Abstract
Planktonic archaea are thought to play an important role in ammonia oxidation in marine environments. Data on the distribution, abundance, and diversity of ammonia oxidizers in the coastal sea-surface microlayer (SML) are lacking, despite previous reports of high abundance of Thaumarchaeota in the SML of estuaries and freshwater lakes. Here, we failed to detect the presence of ammonia-oxidizing bacteria in any of our samples taken from a semi-enclosed marine inlet in Japan. Therefore, we shifted our focus to examine the archaeal community composition as well as the Thaumarchaeota marine group I (MG-I) and ammonia monooxygenase subunit A (amoA) gene copy numbers and composition in the SML and corresponding underlying water (UW, 20 cm). amoA gene copy numbers obtained by quantitative PCR were consistent with the typical values observed in the surface waters of oceanic and coastal environments where nitrification activity has been detected, but the copy numbers were two- to three-fold less than those reported from the surface layers and UW of high mountain lakes. Both amoA and MG-I 16S rRNA gene copy numbers were significantly negatively correlated with chlorophyll-a and transparent exopolymer particle concentrations in the SML. Communities of archaea and ammonia-oxidizing archaea in SML samples collected during low wind conditions (≤5 m s–1) differed the most from those in UW samples, whereas the communities in SML samples collected during high wind conditions were similar to the UW communities. In the SML, low ratios of amoA to MG-I 16S rRNA genes were observed, implying that most of the SML Thaumarchaeota lacked amoA. To our knowledge, our results provide the first comparison of ammonia-oxidizing communities in the coastal SML with those in the UW.
Introduction
Nitrification is an important process in the nitrogen cycle. The nitrification process involves a two-step biological conversion of ammonia to nitrate via nitrite. Ammonia oxidation—the first, and rate-limiting, step in nitrification—was once thought to be restricted to ammonia-oxidizing bacteria (AOB) belonging to the phylum Proteobacteria [1,2]. The discovery of the putative archaeal ammonia monooxygenase subunit A (amoA) gene [3] and the subsequent isolation of Nitrosopumilus maritimus, a member of the ammonia-oxidizing archaea (AOA) [4] that belongs to the phylum Thaumarchaeota, have provided new insights into the distribution of AOA and their roles in the nitrogen cycle. In marine environments, AOA are considered to be more abundant, and to play a more important role in ammonia oxidation, than AOB [5].
The sea-surface microlayer (SML) is a thin surface film located at the interfacial point between the sea surface and the atmosphere. SML archaeal communities differ from those in the underlying water, and SML-exclusive archaeal groups have been detected [6,7]. In high mountain lakes, members from the phylum Thaumarchaeota dominate the surface-layer archaeal community, whereas members of the phylum Euryarchaeota were more abundant in the subsurface waters [8–10]. Despite intensive studies of the ubiquitous distribution of AOA in marine environments, in the case of the SML, AOA community structure and abundance have been described only in a high mountain lake [11]. The presence of Marine Group-I Thaumarchaeota (MG-I) and amoA genes in the SML indicates the potential for ammonia oxidation in this layer. Furthermore, ammonia, the substrate needed for ammonia oxidation, is known to accumulate in the SML [12,13]. However, to date, no data are available on the ammonia-oxidizing communities in coastal and oceanic SML environments. Much less is known about the ammonia-oxidizing community in the SML than about ammonia oxidizers in other aquatic systems.
Here, we describe the archaeal community composition, and specifically the composition of Thaumarchaeota MG-I, as well as, MG-I, AOA and AOB gene copy numbers and composition, in the SML and the corresponding underlying water (UW, 20 cm depth) of a semi-enclosed marine inlet in Japan. Our findings regarding the ammonia-oxidizing communities in the SML and UW of this coastal inlet should fill the fundamental gaps in our knowledge of these globally important functional groups of microbes in one of the most under-studied areas of the marine environment.
Materials and methods
Study site
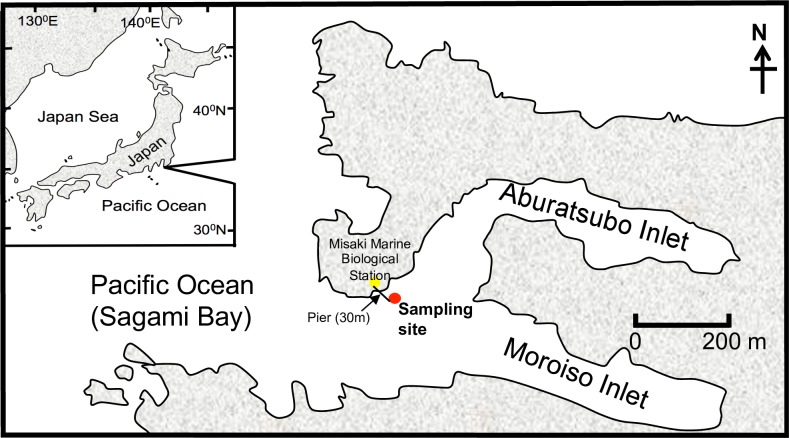
Water samples from Aburatsubo Inlet, in the town of Misaki, in Kanagawa Prefecture, Japan, were obtained from the pier of Misaki Marine Biological Station in September and December 2012 at high tide to avoid resuspended bottom sediments. The sampling site (35°09.5′N, 139°36.5′E) was located approximately 30 m from land, with a maximum water depth of about 5 m (Fig 1). The semi-enclosed inlet is located in a temperate coastal area adjacent to Sagami Bay, which connects to the Pacific Ocean. The topography of the inlet was ideal for SML sampling, as it reduced the effect of oceanic waves from Sagami Bay.
Fig 1. Sampling location.
The location of the Misaki Marine Biological Station is indicated by the yellow dot, and the sampling point is indicated by the red dot.
SML and UW sampling
SML water samples were collected by rotating a Plexiglas drum at a rate of 6 rotations min–1. The rotating drum sampler is widely used to sample large volumes of water from the SML of various environments, including the open ocean and coastal waters [14–16]. Water adhering to the drum was scraped off by using a Teflon scraper into a sterile collection bottle. To condition the drum surface, the first 1 L of SML water sample collected was discarded. Duplicate water samples from the SML were collected by using the drum sampler, and these duplicate samples were pooled for further filtration and analysis. The thickness of the SML sampled by the drum sampler was determined by dividing the volume of water sampled by the surface area of the sampler and the number of drum rotations. The thickness of the sampled SML was 36 ± 4 μm. At the same time as the drum samples were taken, a sterile bottle was submerged to a depth of 20 ± 5 cm to collect UW samples. To prevent contamination with SML water, the cap of the sterile bottle was opened and closed at a depth of 20 cm.
In total, 12 samples (six pooled samples from the SML and six samples from the UW) were collected. The samples were labeled, in sequence, according to the sampling month (September: S; December: W), sample number (e.g. 1, 2) and depth (SML samples collected using drum sampler: D; UW: U). A set of paired samples (SML and UW) was collected daily over 2 or 3 consecutive days from the same sampling point. The temperature and salinity of surface water samples were monitored by using a YSI Model 85 handheld meter (YSI Incorporated, Yellow Springs, USA). The development of a distinct microbial community in the SML requires relatively calm surface conditions and low wind speed [17]. Therefore, the wind speed during sampling was constantly recorded by using a handheld anemometer (GA-06, Be-s Co. Ltd, Osaka, Japan).
Chlorophyll-a and transparent exopolymer particles as proxies for SML formation
Concentrations of transparent exopolymer particles (TEP) and chlorophyll-a (Chl-a) were measured as proxies of organic matter concentrations in the SML. TEP concentrate in the SML and are a major component of it [18]. TEP concentration was quantified as previously described [19,20]. Triplicate 10- to 20-mL subsamples for TEP quantification were filtered onto 0.4-μm-pore-size polycarbonate membrane filters (Isopore HTTP, Millipore, Tokyo, Japan), stained with 500 μL of 0.02% 8GX alcian blue solution (Sigma-Aldrich, Tokyo, Japan) in 0.06% acetic acid (pH 2.5), extracted in 80% sulfuric acid for 2 h, and then measured spectrophotometrically.
For Chl-a analysis, 50 mL each of duplicate subsamples were filtered onto 25-mm GF/F filters (GE Healthcare, Tokyo, Japan) extracted with N,N-dimethylformamide, and stored at −20°C until further analysis [21]. Chl-a concentrations were measured fluorometrically with a model 10-AU Field and Laboratory Fluorometer (Turner Designs, San Jose, USA).
The enrichment factors (EF) of TEP and Chl-a in the SML were calculated as the ratio of the concentration in the SML to that in the UW. These parameters were considered enriched in the SML when the EF value exceeded a ratio of 1.0 [22].
Nucleic acid extraction
Approximately 1 L of water samples collected from the SML and UW were filtered onto 0.22-μm-pore-size Sterivex GP filter units (Millipore). The filters were immediately stored at −80°C until further analysis. DNA extraction was performed with a ChargeSwitch Forensic DNA Purification Kit (Invitrogen, Tokyo, Japan) after bead beating with zirconia beads (FastGene, Tokyo, Japan) at 5,000 rpm for 30 s. Each filter was extracted twice to maximize the DNA yield.
Archaeal 16S rRNA gene pyrosequencing and data processing
The V1 to V3 hyper-variable regions of the archaeal 16S rRNA gene were amplified by using the forward primer A20F [23] and reverse primer 519R [24]. PCRs for each sample were performed in triplicate with 20 μL of mixture consisting of 6 μL of 10-fold diluted template DNA (5 to 8 ng μL–1), 0.13 μM of each primer, 0.2 mM of each dNTP, 2 μL of 10× Ex Taq Buffer (Takara, Shiga, Japan), and 0.5 U of Ex Taq HS (Takara). Touchdown PCR was performed for a total of 35 cycles under the following conditions: initial denaturation at 94°C for 3 min, 10 cycles of denaturation at 98°C for 10 s, annealing at 68 to 59°C for 30 s, elongation at 72°C for 45 s; followed by 25 cycles of denaturation at 98°C for 10 s, annealing at 58°C for 30 s, elongation at 72°C for 45 s; and final elongation at 72°C for 10 min. The prepared PCR products were purified with an AMPure system (Beckman Coulter, Brea, USA) and sequenced at the Atmosphere and Ocean Research Institute, Chiba, on a Roche 454 GS Junior platform (Roche 454 Life Science, Brandford, USA) in accordance with the manufacturer’s instructions.
After the sequencing, the open-source software mothur 1.35.1 [25,26] was used for subsequent analysis in accordance with the 454 standard operating procedure (http://www.mothur.org/wiki/454_SOP; accessed on 23 March 2015). Reads for downstream sequence processing were retained if they had a minimum quality score of 30, and representative sequences were assigned to operational taxonomic units (OTUs) by using the furthest-neighbor clustering algorithm based on 97% identity.
Detection of ammonia-oxidizing bacteria
The betaproteobacterial and gammaproteobacterial amoA gene fragments were amplified by using the amoAr-New/amoA-2R [27] and amoA3F/amoB-4R [28] primer sets, respectively. PCR for each sample was performed in triplicate with 20 μL of reaction mixture consisting of 3 μL of 10-fold-diluted template DNA (5 to 8 ng μL–1), 0.2 μM of each primer, 0.2 μM of each dNTP, 2 μL of 10× Ex Taq Buffer (Takara), and 0.5 U of Ex Taq HS Polymerase (Takara). Thermal cycling was performed under the following conditions: initial denaturation at 95°C for 3 min, 40 cycles of amplification at 94°C for 30 s, annealing at 47°C (amoAr-New/amoA-2R) and 48°C (amoA3F/amoB-4R) for 1 min, elongation at 72°C for 40 s, and a final elongation at 72°C for 10 min. As the amplification of AOB amoA from all samples was unsuccessful, no further analysis of AOB was done.
Detection and quantification of ammonia-oxidizing archaea
The archaeal amoA gene fragments were quantified by using the Arch-amoA-for /Arch-amoA-rev primer set [5], whereas MG-I 16S rRNA gene fragments were quantified by using the GI-751F/GI-956R primer set [29] in triplicate by quantitative PCR (qPCR). Serial dilutions of known plasmid numbers were used to generate an external standard curve for qPCR analysis [amoA: kt162 plasmid, GenBank accession number: AB592102; and MG-I 16S rRNA gene: kt779 plasmid, GenBank accession number: AB919347]. qPCR was performed in reaction mixtures (20 μL) that contained 3 μL of 10-fold-diluted template DNA (5 to 8 ng μL–1), 10 μL of 2× SYBR Premix Ex Taq with Tli RNase H (Takara), and 0.2 μM final concentration of each primer. Assays for both primer sets were performed by using a Roche Light Cycler 480 II Real-Time PCR system (Roche, Sussex, UK) with the following thermal cycling conditions: initial denaturation at 95°C for 30 s; and 45 cycles of amplification at 95°C for 5 s, annealing at 58°C for 30 s, and elongation at 72°C for 30 s, with a detection step at the end of each cycle. After amplification, melting curve analyses were performed at 95°C for 5 s, followed by a temperature gradient of 0.11°C s−1 from 65 to 95°C. The qPCR detection efficiency was 95.1% for MG-I 16S rRNA genes and 90.0% for amoA genes.
Construction of an archaeal amoA clone library and clone library sequence analysis
An approximately 256-bp fragment of archaeal amoA was amplified by using the Arch-amoA-for/Arch-amoA-rev primer set. PCR was performed in triplicate using a 20-μL reaction mixture consisting of 3 μL of 10-fold-diluted template DNA (5 to 8 ng μL–1), 0.2 μM of each primer, 0.2 mM of each dNTP, 2 μL of 10× Ex Taq Buffer, and 0.5 U of Ex Taq HS (Takara). The PCR thermal cycling conditions consisted of initial denaturation at 95°C for 3 min; 40 cycles of amplification at 98°C for 10 s, annealing at 58°C for 30 s, and elongation at 72°C for 1 min; and final elongation at 72°C for 10 min. Amplified PCR products were pooled and purified by using a Qiaquick PCR Purification Kit (Qiagen KK, Tokyo, Japan). Cloning was performed with a TOPO TA Cloning Kit for Sequencing (Invitrogen) and Escherichia coli DH5-α competent cells (Takara). Positive clones with PCR products amplified through colony PCR were purified by using USB ExoSAP-IT product cleanup reagent (Affymetrix Japan, Tokyo, Japan) and sequenced by using an ABI Prism BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Tokyo, Japan) on an ABI 3730xl platform (Applied Biosystems).
A total of 387 sequences obtained from the clone library were aligned by using MEGA 6.0 [30] against reference sequences retrieved from the GenBank database, and sequences with regions that were ambiguously aligned or with ambiguous amino acid sequences were removed. The remaining sequences were clustered into OTUs by using CD-HIT [31] on the basis of 95% nucleotide identity; this yielded 22 representative amoA OTUs. The 22 representative OTU sequences at 95% nucleotide identity (by CD-HIT) and the closest relative reference sequences from GenBank [32] were used to construct a neighbor-joining phylogenetic tree with the Tamura-Nei model [33] of nucleotide substitution and 1000 bootstrap replicates, implemented in MEGA 6.0.
Statistical analysis
Samples from both samplings (September and December) were pooled according to their depth, SML or UW, for statistical analysis using R version 3.2.3 [34]. The correlation between amoA or MG-I 16S rRNA gene copy numbers and Chl-a and TEP concentrations in all pooled samples were calculated in R by using the non-parametric Spearman’s rank coefficient test. Differences in Chl-a and TEP concentrations between the SML and UW were tested by using the non-parametric Mann-Whitney U-test in R. Principal coordinate analysis (PcoA) of archaeal community structure based on weighted and unweighted UniFrac distances and of AOA community structure based on amoA gene clones was conducted in R by using the phyloseq package [35]. Phylogenetic trees used in the UniFrac analysis were generated using the clearcut function [36] in mothur 1.35.1 [25,26]. Differences between the SML and UW archaeal 16S rRNA gene communities and amoA clone libraries were tested by using weighted and unweighted UniFrac and analysis of molecular variance (AMOVA) functions in the mothur 1.35.1 platform according to the default parameters.
Nucleotide sequence accession numbers
The amoA sequences reported here were deposited in GenBank under accession numbers KU216137 to KU216158. The archaeal 16S rRNA gene sequences obtained by pyrosequencing were deposited in the DNA Data Base of Japan Sequence Read Archive (http://trace.ddbj.nig.ac.jp/dra/index_e.html) under the accession number DRA004205.
Results
Environmental conditions and enrichment of organic matter in the SML
Sea-surface temperature in Aburatsubo Inlet peaks (at 25 to 26°C) with low salinity in August and September (summer), and the temperature reaches its lowest (15 to 16°C) with high salinity in December (winter) [37]. The temperature and salinity ranges obtained during our sampling in September (26.2 to 27.7°C; salinity, 31.7 to 32.4) and December (14.3 to 15.9°C, 34.1 to 34.2) corresponded to the summer and winter values previously reported by Yamaguchi [37]. Salinity in the Aburatsubo Inlet remained similar to that of the surrounding ocean, with slight seasonal changes; there is no known direct river input into the inlet [38]. Ammonium and phosphate concentrations in the semi-enclosed inlet are also higher in summer (September) than in winter (December) [38].
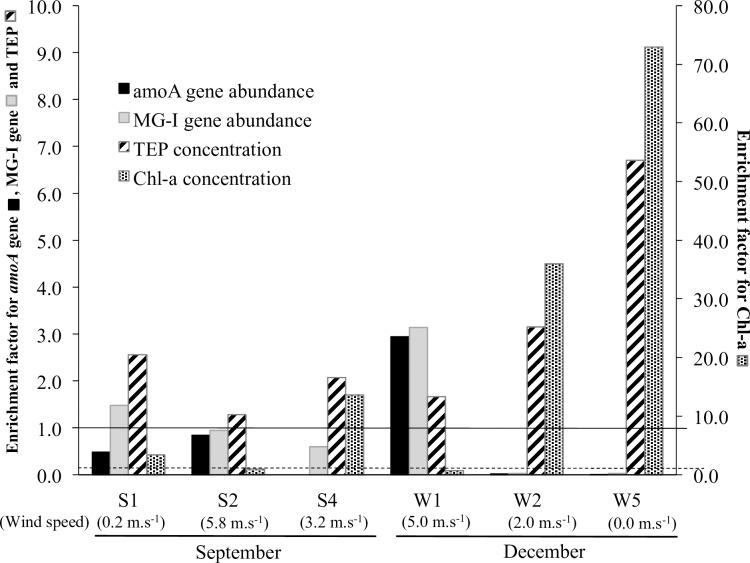
EF ranged from 1.3 to 6.7 for TEP and 0.7 to 72.9 for Chl-a throughout the sampling periods in September and December (Fig 2). Chl-a and TEP concentrations were significantly positively correlated (P < 0.001; S1 Table). When the wind speed was low (≤5.0 m s−1), the EF for all Chl-a samples exceeded 1.0 (SML enriched) whereas the TEP was constantly enriched in the SML regardless of the wind speed (Fig 2). The Chl-a (Mann–Whitney U, P = 0.06) and TEP (Mann-Whitney U, P < 0.01) concentrations were also significantly higher in the SML than in the UW at times of low wind speed. The highest EF values for Chl-a and TEP were observed in December, when the wind speed was the lowest (Fig 2). At high wind speeds (≥5.0 m s−1), the EF values in the SML for Chl-a were lowest and the TEP enrichment in the SML was lower.
Fig 2. Enrichment factors of archaeal ammonia monooxygenase subunit A (amoA) gene copy numbers, archaeal Marine Group-I (MG-I) 16S rRNA gene copy numbers, transparent exopolymer particles (TEP) concentration, and chlorophyll-a (Chl-a) concentration in the sea-surface microlayer (SML) in samples collected in September (S) and December (W).
Enrichment value of 1.0 is indicated by a solid line extending from the first y-axis in the case of amoA copy numbers, MG-I 16S rRNA gene copy numbers, and TEP concentrations and by a dashed line extending from the secondary y-axis in the case of Chl-a concentration. Enrichment >1.0 indicates enrichment in the SML.
Archaeal community structure
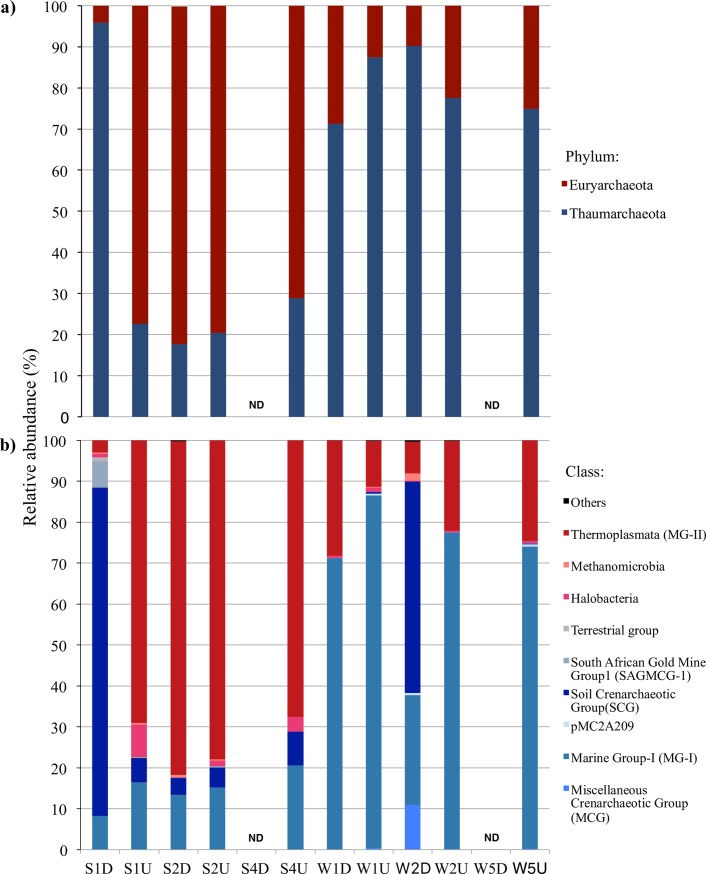
The archaeal community from MG-II belonging to the phylum Euryarchaeota was more abundant in the SML and UW (except in sample S1D) in the September samples than in the December samples, with an average relative abundance of 77.6% ± 4.1% across both the SML and UW combined (Fig 3A). In December, the major archaeal phylum from SML and UW combined samples shifted to Thaumarchaeota, for which the relative abundance ranged from 71.3% to 90.0% (Fig 3A). Phylum Thaumarchaeota accounted for a large fraction of the archaeal community in the SML samples, with an average relative abundance of 68.8% (range: 17.7% to 95.9%) across all SML samples, excluding samples S4D and W5D, for which PCR amplification was unsuccessful (Fig 3A). The relative abundance of the Soil Crenarchaeota Group (SCG) (Fig 3B) was generally 50- to eight-fold higher in the SML samples from S1D and W2D than in the paired UW samples collected from the respective samplings, whereas the Terrestrial group was detected only in the S1D and W1D SML samples (Fig 2B). The PCoA ordination plot based on weighted UniFrac distances showed that the archaeal community composition and relative abundance in SML samples differed from those in the UW samples and SML samples that was collected during high wind speed (S1 Fig). All of the UW archaeal community was clustered closely together by sampling month. SML samples collected during times of high wind speed (≥5 m s–1) (S2D and W1D) were clustered close to the UW samples collected from the same respective samplings.
Fig 3.
Relative abundances of archaeal taxonomic groups, obtained by using 454 pyrosequencing of the 16S rRNA gene, at the a) phylum and b) class levels for surface microlayer (D) and underlying water (U) samples obtained in September (S) and December (W). Abundances are presented in terms of percentage of total archaeal operational taxonomic units at a 97% identity level. “ND” indicates that the sample was not successfully amplified by using the A20F/519R primer set for 454 pyrosequencing.
Archaeal and bacterial amoA and MG-I 16S rRNA gene copy numbers
Interestingly, no beta- or gamma-proteobacterial amoA genes were detected in any of the SML or UW samples.
The average archaeal amoA copy numbers ranged from 0.04 to 11.89 × 105 copies L−1 across all SML samples (with the exception of sample S4D, for which the value fell below the qPCR detection limit) and from 0.31 to 10.71 × 105 copies L−1 across all UW samples. The copy numbers of the MG-I 16S rRNA gene across the SML samples ranged from 0.14 to 24.7 × 105 copies L−1 and from 0.40 to 22.64 × 105 copies L−1 across the UW samples (S2 Fig).
The copy numbers of amoA were significantly higher in UW than in the SML (Mann-Whitney U, P < 0.05) as evidenced by the enrichment values of amoA, which were depleted in the SML (EF = 0.34 ± 0.40), except in the case of sample W1 (EF = 2.9) (Fig 2). No significant differences were found between the SML and UW for MG-I 16S rRNA gene copy numbers from all samples from both samplings (Mann-Whitney U, P = 0.21). MG-I genes in the SML were enriched only in samples S1 and W1 (Fig 2). Significant positive correlations were found between amoA and MG-I 16S rRNA gene copy numbers (S1 Table). The ratio of amoA to MG-I 16S rRNA genes was, on average, 0.4 ± 0.2 in the SML and 0.7 ± 0.1 in the UW.
Both amoA and MG-I 16S rRNA gene copy numbers were negatively correlated with TEP (amoA gene number: P < 0.001; MG-I 16S rRNA gene number: P < 0.01) and Chl-a concentrations (amoA gene number: P < 0.001; MG-I 16S rRNA gene number: P < 0.001; S1 Table). In two samples from the SML (S4D and W5D) they either were present in very low abundance or were undetectable using qPCR primer sets targeting the archaeal amoA and MG-I 16S rRNA genes (S2 Fig). Furthermore, S4D and W5D samples were also unsuccessfully amplified using the current archaeal 16S rRNA gene primer set for pyrosequencing (A20F/519R; Fig 3). The patterns of low gene copy numbers or unsuccessful PCR amplifications from the two samples coincided with the highest peaks of both Chl-a and TEP in the SML during the respective samplings (S2 Fig).
Community structure and diversity of AOA
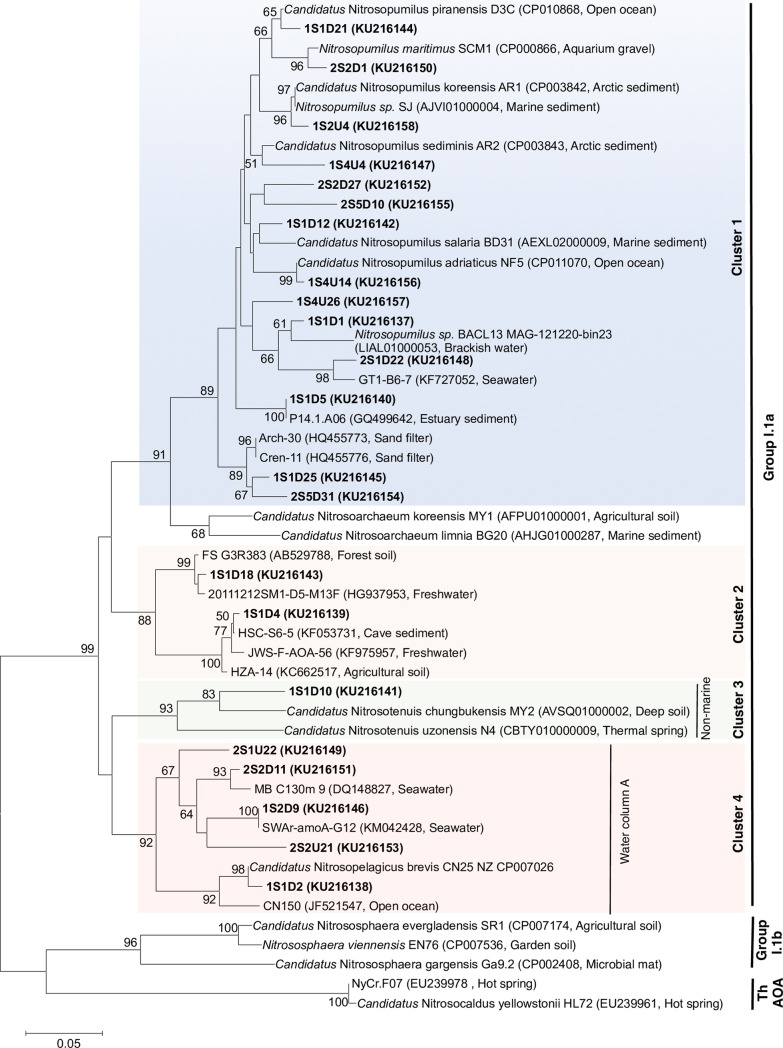
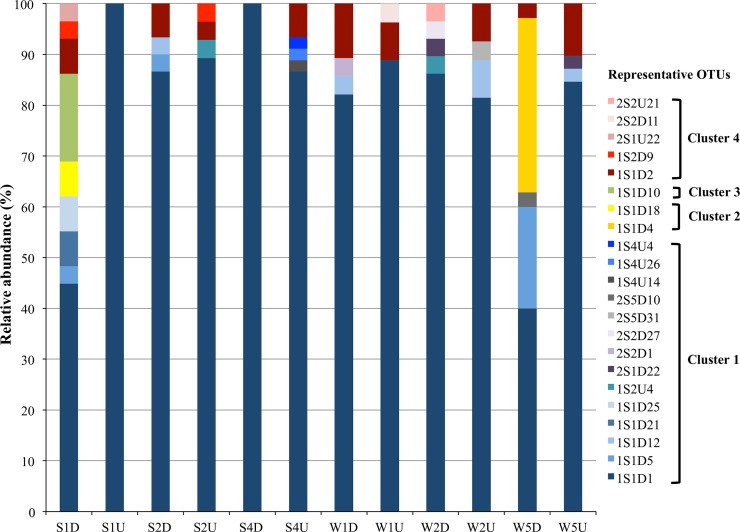
Four major clusters were detected in the SML and UW samples obtained in September and December; 329 of the clone sequences belonged phylogenetically to Cluster 1 (Fig 4), which consisted mainly of sequences that were closely related to the ammonia-oxidizing archaeon Nitrosopumilus maritimus. A majority of the phylotypes obtained from the SML and UW were also ubiquitously distributed within Cluster 4, which consisted of sequences from Water Column Cluster A (WCA) or a surface cluster [29,39]. SML-specific phylotypes represented by 1S1D5, 1S1D25, and 2S5D31 from Cluster 1, as well as 1S1D18 and 1S1D4 from Cluster 2, were found (Figs 4 and 5). These phylotypes were closely related to other clones obtained from sand filters, soil environments, and sediment environments, but not to any closely described species (Fig 5). Clone 1S1D10 from Cluster 3, which was found only in S1D (Fig 5), was clustered close to Candidatus Nitrosotenuis chungbukensis and Candidatus Nitrosotenuis uzonensis, which have been obtained from soil [40] and thermal spring enrichment cultures [41], respectively (Figs 4 and 5).
Fig 4. Consensus neighbor-joining phylogenetic tree of archaeal ammonia monooxygenase subunit A (amoA) gene nucleotide sequences based on Tamura-Nei distances.
The tree contains 22 representative operational taxonomic units (OTUs) from this study (in bold) obtained at a cutoff value of 0.05% by using the CD-HIT program. Bootstrap values (>50%) are indicated at the nodes of the tree. Accession numbers for the representative OTUs and reference sequences are indicated in parentheses. Scale bar represents 0.05 amino acid substitutions per site.
Fig 5. Relative abundances of archaeal ammonia monooxygenase subunit A (amoA) gene clones from surface microlayer (D) and underlying water (U) samples collected in September (S) and December (W).
Clones were grouped into four different clusters based on representative operational taxonomic units with 95% sequence identity, obtained by using the CD-HIT program.
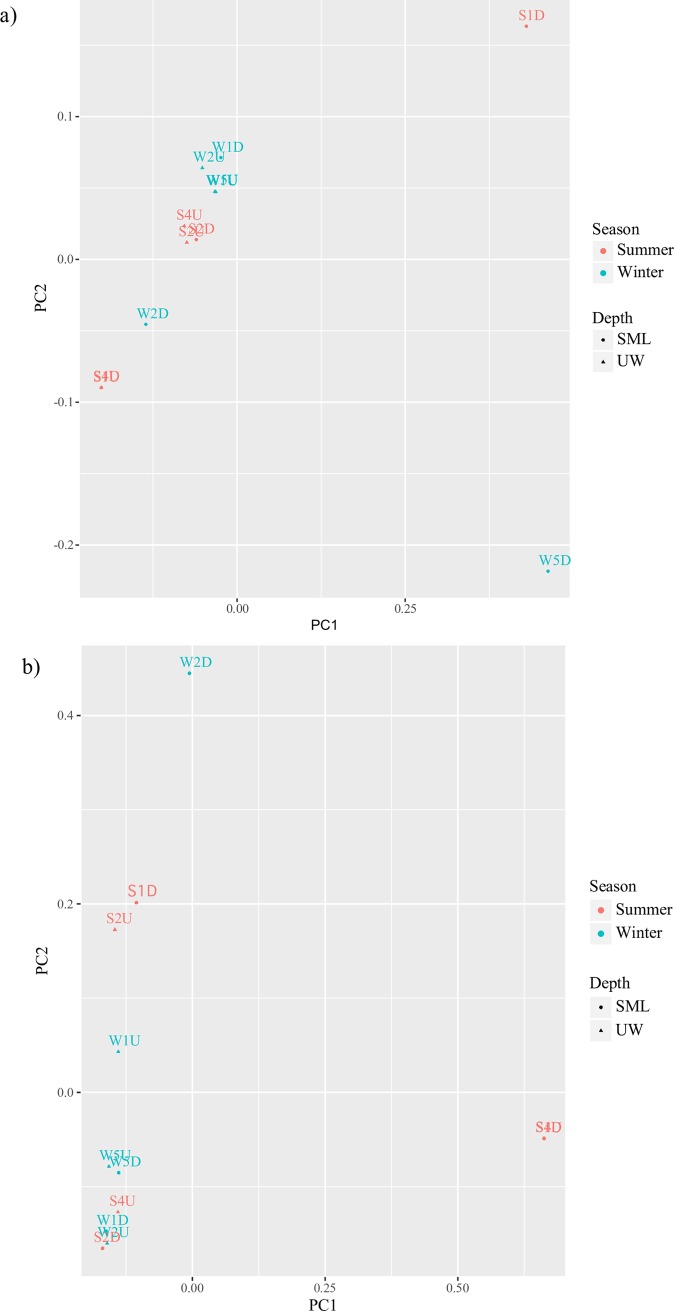
AMOVA tests also showed significant differences between the SML and UW amoA structures (Fs = 3.02, P < 0.05). Additionally, both weighted (P < 0.001) and unweighted (P < 0.01) UniFrac showed that the AOA community structure and composition in the SML differed from those in the UW. All AOA communities from the UW were clustered closely in both September and December samplings, especially in the weighted UniFrac PCoA plot (Fig 6). Weighted and unweighted UniFrac analyses (Fig 6) revealed that AOA communities in the SML samples S2D and W1D, which were collected during high wind speed conditions, were also clustered close to the UW communities. Weighted UniFrac showed that the SML AOA communities from samples S1D, S4D, W2D, and W5D differed from their paired UW samples, whereas samples S1D, S4D, and W2D differed from their paired UW samples in the unweighted UniFrac plots.
Fig 6.
Principal coordinate analysis (PCoA) plots based on a) weighted and b) unweighted UniFrac distances of the amoA community structure from clone libraries.
Discussion
AOA and AOB gene copy numbers in the SML and UW
Our results provide insights into the gene copy numbers and diversity of AOA in the SML in coastal waters compared with those in the UW. In contrast to Zhang et al. [42], we did not amplify any beta- and gamma-proteobacterial amoA from any of our coastal SML and UW samples. Mincer et al. [29] and Beman et al. [43] also found that the gene copy number of AOB tended to show them as absent, or at lower numbers than AOA, in photic zones. Therefore, we surmise that ammonia oxidation in the SML and UW in our samples—if any—was performed mostly by AOA. The amoA and MG-I 16S rRNA gene datasets reported here are consistent with the typical values observed in the surface waters of oceanic and coastal environments [29,43,44] where nitrification activity has been detected [45]. However, they are two- to three-fold less than those reported in the surface layers and UW of high mountain lakes [8].
Our qPCR assays indicated that the amoA to MG-I 16S rRNA gene copy number ratio was within the reference ranges reported in oceanic regions [46–48]. However, the average amoA to MG-I 16S rRNA gene copy number ratio was lower for SML samples than for UW, suggesting that amoA genes in our samples were apparently less abundant than MG-I 16S rRNA genes. Currently, all amoA to MG-I 16S rRNA gene copy number ratios obtained from metagenomic [29] and genomic [49] sequences have been close to 1. However, lower ratios of amoA to MG-I 16S rRNA genes have also been reported previously [46,50]. The lower ratio obtained here could indicate that not all of the MG-I Thaumarchaeota possess amoA, or alternatively that the primer set we used was not able to amplify all of the amoA genes present in our samples. The primer set that we used has nucleotide mismatches, which may have led to spurious qPCR results and the underestimation of amoA copy numbers in the deep meso- and bathypelagic regions of the Pacific Ocean [47,51]. Despite the nucleotide mismatches, this primer set by Wuchter et al. [5] is able to detect amoA clades from both epipelagic and high-latitude regions [52,53].
Low ratios of amoA to MG-I 16S rRNA genes have also been attributed to a lack of amoA genes in the Thaumarchaeota [46,50,54]. Previous isotopic [55,56] and genomic [49,57] studies have shown that members of the phylum Thaumarchaeota are capable of assimilating organic matter to sustain their growth. The high concentrations of organic matter in surface waters—especially the SML—could theoretically attract heterotrophic Thaumarchaeota, thereby increasing the reliance of any Thaumarchaeota present in the SML on chemoorganoheterotrophy rather than chemolithoautotrophy.
Archaeal and AOA community structures in the SML and UW
The Thaumarchaeota detected by amplicon sequencing and in the MG-I 16S rRNA gene qPCR results were more pronounced in the December SML and UW samples than in the samples collected during September. Decline of Thaumarchaeota during spring and summer [54,58,59] has been reported previously, suggesting the sensitivity of this group to photoinhibition or competition with bacteria [58]. Also, light is an important factor structuring the Thaumarchaeota population in the water column [29,47]. Although surface Thaumarchaeota may have developed pathways to help cope with UV-induced damage [60], it is still unclear whether ammonia oxidation is directly affected by photoinhibition.
MG-II from the phylum Euryarchaeota in the photic zone possesses the light-driven proton pump proteorhodopsin, providing an alternative pathway to support a photoheterotrophic lifestyle [61]. The higher relative abundance of MG-II Euryarchaeota in the SML and UW samples collected in September might indicate enhanced phototrophy in these surface waters in response to increased irradiance, as reported elsewhere [62,63]. A time-series study of planktonic archaea in the Santa Barbara Channel (Pacific Ocean off California, USA) has revealed that decreasing chlorophyll-a coincides with sporadic MG-II blooms in the surface waters [64].
Our comparisons between 16S rRNA gene archaeal community structure and amoA diversity and phylogeny showed that a portion of the archaeal communities in the coastal SML could be of sediment or terrestrial origin. In some of our samples—especially those collected when the wind speed was low and formation of the SML was thus maintained—phylotypes belonging to the SCG and Terrestrial groups were found in higher proportions in the SML than in the UW. AOA from soil environments (Clusters 2 and 3) were detected only in SML samples. SML-specific phylotypes closely related to AOA originating from soil, sediments, sand filters, and freshwater environments were present in our samples. This result is consistent with those of Stolle et al. [17], wherein a few bacterial OTUs closely related to sequences found in sediment environments were found specifically in the SML. The presence of these phylotypes could indicate passive transport via sediment resuspension, or intrusion of groundwater into the SML. It has also been speculated that the similarity of the chemical constituents in the SML to that of a solid substratum such as sediment could enable these newly introduced communities to be supported [65,66]. As these communities could have been passively transported into the water column and remained in the SML and UW for a period of time, further studies are needed to determine whether these SML-specific AOA communities are actively oxidizing ammonia and expressing amoA in the SML.
Differences in both total archaeal and AOA communities were observed between the SML and UW and were significantly correlated to the concentrations of organic matter (Chl-a and TEP). amoA and MG-I 16S rRNA gene copy numbers were especially low in our SML samples when Chl-a and TEP concentrations were high, and vice versa. Furthermore, the two SML samples that could not be amplified by the pyrosequencing primer sets had the lowest amoA and MG-I 16S rRNA gene copy numbers, indicating that archaeal abundance in the samples could have been very low. Interestingly, these samples contained the highest Chl-a and TEP concentrations in each sampling period. Negative correlations between amoA [62,67] or MG-I [8] gene copy numbers and Chl-a concentrations have been reported previously. One possible explanation is competition with phytoplankton [68]. It was recently discovered that competition with phytoplankton for ammonium could play a greater role than light intensity in regulating AOA cellular activity and nitrification rates [67]. However, there may be multiple factors regulating the Thaumarchaeota and the amoA composition and abundance: they could be affected by a wide array of environmental parameters, such as temperature [69,70], ammonia concentration [71], and even small changes in pH [72].
Insights into archaeal dynamics in the SML
The presence of amoA and MG-I genes in the SML samples indicates the potential for ammonia oxidation in the top 1 mm of surface waters. However, the gene copy numbers of AOA tended to be low when the SML was well formed during low wind conditions. The lower ratio of amoA to MG-I 16S rRNA genes in the SML than in the UW could indicate that Thaumarchaeota present in the SML lack amoA and might rely on other substrates other than ammonia, such as organic matter, as an energy source. Additionally, the difference in archaeal and AOA community composition could reflect differences in nitrification between the SML and UW, as different AOA communities and clades possess different physiological and metabolic characteristics [45,52]. Because the physiology of AOA and the factors controlling their ammonia oxidation rates are still not well defined, future work should include the nitrification activity and gene transcript contents of AOA in the SML to help elucidate their role and adaptations to ammonia oxidation.
Supporting information
(DOCX)
(PDF)
a) Copy numbers of archaeal ammonia monooxygenase subunit A (amoA) gene and archaeal Marine Group-I (MG-I) 16S rRNA gene; b) chlorophyll-a (Chl-a) and c) transparent exopolymer particles (TEP) concentrations from surface microlayer (D) and underlying water (U) samples collected in September (S) and December (W). * indicates that the concentration was below the qPCR detection limit and ** indicates that the sample was not amplified successfully using the A20F/519R primer set for 454 pyrosequencing. Error bars represent standard deviations of technical triplicates.
(PDF)
Acknowledgments
We thank S. Suzuki, Y. Cui, and the staff of the Misaki Marine Biological Station for their help during sampling. We also thank Y. Yamada and R. Shishikura for their help in the laboratory analyses.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Japan Society for the Promotion of Science (KAKENHI Grant Numbers 24121004, 24241002 and 18K14787). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Teske A, Alm E, Regan JM, Toze S, Rittmann BE, Stahl DA. Evolutionary relationships among ammonia- and nitrite-oxidizing bacteria. J Bacteriol. 1994; 176(21): 6623–6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ward BB, Capone DG, Zehr JP. What’s New in the Nitrogen Cycle? Oceanography. 2007; 20: 101–109. [Google Scholar]
- 3.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004; 304: 66–74. 10.1126/science.1093857 [DOI] [PubMed] [Google Scholar]
- 4.Konneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl, DA. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005; 437: 543–546. 10.1038/nature03911 [DOI] [PubMed] [Google Scholar]
- 5.Wuchter C, Abbas B, Coolen MJL, Herfort L, van Bleijswijk J, Timmers P, et al. Archaeal nitrification in the ocean. Proc Natl Acad Sci USA. 2006; 103: 12317–12322. 10.1073/pnas.0600756103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cunliffe M, Schafer H, Harrison E, Cleave S, Upstill-Goddard R, Murrell JC. Phylogenetic and functional gene analysis of the bacterial and archaeal communities associated with the surface microlayer of an estuary. ISME J. 2008; 2: 776–789. 10.1038/ismej.2008.28 [DOI] [PubMed] [Google Scholar]
- 7.Cunliffe M, Harrison E, Salter M, Schaefer H, Upstill-Goddard RC, Murrell JC. Comparison and validation of sampling strategies for the molecular microbial analysis of surface microlayers. Aquat Microb Ecol. 2009; 57: 69–77. [Google Scholar]
- 8.Auguet JC, Casamayor EO. A hotspot for cold crenarchaeota in the neuston of high mountain lakes. Environ Microbiol. 2008; 10: 1080–1086. 10.1111/j.1462-2920.2007.01498.x [DOI] [PubMed] [Google Scholar]
- 9.Auguet JC, Nomokonova N, Camarero L, Casamayor EO. Seasonal changes of freshwater ammonia-oxidizing archaeal assemblages and nitrogen species in oligotrophic Alpine lakes. Appl Environ Microbiol. 2011; 77(6): 1937–1945. 10.1128/AEM.01213-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villa-Costa M, Barberan A, Auguet JC, Sharma S, Moran MA, Casamayor EO. Bacterial and archaeal community structure in the surface microlayer of high mountain lakes examined under two atmospheric aerosol loading scenarios. FEMS Microbiol Ecol. 2013; 84: 387–397. 10.1111/1574-6941.12068 [DOI] [PubMed] [Google Scholar]
- 11.Auguet JC, Triadó-Margarit X, Nomokonova N, Camarero L, Casamayor EO. Vertical segregation and phylogenetic characterization of ammonia-oxidizing archaea in a deep oligotrophic lake. ISME J. 2012; 6: 1786–1797 10.1038/ismej.2012.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cunliffe M, Engel A, Frka S, Gašparović B, Guitart C, Murrell JC, et al. Sea surface microlayers: A unified physicochemical and biological perspective of the air-ocean interface. Prog Oceanogr. 2013; 109: 104–116. [Google Scholar]
- 13.Reinthaler T, Sintes E, Herndl GJ. Dissolved organic matter and bacterial production and respiration in the sea-surface microlayer of the open Atlantic and the western Mediterranean Sea. Limnol Oceanogr. 2008, 53: 122–136. [Google Scholar]
- 14.Harvey GW. Microlayer collection from the surface of the sea. Limnol Oceanogr. 1966; 10: 602–605. [Google Scholar]
- 15.Kutznetsova M, Lee C, Aller J. Enrichment of amino acids in the sea surface microlayer at coastal and open ocean sites in the North Atlantic Ocean. Limnol Oceanogr. 2004; 49(5): 1605–1619. [Google Scholar]
- 16.Bureekul S, Murashima Y, Furutani H, Uematsu M. Enrichment of particulate phosphorus in a sea-surface microlayer over the Eastern Equatorial Pacific Ocean. Geochem J. 2012; 48(3): e1–e7. [Google Scholar]
- 17.Stolle C, Labrenz M, Meeske C, Jürgens K. Bacterioneuston community structure in the Southern Baltic Sea and its dependence on meteorological conditions. Appl Environ Microbiol. 2011; 77: 3726–3733. 10.1128/AEM.00042-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wurl O, Holmes M. The gelatinous nature of the sea-surface microlayer. Mar Chem. 2008; 110: 89–97. [Google Scholar]
- 19.Alldredge AL, Passow U, Logan BE. The abundance and significance of a class of large, transparent organic particles in the ocean. Deep-Sea Res Pt. I. 1993; 40: 1131–1140. [Google Scholar]
- 20.Passow U, Alldredge AL. A dye-binding assay for the spectrophotometric measurement of transparent exopolymer particles (TEP). Limnol Oceanogr. 1995; 40: 1326–1335. [Google Scholar]
- 21.Suzuki R, Ishimaru T. An improved method for the determination of phytoplankton chlorophyll using N,N-dimethylformamide. J Oceanogr Soc Japan. 1990; 46: 190–194. [Google Scholar]
- 22.Wurl O, Miller L, Vagle S. Production and fate of transparent exopolymer particles in the ocean. J Geophys Res. 2011; 116: C00H13. [Google Scholar]
- 23.Massana R, Taylor LT, Murray AE, Wu KY, Jeffrey WH, DeLong EF. Vertical distribution and temporal variation of marine planktonic archaea in the Gerlache Strait, Antartica, during early spring. Limol Oceanogr. 1998; 43: 607–617. [Google Scholar]
- 24.Reysenbach A-L, Longnecker K, Kirshstein J. Novel bacterial and archaeal lineages from an in-situ growth chamber deployed at a Mid-Atlantic Ridge hydrothermal vent. Appl Environ Microbiol. 2000; 66: 3798–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl Environ Microbiol. 2009; 75: 7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schloss PD, Westcott SL. Assessing and Improving Methods Used in Operational Taxonomic Unit-Based Approaches for 16S rRNA Gene Sequence Analysis. Appl Environ Microbiol. 2011; 77: 3219–3226. 10.1128/AEM.02810-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hornek R, Pommerening-Röser A, Koops H-P, Farnleitner AH, Kreuzinger N, Kirschner A, et al. Primers containing universal bases reduce multiple amoA gene specific DGGE band patterns when analysing the diversity of beta-ammonia oxidizers in the environment. J Microbiol Methods. 2006; 66: 147–155. 10.1016/j.mimet.2005.11.001 [DOI] [PubMed] [Google Scholar]
- 28.Purkhold U, Pommerening-Röser A, Juretschko S, Schmid MC, Koops H-P, Wagner M. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl Environ Microbiol. 2000; 66: 5368–5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mincer TJ, Church MJ, Taylor LT, Preston C, Kar DM, DeLong EF. Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ Microbiol. 2007; 9: 1162–1175. 10.1111/j.1462-2920.2007.01239.x [DOI] [PubMed] [Google Scholar]
- 30.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013; 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang Y, Niu B, Gao Y, Fu L, Li W. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010; 26: 680–682. 10.1093/bioinformatics/btq003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2015;43(Database issue):D30–D35. 10.1093/nar/gku1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10: 512–526. 10.1093/oxfordjournals.molbev.a040023 [DOI] [PubMed] [Google Scholar]
- 34.R Core Development Team. R: a language and environment for statistical computing (version 3.2.3). Vienna: R Foundation for Statistical Computing, 2015. [Google Scholar]
- 35.McMurdie PJ, Holmes S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013; 8(4): e661217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheneman L, Evans J, Foster JA. Clearcut: a fast implementation of relaxed neighbor joining. Bioinformatics. 2006; 22(22): 2823–2824. 10.1093/bioinformatics/btl478 [DOI] [PubMed] [Google Scholar]
- 37.Yamaguchi M. Growth and reproductive cycles of marine fouling ascidians Ciona intestinalis, Styela plicata, Botrylloides violaceus and Leptoclinum mitsukuri at Aburatsubo-Moroiso Inlet (Central Japan). Mar Biol. 1975; 29: 253–259. [Google Scholar]
- 38.Takayanagi K, Yamada H. Effects of Benthic flux on short term variations of nutrients in Aburatsubo Bay. J Oceanogr. 1999; 463–469. [Google Scholar]
- 39.Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci USA. 2005; 102: 14683–14688. 10.1073/pnas.0506625102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jung M–Y, Park S–J, Kim S–J, Kim J–G, Damsté JSS, Jeon CO, Rhee S–K. A mesophilic, autotrophic, ammonia-oxidizing archaeon of Thaumarchaeotal Group I.1a cultivated from a deep oligotrophic soil horizon. Appl Environ Microbiol. 2014; 80(12): 3465–3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lebedeva EV, Hatzenpichler R, Pelletier E, Schuster N, Hauzmayer S, Bulaev A, et al. Enrichment and Genome Sequence of the Group I.1a Ammonia-Oxidizing Archaeon “Ca. Nitrosotenuis uzonensis” Representing a Clade Globally Distributed in Thermal Habitats. PLoS ONE. 2013; 8(11): e80835 10.1371/journal.pone.0080835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Q, Tang F, Zhou Y, Xu J, Chen H, Wang M, Laanbroek HJ. Shifts in the pelagic ammonia-oxidizing microbial communities along the eutrophic estuary of Yong River in Ningbo City, China. Front Microbiol. 2015; 6: 1180 10.3389/fmicb.2015.01180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beman JM, Popp BN, Francis CA. Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J. 2008; 2: 429–441. 10.1038/ismej.2007.118 [DOI] [PubMed] [Google Scholar]
- 44.Wietz M, Gram L, Jørgensen B, Schramm A. Latitudinal patterns in the abundance of major marine bacterioplankton groups. Aquat Microbiol Ecol. 2010; 61: 179–189. [Google Scholar]
- 45.Smith JM, Casciotti KL, Chavez FP, Francis CA. Differential contributions of archaeal ammonia oxidizer ecotypes to nitrification in coastal surface waters. ISME J. 2014; 8(8): 1704–1714. 10.1038/ismej.2014.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Agogué H, Brink M, Dinasquet J, Herndl GJ. Major gradients in putatively nitrifying and non-nitrifying Archaea in the deep North Atlantic. Nature. 2008; 456: 788–792. 10.1038/nature07535 [DOI] [PubMed] [Google Scholar]
- 47.Church MJ, Wai B, Karl DM, DeLong EF. Abundances of crenarchaeal amoA genes and transcripts in the Pacific Ocean. Environ Microbiol. 2010; 12: 679–688. 10.1111/j.1462-2920.2009.02108.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pedneault E, Galand PE, Potvin M, Tremblay J-E, Lovejoy C. Archaeal amoA and ureC genes and their transcriptional activity in the Arctic Ocean. Sci Rep. 2014; 4:4661 10.1038/srep04661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hallam SJ, Konstantinos TK, Putnam N, Schelper C, Watanabe Y, Sugahara J, Preston C, de la Torre J, Richardson PM, Delong EF. Genomic analysis of the uncultivated marine crenarcheote Crenarchaeum symbiosium. Proc Natl Acad Sci USA. 2006; 103(48): 18296–18301. 10.1073/pnas.0608549103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Corte D, Yokokawa T, Varela MM, Agogué H, Herndl GJ. Spatial distribution of bacteria and archaea and amoA gene copy numbers throughout the water column of the Eastern Mediterranean Sea. ISME J. 2008; 3(2): 147–158. 10.1038/ismej.2008.94 [DOI] [PubMed] [Google Scholar]
- 51.Konstantinidis KT, Braff J, Karl DM, DeLong E. Comparative metagenome analysis of a microbial community residing at a depth of 4,000 meters at Station ALOHA in the North Pacific Subtropical Gyre. Appl Environ Microbiol. 2009; 75(16): 5345–5355. 10.1128/AEM.00473-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sintes E, Bergauer K, Corte DD, Yokokawa T, Herndl GJ. Archeal amoA gene diversity points to distinct biogeography of ammonia-oxidizing Crenarchaeota in the ocean. Environ Microbiol. 2012; 15(5): 1647–1658. 10.1111/j.1462-2920.2012.02801.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sintes E, Corte DD, Haberleitner E, Herndl GJ. Geographic distribution of archaeal ammonia oxidizing ecotypes in the Atlantic Ocean. Front Microbiol. 2016; 7: 77 10.3389/fmicb.2016.00077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kalanetra KM, Bano N, Hollibaugh JT. Ammonia-oxidizing archaea in the Arctic Ocean and Antarctic coastal waters. Environ Microbiol. 2009; 11(9): 2434–2445. 10.1111/j.1462-2920.2009.01974.x [DOI] [PubMed] [Google Scholar]
- 55.Herndl GJ, Reinthaler T, Teira E, van Aken H, Veth C, Pernthaler A et al. Contribution of Archaea to total prokaryotic production in the deep Atlantic Ocean. Appl Environ Microbiol. 2005; 71: 2303–2309. 10.1128/AEM.71.5.2303-2309.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teira E, Lebaron P, van Aken H, Herndl GJ. Distribution and activity of Bacteria and Archaea in the deep water masses of the North Atlantic. Limnol Oceanogr. 2006; 51: 2131–2144. [Google Scholar]
- 57.Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinela N, Arp DJ et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA. 2010; 107: 8818–8823. 10.1073/pnas.0913533107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Massana R, Murray AE, Preston CM, DeLong EF. Vertical distributiona nd phylogenetic chracterization of marine planktonic Archaea in the Santa Barbara Channel. Appl Environ Microbiol. 1997; 63: 50–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Church MJ, DeLong EF, Ducklow HW, Karner MB, Preston CM, Karl DM. Abundance and distribution of planktonic Archaea and Bacteria in the waters west of the Antarctica Peninsula. Limnol Oceanogr. 2003; 48(5): 1893–1902. [Google Scholar]
- 60.Luo H, Tolar BB, Swan BK, Zhang CL, Stepanauskas R, Moran MA, Hollibaugh JT. Single-cell genomics shedding light on marine Thaumarchaeota diversification. ISME J. 2014; 8(3): 732–736. 10.1038/ismej.2013.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frigaard NU, Martinez A, Mincer TJ, DeLong EF. Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea. Nature. 2006; 439: 847–850. 10.1038/nature04435 [DOI] [PubMed] [Google Scholar]
- 62.Herfort L, Schouten S, Abbas B, Veldhuis MJW, Coolen MJL, Wuchter C, Boon JP, Herndl GJ, Damsté JSS. Variations in spatial and temporal distribution of Archaea in the North Sea in relation to environmental variables. FEMS Microbiol Ecol. 2007; 62: 242–257. 10.1111/j.1574-6941.2007.00397.x [DOI] [PubMed] [Google Scholar]
- 63.Hugoni M, Taib N, Debroas D, Domaizon I, Dufournel IJ, Bronner G, Agogué H, Mary I, Galand PE. Structure of the rare archaeal biosphere and seasonal dynamics of active ecotypes in surface coastal waters. Proc. Natl. Acad. Sci. U.S.A. 2013; 110: 6004–6009. 10.1073/pnas.1216863110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Murray AE, Blakis A, Massana R, Strawzewski S, Passow A, Alldredge A, DeLong EF. A time series assessment of planktonic archaeal variability in the Santa Barbara Channel. Aquat Microb Ecol. 1999; 20: 129–145. [Google Scholar]
- 65.Baier RE. “Applied chemistry at protein interfaces”, in: Applied Chemistry at Protein Interfaces, ed. Gould R. F., (Washington D.C: American Chemical Society; ); 1975. pp. 1–25. [Google Scholar]
- 66.Kjelleberg S. “Mechanisms of bacterial adhesion at gas-liquid interfaces” in Bacterial Adhesion, eds. Savage D. C. and Fletcher M. (New York, NY: Plenum Press; ); 1985. pp. 163–194. [Google Scholar]
- 67.Smith JM, Chavez FP, Francis CA. Ammonium uptake by phytoplankton regulates nitrification in the sunlit ocean. PLoS ONE, 2014; e108173 10.1371/journal.pone.0108173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Christman GD, Cottrell MT, Popp BN, Gier E, Kirchman DL. Abundance, diversity and activity of ammonia-oxidizing prokaryotes in the coastal Arctic Ocean in summer and winter. Appl Environ Microbiol. 2011; 77(6): 2016–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Urakawa H, Tajima Y, Numata Y, Tsuneda S. Low temperature decreases the phylogenetic diversity of ammonia-oxidizing archaea and bacteria in aquarium biofiltration systems. Appl Environ Microbiol. 2008; 74: 894–900. 10.1128/AEM.01529-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ijichi M, Hamasaki K. Community structure of ammonia-oxidizing marine archaea differs by depth of collection and temperature of cultivation. J Oceanogr. 2011; 68: 369–386. [Google Scholar]
- 71.Urakawa H, Martens-Habbena W, Huguet C, de la Torre JR, Ingalls AE, Devol AH, Stahl DA. Ammonia availability shapes the seasonal distribution and activity of archaeal and bacterial ammonia oxidizers in the Puget Sound Estuary. Limnol Oceanogr. 2014; 59: 1321–1335. [Google Scholar]
- 72.Beman JM, Chow CE, King AL, Feng Y, Fuhrman JA, Andersson A, et al. Global declines in oceanic nitrification rates as a consequence of ocean acidification. Proc Natl Acad Sci U.S.A. 2011; 108: 208–213. 10.1073/pnas.1011053108 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(PDF)
a) Copy numbers of archaeal ammonia monooxygenase subunit A (amoA) gene and archaeal Marine Group-I (MG-I) 16S rRNA gene; b) chlorophyll-a (Chl-a) and c) transparent exopolymer particles (TEP) concentrations from surface microlayer (D) and underlying water (U) samples collected in September (S) and December (W). * indicates that the concentration was below the qPCR detection limit and ** indicates that the sample was not amplified successfully using the A20F/519R primer set for 454 pyrosequencing. Error bars represent standard deviations of technical triplicates.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.