Abstract
Amyloplast is the site of starch synthesis in the storage tissue of maize (Zea mays). The amyloplast stroma contains an enriched group of proteins when compared with the whole endosperm. Proteins with molecular masses of 76 and 85 kD have been identified as starch synthase I and starch branching enzyme IIb, respectively. A 112-kD protein was isolated from the stromal fraction by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subjected to tryptic digestion and amino acid sequence analysis. Three peptide sequences showed high identity to plastidic forms of starch phosphorylase (SP) from sweet potato, potato, and spinach. SP activity was identified in the amyloplast stromal fraction and was enriched 4-fold when compared with the activity in the whole endosperm fraction. Native and sodium dodecyl sulfate-polyacrylamide gel electrophoresis analyses showed that SP activity was associated with the amyloplast stromal 112-kD protein. In addition, antibodies raised against the potato plastidic SP recognized the amyloplast stromal 112-kD protein. The amyloplast stromal 112-kD SP was expressed in whole endosperm isolated from maize harvested 9 to 24 d after pollination. Results of affinity electrophoresis and enzyme kinetic analyses showed that the amyloplast stromal 112-kD SP preferred amylopectin over glycogen as a substrate in the synthetic reaction. The maize shrunken-4 mutant had reduced SP activity due to a decrease of the amyloplast stromal 112-kD enzyme.
In the developing endosperm of monocotyledonous plants, starch granules are synthesized and deposited within the amyloplast (Lopes and Larkins, 1993; Nelson and Pan, 1995). The amyloplast is a specialized plastid comprised of three distinct components, the envelope, the starch granule, and a soluble compartment known as the stroma (Lopes and Larkins, 1993; Nelson and Pan, 1995). The identification of the enzymes involved in the synthesis of the starch granule is essential to understanding the structure and functionality of starch. The amyloplast stroma of maize (Zea mays) contains a unique group of proteins, some of which are enzymes involved in starch biosynthesis (Yu et al., 1998). These enzymes include starch synthase, starch branching enzyme, and starch debranching enzyme (Yu et al., 1998). One of the unique proteins in the stroma, whose abundance is second only to the starch branching enzyme IIb, has a subunit molecular mass of 112 kD (Yu et al., 1998). The identification of this 112-kD protein was the focus of the present investigation. Amino acid sequence analysis of the amyloplast stromal 112-kD protein revealed that this protein was a putative starch phosphorylase (SP).
SP catalyzes reaction 1 (Hanes, 1940a, 1940b). In the synthetic direction, a Glc unit is transferred from Glc-1-P to a growing primer, with release of inorganic phosphate.
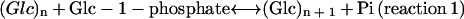 |
In the phosphorolytic direction, addition of inorganic phosphate liberates a molecule of Glc-1-P. Higher plant SP enzymes are classified into two types based upon their molecular mass, subcellular localization, and affinity for various α-glucans (Shimomura et al., 1982; Steup, 1988). One type of SP is localized in plastids (e.g. amyloplast and chloroplast) and has a subunit molecular mass >100 kD. The plastidic SP prefers α-glucans possessing long linear glucan chains, such as amylopectin or maltodextrins. The other type of SP is localized in the cytosol and has a subunit molecular mass of approximately 90 kD. The cytosolic SP exhibits a high affinity for highly branched glucans such as glycogen.
Four different forms of SP have been characterized from maize by Nelson and coworkers (Tsai and Nelson, 1968, 1969a). The four forms of SP differ in their pH optima, primer dependence, and developmental expression (Tsai and Nelson, 1968, 1969a). The major form of SP has been purified to apparent homogeneity from maize kernels by Burr and Nelson (1975). The purified enzyme is a dimer composed of identical subunits having a molecular mass of 112 kD (Burr and Nelson, 1975). The purified enzyme has been characterized with respect to its amino acid composition, pyridoxal-5-phosphate content, and enzymological and kinetic properties (Burr and Nelson, 1975). In this paper we presented biochemical data showing that the amyloplast stromal 112-kD protein is indeed a SP enzyme.
RESULTS
Identification of the 112-kD Amyloplast Stroma Protein as an SP
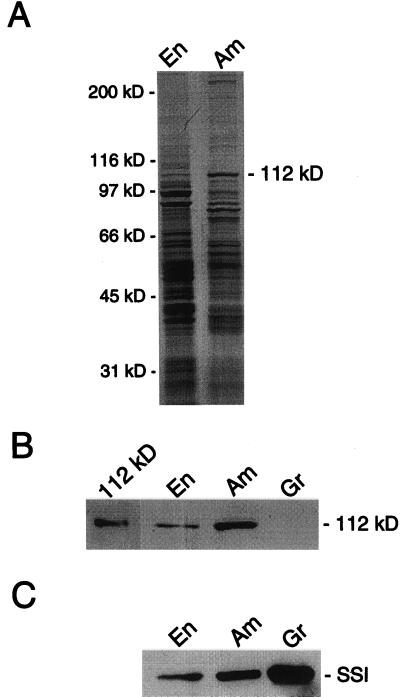
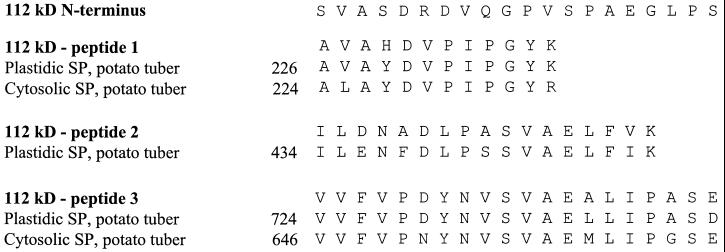
The maize amyloplast stroma contains a characteristic group of proteins that are enriched when compared with whole endosperm (Yu et al., 1998; Fig. 1). The 76- and 85-kD proteins have been identified as starch synthase I and starch branching enzyme IIb, respectively, and the 81-kD protein has been identified as a heat shock cognate (Yu et al., 1998). The identity of the 112-kD protein, the abundance of which is second only to the 85-kD protein, was the focus of this work. The amyloplast stromal fraction was isolated from maize endosperm and subjected to SDS-PAGE. The 112-kD protein was isolated by electroelution from the polyacrylamide gel and subjected to N-terminal and internal amino acid sequence analyses. Unambiguous amino acid sequences were obtained for the N terminus and for three internal peptide fragments (Fig. 2). These sequences were compared with sequences of the plastidic and cytosolic forms of SP from potato, the only species where sequences of both SP forms have been published. The amino acid sequences of the three internal fragments derived from the maize 112-kD protein closely aligned with internal sequences of the plastidic form of SP from potato tuber (Fig. 2). On the other hand, only peptides 1 and 3 of the 112-kD protein aligned with sequences of the cytosolic form of SP from potato tuber (Fig. 2). The sequence of peptide 2 of the 112-kD protein is found in the central portion of plastidic SP enzymes commonly referred to as the insertion sequence (Lin et al., 1991; Mori et al., 1991). The insertion sequence is thought to reflect the affinity of plastidic SP enzymes for α-glucan substrates (Mori et al., 1993). The three peptides of the 112-kD protein also aligned closely with sequences from the plastidic SP enzymes from potato leaf (Sonnewald et al., 1995), sweet potato (Lin et al., 1991), and spinach leaf (Duwenig et al., 1997a). The N-terminal amino acid sequence of the 112-kD protein was unique and did not align with the N terminus of the plastidic SP enzymes or with any sequences in the GenBank database.
Figure 1.
SDS-PAGE analysis of the endosperm and amyloplast stromal fraction and immunoblot analysis of endosperm, amyloplast stromal, and granule fractions. A, Samples (30 μg) of the endosperm (En) and amyloplast stromal fraction (Am) were subjected to SDS-PAGE followed by Coomassie Blue staining. The molecular mass standards from top to bottom are myosin (200 kD), β-galactosidase (116.2 kD), phosphorylase b (97.4 kD), bovine serum albumin (66.2 kD), ovalbumin (45 kD), and carbonic anhydrase (31 kD). B, A sample (0.1 μg) of the isolated 112-kD protein and samples (30 μg) of the endosperm, amyloplast stromal, and granule fractions were subjected to immunoblot analysis using anti-SP antibodies. C, Samples (30 μg) of the endosperm, amyloplast stromal, and granule fractions were subjected to immunoblot analysis using anti-starch synthase I antibodies. A portion of the immunoblots is shown in B and C. The position of the 112-kD protein is indicated in A and B, and the position of starch synthase I (SSI) is indicated in C. The data shown in each A through C is representative of two independent experiments.
Figure 2.
Amino acid sequences of the amyloplast stromal 112-kD protein and alignment with amino acid sequences of plastidic and cytosolic forms of potato SP. The isolated 112-kD protein was subjected to SDS-PAGE. The protein was then transferred to polyvinylidene difluoride paper and subjected to N-terminal amino acid sequence analysis. Another sample of the protein was digested with trypsin, three peptides were isolated, and subjected to amino acid sequence analysis. Sequences of the N terminus and from three internal peptides of the 112-kD protein were aligned with published sequences of plastidic SP (Nakano and Fukui, 1986; Nakano et al., 1989) and cytosolic SP (Mori et al., 1991) from potato tuber. The numbers in the figure represent the residue numbers that begin with the indicated published sequences.
To examine the hypothesis that the 112-kD protein was a SP enzyme, the protein was isolated from an SDS-polyacrylamide gel and subjected to immunoblot analysis using anti-SP antibodies raised against the plastidic SP of potato (Brisson et al., 1989). Indeed these antibodies recognized the isolated 112-kD protein (Fig. 1B). Moreover, the anti-SP antibodies recognized the 112-kD protein in the endosperm and amyloplast stromal fraction (Fig. 1B). The relative abundance of the immunoreactive 112-kD protein in the endosperm and amyloplast stromal fraction was similar to that observed for the Coomassie Blue-stained 112-kD protein in these fractions (Fig. 1A). These antibodies did not recognize any other proteins in the endosperm and amyloplast stromal fraction of maize.
A protein extract was prepared from starch granules isolated from the amyloplast fraction and subjected to immunoblot analysis using the anti-SP antibodies. These antibodies did not recognize any proteins in the granule fraction (Fig. 1B). Immunoblot analysis of the endosperm, amyloplast stromal, and granule fractions using anti-starch synthase I antibodies was performed as positive control. As described previously (Mu et al., 1994; Mu-Forster et al., 1996; Yu et al., 1998), starch synthase I was enriched in the amyloplast stroma when compared with the whole endosperm and also associated with the starch granule (Fig. 1C).
The whole endosperm and amyloplast stromal fractions were assayed for SP activity. The specific activity of the enzyme in the amyloplast stromal fraction (0.6 μmol min−1 mg−1) was 4-fold greater than the activity in the whole endosperm fraction (0.15 μmol min−1 mg−1). These two fractions were also subjected to native PAGE in the presence of either 12 mg/mL amylopectin or 24 mg/mL glycogen. Following electrophoresis, the SP activity in the polyacrylamide gel was measured by iodine staining. A major positively stained band was observed in gels containing amylopectin or glycogen from each fraction. The SP activity band was enriched in the amyloplast stromal fraction when compared with the whole endosperm fraction. A duplicate polyacrylamide gel was not stained with iodine. A gel slice corresponding to the position of the major iodine-stained band was minced with a razor blade into small pieces. This sample was then subjected to SDS-PAGE followed by immunoblotting with anti-SP antibodies. This analysis showed that the anti-SP antibodies recognized a 112-kD protein that was present in the native polyacrylamide gel band that possessed SP activity. Taken together, these data provided evidence that the 112-kD protein that was enriched in the amyloplast stromal fraction was a plastidic SP.
Levels of the 112-kD SP during Maize Endosperm Development
We examined the temporal expression of the 112-kD amyloplast stroma SP during endosperm development. Whole-endosperm fractions were isolated from maize harvested 9 to 24 d after pollination (DAP). This time interval corresponds with the time when starch is synthesized in maize endosperm (Tsai and Nelson, 1968; Ozbun et al., 1973). SP activity was measured within native polyacrylamide gels containing glycogen by iodine staining and the 112-kD SP protein was analyzed by immunoblotting using the anti-SP antibodies. SP activity and the amyloplast stromal 112-kD protein were present 9 DAP and persisted through 24 DAP. As a positive control, immunoblot analysis was used to examine the levels of the 76-kD starch synthase I during endosperm development. As described previously (Mu et al., 1994), the levels of starch synthase I did not change significantly from 9 to 24 DAP.
Affinity of the 112-kD Stroma SP for Amylopectin and Glycogen
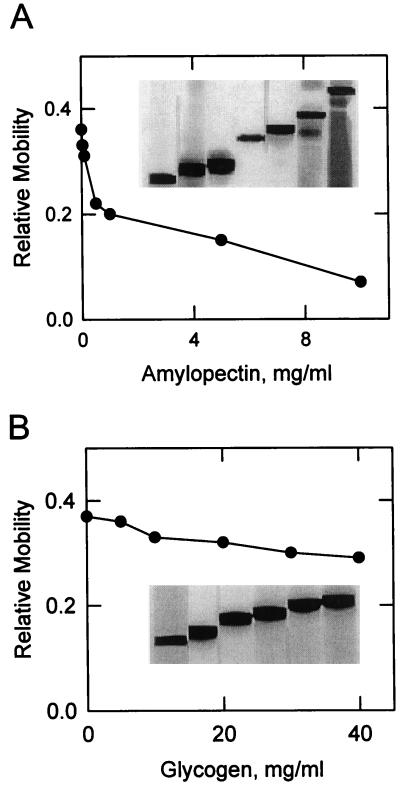
The affinity of the 112-kD amyloplast stromal SP for α-glucans was examined by affinity electrophoresis (Takeo, 1984). The amyloplast stromal fraction was subjected to native PAGE in the presence of varying concentrations of α-glucan immobilized within the gel. Following electrophoresis, SP activity was measured by iodine staining. Immunoblot analysis confirmed the identity of the SP activity bands as the amyloplast stromal 112-kD SP enzyme. The addition of increasing concentrations of amylopectin to polyacrylamide gels resulted in a dose-dependent decrease in the mobility of SP activity (Fig. 3A). This demonstrated that the enzyme bound to its α-glucan substrate within the polyacrylamide gel (Shimomura and Fukui, 1980; Takeo, 1984). The dissociation constant (Kd) of the enzyme for amylopectin was calculated to be 0.43 mg/mL. The addition of glycogen to polyacrylamide gels also resulted in a dose-dependent decrease in the electrophoretic mobility of SP activity (Fig. 3B). The dissociation constant for glycogen was 17 mg/mL. The decrease in electrophoretic mobility of the SP enzyme in the presence of amylopectin and glycogen was specific. These α-glucans did not affect the electrophoretic mobility of 0.2 mg/mL bovine serum albumin.
Figure 3.
Dependence of the relative mobility of the 112-kD stromal SP on the concentration of amylopectin and glycogen upon affinity electrophoresis. Samples (30 μg) of the amyloplast stromal fraction were subjected to native PAGE in the presence of the indicated concentrations of amylopectin (A) and glycogen (B). Following electrophoresis, SP activity was measured by iodine staining. The relative mobility was calculated by dividing the migration of the activity band by the migration of the dye front. The inset of A and B contains a portion of the native gels showing the relative mobility of SP. The data shown in A and B is representative of two independent experiments.
Dependence of the 112-kD Stroma SP Activity on Amylopectin and Glycogen
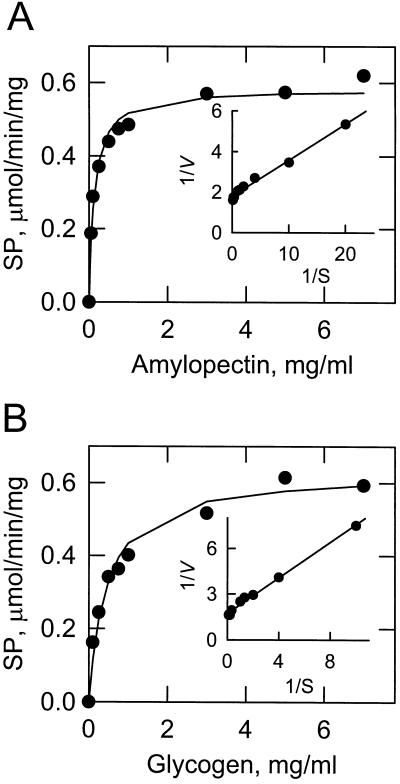
The dependence of the 112-kD amyloplast stroma SP activity on amylopectin and glycogen was examined using a saturating concentration of Glc-1-P (20 mm). SP exhibited typical saturation kinetics with respect to amylopectin (Fig. 4A). The Vmax was 0.58 μmol min−1 mg−1, and the apparent Km value for amylopectin was 0.13 mg/mL. The enzyme displayed saturation kinetics for glycogen (Fig. 4B). The Vmax was 0.63 μmol min−1 mg−1 and the apparent Km value for glycogen was 0.45 mg/mL.
Figure 4.
Dependence of SP activity on the concentration of amylopectin and glycogen. SP activity was measured as a function of the indicated concentrations of amylopectin (A) and glycogen (B). The concentration of Glc-1-P was maintained at 20 mm. The curves drawn were the result of the analysis of the data according to the Michaelis-Menten equation. The insets shown in A and B are double reciprocal plots of the data. The lines drawn in the insets are the result of a least-squares analysis of the data. The data shown in A and B is representative of two independent experiments carried out in duplicate.
Analysis of the 112-kD Stroma SP in the shrunken-4 Mutant
The maize shrunken-4 mutant kernel contains one-third as much starch as the wild-type kernel, resulting in its shriveled, opaque phenotype (Tsai and Nelson, 1969b; Burr and Nelson, 1973). We prepared extracts from wild-type and shrunken-4 mutant endosperms of 30 DAP and measured SP activity. As described previously (Tsai and Nelson, 1969b), the specific activity of SP in the extract of the shrunken-4 mutant was reduced (66%) when compared with the SP activity from wild-type endosperm. The reduced SP activity in the shrunken-4 mutant has been attributed to a deficiency for pyridoxal-5-phosphate within the cell (Burr and Nelson, 1973). Pyridoxal-5-phosphate is a cofactor of SP (Burr and Nelson, 1975). The addition of pyridoxal-5-phosphate to the assay mixture of the shrunken-4 mutant did not affect the level of SP activity. We questioned if the reduction in SP activity in the extract derived from the shrunken-4 mutant was due to the presence of an enzyme inhibitor or the loss of an enzyme activator. Extracts from the endosperms of wild-type and shrunken-4 mutant were mixed, incubated for 20 min, and then assayed for SP activity. The SP activity of the mixture was the average of the specific activities of the SP assayed from each extract separately. These data suggested that the lower SP activity in the shrunken-4 mutant endosperm was not due to effector molecules.
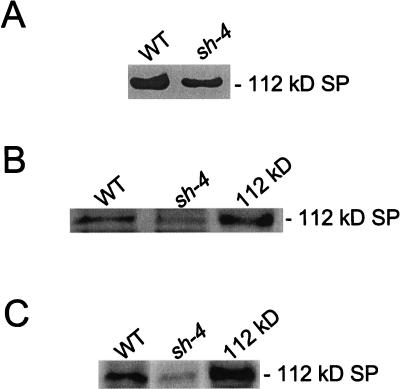
We questioned whether the reduction of the SP activity in the shrunken-4 mutant was due to the level of the 112-kD stromal SP. Equal amounts of the endosperm fractions of the wild-type and shrunken-4 mutant were subjected to native PAGE followed by the measurement of SP activity by iodine staining. A positively stained band was observed in the gel containing the sample that was derived from the shrunken-4 mutant. This activity band migrated to the same position as the 112-kD stromal SP from the wild-type control (Fig. 5A). Scanning densitometry of the SP bands on the polyacrylamide gels showed that the activity in the shrunken-4 mutant was reduced by 50%. The amount of the 112-kD stroma SP in the shrunken-4 mutant endosperm was also examined by SDS-PAGE followed by Coomassie Blue staining (Fig. 5B) and by immunoblot analysis using anti-SP antibodies (Fig. 5C). This analysis showed that the level of the 112-kD stroma SP was reduced by 50% in the shrunken-4 mutant when compared with the wild-type control.
Figure 5.
Levels of the 112-kD stromal SP in the shrunken-4 mutant. A, Samples (30 μg) of the endosperm fraction from wild-type (WT) and the shrunken-4 (sh-4) mutant were subjected to native PAGE in the presence of 24 μm glycogen. Following electrophoresis, SP activity was measured by iodine staining. B, The isolated 112-kD protein (0.1 μg) and samples (60 μg) of the endosperm fraction from wild-type (WT) and the shrunken-4 (sh-4) mutant were subjected to SDS-PAGE followed by Coomassie Blue staining. C, The isolated 112-kD protein (0.1 μg) and samples (30 μg) of the endosperm fraction from wild-type and the shrunken-4 mutant were subjected to immunoblot analysis using anti-SP antibodies. A portion of the polyacrylamide gels (A and B) and the immunoblot (C) is shown, and the position of the 112-kD stromal SP is indicated in the figure. The data shown in A through C is representative of two independent experiments.
DISCUSSION
In maize non-photosynthetic sink tissue, starch biosynthesis occurs in the amyloplast (Lopes and Larkins, 1993; Nelson and Pan, 1995). Some starch biosynthetic enzymes are localized exclusively in the starch granule (e.g. granule bound starch synthase I; Nelson and Chourey, 1978; Macdonald and Preiss, 1985), whereas some are localized exclusively in the amyloplast stroma (e.g. starch debranching enzyme; Yu et al., 1998). In addition, some starch biosynthetic enzymes are localized in both locations (e.g. starch synthase I and starch branching enzyme IIb; Mu-Forster et al., 1996; Yu et al., 1998). In this study we identified the maize amyloplast stromal 112-kD protein as a plastidic SP enzyme localized in the stroma. The classification of this SP as a plastidic enzyme was based on the following data. SP activity was associated with the 112-kD protein enriched in the amyloplast stroma. The 112-kD protein exhibited amino acid sequence identity with sequences of known plastidic SP enzymes. The 112-kD protein reacted with specific antibodies raised against the potato plastidic SP. We hypothesize that the amyloplast stromal 112-kD plastidic SP is the same enzyme originally purified from maize kernels by Burr and Nelson (Burr and Nelson, 1975). At the time of this earlier work the localization of the SP enzyme purified by Burr and Nelson (Burr and Nelson, 1975) was not known, nor were SP enzymes classified as being plastidic or cytosolic.
A characteristic property of plastidic SP enzymes is their preference for α-glucans having long linear glucan chains. For example, the plastidic SP enzymes from potato tuber (Liddle et al., 1961), spinach leaf (Shimomura et al., 1982), and sweet corn (Lee and Braun, 1973) prefer amylopectin as a substrate to glycogen, which is a more highly branched α-glucan. Indeed the maize amyloplast stromal 112-kD SP preferred amylopectin as a substrate when compared with glycogen. The Km value for amylopectin in the synthetic direction of the SP reaction was 3.4-fold lower than that of glycogen. Moreover, the Kd for amylopectin as determined by affinity electrophoresis was 40-fold lower than that of glycogen.
The maize shrunken-4 mutant is characterized by having one-third as much starch and one-third as much soluble protein as the wild-type kernel (Tsai and Nelson, 1969b; Burr and Nelson, 1973). Nelson and coworkers (Tsai and Nelson, 1969b; Burr and Nelson, 1973) have shown that the SP activity in maize endosperm is reduced in the shrunken-4 mutant. Moreover, the activities of other starch biosynthetic enzymes, including ADP-Glc pyrophosphorylase and starch synthase, are also reduced in the shrunken-4 mutant (Akatsuka and Nelson, 1966). The total amount of pyridoxal-5-phosphate in the endosperm of the shrunken-4 mutant is reduced 8-fold when compared with wild-type endosperm (Burr and Nelson, 1973). Burr and Nelson (1973) have suggested that the decrease in SP activity in the mutant is due to a deficiency of its cofactor pyridoxal-5-phosphate. In the present work the addition of pyridoxal-5-phosphate to the assay system for SP activity of the mutant did not affect the activity. The fact the enzyme activity could not be restored with pyridoxal-5-phosphate could result from the instability and/or degradation of SP in the extract due to the deficiency of the cofactor. Indeed the reduced SP activity in the shrunken-4 mutant could be attributed to a decrease of the amyloplast stromal 112-kD SP enzyme. Additional studies will be required to determine whether the decrease of the amyloplast stromal 112-kD SP in the shrunken-4 mutant was due to an increase in enzyme turnover or due to a decrease in enzyme expression. The decrease in SP activity in the shrunken-4 mutant did not appear to result from the presence or absence of effector molecules based on results of mixing extracts of mutant and wild-type endosperms.
SP was once considered the predominant enzyme catalyzing starch chain elongation in maize endosperm (Tsai and Nelson, 1968, 1969a). However, interest in SP as a major starch biosynthetic enzyme had declined with the discovery of ADP-Glc pyrophosphorylase (Akatsuka and Nelson, 1966; Ozbun et al., 1973). Over the past three decades most efforts to characterize the flow of carbon from Glc-1-P to starch have focused on ADP-Glc pyrophosphorylase, starch synthase, starch branching enzyme, and starch debranching enzyme (Smith et al., 1997). Antisense experiments used to reduce plastidic SP in potato leaf (Sonnewald et al., 1995) and cytosolic SP in potato tuber (Duwenig et al., 1997b) have shown little effect on starch synthesis or degradation. Thus the major route of starch synthesis occurs via the ADP-Glc pyrophosphorylase-starch synthase pathway (Smith et al., 1997).
There has been a renewed interest in the SP enzyme. cDNAs encoding the plastidic forms of SP enzymes have been isolated and characterized from a variety of higher plants (Steup, 1988). Studies with potato (Brisson et al., 1989; St-Pierre and Brisson, 1995), spinach (Duwenig et al., 1997a), and pea (van Berkel J et al., 1991) have shown that the expression of plastidic SP genes correlates with starch biosynthesis. The fact that the 112-kD SP enzyme was enriched in the amyloplast stroma where other starch synthetic enzymes are localized supports the hypothesis that SP may play some role in starch biosynthesis. Takaha et al. (1998) have recently suggested that disproportionating enzymes may work in conjunction with SP for starch synthesis via the SP phosphorolytic reaction in the context of the “discontinuous synthesis model” or the “glucan-trimming model” (Ball et al., 1996; Myers et al., 2000). According to the model (Ball et al., 1996; Myers et al., 2000) pre-amylopectin molecules are trimmed by the starch debranching enzyme. The short chain malto-oligo-saccharides liberated in the trimming reaction are converted to longer chain glucan molecules by the action of disproportionating enzymes (Takaha et al., 1998). The longer-chain glucan molecules can then be utilized by SP via the phosphorolytic reaction to generate Glc-1-P. In turn, the Glc-1-P may be utilized by ADP-Glc pyrophosphorylase for the synthesis of starch. Recent studies have shown that the phosphorolytic reaction of SP is in fact stimulated by the presence of disproportionating enzymes (Colleoni et al., 1999). It is clear that additional studies are needed to establish the physiological role of SP enzymes in starch metabolism. The identification of the maize amyloplast stromal 112-kD protein as a plastidic SP enzyme provides a foundation for future studies.
MATERIALS AND METHODS
Materials
All chemicals were reagent grade. Amylopectin, glycogen, bovine serum albumin, Glc-1-P, 2-(N-morpholino)-ethanesulfonic acid, Triton X-100, ferrous molybdate, and pyridoxal-5-phosphate were from Sigma (St. Louis). Protein assay reagent, electrophoresis reagents, molecular mass standards, polyvinylidene difluoride paper for protein sequencing, and immunochemical reagents were from Bio-Rad (Hercules, CA). Nitrocellulose paper for immunoblotting was from Schleicher & Schuell (Keene, NH). The enhanced chemiluminescence direct labeling and detection system was from Amersham Pharmacia Biotech (Uppsala). Pro-Blue PAGE stain was from Owl Separation Systems (Woburn, MA). The shrunken-4 mutant maize ears of 30 DAP were provided by ExSeed Genetics (Ames, IA).
Preparation of Endosperm and Amyloplast Fractions
The endosperm fraction was prepared from Dent inbred maize (Zea mays cv B73) that were field grown and harvested at 9, 12, 15, 18, 21, and 24 DAP. Endosperms were obtained from maize ears by manual removal of embryos and pericarp. Cell extracts were prepared from endosperms as described by Yu et al. (1998). The amyloplast stromal fraction was isolated from endosperms of 15 DAP (Yu et al., 1998). The purity of the amyloplast stromal fraction was assessed using appropriate marker enzymes (Yu et al., 1998). Starch granules were isolated from amyloplasts as described previously (Yu et al., 1998). Granule-associated proteins were isolated by extracting starch granules with SDS-PAGE sample buffer (20 μL buffer mg −1 dry weight granule). Mixtures were then boiled for 15 min and cooled to room temperature. Annealed starch was removed by centrifugation at 13,000g for 15 min (Mu et al., 1994; Mu-Forster et al., 1996).
Electrophoresis, Amino Acid Sequence Analyses, and Immunoblotting
Affinity electrophoresis (Shimomura and Fukui, 1980; Takeo, 1984; Duwenig et al., 1997a) was performed with 7.5% (w/v) polyacrylamide tube gels in the absence or presence of the indicated glucan substrates at 4°C. Electrophoresis was performed for 1 h at 3 mA per tube, and then for 3 h at 5 mA per tube. Following electrophoresis, gels were soaked in 100 mm citrate-NaOH buffer (pH 6.0) for 1 h. The tube gels were then incubated for 2 h at 37°C in a reaction mixture containing 100 mm citrate-NaOH buffer (pH 6.0) and 20 mm Glc-1-P. SP activity was identified as blue-staining bands in the tube gels after a 3- to 5-min incubation with 10 mm I2 and 14 mm KI. In an alternate manner, electrophoresis was carried out in 8% (w/v) polyacrylamide slab gels.
SDS-PAGE (Laemmli, 1970; Porzio and Pearson, 1976) was performed with 9% to 18% (w/v) gradient slab gels. Proteins on SDS-polyacrylamide gels were visualized with Pro-Blue stain. Molecular mass standards were myosin (200 kD), β-galactosidase (116.2 kD), phosphorylase b (97.4 kD), bovine serum albumin (66.2 kD), ovalbumin (45 kD), and carbonic anhydrase (31 kD).
The 112-kD amyloplast stromal protein was eluted from SDS-polyacrylamide gel slices using an electroeluter (model 422, Bio-Rad) according to the instructions provided by the manufacturer. The electroeluted protein was then subjected to SDS-PAGE and transferred to polyvinylidene difluoride paper (Matsudaira, 1987). The N-terminal amino acid sequence of a protein sample was determined by automated Edman degradation. Another sample of the protein on the polyvinylidene difluoride paper was digested with trypsin and the resulting peptide fragments were purified by HPLC. Selected peptides were subjected to amino acid sequence analysis. The trypsin digestion and the amino acid sequence analyses were performed at the Macromolecular Structure Facility of Michigan State University.
Immunoblot analyses (Haid and Suissa, 1983) were performed with anti-SP antibodies raised against the plastidic form of potato SP (Brisson et al., 1989) and with anti-starch synthase I antibodies raised against maize soluble starch synthase I (Mu et al., 1994). The anti-SP antibodies and anti-starch synthase antibodies were used at dilutions of 1:5,000 and 1:100,000, respectively. Protein bands were identified on immunoblots using the enhanced chemiluminescence direct labeling and detection kit. The density of bands was quantified by scanning densitometry. Immunoblot signals were in the linear range of detection.
Enzyme Assays and Protein Determination
SP activity was measured in the synthetic direction. The reaction mixture contained 100 mm 2-(N-morpholino)-ethanesulfonic acid buffer (pH 6.0), 20 mm Glc-1-P, the indicated concentration of glucan substrate, and enzyme protein in a total volume of 0.1 mL. The reaction mixtures were incubated at 37°C for 45 min and were terminated by addition of 0.05 mL 0.4 n H2SO4. The amount of inorganic phosphate released from Glc 1-P was determined by a molybdate-based colorimetric assay (Fiske and Subbarow, 1925). All assays were conducted in duplicate and were linear with time and protein concentration. A unit of SP activity was defined as the amount of enzyme that catalyzed the formation of 1 μmol of product/min. Specific activity was defined as units per milligram of protein. Protein concentration was determined by the method of Bradford (1976) using bovine serum albumin as the standard. Buffers (e.g. containing Triton X-100) that were identical to those containing protein samples were used as blanks.
Calculation of Kd and Analysis of Kinetic Data
The Kd of SP for amylopectin and glycogen were based on the relative mobility of the enzyme after affinity electrophoresis (Takeo, 1984). Kinetic data were analyzed with the EZ-FIT enzyme kinetic model-fitting program according to the Michaelis-Menten equation. EZ-FIT uses the Nelder-Mead Simplex and Marquardt/Nash nonlinear regression algorithms sequentially and test for the best fit of the data among different kinetic models (Perrella, 1988).
ACKNOWLEDGMENTS
We thank Peter Keeling and Ed Wilhelm for providing maize samples and Normand Brisson for providing the anti-SP antibodies. We thank Connie Shih and Justin Belles for their help with amyloplast preparations and Makoto Yamamori for advice on affinity electrophoresis. We also acknowledge Peter Keeling for many helpful discussions.
Footnotes
This work was supported by the U.S. Department of Agriculture National Research Initiative (grant no. 95–02531) and by ExSeed Genetics.
This paper is dedicated to the memory of our friend and colleague Bruce P. Wasserman, who passed away on August 26, 1998.
LITERATURE CITED
- Akatsuka T, Nelson OE. Starch granule-bound adenosine diphosphate Glc-starch glucosyltransferases of maize seeds. J Biol Chem. 1966;241:2280–2286. [PubMed] [Google Scholar]
- Ball S, Guan H, James M, Myers A, Keeling P, Mouille G, Buleon A, Colonna P, Preiss J. From glycogen to amylopectin: a model for the biogenesis of the plant starch granule. Cell. 1996;86:349–352. doi: 10.1016/s0092-8674(00)80107-5. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein using the principles of protein dye-binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brisson N, Giroux H, Camirand A, Simard C. Maturation and subcellular compartmentation of potato starch phosphorylase. Plant Cell. 1989;1:559–566. doi: 10.1105/tpc.1.5.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr B, Nelson OE. The phosphorylases of developing maize seeds. Ann NY Acad Sci. 1973;210:129–138. doi: 10.1111/j.1749-6632.1973.tb47567.x. [DOI] [PubMed] [Google Scholar]
- Burr B, Nelson OE. Maize α-glucan phosphorylase. Eur J Biochem. 1975;56:539–546. doi: 10.1111/j.1432-1033.1975.tb02260.x. [DOI] [PubMed] [Google Scholar]
- Colleoni C, Dauvillee D, Mouille G, Morell M, Samuel M, Slomiany M, Lienard L, Wattebled F, D'Hulst C, Ball S. Biochemical characterization of the Chlamydomonas reinhardtii α-1,4 glucanotransferase supports a direct function in amylopectin biosynthesis. Plant Physiol. 1999;120:1005–1014. doi: 10.1104/pp.120.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duwenig E, Steup M, Kossmann J. Induction of genes encoding plastidic phosphorylase from spinach (Spinacia oleracea L.) and potato (Solanum tuberosum L.) by exogenously supplied carbohydrates in excised leaf discs. Planta. 1997a;203:111–120. doi: 10.1007/s004250050171. [DOI] [PubMed] [Google Scholar]
- Duwenig E, Steup M, Willmitzer L, Kossmann J. Antisense inhibition of cytosolic phosphorylase in potato plants (Solanum tuberosum L.) affects tuber sprouting and flower formation with only little impact on carbohydrate metabolism. Plant J. 1997b;12:323–333. doi: 10.1046/j.1365-313x.1997.12020323.x. [DOI] [PubMed] [Google Scholar]
- Fiske CH, Subbarow Y. The colorimetric determination of phosphorus. J Biol Chem. 1925;66:375–400. [Google Scholar]
- Haid A, Suissa M. Immunochemical identification of membrane proteins after SDS-PAGE. Methods Enzymol. 1983;96:192–205. doi: 10.1016/s0076-6879(83)96017-2. [DOI] [PubMed] [Google Scholar]
- Hanes CS. The breakdown and synthesis of starch by an enzyme system from pea seeds. Proc Roy Soc (London) 1940a;B128:421–500. [Google Scholar]
- Hanes CS. The reversible formation of starch from Glc-1-phosphate catalyzed by potato phosphorylase. Proc R Soc Lond. 1940b;B129:174–208. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee EY, Braun JJ. Sweet corn phosphorylase: purification and properties. Arch Biochem Biophys. 1973;156:276–286. doi: 10.1016/0003-9861(73)90366-4. [DOI] [PubMed] [Google Scholar]
- Liddle AM, Manners DJ, Wright A. Studies on carbohydrate-metabolizing enzymes: VI. The action of potato phosphorylase on starch-type polysaccharides. Biochem J. 1961;80:304–309. doi: 10.1042/bj0800304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CT, Yeh KW, Lee PD, Su JC. Primary structure of sweet potato starch phosphorylase deduced from its cDNA sequence. Plant Physiol. 1991;95:1250–1253. doi: 10.1104/pp.95.4.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes MA, Larkins BA. Endosperm origin, development, and function. Plant Cell. 1993;5:1383–1389. doi: 10.1105/tpc.5.10.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald FD, Preiss J. Partial purification and characterization of granule-bound starch synthases from normal and waxy maize. Plant Physiol. 1985;78:849–852. doi: 10.1104/pp.78.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- Mori H, Tanizawa K, Fukui T. Potato tuber type H phosphorylase isozyme: molecular cloning, nucleotide sequence, and expression of a full-length cDNA in Escherichia coli. J Biol Chem. 1991;266:18446–18453. [PubMed] [Google Scholar]
- Mori H, Tanizawa K, Fukui T. A chimeric α-glucan phosphorylase of plant type L and H isozymes: functional role of 78-residue insertion in type L isozyme. J Biol Chem. 1993;268:5574–5581. [PubMed] [Google Scholar]
- Mu C, Harn C, Ko YT, Singletary GW, Keeling PL, Wasserman BP. Association of a 76-kD polypeptide with soluble starch synthase I activity in maize (cv73) endosperm. Plant J. 1994;6:151–159. [Google Scholar]
- Mu-Forster C, Huang R, Powers JR, Harriman RW, Knight M, Singletary GW, Keeling PL, Wasserman BP. Physical association of starch biosynthetic enzymes with starch granules of maize endosperm: granule-associated forms of starch synthase I and starch branching enzyme II. Plant Physiol. 1996;111:821–829. doi: 10.1104/pp.111.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AM, Morell MK, James MG, Ball SG. Recent progress toward understanding biosynthesis of the amylopectin crystal. Plant Physiol. 2000;122:989–998. doi: 10.1104/pp.122.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K, Fukui T. The complete amino acid sequence of potato α-glucan phosphorylase. J Biol Chem. 1986;261:8230–8236. [PubMed] [Google Scholar]
- Nakano K, Mori H, Fukui T. Molecular cloning of cDNA encoding potato amyloplast α-glucan phosphorylase and the structure of its transit peptide. J Biochem. 1989;106:691–695. doi: 10.1093/oxfordjournals.jbchem.a122918. [DOI] [PubMed] [Google Scholar]
- Nelson O, Pan D. Starch synthesis in maize endosperms. Annu Rev Plant Physiol Mol Biol. 1995;46:475–496. [Google Scholar]
- Nelson OE, Chourey PS. Nucleoside diphosphate sugar-starch glucosyl transferase activity of wx starch granules. Plant Physiol. 1978;62:383–386. doi: 10.1104/pp.62.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbun JL, Hawker JS, Greenberg E, Lammel C, Preiss J, Lee EYC. Starch synthetase, phosphorylase, ADP-Glc pyrophosphorylase and UDP-Glc pyrophosphorylase in developing maize kernels. Plant Physiol. 1973;51:1–5. doi: 10.1104/pp.51.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrella F. A practical curve-fitting microcomputer program for the analysis of enzyme kinetic data on IBM-PC compatible computers. Anal Biochem. 1988;174:437–447. doi: 10.1016/0003-2697(88)90042-5. [DOI] [PubMed] [Google Scholar]
- Porzio MA, Pearson AM. Improved resolution of myofibrillar proteins with SDS-PAGE. Biochim Biophys Acta. 1976;490:27–34. doi: 10.1016/0005-2795(77)90102-7. [DOI] [PubMed] [Google Scholar]
- Shimomura S, Fukui T. A comparative study on α-glucan phosphorylases from plant and animal: interrelationship between the polysaccharide and pyridoxal phosphate binding sites by affinity electrophoresis. Biochemistry. 1980;19:2287–2294. doi: 10.1021/bi00552a001. [DOI] [PubMed] [Google Scholar]
- Shimomura S, Nagai M, Fukui T. Comparative glucan specificities of two types of spinach leaf phosphorylase. J Biochem. 1982;91:703–717. doi: 10.1093/oxfordjournals.jbchem.a133743. [DOI] [PubMed] [Google Scholar]
- Smith AM, Denyer K, Martin C. The synthesis of the starch granule. Annu Rev Plant Physiol Mol Biol. 1997;48:65–87. doi: 10.1146/annurev.arplant.48.1.67. [DOI] [PubMed] [Google Scholar]
- Sonnewald U, Basner A, Greve B, Steup M. A second L-type isozyme of potato glucan phosphorylase: cloning, antisense inhibition and expression analysis. Plant Mol Biol. 1995;27:567–576. doi: 10.1007/BF00019322. [DOI] [PubMed] [Google Scholar]
- Steup M. Starch degradation. In: Preiss J, editor. The Biochemistry of Plants. Vol. 14. San Diego, CA: Academic Press; 1988. pp. 255–296. [Google Scholar]
- St-Pierre B, Brisson N. Induction of the plastidic starch-phosphorylase gene in potato storage sink tissue. Planta. 1995;195:339–344. [Google Scholar]
- Takaha T, Critchley J, Okada S, Smith SM. Normal starch content and composition in tubers of antisense potato plants lacking D-enzyme (4-α-glucanotransferase) Planta. 1998;205:445–451. [Google Scholar]
- Takeo K. Affinity electrophoresis: principles and applications. Electrophoresis. 1984;5:187–195. [Google Scholar]
- Tsai CY, Nelson OE. Phosphorylases I and II of maize endosperm. Plant Physiol. 1968;43:103–112. doi: 10.1104/pp.43.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CY, Nelson OE. Two additional phosphorylases in developing maize seeds. Plant Physiol. 1969a;44:159–167. doi: 10.1104/pp.44.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CY, Nelson OE. Mutations at the shrunken-4 locus in maize that produce three altered phosphorylases. Genetics. 1969b;61:813–821. doi: 10.1093/genetics/61.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berkel J, Conrads-Strauch J, Steup M. Glucan-phosphorylase forms in cotyledons of Pisum sativum L.: localization, developmental change, in vitro translation, and processing. Planta. 1991;185:432–439. doi: 10.1007/BF00201068. [DOI] [PubMed] [Google Scholar]
- Yu Y, Mu HH, Mu-Forster C, Wasserman BP. Polypeptides of the maize amyloplast stroma: stromal localization of starch biosynthetic enzymes and identification of an 81-kilodalton amyloplast stromal heat shock cognate. Plant Physiol. 1998;116:1451–1460. doi: 10.1104/pp.116.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]