Abstract
β-amyloid (Aβ) is a hydrophobic peptide with an intrinsic tendency to self-assemble into aggregates. Among various aggregates, Aβ oligomer is widely accepted as the leading neurotoxin in the progress of Alzheimer's disease (AD) and is considered to be the crucial event in the pathogenesis of AD. Therefore, Aβ oligomer inhibitors might prevent neurodegeneration and have the potential to be developed as disease-modifying treatments of AD. However, different formation protocols of Aβ oligomer might lead to oligomers with different characteristics. Moreover, there are not many methods to effectively screen Aβ1-42 oligomer inhibitors. An A11 antibody can react with a subset of toxic Aβ1-42 oligomer with anti-parallel β-sheet structures. In this protocol, we describe how to prepare an A11-positive Aβ1-42 oligomer-rich sample from a synthetic Aβ1-42 peptide in vitro and to evaluate relative amounts of A11-positive Aβ1-42 oligomer in samples by a dot blotting analysis using A11 and Aβ1-42-specific 6E10 antibodies. Using this protocol, inhibitors of A11-positive Aβ1-42 oligomer can also be screened from semi-quantitative experimental results.
Keywords: Biochemistry, Issue 135, Alzheimer's disease, Aβ, oligomer, dot blotting analysis, transmission electronic microscopy, curcumin
Introduction
Alzheimer's disease (AD) is one of the most important neurodegenerative diseases affecting elderly people worldwide1. It is widely accepted that the abnormal aggregation of β-amyloid (Aβ) may be the leading pathological factor of AD. Aβ aggregates are the main components of the senile plaques, one of the biological markers in the brains of AD patients. Moreover, Aβ aggregates, including oligomers in particular, produce potent neurotoxicity, which might be the cause of neuronal death as AD progresses. Therefore, the inhibition of Aβ oligomer formation might prevent neurodegeneration, and Aβ oligomer inhibitors could be developed as disease-modifying treatments of AD. Many studies have used a synthetic Aβ peptide to form oligomers in vitro, explore morphologies and structures of artificial Aβ oligomers, and investigate the inhibitors of Aβ oligomer using in vitro models2,3,4. However, different in vitro formation protocols of Aβ oligomer could lead to oligomer with different morphological characteristics, which might cause the incomparable results among different research groups. Therefore, a standard formation protocol for Aβ oligomer is urgently needed.
Until now, not many methods have been reported to directly detect Aβ oligomers. Transmission electronic microscopy (TEM), non-denaturing gel electrophoresis, enzyme-linked immunosorbent assay (ELISA), and dot blotting analysis can be used to examine the amount and/or morphology of Aβ oligomer in vitro5,6. For example, the morphology and structure of Aβ oligomer can be observed in TEM. The relative amounts and molecular size of Aβ aggregations could be measured by non-denaturing gel electrophoresis. ELISA could be used to determine Aβ oligomer in serum, plasma, and extracts from brain tissue. Lastly, dot blotting analysis, a technique used for detecting, analyzing, and identifying proteins, could be used to evaluate the relative concentration of Aβ oligomer in different samples with the help of oligomer-specific and Aβ-specific antibodies. Moreover, a dot blotting assay offers significant time savings, as gel electrophoresis and the blotting procedures for gels are not required. Therefore, this assay is normally used to screen potential Aβ oligomer inhibitors. The overall goal of this protocol is to describe a relatively simple, reliable, and reproducible method to prepare an Aβ1-42 oligomer-rich sample, to analyze the amounts of Aβ1-42 oligomer by dot blotting analysis, and to screen Aβ oligomer inhibitors using semi-quantitative experimental results.
Protocol
1. Solution Preparation
NOTE: See Table of Materials for reagent sources.
Prepare a 5% bovine serum albumin (BSA) solution by adding 5 g of BSA to 100 mL of double-distilled water. Mix them completely by vortexing them. Store the solution at 4 °C for up to 1 month.
Prepare an anti-oligomer antibody A11 solution (1:1,000) by adding 10 µL of antibody stock solution to 10 mL of the 5% BSA solution. Mix them completely by vortexing them. Store the solution at 4 °C for up to 1 month.
Prepare an anti-Aβ antibody 6E10 solution (1:1,000) by adding 10 µL of antibody stock solution to 10 mL of 5% BSA solution. Mix them completely by vortexing. Store the solution at 4 °C for up to 1 month.
Prepare an anti-fibrillar oligomer antibody OC solution (1:1,000) by adding 10 µL of antibody stock solution to 10 mL of 5% BSA solution. Mix them completely by vortexing. Store the solution at 4 °C for up to 1 month.
Prepare a Tris-buffered saline (TBS) stock solution by adding 24 g of Tris base and 88 g of NaCl to 1,000 mL of double-distilled water. Adjust the pH to 7.4. Store the solution at 4 °C for up to 3 months.
Prepare a Tris-buffered saline and Tween-20 (TBST) solution by adding 1 mL of Tween-20 to 100 mL of TBS stock solution and 900 mL double-distilled water.
Prepare a secondary antibody solution by adding 10 µL of horseradish peroxidase (HRP) Goat anti-Rabbit IgG (H + L) to 10 mL of TBST solution. Mix them completely by vortexing them. Store the solution at 4 °C for up to 1 month.
Dissolve 3.68 g of curcumin in 1 mL of dimethyl sulfoxide (DMSO) to form a 10-mM curcumin stock solution.
Dissolve 5 mg of synthetic Aβ1-42 in 2 mL of 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP) to form a 2.5-mg/mL Aβ monomer solution. Place it at room temperature (25 °C) for 20 min. Make 100 µL aliquots and store at -20 °C for up to 6 months. NOTE: This procedure should be run as fast as possible. The pipette tips should be cut off to ensure accurate pipetting.
Prepare electrochemiluminescence (ECL) fluid by mixing ECL fluid A and B with a volume ratio of 1:1. This solution should be prepared just before use.
2. Sample Preparation
NOTE: Perform the sample preparation 2 days before the dot blotting analysis.
Add 900 µL of double-distilled water to a tube of Aβ1-42 monomer solution; the concentration of Aβ1-42 will be 0.25 mg/mL. Place the mixture at room temperature for 20 min.
Evaporate the solution with high-purity nitrogen gas until its volume is about 850 µL. The concentration of the Aβ1-42 solution will be about 0.29 mg/mL. NOTE: The solution should be shaken from time to time to ensure the HFIP is fully evaporated. Normally, after 30 min of evaporation, the volume of the residual solution is about 850 µL.
Dilute the curcumin stock solution to a curcumin working solution (0.2 and 2 µM) with double-distilled water. Mix the Aβ1-42solution and curcumin working solution with a volume ratio of 1:1. The final concentrations of curcumin will be 0.1 and 1 µM. NOTE: Potential oligomer inhibitors can be mixed with the Aβ1-42 solution in any ratio if needed.
- Shake the solution continuously in a magnetic agitator (see Table of Materials).
- Fix a plastic divider box to the magnetic agitator. Place 2 magnetic stir bars at 2 corners of the box and place the sample tubes in the center of the box.
- Shake the box at room temperature (25 °C) for 48 h. NOTE: The speed of the magnetic agitator is around 60 rpm.
Centrifuge the tubes for 15 min at 4 °C and 18,000 x g. Collect the supernatant.
3. Dot Blotting Analysis
NOTE: All incubations are performed on a horizontal shaker.
Cut nitrocellulose membrane into 1-cm wide strips. NOTE: The width of the nitrocellulose membrane can be adjusted according to experimental needs.
Place 2-µL samples evenly on 2 strips (strip 1 and 2) with each point interval at 0.5 cm.
Place the strips at room temperature for 5 min until the droplet on the band is dry.
Incubate the strips with a 5% BSA solution for 30 min at room temperature.
Rinse the strips with a TBST solution for 5 min at room temperature.
Aspirate the TBST solution. For 1 h at room temperature, incubate strip 1 with an anti-oligomer antibody A11 solution and strip 2 with an anti-Aβ antibody 6E10 solution.
Rinse the strips with a TBST solution three times, each time for 5 min at room temperature.
Aspirate the TBST solution. Incubate the strips with a secondary antibody solution for 40 min at room temperature.
Rinse the strips with a TBST solution three times, each time for 5 min at room temperature.
Evenly apply the mixed ECL fluid to the surface of the strips. Expose the strips in an automatic chemiluminescence imaging system (see Table of Materials). NOTE: Normally, 300 µL of ECL fluid is enough for one membrane. The membrane must be kept moist during the exposure. The exposure time is automatically calculated by the imaging system.
Do a grayscale analysis using ImageJ (National Institutes of Health) or another image-processing software to obtain semi-quantitative results.
Representative Results
To investigate whether an Aβ1-42 monomer can form an Aβ1-42 oligomer after preparation, TEM analysis was used. No visible aggregates were observed in the HFIP-dissolved Aβ1-42 monomer sample (Figure 1A). Moreover, mainly globular aggregates with a diameter of around 10 - 80 nm were observed in the Aβ1-42 sample after 48 h of shaking, suggesting that Aβ1-42 forms oligomers after preparation (Figure 1B).
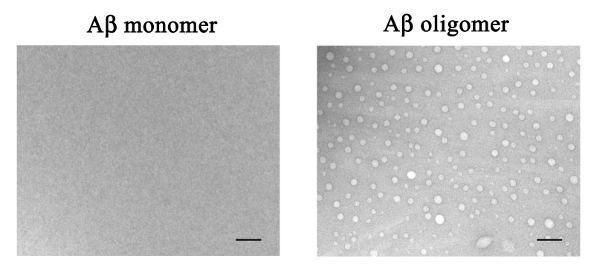
Figure 1. TEM analysis of Aβ1-42 monomer and Aβ1-42 oligomer-rich samples. The HFIP-dissolved Aβ1-42 monomer sample (10 µM) and the Aβ1-42 oligomer-rich sample (10 µM) prepared according to this protocol were examined by TEM. The scale bar = 200 nm. Please click here to view a larger version of this figure.
Additionally, dot blotting analysis was used to evaluate the relative amounts of Aβ1-42 oligomer in the samples. An A11 antibody could react with a subset of toxic Aβ oligomer with anti-parallel β-sheet structures. An OC antibody reacts with fibrillar aggregates with parallel in-register β-sheet structures. By using these antibodies, we demonstrated that A11-positive Aβ1-42 oligomers, but not OC-positive Aβ1-42 fibrillar aggregates, mainly appeared in the Aβ1-42 oligomer-rich samples that were prepared according to the protocol (Figure 2). By using the anti-Aβ antibody 6E10, we observed that the number of Aβ1-42 peptides was similar in the HFIP-dissolved Aβ1-42 monomer sample and the Aβ1-42 oligomer-rich sample (Figure 2).
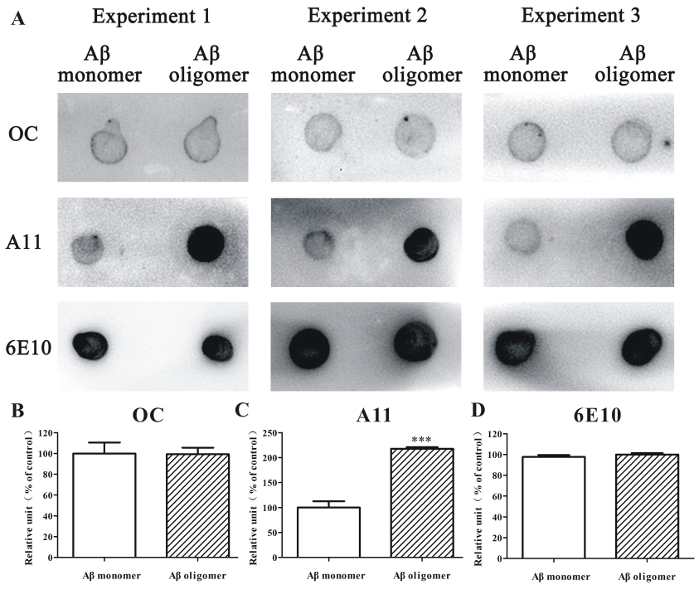
Figure 2. Dot blotting analysis of Aβ1-42 monomer and Aβ1-42 oligomer-rich samples. (A) The HFIP-dissolved Aβ1-42 monomer sample (10 µM) and the Aβ1-42 oligomer-rich sample (10 µM) prepared according to this protocol were examined by dot blotting analysis using the anti-oligomer antibody A11, anti-fibrillar oligomer antibody OC, and anti-Aβ antibody 6E10. (B - D) The semi-quantitative analysis of grayscale was performed by ImageJ. The data, expressed as a percentage of control, were the mean ± SEM of 3 independent experiments; ***p <0.001 vs. the Aβ1-42 monomer group (ANOVA and t-test). Please click here to view a larger version of this figure.
To further investigate if this protocol could be used to screen inhibitors of Aβ1-42 oligomers, curcumin, a known Aβ oligomer inhibitor, was used. We found that a co-incubation with curcumin (0.1 - 1 µM) significantly reduced the relative amounts of A11-positive Aβ1-42 oligomer, as evidenced by the decreased staining of A11 in the curcumin co-incubated Aβ1-42 oligomer sample than in the normal incubated Aβ1-42 oligomer-rich sample (Figure 3). At the same condition, curcumin (0.1 - 1 µM) did not alter the number of Aβ1-42 peptides, as the staining of 6E10 was even in all samples (Figure 3).
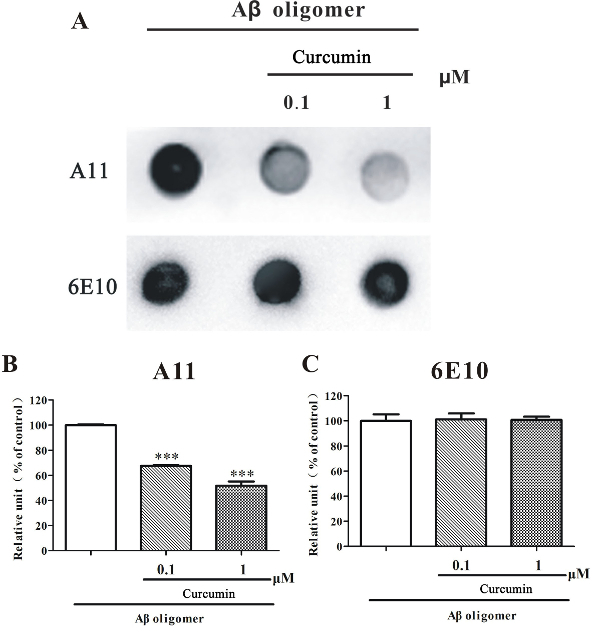
Figure 3. Dot blotting analysis of curcumin co-incubated Aβ1-42 oligomer-rich samples. (A) HFIP-dissolved Aβ1-42 monomers (10 µM) were prepared according to this protocol and incubated with or without curcumin (0, 0.1, 1 µM) under shaking for 48 h. The samples were examined by a dot blotting analysis using A11 and 6E10. (B) Semi-quantitative grayscale analysis was performed by ImageJ. The data, expressed as a percentage of control, were the mean ± SEM of 3 independent experiments; ***p <0.001 vs. the Aβ1-42 alone group (ANOVA and Tukey's test). Please click here to view a larger version of this figure.
To verify oligomer formation, we have also used a non-denaturing gel. We found that oligomer (>4 KD) was present in the Aβ1-42 oligomer-rich sample, while most monomer (4 KD) was found in the Aβ1-42 monomer sample (Figure S1).
We have investigated Aβ aggregates in the supernatant and pellet of the Aβ1-42 sample. After 48 h of incubation, Aβ1-42 oligomer but not Aβ1-42 fibrillar aggregates were found in the supernatant of the sample as determined by A11, OC, and 6E10 antibodies (Figure S2). At the same condition, Aβ1-42 fibrillar aggregates but not Aβ1-42 oligomers were found in the pellet of the sample (Figure S2).
We have also examined the morphology of the curcumin-treated Aβ1-42 sample by using TEM. The curcumin-treated sample contains a few spherical spots, suggesting that curcumin could reduce the amount of Aβ1-42 oligomer (Figure S3).
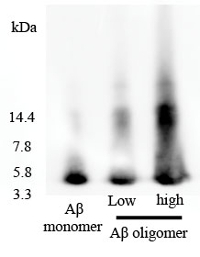 Figure S1. Non-denaturing gel electrophoresis analysis of Aβ1-42 monomer and Aβ1-42 oligomer-rich samples. The HFIP-dissolved Aβ1-42 monomer sample (10 µM) and the Aβ1-42 oligomer-rich sample (5 µM, low; 10 µM, high) prepared according to this protocol were examined by non-denaturing gel electrophoresis using the anti-Aβ antibody 6E10. Please click here to view a larger version of this figure.
Figure S1. Non-denaturing gel electrophoresis analysis of Aβ1-42 monomer and Aβ1-42 oligomer-rich samples. The HFIP-dissolved Aβ1-42 monomer sample (10 µM) and the Aβ1-42 oligomer-rich sample (5 µM, low; 10 µM, high) prepared according to this protocol were examined by non-denaturing gel electrophoresis using the anti-Aβ antibody 6E10. Please click here to view a larger version of this figure.
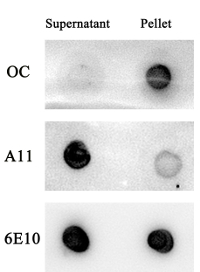 Figure S2. The supernatant of Aβ1-42 solution contains mainly oligomer, but not fibrillar aggregates. An Aβ1-42 solution (50 µL), after shaking for 48 h, was centrifuged at 18,000 x g for 10 min. The supernatant was collected and the pellet was re-dissolved in 850 µL of double-distilled water. The supernatant and pellet were further examined by a dot blot assay using OC, A11, and 6E10 antibodies. Please click here to view a larger version of this figure.
Figure S2. The supernatant of Aβ1-42 solution contains mainly oligomer, but not fibrillar aggregates. An Aβ1-42 solution (50 µL), after shaking for 48 h, was centrifuged at 18,000 x g for 10 min. The supernatant was collected and the pellet was re-dissolved in 850 µL of double-distilled water. The supernatant and pellet were further examined by a dot blot assay using OC, A11, and 6E10 antibodies. Please click here to view a larger version of this figure.
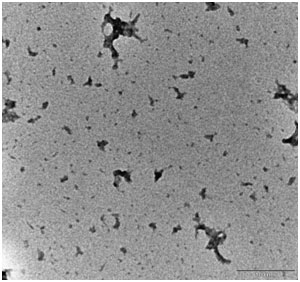 Figure S3. TEM analysis of curcumin co-incubated Aβ1-42 oligomer-rich samples. HFIP-dissolved Aβ1-42 monomers (10 µM) were prepared according to this protocol and incubated with 1 µM curcumin under shaking for 48 h. The samples were examined by TEM. Scale bar = 100 nm. Please click here to view a larger version of this figure.
Figure S3. TEM analysis of curcumin co-incubated Aβ1-42 oligomer-rich samples. HFIP-dissolved Aβ1-42 monomers (10 µM) were prepared according to this protocol and incubated with 1 µM curcumin under shaking for 48 h. The samples were examined by TEM. Scale bar = 100 nm. Please click here to view a larger version of this figure.
Discussion
In this protocol, we have reported a method to prepare samples containing Aβ1-42 oligomer, and to analyze the amounts of A11-positive Aβ1-42 oligomer by a dot blotting analysis. Although our methods for the preparation of Aβ1-42 oligomer-rich samples are quite simple, reliable, and reproducible, there are still some points to be noticed. Firstly, HFIP is used to dissolve the synthetic Aβ1-42 peptide. An aggregated Aβ1-42 peptide can disassemble into monomer in the HFIP solution. However, HFIP is easy to volatilize, and the viscosity of HFIP is very low. Therefore, the Aβ1-42 peptide should be dissolved into HFIP and the HFIP solution should be aliquoted as fast as possible to reduce the potential loss of HFIP during the operation. Moreover, the pipette tips should be cut off to ensure accurate pipetting in this procedure. Secondly, high-purity nitrogen gas is used to evaporate the HFIP from the solution. During this procedure, the Aβ1-42 solution should be shaken from time to time to ensure that the HFIP can be fully evaporated. At the end of this procedure, the smell of HPIF should not be detected in the solution. Normally, 30 min of evaporation reduces the concentration of HFIP to less than 0.1%. At this concentration, HFIP is reported not to alter the conformation transition of Aβ1-427. Thirdly, when preparing samples containing Aβ1-42 oligomer, the minimum volume of the liquid should be greater than 50 µL. Otherwise, the samples are likely to scatter into small droplets, attaching to the wall of the tubes. Also, Aβ1-42 monomer cannot form oligomer without a thorough mixing.
The protocol to prepare an Aβ1-42 oligomer-rich sample as described here is slightly different from some protocols used in other groups. For example, in some studies, HFIP was evaporated before adding double-distilled water into the solution8,9. However, in such a condition, the Aβ1-42 peptide may form small sheets, which are hard to dissolve into the solution. Therefore, we chose to add double-distilled water first, and then evaporate the HFIP by nitrogen gas. Most importantly, oligomeric shapes of aggregates prepared by this protocol are confirmed by the TEM. Moreover, Aβ1-42 oligomer formed by this protocol could induce neurotoxicity in SH-SY5Y cells and primary hippocampal neurons and reduce cognitive performance after injection into the hippocampal region of mice, suggesting that the Aβ1-42 oligomer prepared is quite toxic5,10. These results are consistent with previous studies11.
This protocol to evaluate A11-positive Aβ1-42 oligomer by a dot blotting analysis still has some shortcomings. Firstly, by using manual sampling, it is not easy to keep the droplets uniform in size. Secondly, a dot blotting analysis is a semi-quantitative but not quantitative method to measure the amounts of A11-positive Aβ1-42 oligomer, suggesting that other methods, such as ELISA, are required if the accurate evaluation of amounts of Aβ1-42 oligomer is necessary. Thirdly, some drugs may react to the antibodies, leading to false positive results.
In general, we have provided a reliable protocol to prepare samples containing A11-positive Aβ1-42 oligomer and to analyze the amounts of A11-positive Aβ1-42 oligomer by using a dot blotting assay. By using this protocol, potential Aβ1-42 oligomer inhibitors with the potential to treat AD might be screened effectively.
Disclosures
The authors have nothing to disclose.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (U1503223, 81673407, 21475131), the Applied Research Project on Nonprofit Technology of Zhejiang Province (2016C37110), the Ningbo International Science and Technology Cooperation Project (2014D10019), the Ningbo Municipal Innovation Team of Life Science and Health (2015C110026), the Ningbo Sci & Tech Project for Common Wealth (2017C50042), the Li Dak Sum Yip Yio Chin Kenneth Li Marine Biopharmaceutical Development Fund, and the K. C. Wong Magna Fund at Ningbo University.
References
- Lauren J. Cellular prion protein as a therapeutic target in Alzheimer's disease. Journal of Alzheimer's Disease. 2014;38(2):227–244. doi: 10.3233/JAD-130950. [DOI] [PubMed] [Google Scholar]
- De Maio A, Rivera I, Cauvi DM, Arispe N. Modulation of amyloid peptide oligomerization and toxicity by extracellular Hsp70. Biophysical Journal. 2017;112(3):444. doi: 10.1007/s12192-017-0839-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Scala C, Chahinian H, Yahi N, Garmy N, Fantini J. Interaction of Alzheimer's beta-amyloid peptides with cholesterol: mechanistic insights into amyloid pore formation. Biochemistry. 2014;53(28):4489–4502. doi: 10.1021/bi500373k. [DOI] [PubMed] [Google Scholar]
- Xiang SY, et al. Fucoxanthin inhibits beta-amyloid assembly and attenuates beta-amyloid oligomer-induced cognitive impairments. Journal of Agricultural and Food Chemistry. 2017;65(20):4092–4102. doi: 10.1021/acs.jafc.7b00805. [DOI] [PubMed] [Google Scholar]
- Chang L, et al. Protection against beta-amyloid-induced synaptic and memory impairments via altering beta-amyloid assembly by bis(heptyl)-cognitin. Scientific Reports. 2015;5:10256. doi: 10.1038/srep10256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam GHF, et al. In vitro amyloid aggregate forming ability of TGFBI mutants that cause corneal dystrophies. Investigative Ophthalmology & Visual Science. 2012;53(9):5890–5898. doi: 10.1167/iovs.11-9068. [DOI] [PubMed] [Google Scholar]
- Tomaselli S, et al. The alpha-to-beta conformational transition of Alzheimer's A beta-(1-42) peptide in aqueous media is reversible: A step by step conformational analysis suggests the location of beta conformation seeding. ChemBioChem. 2006;7(2):257–267. doi: 10.1002/cbic.200500223. [DOI] [PubMed] [Google Scholar]
- Shigemitsu Y, et al. Nuclear magnetic resonance evidence for the dimer formation of beta amyloid peptide 1-42 in 1,1,1,3,3,3-hexafluoro-2-propanol. Analytical Biochemistry. 2016;498:59–67. doi: 10.1016/j.ab.2015.12.021. [DOI] [PubMed] [Google Scholar]
- Khan MV, Rabbani G, Ahmad E, Khan RH. Fluoroalcohols-induced modulation and amyloid formation in conalbumin. International Journal of Biological Macromolecules. 2014;70:606–614. doi: 10.1016/j.ijbiomac.2014.07.027. [DOI] [PubMed] [Google Scholar]
- Fang F, et al. 5-hydroxycyclopenicillone, a new beta-amyloid fibrillization inhibitor from a sponge-derived fungus trichoderma sp HPQJ-34. Marine Drugs. 2017;15(8):260. doi: 10.3390/md15080260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiao YJ, Su MH, Lin HC, Wu CR. Echinacoside ameliorates the memory impairment and cholinergic deficit induced by amyloid beta peptides via the inhibition of amyloid deposition and toxicology. Food & Function. 2017;8(6):2283–2294. doi: 10.1039/c7fo00267j. [DOI] [PubMed] [Google Scholar]


