Abstract
The pancreas is a complex organ composed of many different cell types that work together to regulate blood glucose homeostasis and digestion. These cell types include enzyme-secreting acinar cells, an arborized ductal system responsible for the transportation of enzymes to the gut, and hormone-producing endocrine cells.
Endocrine beta-cells are the sole cell type in the body that produce insulin to lower blood glucose levels. Diabetes, a disease characterized by a loss or the dysfunction of beta-cells, is reaching epidemic proportions. Thus, it is essential to establish protocols to investigate beta-cell development that can be used for screening purposes to derive the drug and cell-based therapeutics. While the experimental investigation of mouse development is essential, in vivo studies are laborious and time-consuming. Cultured cells provide a more convenient platform for screening; however, they are unable to maintain the cellular diversity, architectural organization, and cellular interactions found in vivo. Thus, it is essential to develop new tools to investigate pancreatic organogenesis and physiology.
Pancreatic epithelial cells develop in the close association with mesenchyme from the onset of organogenesis as cells organize and differentiate into the complex, physiologically competent adult organ. The pancreatic mesenchyme provides important signals for the endocrine development, many of which are not well understood yet, thus difficult to recapitulate during the in vitro culture. Here, we describe a protocol to culture three-dimensional, cellular complex mouse organoids that retain mesenchyme, termed pancreatoids. The e10.5 murine pancreatic bud is dissected, dissociated, and cultured in a scaffold-free environment. These floating cells self-assemble with mesenchyme enveloping the developing pancreatoid and a robust number of endocrine beta-cells developing along with the acinar and the duct cells. This system can be used to study the cell fate determination, structural organization, and morphogenesis, cell-cell interactions during organogenesis, or for the drug, small molecule, or genetic screening.
Keywords: Developmental Biology, Issue 136, Pancreas, Organoids, Beta Cells, Development, Mesenchyme, Differentiation, Diabetes, 3D Culture, Pancreatoids
Introduction
Delineating the mechanisms of the normal development and the physiology is paramount to understand disease etiology and ultimately cultivate treatment methods. While culturing and differentiating stem cells enables quick and high-throughput analysis of development, it is limited by the existing body of knowledge regarding mechanisms regulating cell fate and artificially recapitulates development in a relatively homogenous, two-dimensional state1,2. Not only is in vivo development affected by extrinsic influences, with different cell types in the niche and milieu providing paracrine signals and organizational support to guide organogenesis, but the function of these cells also relies on their surroundings for guidance3,4,5. Given the importance of these external cues, the limitations of differentiation protocols, and the laborious nature of in vivo mouse models, new systems are needed to experimentally investigate basic developmental processes and physiology.
The emergence of protocols to generate three-dimensional, complex organoids provides a convenient and congruent system to study organogenesis, physiology, drug efficacy, and even pathogenesis. Establishing murine organoids for different systems such as the stomach6 and intestine7 have expanded our understanding of organogenesis, providing a tool to study developmental complexities with fewer restrictions than in vivo and in vitro models. Due to these advances in the murine organoid formation and the advent of human pluripotent stem cells, human intestinal8, retinal9, renal10,11, and cerebral12 organoids have been produced, and this repertoire is only limited by the existing knowledge regarding mechanisms of development.
Of particular interest is the generation of pancreatic organoids, as a myriad of diseases plagues different pancreatic cell types, including acinar cells and ducts in exocrine pancreatic insufficiency13, acinar cells in pancreatitis14, and beta cells in diabetes15. Gaining knowledge regarding the development of these different cell types could aid in understanding their pathology and can, also, act as a platform for personalized drug screening or transplantation. Previously, Greggio et al. developed a method to create murine pancreatic organoids that recapitulate in vivo morphogenesis and develop organized, three-dimensional, complex structures composed of all major pancreatic epithelial cell types16,17. This is a major step forward in the pancreatic field, especially as making cells in vitro can enable biological investigation of beta-cell development. However, a scarcity of endocrine cells formed in this protocol unless the organoids were transplanted into tissue, where the niche could interact and provide instructional cues17. The mesenchyme constitutes the largest portion of the niche, heavily enveloping the developing epithelium from early stages of organogenesis to later stages including endocrine delamination and differentiation3,4,18. The interaction of the mesenchyme with the developing pancreas is yet another example of extrinsic signaling and the importance of maintaining in vivo cellular complexity to study organogenesis.
Here, we describe how to generate three-dimensional pancreatic organoids, termed pancreatoids, from dissociated e10.5 murine pancreatic progenitors. These pancreatoids retain native mesenchyme, self-assemble in free-floating conditions, and generate all major pancreatic cell types, including a robust number of endocrine beta cells19. This approach is best suited for the analysis of endocrine development, as previous protocols lack robust endocrine differentiation. However, using the protocol for pancreatic organoids as described by Greggio et al. is better suited for analysis of pancreatic epithelial branching and morphogenesis, as branching is more limited in pancreatoids.
Protocol
All animal experiments described in this method were approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine.
1. Preparation of Mouse Embryonic Day 10.5 Pancreatic Progenitors
Note: This protocol does not need to be followed under sterile conditions until step 2, however it is optimal to sterilize dissection tools and spray with 70% ethanol prior to use.
- To set up for dissection, fill an ice bucket and place a container of phosphate buffered saline (PBS) in the ice. Clean two fine-tipped forceps and dissection scissors with 70% ethanol. Set near the dissection area, a container with at least three borosilicate capillary tubes, a mouth pipette, a lighter, and filtered 1,000 µL pipette tips.
- Prepare at least 1 mL of 1.25 mg/mL cold dispase diluted in sterilized water and place in the second well of a 12 well plate on ice. Put PBS in the first, third, and fourth wells of the 12 well plate. Place 1 mL of 0.05% Trypsin in a 1.5 mL tube on ice. Pre-warm a shaking incubator to 37 oC.
- Euthanize e10.5 timed-pregnant mice in accordance with IACUC guidelines. Spray the abdomen with 70% ethanol to clean the region before making a V-shaped incision at the genital area of the mouse using scissors and forceps and continue it up to the diaphragm. NOTE: Appearance of a vaginal plug in this protocol indicates day 0.5. Using 5 - 6-week-old mice, a weight gain of more than 2 g acts as a secondary measure of successful impregnation.
- Carefully excise the uterus by making an incision at the uppermost lateral portions of the uterus, pulling the organ upwards away from the mouse, and following this incision down to the genital region before repeating on the opposite side. Place it in a 10 cm Petri dish with the cold PBS. NOTE: It is possible to obtain organoids from e11.5 dissociated cells, however unlike the e10.5 pancreatoids, these organoids are not yet characterized and thus may not retain the capacity to differentiate into all three major pancreatic lineages.
Under a light dissection microscope, place the Petri dish and carefully open the uterine tissue by placing two forceps between each embryo and peeling tissue away from the yolk sac. Transfer embryos by grasping the yolk sac gently with forceps and place in a 10 cm Petri dish with fresh, cold PBS. Place the Petri dish on ice. NOTE: It is important to heedfully remove embryos, preferably maintained within the yolk sac, as forceful removal can pull the umbilical cord, ripping tissue of the embryo and compromising the structural integrity.
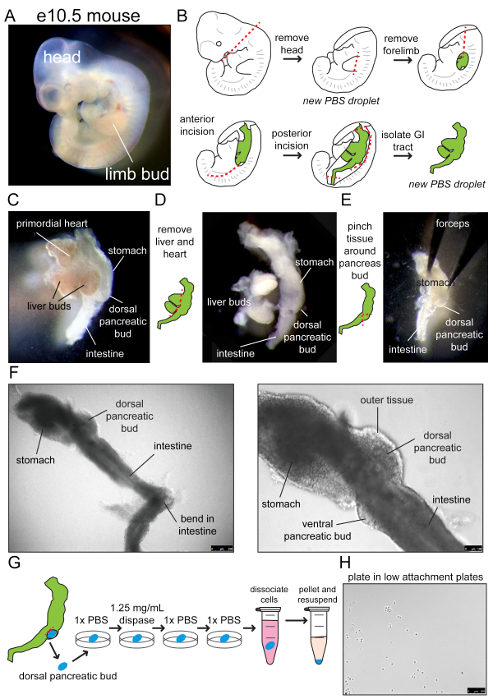
- Place multiple PBS drops on a 10 cm Petri dish lid. Use these droplets to transfer the embryo during dissection as extraembryonic tissues are removed to ensure visibility. Transfer an embryo to a PBS drop and gently remove from the yolk sac using forceps, while the remaining embryos stay in PBS on ice (Figure 1A). Remove the head before moving the embryo to a new PBS drop using forceps to lightly scoop the embryo up, to not damage the tissue (Figure 1B). NOTE: The tissue might need to be transferred more frequently to fresh PBS droplets than described here, depending on the opacity of the liquid.
- In a new PBS drop, remove the forelimb buds using forceps (Figure 1B). Place forceps into the opening where limb bud was present and gently tear only the most external tissue anteriorly (Figure 1B). Rotate embryo and repeat in the posterior direction, stopping at the hindlimb bud. At this point, the gastrointestinal tract should be visible (Figure 1B).
- The posterior region of the gastrointestinal tract has a slight bend (Figure 1F). Insert forceps behind this bend in the opening between the gastrointestinal tract and the spinal region of the body wall. Detach the gastrointestinal tract, working slowly upwards until the most anterior region is reached where the cardiac region connects (Figure 1B-E). OPTIONAL: Remove the primordial heart and the liver buds from the ventral regions of the gastrointestinal tract, leaving only the continuous gut tube. The first few times this protocol is performed it might be easier to leave the heart and liver as ventral landmarks until the morphology of the gastrointestinal tract and the dorsal pancreatic bud are more familiar (Figure 1C and D).
- Transfer the gastrointestinal tract to the fresh PBS droplet.
- Take forceps and gently pinch the tissue beneath the protruding bud and the intestine and slightly lift the outer tissue off (Figure 1D-F). NOTE: The dorsal pancreatic bud is located between the stomach and intestine (Figure 1C-G). The pancreatic bud beneath should look like a round, knobby structure (Figure 1G).
- Place forceps where the bud connects to the intestine and pinch upward to loosen the bud (Figure 1G). Wash one time in a new PBS bubble before transferring the bud into the first well of the 12 well plate in cold PBS on ice. Repeat until all buds are collected from embryos and placed in the first well of the 12 well plate.
- Place the 12 well dish under the light dissection microscope. Count the number of pancreatic buds successfully dissected to later calculate the split ratio.
- Place a filtered 1,000 µL pipette tip into the mouth pipette with a capillary tube attached. Flame sterilize the capillary tube and create a bend in the tube for the ease of use. Use the capillary to transfer buds to the second well containing cold dispase solution for 2 min before transferring to the third well with the clean PBS (Figure 1G). NOTE: While transferring buds from dispase to PBS, avoid touching the tissue to the tip of the capillary tube. If the tissue gets stuck at the tip of the tube, pipette up and down or shake in the clean PBS solution until loosened.
- Pipette buds up and down, and transfer to the fourth well with clean PBS. Finally, using the capillary device transfer buds to the 1.5 mL tube with 0.05% Trypsin. Place the tube in the pre-warmed 37 oC shaker and shake at 1,500 rpm for 4 min.
- Vortex for approximately 10 s then immediately place the tube in a centrifuge and spin for 5 min at 200 g. Remove all but approximately 50 µL of solution cautiously as to not disturb the centrifuged cells (Figure 1G).
2. Culturing Dissociated Progenitors to Form Pancreatoids
Note: The following should be performed in a sterile atmosphere in a standard tissue culture hood, using standard sterile procedures.
For every bud collected, add 400 µL organogenesis media16,17 (Table 1) to the centrifuged cells. Pulse vortex three times and plate 100 µL per well of a low attachment 96 well plate, splitting buds at a 1:4 ratio (Figure 1G).
Check under the microscope to visualize dissociated cells freely floating (Figure 1H).Place the culture dish in a 37 oC incubator with 5% CO2 on a rocker at medium speed.
Monitor the progress of pancreatoids daily. Replace with 100 µL of fresh media every 3 days, carefully pipetting media under microscopic control in the hood to remove while leaving pancreatoid in the well. Alternatively, fresh 100 µL of media can be added to a new well and pancreatoid transferred using the borosilicate capillary tube attached to mouth pipette and 1000 µL filtered pipette tip.
3. Processing Pancreatoids for Immunofluorescent Imaging
- To process pancreatoids for immunofluorescent images, first prepare 4% paraformaldehyde/PBS, pH7.4 (PFA) solution. Place 50 µL of 4% PFA in wells of a 96 well plate.
- Using the borosilicate capillary tubes and mouth pipette with the 1,000 µL filtered pipette tip, transfer pancreatoids from culture medium taking as little media as possible into the fresh well with 4% PFA. Set on the rocker at room temperature for 15 min.
- Transfer pancreatoids from 4% PFA into a well with 100 µL of fresh PBS, rock at room temperature for 5 - 10 min. Repeat for a total of three fresh PBS washes. NOTE: The protocol can be paused here, with pancreatoids stored in 100 µL of PBS at 4 oC for up to one week.
- For wholemount imaging (recommended if antibodies are suitable, alternative approach in section 3.3. and microscopy in 3.4.), transfer from PBS into 50 µL 5% donkey serum in 1x PBS with 0.1% nonionic detergent (PBST) to block. Rock overnight at 4 oC or alternatively for 4 h at room temperature.
- Transfer pancreatoids to a new well containing primary antibodies diluted in 50 µL of 5% donkey serum in 1x PBST. Optimally, rock for 24 h at 4 oC (at least 12 h but up to 36 h). NOTE: Antibodies listed in the Table of Materials against Chga (1:100), DBA (1:400), Vim (1:400), and Pdx1 (1:100) are suitable for wholemount staining; other antibodies may require optimization.
- Transfer pancreatoids to a well containing 100 µL of fresh PBST to wash and rock for 30 min at room temperature. Repeat this step for a total of three fresh PBST washes.
- Transfer pancreatoids to a new well containing secondary antibodies diluted in 50 µL of 5% donkey serum in 1x PBST. Protect the plate from light and place in 4 oC, rocking for optimally 24 h (at least 12 h but up to 36 h). NOTE: All secondary antibodies listed in the Table of Materials and DAPI nuclear counterstain are suitable for wholemount staining.
- Transfer pancreatoids to a new well containing 100 µL of 1x PBST and rock at room temperature for 30 min. Repeat for a total of three washes in 1x PBST. In the third and final wash, add 300 nM DAPI.
- Finally, place into 100 µL of clean PBS and store at 4 oC protected from light for up to a week before imaging by confocal microscopy, although best results come from imaging immediately after staining. To prevent movement of pancreatoids during imaging but prevent drying out, place each pancreatoid into a 20 µL droplet of PBS. If pancreatoids do not remain still, reduce PBS volume.
- For frozen sections, transfer pancreatoids from PBS into 30% sucrose solution overnight at 4 oC.
- Add embedding media in a bubble under a dissection microscope. Using forceps under microscopic control, soak pancreatoids by gently pushing through the embedding media several times to remove residual sucrose before transferring to a tissue block with frozen embedding media.
- Place the tissue block on dry ice until frozen and section on the cryostat at 8 µm. NOTE: Sections can be stored at -80 oC or immunostained immediately.
- Allow sections from -80 oC to thaw at room temperature for approximately 5 min until dry. Draw a hydrophobic barrier using the hydrophobic pap pen around the tissue and the block using 5% donkey serum diluted in 1x PBST for 30 min at room temperature.
- Remove media and replace immediately with primary antibodies diluted in 5% donkey serum diluted in 1x PBST overnight at 4 oC.
- Remove the primary solution and wash tissues three times with 1x PBST for 10 min each. Following this, add secondary antibody solution diluted in 5% donkey serum diluted in 1x PBST for 1 h at room temperature and protect slides from light.
- Remove the secondary solution and wash slides three times in 1x PBST for 10 min each. In the final wash, add 300 nM DAPI.
- Remove media and add mounting media and a coverslip. Store slides away from light in 4 oC until ready to image.
4. Isolation of RNA from Pancreatoids for Transcript Analysis
Collect pancreatoids using borosilicate capillary attached to a mouth pipette with a filtered 1,000 µL pipette tip for sterility and place into 500 µL of acid guanidinium thiocyanate-phenol-chloroform solution in a 1.5 mL tube. Store at -80 oC or proceed immediately to RNA isolation.
- For RNA isolation, thaw samples on ice if necessary and add 100 µL of chloroform. Vortex well and place in 4 oC for 15 min. Pre-cool a centrifuge to 4 oC.
- Place tubes into centrifuge and spin for 20 min at 12,000 g at 4 oC.
- Carefully remove tubes to not disturb the separation of layers. Using a 200 µL pipette, collect the clear aqueous solution and place into a new 1.5 mL tube, leaving behind a small amount to buffer from the white protein layer and pink DNA layer. Do not touch the tip of the pipette to anything in this process and do not touch the walls of the 1.5 mL tube.
- Add 500 µL of 100% isopropanol to the new tube containing the aqueous layer and vortex. Let sit at room temperature for 20 min, longer on the ice, or for maximum precipitation leave overnight at -20 oC.
- Centrifuge at 12,000 g for 15 min at 4 oC to pellet the RNA. Remove the isopropanol without disturbing the pellet. NOTE: As pancreatoids are small, it is likely the pellet will not be visible.
- Add cold, 75% ethanol and invert the tube several times. Place in the centrifuge and spin at 9,000 x g for 10 min at 4 oC. Remove the ethanol and spin for 2 min at 9,000 x g.
- Use a 200 µL pipette to remove any remaining ethanol in the tube, without contacting the region the pellet resides in. Leave the cap open on the tube for 5 - 10 min at room temperature to ensure evaporation of residual ethanol.
- Resuspend pellet in by vortexing in 20 µL of clean, nuclease-free water. Optional: Heat the RNA at 65 oC for 3 min and immediately return to the ice. RNA can be stored at -80 oC or immediately used for reverse transcription and qPCR.
Representative Results
Careful dissection of mouse embryos at e10.5 from the uterine horn should yield undamaged embryos in PBS for further dissection (Figure 1A). The gastrointestinal tract can be efficiently removed from the embryo (Figure 1B), permitting discernment of the dorsal pancreatic bud at the junction of the intestine and stomach (Figure 1C-F). The e10.5 pancreatic bud has previously been characterized; progenitors should express Pdx1, Sox9, Ptf1a, and Hes120,21,22,23. Following tissue processing steps, dissociated pancreatic single cells or small groups of cells can be visualized by light microscopy (Figure 1G-H).
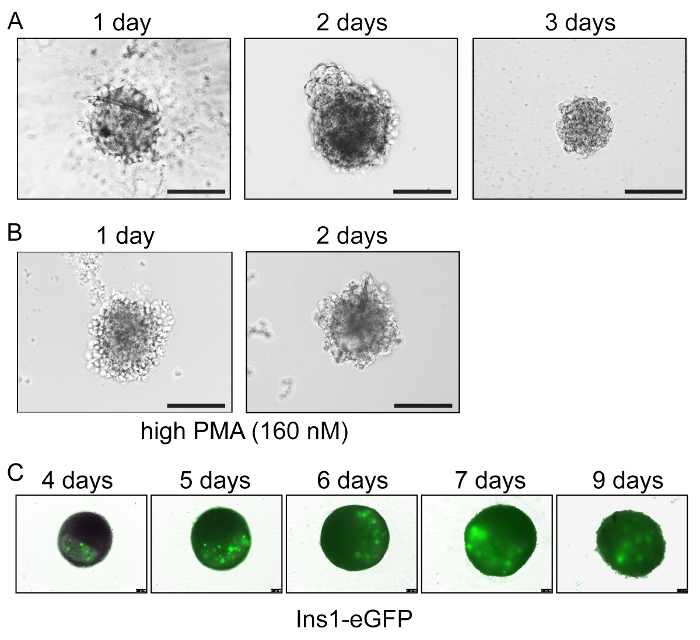
In organogenesis media, these free-floating, scaffold-free cells self-assemble and organize into three-dimensional pancreatoids that grow and persist for at least ten days in culture (Figure 2A). The pancreatoids have morphological similarities to the in vivo pancreas, with branching morphogenesis occurring. Using transgenic mice, different cell types or processes can be monitored in real time. For example, using Ins1-eGFP24 mice to mark the formation of endocrine beta cells, pancreatoids can be imaged in daily to visualize beta cell development (Figure 2B). Further, by altering culture conditions, adding small molecules, drugs, or manipulating the genome, not only can the cell fate determination be assessed but changes in structure and morphology can also be investigated. Here we show the application of protein kinase C activator phorbol 12-myristate 13-acetate (PMA) at high concentrations (160 nM), which alters the morphology of developing pancreatoids, leading to loosely associated epithelial cells and increased branching19 (Figure 2C).
To assess the association of pancreatic mesenchyme tissue with pancreatic epithelial cells, immunostaining can be performed for markers of each tissue. Immunostaining of duct marker, DBA25,26, with an endocrine marker, Chga27, and nuclei marked by DAPI reveal multi-lineage formation in pancreatoids (Figure 3A). A mesenchyme marker, Vimentin, co-stained with a pancreatic progenitor marker (from e9.0 to approximately e15.5) Pdx1, shows that the mesenchyme envelops the pancreatoid (Figure 3B). Immunostaining can also be used to examine morphology, with Pdx1 revealing branching structures, as well as different markers of cell types of interest. In Figure 3C, we visualize beta cell development by staining for the epithelial marker Pdx1 and the beta cell marker Ins. Using qPCR analysis, transcripts of progenitors and differentiating cells an be assessed, such as progenitor genes Pdx1 and Hes1, and differentiated markers Prss1, Prss3, Hnf6, Isl1, Nkx6-1, and Ins1 (Figure 4).
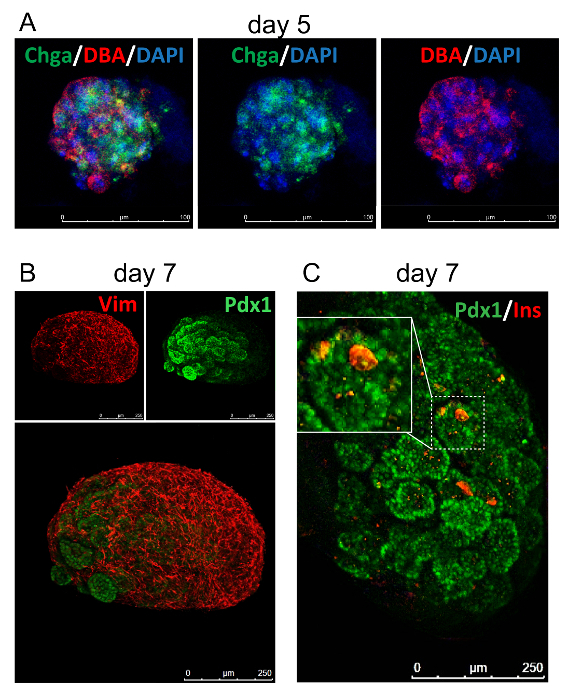
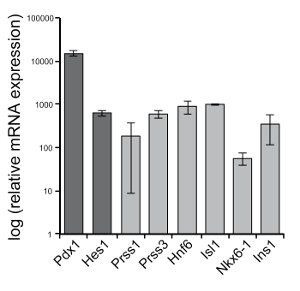
Figure 1: Outline of dissection, dissociation, and plating of e10.5 pancreatic progenitors for the pancreatoid generation. (A) The brightfield image of e10.5 embryo. (B) The schematic of dissection procedure, starting at the top left. The red, dashed lines indicate regions to cut, while the green region is the gastrointestinal tract. First, the head is removed followed by the removal of forelimb buds and the opening of the side of the organism. (C) The gastrointestinal tract is carefully removed, with the heart and liver buds protruding from the ventral region of the tract. The stomach, dorsal pancreatic bud, and intestine can be visualized, as shown in the brightfield image. (D) Removal of the heart and liver buds. Schematic is on the left and the brightfield image is on the right. (E) Pinching of the tissue around the dorsal pancreatic bud to expose the bud for dissection. Schematic is on the left and the brightfield image is on the right. (F) The gastrointestinal tract with the dorsal pancreatic bud under brightfield microscopy. In the image on the left, the bend in the intestine is visible, where forceps can be placed when detaching the gastrointestinal tract from the spinal region. On the right, the high magnification brightfield image shows both the dorsal and ventral pancreatic buds. (G) Removal of the dorsal pancreatic bud and processing of tissue before dissociation, resuspension in organogenesis media, and plating. (H) the brightfield image shows dissociated cells immediately after plating. Please click here to view a larger version of this figure.
Figure 2: Monitoring pancreatoids over time. (A) Brightfield images of pancreatoids at day 1, day 2, and day 3. Scale bars=100 um. (B) Manipulation of culture conditions to investigate pancreatic morphogenesis. Here, the addition of high levels of PMA leads to loosened epithelial structure and increased branching, as visualized by brightfield microscopy at day 1 and 2. Scale bars = 100 µm. (C) Ins1-eGFP mouse pancreatoids at day 4, day 5, day 6, day 7, and day 9, with the development of beta cells indicated by eGFP in green. Scale bars = 100 µm. Please click here to view a larger version of this figure.
Figure 3: Immunostaining of pancreatoids.(A) Immunostaining pancreatoid at day 5 to mark ducts by DBA in red, endocrine cells by Chga in green, and nuclei marked by DAPI in blue. (B) Immunostaining pancreatoid at day 7 to mark mesenchyme by Vimentin in red and pancreatic progenitors by Pdx1 in green. (B) Immunostaining pancreatoid at day 7 to mark beta cells by Insulin in red and pancreatic progenitors by Pdx1 in green. Please click here to view a larger version of this figure.
Figure 4: Transcript analysis. Quantitative PCR of day 7 pancreatoids for markers of progenitors and differentiating cells. Comparison to in vivo murine tissue is shown in Scavuzzo et al. (2017). Results are normalized to Gapdh. N = 2, scale bars are SEM. Please click here to view a larger version of this figure.
| Component | Abbreviation | Final concentration |
| Penicillin-Streptomycin | P/S | 1% |
| FBS-free media supplement | 10% | |
| Beta-Mercaptoethanol | bME | 0.1 mM |
| Phorbol 12-Myristate 13-Acetate | PMA | 16 nM |
| Y-27632 or ROCK inhibitor | RI | 10 uM |
| Epidermal Growth Factor | EGF | 25 ng/mL |
| R-Spondin1 | - | 500 ng/mL |
| acidic Fibroblast growth factor, Fibroblast growth factor 1 | aFGF, FGF1 | 25 ug/mL |
| Heparin sodium salt | Heparin | 2 U/mL |
| Fibroblast growth factor 10 | FGF10 | 100 ng/mL |
| Dulbecco's Modified Eagle Medium/Nutrient Mixture F-12 | DMEM/F12 | to 5 mL |
Table 1. Organogenesis media.
Discussion
The progression of cell culture models is critical to properly model development, produce clinically relevant cell types, test drug efficacy, or even transplant to patients. However, artificially recapitulating development in a dish is challenging as we are still far from understanding the mechanisms of organogenesis and physiology in vivo. Thus in vitro cells are inefficiently generated, not fully functional, unable to be maintained for long periods of time, or harbor other abnormalities from comparable cells in the body. This is because many different cell types interact while complex morphogenetic changes occur to influence development and physiology. Gaining an understanding of how development proceeds by combining the convenience of in vitro systems while retaining the complexity of in vivo development would impart a valuable tool for the biomedical research.
The development of organoids is a promising avenue towards modeling the complexities of development. In this protocol, we outline how to make pancreatic organoids, termed pancreatoids, which retain native mesenchyme and robustly generate endocrine cells, albeit pancreatoids do not exhibit glucose responsiveness. This tool can be used to investigate mechanisms of development as well as functionality in a culture system that maintains the heterogeneity of tissue found in vivo. Further, this can be used for genetic screens or to screen small molecules or drugs. This is particularly interesting, as testing candidate drugs in relatively homogenous cell culture systems may circumvent effects of these compounds on other closely related cell types.
There are several critical steps during this protocol. First, the careful removal of embryos from the uterus is important as pulling the embryos out forcefully can rip the abdominal region and make it difficult to discern the gastrointestinal tract to obtain the pancreatic bud (step 1.3). Second, clean dissection of the pancreatic bud from the gastrointestinal tract is critical, as taking excess tissue may lead to differentiation of other cell types (step 1.4.4). To do this, it is important to pinch below the pancreatic bud and lift the overlaying tissue prior to isolating the pancreatic bud. Finally, when moving buds from the dispase back into the PBS to wash, it is imperative to use a borosilicate capillary tube and to not let the tissue touch the edge of the tube opening, otherwise, the tissue will stick to the tip of the tube (step 1.5.1). To avoid this, begin mouth pipetting solution before moving towards the bud, allowing the bud to go into the tube rather than on the outside.
This method permits the formation of endocrine cells in a 3D pancreatoid, however, the epithelial branching is more limited than other existing protocols16,17. Further, while insulin-producing endocrine cells form, they do not exhibit glucose responsiveness. Thus, further investigation into the maturation of these cells will provide valuable information both in the generation of functional pancreatoids as well as to the field of beta cell generation in general.
The generation of pancreatoids that develop endocrine cells, and, in the future, that obtain glucose responsiveness, has potential implications for the treatment of diabetes. Diabetes is a prime candidate for regenerative therapy, as pancreatic beta cells are lost or dysfunctional and replacing these cells can potentially alleviate disease complications. Progress has been made in differentiating hPSCs into beta cells, however, in diabetes, there are often dysfunctions to other cell types in the pancreas along with beta cells, including alpha cells or acinar cells28,29,30,31,32. Thus, generating new pancreatic tissue for transplantation can fully replace the afflicted tissue. With the additional investigation of murine pancreatoid formation and functioning, the developmental trajectory can be mimicked to generate human pancreatoids out of hPSCs. These human pancreatoids can be used for the personalized medicine to screen for responsiveness to drugs in a patient-specific context, and for regenerative therapy.
Disclosures
The authors have nothing to disclose.
Acknowledgments
We thank Jolanta Chmielowiec for helpful discussion regarding the protocol and manuscript. We also thank Benjamin Arenkiel for access to confocal microscope. This work was supported by the NIH (P30-DK079638 to M.B. and T32HL092332-13 to M.A.S. and M.B.), the McNair Medical Foundation (to M.B.), and the confocal core at the BCM Intellectual and Developmental Disabilities Research Center (NIH U54 HD083092 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development).
References
- Clevers H. Modeling Development and Disease with Organoids. Cell. 2016;165(7):1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- Akkerman N, Defize LH. Dawn of the organoid era: 3D tissue and organ cultures revolutionize the study of development, disease, and regeneration. Bioessays. 2017;39(4) doi: 10.1002/bies.201600244. [DOI] [PubMed] [Google Scholar]
- Guo T, Landsman L, Li N, Hebrok M. Factors expressed by murine embryonic pancreatic mesenchyme enhance generation of insulin-producing cells from hESCs. Diabetes. 2013;62(5):1581–1592. doi: 10.2337/db12-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsman L, et al. Pancreatic mesenchyme regulates epithelial organogenesis throughout development. PLoS Biology. 2011;9(9):1001143. doi: 10.1371/journal.pbio.1001143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammert E, Cleaver O, Melton D. Induction of pancreatic differentiation by signals from blood vessels. Science. 2001;294(5542):564–567. doi: 10.1126/science.1064344. [DOI] [PubMed] [Google Scholar]
- Barker N, et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6(1):25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Spence JR, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470(7332):105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiraku M, et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 2011;472(7341):51–56. doi: 10.1038/nature09941. [DOI] [PubMed] [Google Scholar]
- Xia Y, et al. The generation of kidney organoids by differentiation of human pluripotent cells to ureteric bud progenitor-like cells. Nature Protocols. 2014;9(11):2693–2704. doi: 10.1038/nprot.2014.182. [DOI] [PubMed] [Google Scholar]
- Takasato M, et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature. 2015;526(7574):564–568. doi: 10.1038/nature15695. [DOI] [PubMed] [Google Scholar]
- Lancaster MA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501(7467):373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus SK, et al. Acinar cell apoptosis in Serpini2-deficient mice models pancreatic insufficiency. PLoS Genetics. 2005;1(3):38. doi: 10.1371/journal.pgen.0010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleeff J, et al. Chronic pancreatitis. Nat Rev Dis Primers. 2017;3:17060. doi: 10.1038/nrdp.2017.60. [DOI] [PubMed] [Google Scholar]
- Murtaugh LC, Melton DA. Genes, signals, and lineages in pancreas development. Annual Review of Cell and Developmental Biology. 2003;19:71–89. doi: 10.1146/annurev.cellbio.19.111301.144752. [DOI] [PubMed] [Google Scholar]
- Greggio C, De Franceschi F, Figueiredo-Larsen M, Grapin-Botton A. In vitro pancreas organogenesis from dispersed mouse embryonic progenitors. Journal of Visualized Experiments. 2014. p. 51725. [DOI] [PMC free article] [PubMed]
- Greggio C, et al. Artificial three-dimensional niches deconstruct pancreas development in vitro. Development. 2013;140(21):4452–4462. doi: 10.1242/dev.096628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneddon JB, Borowiak M, Melton DA. Self-renewal of embryonic-stem-cell-derived progenitors by organ-matched mesenchyme. Nature. 2012;491(7426):765–768. doi: 10.1038/nature11463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scavuzzo MA, Yang D, Borowiak M. Organotypic pancreatoids with native mesenchyme develop Insulin producing endocrine cells. Scientific Reports. 2017;7(1):10810. doi: 10.1038/s41598-017-11169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(25):14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SB, Schaffer AE, Sander M. The transcription factors Nkx6.1 and Nkx6.2 possess equivalent activities in promoting beta-cell fate specification in Pdx1+ pancreatic progenitor cells. Development. 2007;134(13):2491–2500. doi: 10.1242/dev.002691. [DOI] [PubMed] [Google Scholar]
- Seymour PA, et al. SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(6):1865–1870. doi: 10.1073/pnas.0609217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, et al. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nature Genetics. 2002;32(1):128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- Hara M, et al. Transgenic mice with green fluorescent protein-labeled pancreatic beta -cells. American Journal of Physiology: Endocrinology and Metabolism. 2003;284(1):177–183. doi: 10.1152/ajpendo.00321.2002. [DOI] [PubMed] [Google Scholar]
- Holthofer H, Schulte BA, Spicer SS. Expression of binding sites for Dolichos biflorus agglutinin at the apical aspect of collecting duct cells in rat kidney. Cell and Tissue Research. 1987;249(3):481–485. doi: 10.1007/BF00217319. [DOI] [PubMed] [Google Scholar]
- Reichert M, et al. Isolation, culture and genetic manipulation of mouse pancreatic ductal cells. Nature Protocols. 2013;8(7):1354–1365. doi: 10.1038/nprot.2013.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler H, Fischer-Colbrie R. The chromogranins A and B: the first 25 years and future perspectives. Neuroscience. 1992;49(3):497–528. doi: 10.1016/0306-4522(92)90222-N. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burcelin R, Knauf C, Cani PD. Pancreatic alpha-cell dysfunction in diabetes. Diabetes and Metabolism. 2008;34:49–55. doi: 10.1016/S1262-3636(08)73395-0. Suppl 2. [DOI] [PubMed] [Google Scholar]
- Del Prato S, Marchetti P. Beta- and alpha-cell dysfunction in type 2 diabetes. Hormone and Metabolic Research. 2004;36(11-12):775–781. doi: 10.1055/s-2004-826163. [DOI] [PubMed] [Google Scholar]
- Piciucchi M, et al. Exocrine pancreatic insufficiency in diabetic patients: prevalence, mechanisms, and treatment. International Journal of Endocrinology. 2015;2015:595649. doi: 10.1155/2015/595649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell-Thompson M, Rodriguez-Calvo T, Battaglia M. Abnormalities of the Exocrine Pancreas in Type 1 Diabetes. Current Diabetes Reports. 2015;15(10):79. doi: 10.1007/s11892-015-0653-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad C, Pulikkal AA, Kumar KM. Pancreatic exocrine insufficiency in type 1 and type 2 diabetics of Indian origin. Pancreatology. 2015;15(6):616–619. doi: 10.1016/j.pan.2015.09.018. [DOI] [PubMed] [Google Scholar]


