-293 cells on the culture dish.
After the transfection, allow the transfectants to grow and express the puromycin-resistance gene products under non-selective culture medium (without puromycin) for 24 - 48 h at 37 °C.
Dissociate the cells from the culture dishes by trypsinization. After discarding the culture medium, rinse the transfectants with pre-warmed PBS (37 °C), add 1 mL of 0.25% trypsin/ethylenediaminetetraacetic acid (EDTA), and incubate for 1 min at 37 °C. After quenching the trypsin/EDTA by adding pre-warmed culture medium (10 mL, 37 °C), harvest the cells using a cell scraper, followed by centrifugation in a 15 mL conical tube at 21,000 x g for 2 min at 18-25°C.
Re-suspend the transfectants in 50 mL of pre-warmed culture medium (37 °C), and transfer 10 mL of the cells into 90 mm fresh cell culture dishes. Culture the cells at 37 °C until 50 - 60% confluency before the puromycin treatment (in general, this takes approximately 24 - 48 h).
Cultivate the transfectants at 37 °C in cell culture medium supplemented with 3 µg/mL puromycin for 3 - 4 weeks. During the selection period, change the cell culture medium containing puromycin (3 µg/mL) every 2 - 3 days to maintain selection pressure. When the cells become fully confluent, split them following the procedures in steps 3.3 - 3.4, with puromycin-containing culture medium. Discard the remaining cells in the culture medium, if not needed. NOTE: Puromycin selection conditions must be established experimentally for specific cell types. We used 3 µg/mL of puromycin in cell culture medium; this was enough to induce cell death in non-transfected HEK293.
- Harvest the cells from individual colonies, using a cloning cylinder, and follow the manufacturer's instructions.
- Discard the cell culture medium. Rinse the plate twice with pre-warmed (37 °C) PBS to remove floating cells.
- Using sterile forceps, set the cloning cylinder over the individual colonies.
- Add 0.2 mL of 0.35% trypsin/EDTA to each cylinder.
- Incubate the dish at 37 °C for 1 min until the cells are detached, and then add a few drops of pre-warmed (37 °C) culture medium to the cylinders.
- Transfer the cells from each cylinder to individual wells of a 6-well plate, with 2 mL of culture medium and 1 µg/mL of puromycin in each well. NOTE: When the cells become confluent, scale up the culture. Now clone(s) may be established.
Maintain individual clones in cell culture medium supplemented with 1 µg/mL of puromycin.
Validate individual colonies by testing their sensitivity to Teb treatment (final concentration, 10 µM) for 24 h. Test the responsiveness of the clones to the Teb by analyzing the expression level of the transgene with immunoblotting. Select a desired clone (or clones). Cultivate a Teb-treated and an untreated sample in culture medium for 12 - 48 h at 37 °C before analyzing the induction level of the transgene. The expression level of transgene can be measured through immunoblotting, following the procedures in steps 2.5.1 - 2.5.7.
4. Depletion of Teb
- Quench the Teb stimuli by replacing Teb containing cell culture medium with Teb-free medium. NOTE: It is recommended to replace the growth media without first reseeding the cells onto a new culture dish. Residual Teb appears to cling to the plastic dish and will elicit a strong background expression of the transgene. Before reseeding the cells on a new culture plate, rinse the harvested cells several times with either culture medium or PBS.
- Dissociate the cells from the culture dishes by trypsinization. After discarding the Teb containing culture medium, rinse the Teb-stimulated cells twice with pre-warmed PBS (37 °C), add 1 mL of 0.25% trypsin/EDTA, and incubate for 1 min at 37 °C. After quenching the trypsin/EDTA by adding pre-warmed Teb-free culture medium (10 mL, 37 °C), harvest the cells using a cell scraper, followed by centrifugation in a 15 mL conical tube at 21,000 x g for 2 min at 18-25 °C.
- Re-suspend the cells in 50 mL of pre-warmed Teb-free culture medium (37 °C), and transfer 5 mL of the cells into three 60 mm fresh cell culture dishes. Culture the cells at 37 °C for 24, 48, and 72 h respectively.
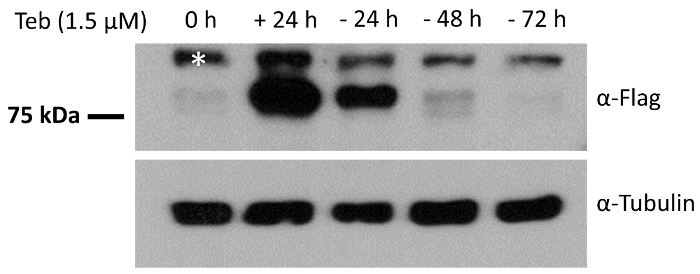
- Harvest the cells at different time points and measure the expression level of the transgene by using immunoblotting. NOTE: Under these experimental conditions, the transgene expression became undetectable approximately 3 days post-culture of the cells in the Teb-free cultural medium (Figure 3). The expression level of transgene can be measured by immunoblotting, as per steps 2.5.1 - 2.5.7.
Representative Results
The GvEcR-based singular gene switch is depicted in Figure 1A. The pEUI(+) vector was optimized for regulated transgene expression by the treatment with Teb. The effector region of pEUI(+) comprises a 10xUAS and an E1b minimal promoter followed by an MCS containing EcoRI, PmlI, NheI, BmtI, AfeI, and AflII restriction enzyme recognition sites. An SV40 poly(A) signal was added behind the MCS (Figure 1B). A PAC gene was inserted between the driver and effector regions of pEUI(+) to confer resistance to puromycin.
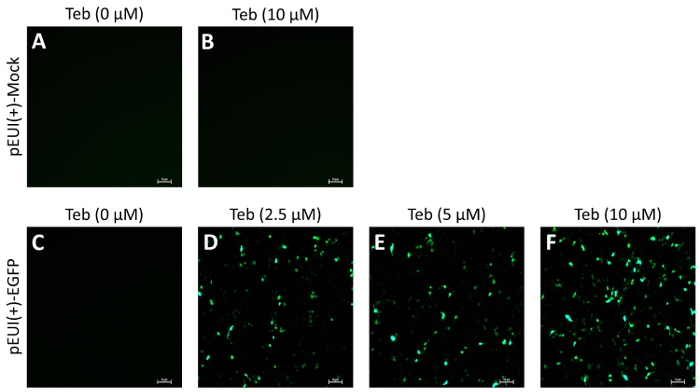
To evaluate the activity and leakiness of pEUI(+), we subcloned EGFP into the EcoRI site of the MCS (pEUI(+)-EGFP), transiently transfected the plasmid construct into HEK293 cells, and administered Teb at different concentrations for 24 h (Figure 2). While mock vector transfectants did not show fluorescent signals regardless of Teb treatment (Figure 2A, B), pEUI(+)-EGFP transfectants became progressively sensitized to the increased amount of Teb (Figure 2C-F). More importantly, pEUI(+)-EGFP transfectants without treatment of Teb did not elicit any detectable EGFP signals under a fluorescent microscope (Figure 2C).
To further validate the responsiveness of pEUI(+) to Teb, we subcloned a FLAG epitope-tagged ANKRD13A gene into the EcoRI site of pEUI(+), transferred the construct into HEK293 cells, and then subjected them to puromycin selection for 3 weeks to establish a desired cell line with the ANKRD13A transgene. We analyzed the expression of the ANKRD13A transgene after a 24 h treatment with Teb and the established cell line responded well (Figure 3). We further demonstrated the reversibility of the pEUI(+) gene switch by depleting Teb from the cultural media. As shown in Figure 3, ANKRD13A expression was no longer detectable when we cultured the cells in Teb-free media.
Figure 1. Schematic diagram of pEUI(+). (A) The vector map of pEUI(+) was constructed. A CMV promoter drives constitutive expression of GvEcR. The Teb-bound GvEcR will translocate into the nucleus and bind the 10xUAS cis-acting regulatory sequences to stimulate expression of the transgene inserted into the MCS. (B) The sequence information between the square brackets in (A) contains the entire effector sequences of pEUI(+). Arrows represent the individual restriction enzyme recognition sites included in the MCS. Please click here to view a larger version of this figure.
Figure 2. GvEcR-based singular gene switch responds to the treatment of Teb. HEK293 cells were transfected with the specified DNA constructs. After 24 h of transfection, the cells were stimulated for 24 h with Teb at the indicated concentration. EGFP transgene activity was observed under a fluorescent microscope. Mock transfectants with pEUI(+) did not show any detectable fluorescence with (A) or without (B) Teb treatment. (C-E) The fluorescent intensity in pEUI(+)-EGFP-transfected cells increases in a Teb dosage-dependent manner. Scale bar, 10 µm. Please click here to view a larger version of this figure.
Figure 3. HEK293 cells stably harboring pEUI(+)/FLAG-ANKRD13A respond reversibly to the administration of Teb. FLAG epitope-tagged ANKRD13A expression was triggered by Teb (1.5 µM) treatment. After 24 h of treatment with Teb, the cells were switched to Teb drop-out media for the indicated amount of time. The relative amount of ANKRD13A was measured by Western blotting with anti-FLAG antibody. The endogenous expression level of α-tubulin was measured with anti-α-tubulin antibody as a control. The white asterisk indicates an unidentified cross-reactive species. Please click here to view a larger version of this figure.
Discussion
The main drawback of using a binary gene expression switch is the need to simultaneously deliver two separate plasmids (driver and effector) into the target cell. This can lead to an unequal distribution of plasmids and result in an inconsistent response of the switch to the inducers1,2,3. Another shortcoming of the two-vector system is that a minimum of two rounds of antibiotic selection are required to identify promising cell lines. Therefore, a gene-inducible cassette comprising one unit has long been sought for use in biological research. The pEUI(+) singular transgene expression switch is contained in a single vector equipped with all regulatory units required for transgene stimulation. Thus, the induction level of the transgene, subcloned in pEUI(+), is dependent on the amount of DNA transfected in the transient transfection experiment and dosage of the chemical inducer. In addition, we further tested the usage of pEUI(+) vector by generating a stable cell line containing a single copy of the ANKRD13A transgene15. The established cell line showed high sensitivity to Teb treatment (Figure 3). Collectively its high sensitivity, reversibility, and low leakiness15, along with limited variation in expression level make pEUI(+) a valuable molecular and cellular tool for manipulating target gene expression.
Among various inducible gene systems, we selected the GvEcR-dependent gene induction cassette. The GvEcR has several advantages over other systems: first, the lipophilic nature of several ecdysone analogs used as chemical inducers (including Teb) is advantageous for efficient penetration of cellular membranes21; second, there are no EcR orthologs in vertebrates, so ecdysteroid agonists do not influence the function of endogenously expressed nuclear receptors21,22; third, ecdysteroids are innocuous in vertebrates and rapidly cleared from the circulation system in vivo21,23; and fourth, since numerous ecdysone agonists have been developed thus far, by choosing optimally-sized small molecules, researchers can avoid undesirable complications13,22,24. Although the pEUI(+) shows high sensitivity to Teb, testing the responsiveness of pEUI(+) to other ecdysteroid agonists may prove valuable. The existence of non-steroidal EcR agonists, such as methoxyfenozide, that are more soluble in water than Teb24 may widen the scope of applications of pEUI(+). Though the expression of transgene in pEUI(+) is relatively weaker than that in a Tet-On system15, the level of transgene induction could be augmented by the addition of a multi-copy of the VP16 transactivation domain (data not shown). Recent gene therapy applications have focused on the safe delivery of a target gene into cells or tissues that lack the respective functional gene product owing to genetic disease25,26,27. However, non-regulated transgene under the control of tissue-specific or viral constitutive promoters can elicit hyper-expression of the target gene, which elevates the possibility of local tissue damage28,29. Thus, the application of a reliable gene switch for gene therapy could avoid unwanted complications. The pEUI(+) vector provides a convenient and powerful tool to control transgene expression in biological and clinical studies. Currently, we are developing a singular lentiviral vector based on GvEcR to increase the application range of our system.
Disclosures
We have no conflicts of interest related to this report.
Acknowledgments
The authors thank S-Y Choi (Chonnam National University) for valuable comments and thorough reading of our manuscript. This work was supported by a research fund of Chungnam National University.
References
- Rossi FM, Blau HM. Recent advances in inducible gene expression systems. Curr. Opin. Biotechnol. 1998;9(5):451–456. doi: 10.1016/s0958-1669(98)80028-1. [DOI] [PubMed] [Google Scholar]
- Clackson T. Regulated gene expression systems. Gene Ther. 2000;7(2):120–125. doi: 10.1038/sj.gt.3301120. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Suzuki Y. Gene regulatable lentiviral system. In: Xu K, editor. Viral Gene Therapy. InTech; 2011. pp. 285–309. Available from: http://www.intechopen.com/books/viralgene-therapy/gene-regulatable-lentiviral-vector-system. [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA. 1992;89(12):5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron U, Bujard H. Tet repressor-based system for regulated gene expression in eukaryotic cells: principles and advances. Methods Enzymol. 2000;327:401–421. doi: 10.1016/s0076-6879(00)27292-3. [DOI] [PubMed] [Google Scholar]
- Hentges KE, et al. FRAP/mTOR is required for proliferation and patterning during embryonic development in the mouse. Proc Natl Acad Sci U S A. 2001;98(24):13796–13801. doi: 10.1073/pnas.241184198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaszynski LA, Liu CW, Wandless TJ. Characterization of the FKBP.rapamycin.FRB ternary complex. J Am Chem Soc. 2005;127(13):4715–4721. doi: 10.1021/ja043277y. [DOI] [PubMed] [Google Scholar]
- Seto B. Rapamycin and mTOR: a serendipitous discovery and implications for breast cancer. Clin Transl Med. 2012;1(1):29. doi: 10.1186/2001-1326-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmann A, Peyron R, Silvestre L. Clinical uses of mifepristone (MFP) Ann N Y Acad Sci. 1995;761:248–260. doi: 10.1111/j.1749-6632.1995.tb31382.x. [DOI] [PubMed] [Google Scholar]
- Sitruk-Ware R, Spitz IM. Pharmacological properties of mifepristone: toxicology and safety in animal and human studies. Contraception. 2003;68(6):409–420. doi: 10.1016/s0010-7824(03)00171-9. [DOI] [PubMed] [Google Scholar]
- Yao TP, et al. Functional ecdysone receptor is the product of EcR and ultraspiracle genes. Nature. 1993;366(6454):476–479. doi: 10.1038/366476a0. [DOI] [PubMed] [Google Scholar]
- Riddiford LM, Cherbas P, Truman JW. Ecdysone receptors and their biological actions. Vitam Horm. 2000;60(2000):1–73. doi: 10.1016/s0083-6729(00)60016-x. [DOI] [PubMed] [Google Scholar]
- Saez E, Nelson MC, Eshelman B, Banayo E, Koder A, Cho GJ. Identification of ligands and coligands for the ecdysone-regulated gene switch. Proc Natl Acad Sci USA. 2000;97(26):14512–14517. doi: 10.1073/pnas.260499497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karzenowski D, Potter DW, Padidam M. Inducible control of transgene expression with ecdysone receptor: gene switches with high sensitivity robust expression, and reduced size. Biotechniques. 2005;39(2):191–200. doi: 10.2144/05392ST01. [DOI] [PubMed] [Google Scholar]
- Lee S, et al. Ecdysone receptor-based singular gene switches for regulated transgene expression in cells and adult rodent tissues. Mol. Ther. Nucleic Acids. 2016;5(9):e367. doi: 10.1038/mtna.2016.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esengil H, Chang V, Mich JK, Chen JK. Small-molecule regulation of zebrafish gene expression. Nat Chem Biol. 2007;3(3):154–155. doi: 10.1038/nchembio858. [DOI] [PubMed] [Google Scholar]
- Padidam M, Gore M, Lu DL, Smirnova O. Chemical-inducible, ecdysone receptor-based gene expression system for plants. Transgenic Res. 2003;12(1):101–109. doi: 10.1023/a:1022113817892. [DOI] [PubMed] [Google Scholar]
- Knopf F, Schnabel K, Haase C, Pfeifer K, Anastassiadis K, Weidinger G. Dually inducible TetOn systems for tissue-specific conditional gene expression in zebrafish. Proc Natl Acad Sci USA. 2010;107(46):19933–19938. doi: 10.1073/pnas.1007799107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong JY, et al. One-step sequence- and ligation-independent cloning as a rapid and versatile cloning method for functional genomics studies. Appl Environ Microbiol. 2012;78(15):5440–5443. doi: 10.1128/AEM.00844-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Cai G, Hall EO, Freeman GJ. In-fusion assembly: seamless engineering of multidomain fusion proteins, modular vectors, and mutations. Biotechniques. 2007;43(3):354–359. doi: 10.2144/000112536. [DOI] [PubMed] [Google Scholar]
- No D, Yao TP, Evans RM. Ecdysone-inducible gene expression in mammalian cells and transgenic mice. Proc Natl Acad Sci U S A. 1996;93(8):3346–3351. doi: 10.1073/pnas.93.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada Y, et al. Synthesis and insecticidal activity of benzoheterocyclic analogues of N'-benzoyl-N-(tert-butyl)benzohydrazide: Part 1. Design of benzoheterocyclic analogues. Pest Manag Sci. 2003;59(1):25–35. doi: 10.1002/ps.604. [DOI] [PubMed] [Google Scholar]
- Lafont R, Girault JP, Kerb U. Excretion and metabolism of injected ecdysone in the white mouse. Biochem Pharmacol. 1988;37(6):1174–1177. doi: 10.1016/0006-2952(88)90529-1. [DOI] [PubMed] [Google Scholar]
- Carlson GR, et al. The chemical and biological properties of methoxyfenozide, a new insecticidal ecdysteroid agonist. Pest Manag Sci. 2001;57(2):115–119. doi: 10.1002/1526-4998(200102)57:2<115::AID-PS245>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- McConnell MJ, Imperiale MJ. Biology of adenovirus and its use as a vector for gene therapy. Hum Gene Ther. 2004;15(11):1022–1033. doi: 10.1089/hum.2004.15.1022. [DOI] [PubMed] [Google Scholar]
- Gorell E, Nguyen N, Lane A, Siprashvili Z. Gene therapy for skin diseases. Cold Spring Harb Perspect Med. 2014;4(4):a015149. doi: 10.1101/cshperspect.a015149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotterman MA, Chalberg TW, Schaffer DV. Viral Vectors for Gene Therapy: Translational and Clinical Outlook. Annu Rev Biomed Eng. 2015;17:63–89. doi: 10.1146/annurev-bioeng-071813-104938. [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Yamaguchi M, Kuzume M, Matsumiya A, Kumada K. Insulin gene transfer with adenovirus vector via the spleen safely and effectively improves posthepatectomized conditions in diabetic rats. J Surg Res. 2003;110(1):228–234. doi: 10.1016/s0022-4804(03)00023-4. [DOI] [PubMed] [Google Scholar]
- Han J, McLane B, Kim EH, Yoon JW, Jun HS. Remission of diabetes by insulin gene therapy using a hepatocyte-specific and glucose-responsive synthetic promoter. Mol Ther. 2011;19(3):470–478. doi: 10.1038/mt.2010.255. [DOI] [PMC free article] [PubMed] [Google Scholar]


