Abstract
The abundance of plant nucleolin mRNA is regulated during de-etiolation by phytochrome. A close correlation between the mRNA abundance of nucleolin and mitosis has also been previously reported. These results raised the question of whether the effects of light on nucleolin mRNA expression were a consequence of light effects on mitosis. To test this we compared the kinetics of light-mediated increases in cell proliferation with that of light-mediated changes in the abundance of nucleolin mRNA using plumules of dark-grown pea (Pisum sativum) seedlings. These experiments show that S-phase increases 9 h after a red light pulse, followed by M-phase increases in the plumule leaves at 12 h post-irradiation, a time course consistent with separately measured kinetics of red light-induced increases in the expression of cell cycle-regulated genes. These increases in cell cycle-regulated genes are photoreversible, implying that the light-induced increases in cell proliferation are, like nucleolin mRNA expression, regulated via phytochrome. Red light stimulates increases in the mRNA for nucleolin at 6 h post-irradiation, prior to any cell proliferation changes and concurrent with the reported timing of phytochrome-mediated increases of rRNA abundance. After a green light pulse, nucleolin mRNA levels increase without increasing S-phase or M-phase. Studies in animals and yeast indicate that nucleolin plays a significant role in ribosome biosynthesis. Consistent with this function, pea nucleolin can rescue nucleolin deletion mutants of yeast that are defective in rRNA synthesis. Our data show that during de-etiolation, the increased expression of nucleolin mRNA is more directly regulated by light than by mitosis.
As part of the characterization of a pea (Pisum sativum) cDNA with homology to yeast and animal nucleolin, Tong et al. (1997) found that the abundance of the pea nucleolin mRNA was regulated by phytochrome in etiolated pea shoots. The exposure of a dark-grown seedling to light leads to a major developmental transformation known as de-etiolation, which is in part mediated by the red light- (R) sensitive photoreceptor, phytochrome. Relative to light-grown plants, etiolated plants are thinner, taller, and lack photosynthetic pigments. Phytochrome mediation of de-etiolation begins in the first few seconds after exposure to R and continues for up to a day or more. Phytochrome effects that occur within the first few seconds after R irradiation include changes in electrical potentials (Smith, 1975) and changes in cytosolic free calcium levels (Shacklock et al., 1992). Some R-induced gene expression changes occur in minutes, and other photomorphogenic responses take a few hours to be detected. For instance, increases in leaf growth rate and decreases in stem growth rate have been measured in peas 4 h after R irradiation (Smith, 1975). Full greening and leaf formation may not occur until 12 to 24 h after R and require constant light.
In etiolated plants, changes in growth are among the earliest measurable whole-plant responses mediated by phytochrome. Phytochrome activation causes a decrease in the elongation rate of epicotyls and an increase in the growth rate of leaves. These changes in growth are known to involve shifts in elongation rates, as well as long-term changes in mitotic rates (Cosgrove, 1994). Changes in elongation and cell division occur at two distinct regions of the plant. Elongation rates are mediated in cells that have already divided and are in the process of maturing, whereas changes in the rate of cell division occur in the meristems. For de-etiolation, the elongation response has been studied most intensely. In peas, inhibition of epicotyl elongation (as measured by changes in the zone of elongation) has been detected as rapidly as 1 h after R exposure (Behringer et al., 1990). Studies of changes in cell division induced by phytochrome are much less exact. What is known up to now is that phytochrome does cause changes in cell division rates. The data about R-induced cell division changes in flowering plants come from studies measuring the differences in cell size many hours after irradiation (Cosgrove, 1994). The only detailed study of phytochrome-mediated cell division kinetics is in germinating fern spores (Furuya et al., 1997). Prior to the work presented here, the relative kinetics of light-induced changes in cell division that occur during de-etiolation were not well documented.
In contrast, differential gene expression during de-etiolation has received much attention. The regulation of genes that are required for greening and photosynthesis have been well characterized. There are many genes that have been identified as being up-regulated by the activation of phytochrome in etiolated seedlings (Batschauer et al., 1994). One of the earliest studied phytochrome regulated genes was rDNA. In higher plants the photoreversible increase in accumulation of rRNA has been measured at 2 to 6 h after R irradiation (Tobin and Silverthorne, 1985). In peas specifically, Koller and Smith (1972) showed that plumules reacted to an R pulse by transiently increasing rRNA synthesis beginning 2 to 3 h post-irradiation. In addition, phytochrome mediates an increase in polysomes (Pine and Klein, 1972). This phytochrome-mediated increase in ribosomes and protein synthesis is presumably acting to increase the population of regulatory, enzymatic, and structural proteins as the plant undergoes de-etiolation. To up-regulate ribosome levels, proteins involved in ribosome synthesis must also be up-regulated.
Nucleolin is one of the best-characterized proteins involved in ribosome synthesis (Tuteja and Tuteja, 1998; Ginisty et al., 1999). The initial steps in ribosome synthesis, such as the transcription of the rRNA and its subsequent splicing, occur in the nucleolus. Although the involvement of nucleolin in the process of ribosome synthesis has been established, the exact step or steps in which it is involved are still unclear. In an in vitro experiment using mouse cell extracts, nucleolin was shown to be a vital component for splicing of the 5′-external transcribed spacer region of the initial rRNA transcript (Ginisty et al., 1998). Other studies using yeast mutants deleted in the gene encoding the yeast nucleolin homolog, NSR1, have shown nucleolin to be involved in the maturation of the rRNA from a single transcript to the individual subunits (Kondo and Inouye, 1992; Lee et al., 1992). Although participation of nucleolin in ribosome synthesis appears to be its primary function, other functions such as helicase activity, binding to laminin, transcriptional repression of the AGP gene (Tuteja and Tuteja, 1998; Ginisty et al., 1999), binding to the Myb transcription factor (Ying et al., 2000), and binding to telomeres (Pollice et al., 2000) have been proposed. In addition to functional data about nucleolin, in animals (Bugler et al., 1982), yeast (Kondo et al., 1992), and plants (Bögre et al., 1996), nucleolin has been characterized as being up-regulated during times of increased cell proliferation.
The correlation of increased nucleolin expression with increased cell proliferation led to the postulate that the up-regulation of nucleolin could serve as a molecular marker for the onset of mitosis in plants (Bögre et al., 1996). We wondered whether this correlation would hold in the case of de-etiolation, in which phytochrome regulates nucleolin expression (Tong et al., 1997) and mitosis (Cosgrove, 1994). Using the nucleolin-like cDNA described in Tong et al. (1997), we performed experiments designed to determine the relationship between nucleolin and mitosis, as well as to learn whether plant nucleolin, like animal and yeast nucleolin, functions in ribosome synthesis.
RESULTS
S-Phase Kinetics Show an Increase 9 h after R Irradiation
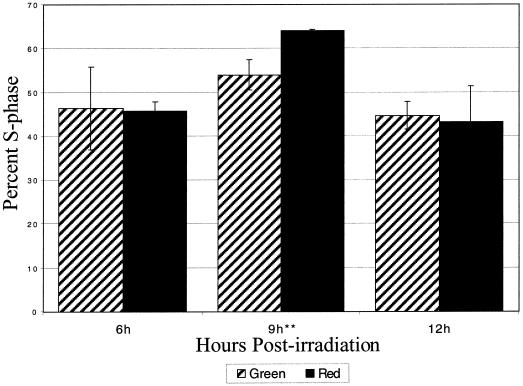
A comparison of the S-phase index of plumules at each time point after R irradiation to its green safe-light (G) control shows that there is a statistically significant (P < 0.05) increase 9 h after the irradiation (Fig. 1). At 6 and 12 h after R irradiation, the S-phase index was lower than at 9 h. In addition, there was no statistically significant difference between R and G plumules at 6 or 12 h post-irradiation. These data indicate that there is an increase in the number of pea plumule cells replicating their DNA 9 h after an R pulse.
Figure 1.
S-phase index of R-irradiated plumules. Seven-day-old etiolated pea plumules were irradiated with an R pulse and returned to darkness. S-phase index was measured by the number of nuclei that had incorporated 5′-bromo-2′-deoxyuridine (BrdU) relative to the total number of nuclei per time point. A significant increase (P < .05) between R and G plumules was found only at 9 h postirradiation, marked with asterisks. Cross-hatched bars represent the G control plumules, and the solid bars represent the R-irradiated plumules. The error bars are ±1 sd.
Measurement of M-Phase Kinetics after an R Pulse
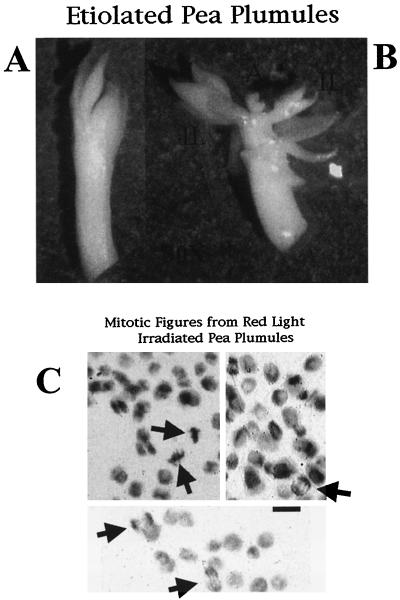
Figure 2A shows a typical excised pea plumule and Figure 2B shows the same plumule after the outer leaves have been removed. The two tissue types are shown with the inner leaves (IL) and the apical region (A) marked. The mass of the plumule is dominated by the leaves so that the A only comprises a small portion of the plumule with the apical meristem being an even smaller region contained within the apex. Figure 2C shows various cells in M-phase with a background of non-mitotic cells. The mitotic cells are indicated by arrows and were counted to obtain the M-phase index.
Figure 2.
Dissected etiolated pea plumules and the mitotic figures obtained by staining the nuclei. A, An etiolated pea plumule after excision from the shoot. The outer leaves surround the IL and the apex. B, The outer leaves have been removed and the inner leaves and apex are ready to be separated and then stained. The mass of the apex is less than one-tenth of the inner leaves. The IL and the apex (A) are marked for reference. C, Example photos of mitotic figures, as visualized by Feulgen staining, from plumules that were counted to obtain the mitotic indices in Figure 3. The arrows point to cells undergoing mitosis. On the left side are two cells in metaphase, and on the right two cells in anaphase. All of these cells were from IL of the plumule. A and B are shown at a magnification of 30×. In C, the bar = 10 μm.
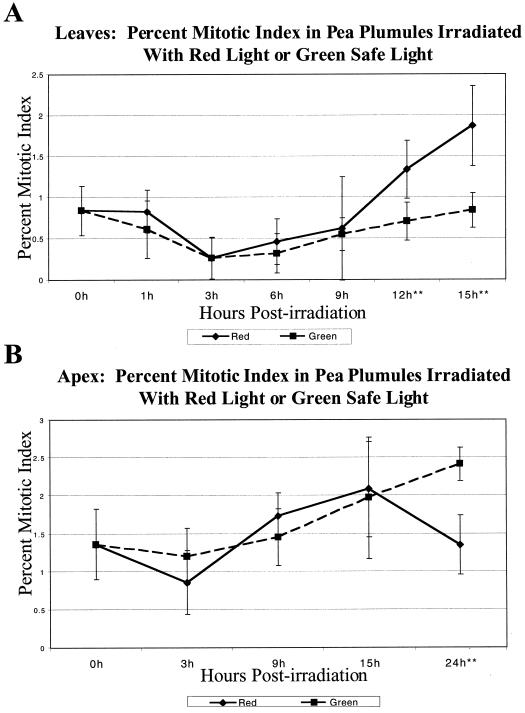
The mitotic index for each time point after R irradiation was compared with a G control to account for any inherent pattern of mitotic rates that might exist in the plumules. Beginning at 12 h post-irradiation and continuing through 15 h post-irradiation there is a significant increase (P < 0.01) in mitotic rates for the R-irradiated leaves (Fig. 3A).
Figure 3.
Mitotic index of R-irradiated pea plumules. A, The mitotic index of the IL of R irradiated plumules. The mitotic index of inner plumule leaves significantly increased (P < 0.01) in comparison to G control IL starting at 12 h postirradiation and continuing through 15 h postirradiation. B, The mitotic index of the As of R-irradiated plumules. The mitotic index of R-irradiated apices significantly decreased (P < 0.001) in comparison to G control apices at 24 h post-irradiation. Each time point shows the mitotic index of R-irradiated plumules and G control plumules picked at the same time. The time course of G is shown in dashed lines, and the time course of R is shown in solid lines. The error bars represent ±1 sd, and the asterisks indicate time points at which G and R are statistically different as noted in the text.
Similar measurements were performed with the apical tissue. Because the A is small and difficult to work with, time points were only taken every 6 h. A significant decrease (P < 0.001) in the mitotic rate of the apex is indicated at 24 h after the R irradiation (Fig. 3B).
mRNA Abundance of Cell Cycle-Regulated Genes
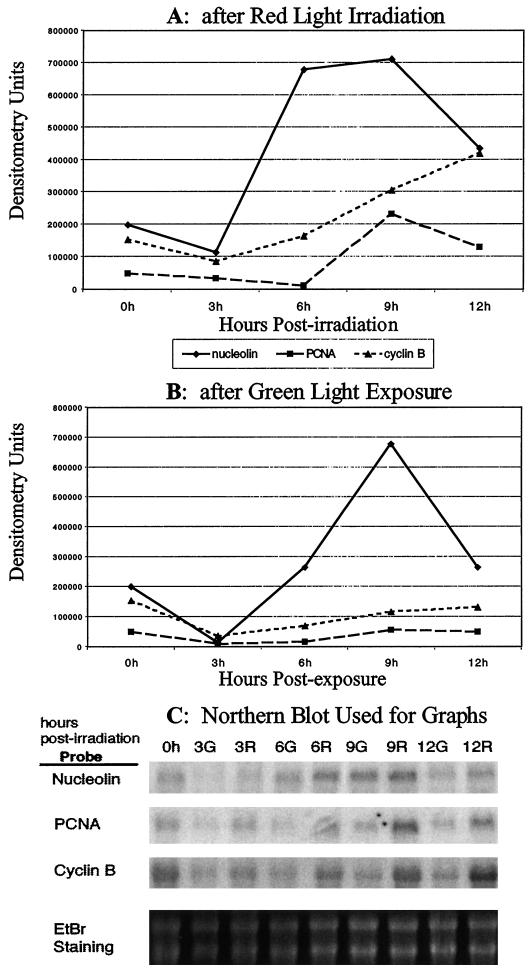
The mRNA abundance of genes known to be regulated in a cell cycle-dependent manner, proliferating cell nuclear antigen (PCNA) and cyclin B, was compared with that of nucleolin after an R pulse (Fig. 4A). Nucleolin rises rapidly at 6 h post-irradiation, stays steady through 9 h, and then begins to decrease at 12 h. The abundance of PCNA stays relatively steady through 6 h, increases at 9 h, and then begins to decrease at 12 h. Cyclin B abundance rises at a relatively constant rate beginningat 6 h post-irradiation and continuing through 12 h.
Figure 4.
The mRNA abundance of cell cycle-regulated genes compared with the mRNA abundance of nucleolin. A, The expression of the mRNA levels of nucleolin, PCNA, and cyclin B after 2 min of R at time zero as measured by northern blots. The densitometry units are values assigned by digitizing the blot on a phosphoImager (Molecular Dynamics). The mRNA abundance of nucleolin increases and rapidly reaches saturation at 6 h postirradiation followed by a gradual increase of cyclin B beginning at 6 h postirradiation and then an increase in PCNA at 9 h postirradiation. Nucleolin and PCNA are decreasing at 12 h postirradiation, whereas cyclin B is continuing its increase. B, The expression of the mRNA levels of nucleolin, PCNA, and cyclin B after an exposure of approximately 10 min of G at time zero as measured by northern blots. This exposure was during the normal handling of the peas in the growth room. The densitometry units are values assigned by digitizing the blot on a phosphoImager (Molecular Dynamics). The mRNA abundance of nucleolin rises at 6 h postirradiation peaking at 9 h and then decreasing at 12 h. Neither cyclin B nor PCNA shows much change when exposed to G. C, Northern blot that was used to obtain the values in A and B. The numbers across the top indicate the hours after exposure to either G or R, and the identification of the probe used for each blot is shown on the left side. Along the bottom is shown the ethidium bromide staining of the large and small subunits of the rRNA that was used as an equal loading control. The RNA at each time point used for these blots was from a single RNA isolation. The lines and points representing the abundance of the various genes are as follows: solid line with diamonds, nucleolin; dashed line with squares, PCNA; and small dashed line with triangles, cyclin B.
Figure 4B shows the pattern of control plants that were exposed only to G. The cell cycle-regulated genes, PCNA and cyclin B, in agreement with the results seen for M-phase and S-phase measurements, showed little or no change in expression when exposed to G. In contrast, nucleolin did increase when exposed to G. Nucleolin mRNA abundance in G-exposed plumules began to rise at 6 h post-irradiation, and reached saturation at 9 h. As with the S-phase index of R-exposed plumules, the abundance of nucleolin mRNA began to markedly decrease at 12 h.
Two patterns emerge from this experiment. When exposed to R, the cell cycle-regulated genes increase their mRNA abundance slowly, peaking at 9 or more hours post-irradiation. In contrast, the pattern of expression for nucleolin is notably different, characterized by a strong initial increase at 6 h post-irradiation. In addition, although the cell cycle-regulated genes show no change when exposed to G, nucleolin mRNA levels do increase, although more slowly than when exposed to R.
Photoreversibility of the R-Induced Increases in mRNA Abundance of Cell Cycle-Regulated Genes
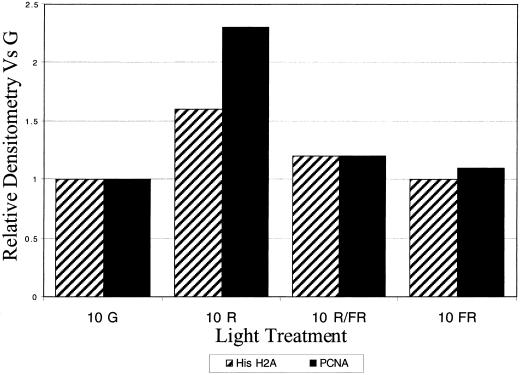
The expression of the cell cycle-regulated genes compared with the S- and M-phase measurements indicated that northern blots could be used as an accurate determination of cell proliferation levels. Therefore, to determine if phytochrome is the photoreceptor for the R-induced increases in cell proliferation that we detected in etiolated pea plants, northern blots of known cell cycle-regulated genes were performed with the following light regimens. Plants were exposed to either R alone, an R pulse followed by a far-R (FR) pulse, or a FR pulse was given without R. PCNA and histone H2A, another cell cycle-regulated gene, show similar increases to R at 10 h as in Figure 4A, and both show little-to-no increase in mRNA abundance after R followed by FR or with FR alone (Fig. 5). Thus changes in the mRNA abundance of cell cycle-regulated genes are photoreversible, implicating phytochrome as the photoreceptor for these changes.
Figure 5.
Photoreversibility of the R induction of the cell cycle-regulated mRNAs. Plumules were irradiated by R alone, R immediately followed by FR, or by FR alone and the accumulation of cell cycle-regulated genes was then measured at 10 h postirradiation. R followed by FR and FR alone did not induce the mRNA accumulation as R did, thus indicating that this response is regulated via phytochrome. Cross-hatched bars indicate histone H2A, and solid bars indicate PCNA.
Rescue of a Yeast nsr1 Mutant with Pea Nucleolin
To determine if the nucleolin-like cDNA from peas was functionally similar to nucleolins from animals and yeast, the pea nucleolin-like cDNA was expressed in a yeast nucleolin mutant (WYL353), which has the yeast nucleolin homolog, nsr1, deleted. The most complete data about the function of nucleolin have come from these yeast deletion mutants. These mutants splice the rRNA transcripts less efficiently than wild type (Lee et al., 1992), and they grow more slowly than wild type (Kondo and Inouye, 1992).
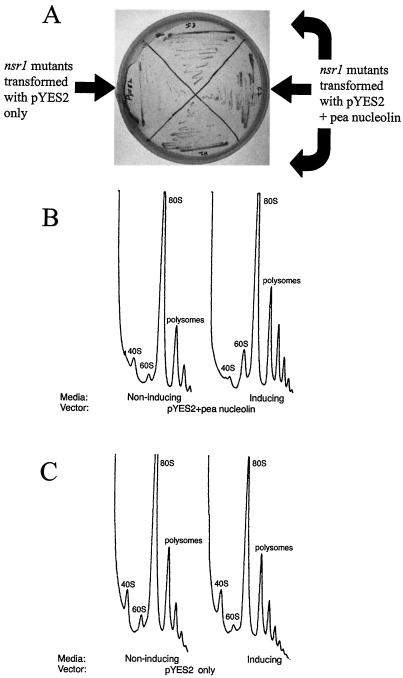
The first indication that the pea nucleolin could rescue the nsr1 mutant was the difference in growth rates of the nsr1 mutant transformed with the pea nucleolin or with vector alone. When grown on plates with inducing media, the nsr1 mutants expressing the pea nucleolin grow faster than those containing the vector only (Fig. 6A). These differences in growth results were demonstrated more quantitatively by growing these yeast strains in liquid inducing media. In this liquid culture assay, the logarithmic phase of growth lasted from 7 to 12 h after inoculation. OD600 readings taken 9, 10, and 11 h after inoculation showed that during logarithmic growth the yeast nsr1 mutant expressing the pea nucleolin gene grew between 1.4- and 1.6-fold more rapidly than the mutant containing vector alone.
Figure 6.
Rescue of the yeast nsr1 mutant with pea nucleolin. A, Yeast transformed with pea nucleolin or vector alone were grown on inducing media plates, and the relative growth rates were observed. Transformed yeast expressing the pea nucleolin grow faster than those with the vector only. B and C, Polysome profiles of yeast cultures in noninducing or inducing media transformed with vector containing pea nucleolin (B) or vector alone (C). The peaks are labeled according to which ribosomal subunits they represent. The profiles read from left to right as from top to bottom of the gradients, and the peaks are in relative OD254 units. When the pea nucleolin is expressed there is an increase in large subunit ribosomes.
Polysome analysis was performed on the same transformed lines (Fig. 6, B and C). When the vector with the pea nucleolin gene was induced, the amount of the 60S subunit increased (Fig. 6B). The vector alone did not show this increase in 60S subunit (Fig. 6C). The polysome analysis was corroborated by densitometry of large subunit rRNA abundance in agarose gels. When the expression of pea nucleolin was induced in the transformed yeast, there was a statistically significant (P < 0.01) increase in large subunit rRNA (data not shown). This increase was not seen in the vector-only controls.
These three experiments show that expressing the pea nucleolin gene in NSR1-deficient yeast cells increases the relative amount of large subunit rRNA and leads to an increased growth rate of the transformed yeast. This increase in large subunit rRNA occurs only when the pea nucleolin cDNA is present and confirms the classification of the pea nucleolin as functioning in ribosome synthesis.
DISCUSSION
Initial experiments performed during the characterization of the pea nucleolin-like cDNA demonstrated that the mRNA abundance of nucleolin in etiolated pea plumules increases following an R pulse and that this increase was photoreversible, implicating phytochrome as the photoreceptor for this response (Tong et al., 1997). This led to the question of whether this light-mediated response was a downstream effect of increased mitotic rates, as might be concluded from previous work on nucleolin (Bugler et al., 1982; Bögre et al., 1996), or whether nucleolin might be regulated independently of proliferation rates.
As etiolated plants proceed through de-etiolation, they shift the focus of their growth from upward to outward. More specifically, the leaves of the plant begin to expand, whereas the upward growth at the shoot apex decreases. These shifts are accomplished by a change in elongation rates and cell division rates (Cosgrove, 1994). Although the kinetics of the shifts in elongation rates has been well studied, the kinetics of cell division changes has not, until this report, been determined. By using several different methods to determine changes in cell proliferation during de-etiolation, the different stages of cell proliferation and their kinetics have been definitively resolved and tracked. The first measurable change in cell proliferation after an R pulse is an increase in cells proceeding through S-phase at 9 h post-irradiation. Next is an increase in M-phase in the inner plumule leaves at 12 h post-irradiation, followed by a decrease in M-phase in the apices at 24 h post-irradiation. The different and opposite reactions to R of the leaves and apices correspond to the opposite directions that their growth rates are changing. By combining these direct measurements of S- and M-phase, the kinetics of cell proliferation changes following an R pulse have been established.
Even though the cell proliferation kinetics were well established by these direct measurements, northern-blot analysis was employed to be able to directly compare R-induced differences in the mRNA abundance of nucleolin with that of other known cell cycle-regulated genes and to more rigorously assess the photoreversibility of these proliferation events. Although each gene has its own pattern of expression after R, the mRNA abundance of the cell cycle-regulated genes increases later and more gradually than that of nucleolin (Fig. 4A). The differences in expression patterns for the cell cycle-regulated genes may be due to the part of the cell cycle in which they are expressed. PCNA is expressed during S-phase (Kodama et al., 1991), it peaks at 9 h postirradiation, and it is noticeably decreasing at 12 h, which mirrors the peak measured for S-phase index (Fig. 1). Cyclin B is expressed during M-phase (Shimizu and Mori, 1998), and its expression increases more gradually and is still increasing at 12 h when M-phase in the leaves is increasing as in Figure 3A. According to these results, during de-etiolation nucleolin is up-regulated prior to increases in cell proliferation changes.
Although nucleolin has been commonly characterized as being regulated in a cell proliferation-dependent manner (Bugler et al., 1982; Bögre et al., 1996), there is at least one other example in which nucleolin is induced by a stimulus without an increase in cell proliferation. Kondo et al. (1992) showed that NSR1, the yeast nucleolin homolog, increased not only with increasing cell proliferation, but also when yeast cells are subjected to a cold shock, a stimulus that suppresses the rate of cell proliferation.
It has been previously shown that increases in the mRNA abundance of nucleolin during de-etiolation are mediated by phytochrome (Tong et al., 1997). In this report we have shown that increases in S-phase, M-phase, and cell cycle-regulated genes are all induced by R and that the direct measurements of S- and M-phase match changes in cell cycle-regulated genes. Therefore, we used northern blots of known cell cycle-regulated genes to determine whether these cell proliferation changes like the mRNA abundance of nucleolin were photoreversible, which implies regulation by phytochrome. Our results (Fig. 5) show that the increases in the cell cycle-regulated genes are photoreversible. So even though the mRNA abundance of nucleolin and changes in cell proliferation have different kinetics during de-etiolation, they are both mediated by phytochrome.
Further analysis of the R-induced cell proliferation data reported here yields additional valuable insights. The first is that these results parallel those of Kaufman et al. (1986). They used whole, etiolated pea plumules and measured the kinetics of fresh weight changes after an R pulse. Their findings show that fresh weight began to significantly increase between 8 and 12 h post-irradiation and that this R-induced effect was reversible by FR. The increase in plumule weight measured by Kaufman et al. (1986), according to our data, would be partially due to the increase in cell division in the plumule leaves. Although elongation rates in pea plumule leaves begin to increase at 4 h post-irradiation (Smith, 1975), significant increases in plumule weight are not recorded until the time that M-phase increases are detected. As Cosgrove (1994) points out, changes in growth cannot involve only changes in cell division, as this would increase the number of cells, but not affect the size of the tissue unless the dividing cells elongate. It is known that the shift in growth that accompanies de-etiolation involves changes in elongation and cell division. But even though elongation rates increase soon after R irradiation, it appears that major growth changes, those that can be measured by an increase in fresh weight, accompany an increase in cell division in the plumule leaves at around 12 h after R irradiation.
Another valuable insight from our data is that R induces some level of synchronization in the number of cells replicating their DNA. There is a significant difference between R- and G-irradiated plants in their S-phase index at 9 h postirradiation, but not before or after. The data presented for the S-phase index are an average of apices and leaves, and, as our M-phase data show, the leaves are increasing their proliferation, whereas the apices are, at a much later time, decreasing their proliferation rate. Although this mixture of apices and leaf tissue could increase background noise, the effect would be small because the mass of the apex is so much smaller than that of the leaves, and the apices are comprised of only a small number of mitotically active cells. Further supporting the idea that light helps to synchronize the number of cells replicating their DNA is the peak in PCNA mRNA abundance at 9 h postirradiation (Fig. 4A). PCNA is known to be up-regulated during S-phase (Kodama et al., 1991), and its peak at 9 h post-irradiation mirrors the peak measured for the S-phase increase. So data from direct measurements and northern blots show some synchrony to the increase in cells entering S-phase following an R pulse.
The fact that R induces an increased abundance of the mRNA for nucleolin somewhat before it stimulates increased rates of cell division could be interpreted to mean that these two processes are regulated differentially by phytochrome, but they do not resolve the question of whether nucleolin expression is independent of cell proliferation. This question is more directly addressed by the results that show G can induce a significant increase in nucleolin expression without having any measurable effect on either cell proliferation or on the expression of cell-cycle related genes. It is clear that in this case, nucleolin gene expression is independent of the onset of cell division.
G is sufficiently actinic to induce the expression of a number of phytochrome regulated genes, via the light hypersensitive-signaling pathway known as the Very Low Fluence response (Thompson et al., 1985; Hsieh et al., 1996). Rigorous demonstration that the effect of G on nucleolin gene expression is mediated through phytochrome would require additional experiments such as repeating these experiments using various phytochrome deficient mutants. If this G induction of nucleolin mRNA expression is via a phytochrome pathway, then because phytochrome can photoreversibly regulate nucleolin expression (Tong et al., 1997) and Very Low Fluence responses are typically not photoreversible, the nucleolin gene would be classified among other genes that can be regulated by R under low fluence (photoreversible) and Very Low Fluence conditions (Thompson et al., 1985). Independent of the question of the photoreceptor for the G response, the fact that G can up-regulate the expression of nucleolin in etiolated pea seedlings without increasing cell division demonstrates conclusively that the expression of nucleolin is regulated differently from cell division.
Phytochrome is known to regulate several different responses such as germination, de-etiolation, shade-avoidance, and flowering. Which response is triggered by phytochrome activation is often controlled by what developmental stage the plant is in. For responses that occur concurrently, however, such as different genes being regulated during de-etiolation, it is known that some of these responses are regulated with very different kinetics and that there are at least two signal transduction pathways that regulate some of these genes (Neuhaus et al., 1993). For instance, phytochrome-mediated up-regulation of the mRNAs for the small subunit of Rubisco and chlorophyll a/b-binding protein requires only a short burst of R, whereas chalcone synthase up-regulation requires a longer R exposure (Peters et al., 1998). Phytochrome can also act through different signal transduction pathways. The mRNA abundance for chlorophyll a/b-binding protein is regulated through a calcium-dependent pathway, whereas chalcone synthase is regulated via a calcium-independent pathway (Neuhaus et al., 1993; Batschauer, 1999). These three genes are all up-regulated during de-etiolation, whereas the mRNA for phytochrome A is down-regulated (Colbert et al., 1983). Thus during the single developmental process of de-etiolation, several different regulatory events are occurring simultaneously, which are all regulated via phytochrome. The different pathways through which phytochrome regulates responses, which are possibly mediated by different members of the phytochrome gene family, can explain how nucleolin and rRNA are up-regulated with different kinetics than the up-regulation of cell proliferation, even though they are all induced by phytochrome.
Knowing when and how nucleolin is regulated during de-etiolation does not resolve what is its function. Much information has been learned about the yeast nucleolin homolog, NSR1, through analysis of deletion mutants. By transforming the pea nucleolin-like cDNA into one of these mutants, we hoped to determine if it had similar functions to other nucleolins. Our findings that yeast deletion mutants expressing the pea nucleolin gene show increases in large rRNA subunit and an increased growth rate demonstrate a rescue of the yeast mutant phenotype. The pea nucleolin shares about the same alignment similarity at the amino acid level with the yeast and animal nucleolins (pea:yeast, 50% similarity, pea:Xenopus, 45% similarity), however, the animal nucleolin does not rescue the yeast nsr1 mutants (Xue et al., 1993). The ability of the plant nucleolin to rescue nsr1 mutants may be related to the fact that the pea and yeast nucleolin share the structural feature of having only two RNA recognition motifs, whereas the animal nucleolins have four (Tong et al., 1997). These RNA recognition motifs are likely responsible for the interaction of nucleolin with rRNA (Serin et al., 1996), and changes in these motifs could be expected to have functional consequences. Although these results do not indicate a precise role for the pea nucleolin in ribosome synthesis, they do demonstrate that the pea nucleolin, like other nucleolins, functions in the synthesis of ribosomes.
The function of nucleolin in ribosome synthesis could help explain why its R-induced expression is coordinated with the light-induced up-regulation of rRNA synthesis that has been repeatedly demonstrated in etiolated seedlings. Several studies have looked at rRNA abundance and polysome formation after phytochrome activation. Increases in rRNA have been measured at around 2 to 6 h post-irradiation, depending on the species (Koller and Smith, 1972; Tobin and Silverthorne, 1985), which is similar to changes in the abundance of the mRNA for pea nucleolin after a pulse of R (Fig. 4A). The observation that nucleolin and rRNA increase with similar kinetics would be predicted by the fact that the primary function of nucleolin has been identified as the processing of rRNA into pre-ribosomes. However, nucleolin is known to be multifunctional (Tuteja and Tuteja, 1998; Ginisty et al., 1999), so this coincidence of up-regulation for rRNA and nucleolin does not preclude other possible functions that nucleolin may have during de-etiolation.
The process of de-etiolation involves a massive change in expressed proteins. This increased need for new protein synthesis mandates precedent up-regulation of nucleolar activity and accumulation of ribosomes for increased translation. Our results indicate that nucleolin up-regulation is likely to be a key event in the overall build-up of molecular machinery leading to increased ribosome synthesis. Because it appears that pea, like yeast and Xenopus, has only one gene encoding nucleolin (Tong et al., 1997), this gene would be a prime target for knockout studies to assess the relative importance of nucleolin in plant rRNA synthesis and in R-induced de-etiolation.
The study of pea nucleolin and its regulation has yielded three main findings. The first is that phytochrome partially synchronizes and alters the time course of cell division during its induction of de-etiolation. Second, the fact that the pea cDNA rescues a yeast nsr1 mutant confirms that the pea gene encodes a functional nucleolin homolog and that the pea nucleolin, like animal and yeast nucleolins, is importantly involved in the synthesis of ribosomes. Third, nucleolin expression, although linked to cell proliferation, is independent of it since it can be induced without an accompanying change in the rate of DNA synthesis or mitosis.
MATERIALS AND METHODS
Plant Material and Irradiations
Pea (Pisum sativum cv Alaska) seedlings were grown in the dark for 7 d at 22°C ± 3°C. They were the source for the pea plumules used in S- and M-phase determinations and for isolation of the RNA used in the northern analyses.
The R irradiations were for 2 min; FR irradiations were for 4 min. The R source produced a fluence of 43 mW/m2 through an interference filter with a λmax of 660 nm and a band width of 20 nm. The FR source produced a fluence of 26 mW/m2 through an interference filter with a λmax of 730 nm and a band width of 20 nm. The dark-grown seedlings were irradiated by R alone, FR alone, or by R followed by FR. All other manipulations of the pea seedlings were done under G, described in Hsieh et al. (1996). After each irradiation treatment, seedlings were returned to the dark and then harvested at different time points as indicated in the figure legends.
Determination of S-Phase Index
After the 7-d-old seedlings were irradiated, an apical section including the plumule + 2 cm of epicotyl was cut off and placed in a Petri dish with 10 nm BrdU (Amersham, Buckinghamshire, UK) for 2 h. At the end of the incubation period, the basal 2 cm of epicotyl was removed and the plumules were fixed in 4% (w/v) paraformaldehyde in Tris buffer (50 mm Tris, 10 mm EDTA, and 100 mm NaCl, pH 7.8) with 0.01% (w/v) Triton X-100 for 1.5 h. The fixative was rinsed out of the plumules by three washes in the Tris buffer. The plumules were then “teased” apart using forceps and a scalpel to release intact cells from the tissue, and the released cells were immobilized on a poly-Lys-coated microscope slide. The cells were washed in the Tris buffer plus 0.01% (w/v) Triton X-100 for 30 min, treated with 2 n HCl for 20 min, and washed twice for 5 min each time in the Tris buffer. The slides were blocked in 1% (w/v) bovine serum albumin in Tris buffer for 20 min followed by a short wash in a buffer (Tris buffer, pH 7.8, 0.1% [w/v] bovine serum albumin, and 0.1% [w/v] Triton X-100). All subsequent washes of the sample used this buffer. The samples were then incubated with mouse anti-BrdU (Amersham) overnight at 4°C. The samples were washed three times for 5 min each prior to incubation with secondary antibody. Goat anti-mouse IgG conjugated to Bodipy was used diluted 1:100. This incubation was performed at room temperature for 2 h. The samples were then washed three times for 5 min each followed by a 10-min incubation with 4′, 6-diamino-phenylindole (to allow identification of nuclei). One final wash was followed by the addition of citifluor-glycerol and overlay of the sample with a coverslip. The cells were then scored for BrdU incorporation by fluorescence of the secondary antibody, and this number was compared with the total number of nuclei to obtain the S-phase index. A minimum of 500 nuclei were counted per data point, and each individual experiment was performed twice. The Student's t test was used to determine statistical significance between light treatments at each time point.
Determination of M-Phase Index
The irradiated plumules were excised from the seedlings and immersed in Farmer's fixative (3 parts ethanol:1 part glacial acetic acid) for 30 to 45 min. The plumules were then washed for 5 min in 100% (w/v) ethanol and stored in 70% (w/v) ethanol at 4°C. The plumules were washed in tap water immediately prior to Feulgen staining. The Feulgen stain binds to DNA, and therefore allows visualization of nuclei. The stained plumules were then prepared for mitotic index counting.
The outer leaves were removed (very little mitosis could be detected in this tissue) and discarded, whereas the IL and A were separated. The As were teased apart as described in the S-phase determination. The IL were teased apart in a small volume of water contained within a microfuge tube using a Teflon grinder. The cells and teased tissue were then placed on a microscope slide, excess water was evaporated on a slide warmer, and the samples were covered with a coverslip. The samples were viewed under a light microscope. The number of cells undergoing cell division was counted by identifying cells in prophase, metaphase, anaphase, or telophase, and this number was compared with the total number of nuclei to obtain the mitotic index. A minimum of 500 nuclei were counted per data point. The Student's t test was used to determine statistical significance between light treatments at each time point.
RNA Isolation and Northern-Blot Analysis
Plumules from the G, R, R/FR, or FR light-irradiated plants were frozen in liquid nitrogen and ground in a microfuge tube with a Teflon grinder. The resulting frozen powder was used for total RNA isolation according to the manufacturer's instructions using TRIzol (Gibco-BRL, Cleveland). The RNA was electrophoretically separated in a 1.2% (w/v) agarose gel containing 6% (w/v) formaldehyde, blotted by capillary action onto Zeta-Probe Nylon membrane (Bio-Rad, Hercules, CA) or Hybond-N+ membrane (Amersham), irradiated with short-wavelength UV for 2 min to crosslink the RNA to the membrane, and incubated in prehybridization solution (0.25 m Na2HPO4, pH 7.2, 1 mm EDTA, and 7% [w/v] SDS). Hybridization to a 32P-labeled probe was performed at 65°C or 60°C for 16 h. The washing procedure was carried out under high stringency conditions described in the supplier's protocol. The wet membrane was wrapped with plastic wrap and placed in a phosphoImager (model no. 445S1, Molecular Dynamics, Sunnyvale, CA). The different probes were used with tissue from same RNA isolation.
Probes Used in Northern Blots
Radioactive probes were made from full-length cDNAs for the various genes using a Decaprime II kit (Ambion, Austin, TX) according to the manufacturer's instructions. The pea nucleolin probe was made from the cDNA of clone NA481–5 (Tong et al., 1997). The pea cyclin B and PCNA cDNAs were obtained from Dr. Hitoshi Mori (Shimizu and Mori, 1998). The pea histone H2A was obtained from Dr. Joel Stafstrom (Devitt and Stafstrom, 1995).
Yeast Strains and Their Growth
The nsr1 mutant (WYL353) was obtained from Dr. Teri Melese (Lee et al., 1992). WYL353 (MATa ade2-1 can1-100 ura3-1 leu2-3, 112 trp1-1 his3-11, 15 nsr1::HIS3) was transformed with pYES2 (Invitrogen, Carlsbad, CA), a yeast expression vector containing the GAL1 promoter. WYL353 was transformed with pYES2 alone or pYES2 containing the full-length pea nucleolin cDNA. All transformed yeast were grown at 30°C on either inducing media (5.7 g/L yeast nitrogen base [Bio 101, Inc., La Jolla, CA], 0.77 g/L CSM-URA [Bio 101, Inc.] 10 mm potassium phosphate, 0.1% [w/v] dextrose, 2% [w/v] Gal, and 5% [w/v] glycerol) or non-inducing media (5.7 g/L yeast nitrogen base [Bio 101, Inc.], 0.77 g/L CSM-URA, 10 mm potassium phosphate, and 2% [w/v] dextrose).
For the liquid growth complementation experiments, WYL353 containing either pYES2 alone or pYES2 with the pea nucleolin cDNA was transferred from noninducing plates and grown in 5 mL of noninducing media overnight (each of these experiments was done in triplicate). Equal amounts of each of these yeast strains, as judged by OD600, were then transferred to another 5 mL of pre-warmed noninducing media and allowed to grow to early log phase. Equal amounts of each of these cultures, as judged by OD600, were then transferred to inducing media and grown to saturation. To monitor the growth rates in liquid inducing media, OD600 readings were taken hourly.
Polysome and Total RNA Isolation from Yeast
Yeast polysomes were isolated by growing cells to mid-log (A600 = approximately 0.5) phase and then adding cycloheximide to a final concentration of 200 μg/mL. The cultures with cycloheximide were placed on ice for 30 min, and then spun at 4,000g for 5 min. These cells were frozen at −70°C. The frozen cells were resuspended in 1 volume of lysis buffer (10 mm Tris, pH 7.5, 50 mm KCl, 8 mm MgCl2, 6 mm 2-mercaptoethanol, and 200 μg/mL cycloheximide) and broken open by vortexing with glass beads. The supernatant was collected and spun 2 × 10 min in a microfuge. This supernatant was collected and quantified by reading A260. Continuous Suc gradients from 7% to 47% were prepared in lysis buffer, and the equivalent of 10 absorbance units were loaded per gradient. The samples were centrifuged at 40,000 rpm in an ultracentrifuge (SW-40, Beckman, Fullerton, CA) for 2.5 h. The gradients were placed in an ISCO 640 density fractionator and the polysome profiles were produced by reading A254.
Total RNA from yeast in liquid culture was isolated by centrifuging the cultures in a table-top centrifuge, pouring off the supernatant, and resuspending the pellet in the leftover media. The pellets were frozen in liquid nitrogen and ground in a microfuge tube with a Teflon grinder. The resulting frozen powder was used for total RNA isolation according to the manufacturer's instructions using TRIzol (Gibco-BRL). The total RNA was separated on a 1.2% (w/v) agarose gel, and the relative densitometry of the large and small rRNA subunits was determined by digitizing the gel image on a gel reader (AlphaImager 2000, Alpha Innotech, San Leandro, CA). The densitometry of the large and small subunits was calculated as a percentage for each subunit of the total densitometry for both subunits. The relative densitometry of the large subunit was used to measure changes in rRNA levels that occurred when expression of the vector was induced. Three independent transformants were used along with three different experiments to generate statistical data. The Student's t test was used to determine significance between samples.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Hitoshi Mori for the pea cyclin B and PCNA cDNAs and Dr. Joel Stafstrom for the pea histone H2A cDNA. We would also like to thank Dr. Arlen Johnson and George Kallstrom for assistance in performing the polysome profiles, and Dorcena Deutsch, Yulin Zhang, William Hanson, and Adam Molofsky for valuable research assistance.
Footnotes
This work was supported by the U.S. National Science Foundation (grant no. IBN–9603884 to S.A.R.) and by the National Aeronautics and Space Administration (grant no. NAG2–1347).
LITERATURE CITED
- Batschauer A. Light perception in higher plants. Cell Mol Life Sci. 1999;55:153–165. doi: 10.1007/s000180050282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batschauer A, Gilmartin P, Nagy F, Schafer E. The molecular biology of photoregulated genes. In: Kendrick RE, Kronenberg GHM, editors. Photomorphogenesis in Plants. Ed 2. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 559–599. [Google Scholar]
- Behringer FJ, Davies PJ, Reid JB. Genetic analysis of the role of gibberellin in the red light inhibition of stem elongation in etiolated seedlings. Plant Physiol. 1990;94:432–439. doi: 10.1104/pp.94.2.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bögre L, Jonak C, Mink M, Meskinen I, Traas J, Ha DTC, Swoboda I, Plank C, Wagner E, Heberle-Bors E, Hirt H. Developmental and cell cycle regulation of alfalfa nucMs1, a plant homolog of the yeast Nsr1 and mammalian nucleolin. Plant Cell. 1996;8:417–428. [PMC free article] [PubMed] [Google Scholar]
- Bugler B, Caizergues-Ferrer M, Bouche G, Bourbon H, Amalric F. Detection and localization of a class of proteins immunologically related to a 100-kD nucleolar protein. Eur J Biochem. 1982;128:475–480. doi: 10.1111/j.1432-1033.1982.tb06989.x. [DOI] [PubMed] [Google Scholar]
- Colbert JT, Hershey HP, Quail PH. Autoregulatory control of translatable phytochrome mRNA levels. Proc Natl Acad Sci USA. 1983;80:2248–2252. doi: 10.1073/pnas.80.8.2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. Photomodulation of growth. In: Kendrick RE, Kronenberg GHM, editors. Photomorphogenesisin Plants. Ed 2. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 631–658. [Google Scholar]
- Devitt ML, Stafstrom JP. Cell cycle regulation during growth-dormancy cycles in pea axillary buds. Plant Mol Biol. 1995;29:255–265. doi: 10.1007/BF00043650. [DOI] [PubMed] [Google Scholar]
- Furuya M, Kanno M, Okamoto H, Fukuda S, Wada M. Control of mitosis by phytochrome and a blue-light receptor in fern spores. Plant Physiol. 1997;113:677–683. doi: 10.1104/pp.113.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginisty H, Amalric F, Bouvet P. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 1998;17:1476–1486. doi: 10.1093/emboj/17.5.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginisty H, Sicard H, Roger B, Bouvet P. Structure and functions of nucleolin. J Cell Sci. 1999;112:761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- Hsieh HL, Tong CG, Thomas C, Roux SJ. Light-modulated abundance of an mRNA encoding a calmodulin-regulated, chromatin-associated NTPase in pea. Plant Mol Biol. 1996;30:135–147. doi: 10.1007/BF00017808. [DOI] [PubMed] [Google Scholar]
- Kaufman LS, Roberts LL, Briggs WR, Thompson WF. Phytochrome control of specific mRNA levels in developing pea buds. Plant Physiol. 1986;81:1033–1038. doi: 10.1104/pp.81.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama H, Ito M, Ohnishi N, Suzuka I, Komamine A. Molecular cloning of the gene for plant proliferating-cell nuclear antigen and expression of this gene during cell cycle in synchronized cultures of Catharaanthus roseus cells. Eur J Biochem. 1991;197:495–503. doi: 10.1111/j.1432-1033.1991.tb15937.x. [DOI] [PubMed] [Google Scholar]
- Koller B, Smith H. Relationship between photomorphogenesis and RNA synthesis in oat and pea seedlings. Phytochemistry. 1972;11:1295–1301. [Google Scholar]
- Kondo K, Inouye M. Yeast NSR1 protein that has structural similarity to mammalian nucleolin is involved in pre-rRNA processing. J Biol Chem. 1992;267:16252–16258. [PubMed] [Google Scholar]
- Kondo K, Kowalski LRZ, Inouye M. Cold shock induction of yeast NSR1 protein and its role in pre-rRNA processing. J Biol Chem. 1992;267:16259–16265. [PubMed] [Google Scholar]
- Lee W-C, Zabetakis D, Melese T. NSR1 is required for pre-rRNA processing and for the proper maintenance of steady-state levels of ribosomal subunits. Mol Cell Biol. 1992;12:3865–3871. doi: 10.1128/mcb.12.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus G, Bowler C, Kern R, Chua NH. Calcium/calmodulin-dependent and -independent phytochrome signal transduction pathways. Cell. 1993;73:937–952. doi: 10.1016/0092-8674(93)90272-r. [DOI] [PubMed] [Google Scholar]
- Peters JL, Szell M, Kendrick RE. The expression of light-regulated genes in the high-pigment-1 mutant of tomato. Plant Physiol. 1998;117:797–807. doi: 10.1104/pp.117.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pine K, Klein AO. Regulation of polysome formation in etiolated bean leaves by light. Dev Biol. 1972;28:280–289. doi: 10.1016/0012-1606(72)90144-3. [DOI] [PubMed] [Google Scholar]
- Pollice A, Zibella MP, Bilaud T, Laroche T, Pulitzer JF, Gilson E. In vitro binding of nucleolin to double-stranded telomeric DNA. Biochem Biophys Res Commun. 2000;268:909–915. doi: 10.1006/bbrc.2000.2237. [DOI] [PubMed] [Google Scholar]
- Serin G, Joseph G, Faucher C, Ghisolfi L, Bouche G, Almaric F, Bouvet P. Localization of nucleolin binding sites on human and mouse pre-ribosomal RNA. Biochimie. 1996;78:530–538. doi: 10.1016/0300-9084(96)84759-6. [DOI] [PubMed] [Google Scholar]
- Shacklock PS, Read ND, Trewavas AJ. Cytosolic free calcium mediates red light-induced photomorphogenesis. Nature. 1992;358:753–755. [Google Scholar]
- Shimizu S, Mori H. Analysis of cycles of dormancy and growth in pea axillary buds based on mRNA accumulation patterns of cell cycle related genes. Plant Cell Physiol. 1998;39:255–262. doi: 10.1093/oxfordjournals.pcp.a029365. [DOI] [PubMed] [Google Scholar]
- Smith H. The photomorphogenic response systems and their photoreceptors. In: Smith H, editor. Phytochrome and Photomorphogenesis. London: McGraw-Hill; 1975. pp. 22–53. [Google Scholar]
- Thompson WF, Kaufman LS, Watson JC. Induction of plant gene expression by light. BioEssays. 1985;3:153–159. [Google Scholar]
- Tobin EM, Silverthorne J. Light regulation of gene expression in higher plants. Annu Rev Plant Physiol. 1985;36:569–593. [Google Scholar]
- Tong C-G, Reichler S, Blumenthal S, Balk J, Hsieh H-L, Roux SJ. Light regulation of the abundance of mRNA encoding a nucleolin-like protein localized in the nucleoli of pea nuclei. Plant Physiol. 1997;114:643–652. doi: 10.1104/pp.114.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja R, Tuteja N. Nucleolin: a multifunctional major nucleolar phosphoprotein. Crit Rev Biochem Mol Biol. 1998;33:407–436. doi: 10.1080/10409239891204260. [DOI] [PubMed] [Google Scholar]
- Xue Z, Shan X, Lapeyre B, Melese T. The amino terminus of mammalian nucleolin specifically recognizes SV40 T-antigen type nuclear localization sequences. Euro J Cell Biol. 1993;62:13–21. [PubMed] [Google Scholar]
- Ying GG, Proost P, van Damme J, Bruschi M, Introna M, Golay J. Nucleolin, a novel partner for the Myb transcription factor family that regulates their activity. J Biol Chem. 2000;275:4152–4158. doi: 10.1074/jbc.275.6.4152. [DOI] [PubMed] [Google Scholar]