Abstract
Clostridium difficile is a major cause of hospital-associated diarrhoea, and in severe cases leads to pseudomembranous colitis and toxic megacolon. The frequency of C. difficile infection (CDI) has increased in recent decades, with 453 000 cases identified in 2011 in the USA. This is related to antibiotic-selection pressure, disruption of normal host intestinal microbiota and emergence of antibiotic-resistant C. difficile strains. The burden of community-acquired CDI has been increasingly appreciated, with disease identified in patients previously considered low-risk, such as young women or patients with no prior antibiotic exposure. C. difficile has been identified in livestock animals, meat products, seafood and salads. It has been postulated that the pool of C. difficile in the agricultural industry may contribute to human CDI. There is widespread environmental dispersal of C. difficile spores. Domestic households, turf lawns and public spaces are extensively contaminated, providing a potential reservoir for community-acquired CDI. In Australia, this is particularly associated with porcine-derived C. difficile UK PCR ribotype 014/020. In this article, the epidemiological differences between hospital- and community-acquired CDI are discussed, including some emerging evidence for community-acquired CDI being a possible zoonosis.
Keywords: Clostridium difficile, healthcare-associated infections, zoonoses
Introduction
Clostridium difficile is a major cause of healthcare-related diarrhoea, in severe cases leading to sepsis, pseudomembranous colitis, toxic megacolon and multiorgan failure. Frequency of C. difficile infection (CDI) has increased, with 453 000 US cases identified in 2011 [1, 2]. Community-acquired CDI (CACDI) is increasingly recognized, with severe disease in low-risk groups, including younger women or patients with no prior antibiotic exposure [3–6]. Clostridium difficile has been identified in livestock animals, meat products, seafood and salads. It has been postulated that this pool of C. difficile in the food and agricultural industries contributes to human CDI [7]. For this review, clinical guidelines and microbiology data were sourced from both national and international societies and regulatory bodies. The emergence of CACDI and the evidence for C. difficile being a zoonotic pathogen were derived from a literature review using the National Center for Biotechnology Information (NCBI) database of articles published in English since 1975.
Normal gut function inhibits CDI
In healthy adults, gastric acid and commensal intestinal flora are protective against pathogenic organisms. Clostridium difficile are endospore-forming, obligate anaerobic bacteria. Their vegetative forms do not survive prolonged exposure to oxygen outside the body [8]. Gastric acid (pH = 1–2) kills ingested vegetative C. difficile cells but not endospores [9, 10]. Clostridium difficile spores begin germination in the duodenum and vegetate in the terminal ileum and colon due to activation of the C. difficile serine protease bile acid receptor (CspC) by primary bile acids, particularly taurocholate [11–13]. Spore germination is also enhanced by less-acidic conditions (pH = 6), presence of phosphate, KCl ions and amino acid nutrients (L-glycine) [12]. Conjugated primary bile acids (taurocholate, glycocholate) are normally deconjugated by bacterial bile salt hydrolases (BSHs) in the small intestine by three main phyla: Firmicutes (30%), Bacteroidetes (14.4%) and Actinobacteria (8.9%). These include Clostridium, Bacteroides, Lactobacillus, Bifidobacterium and Enterococcus genera [14, 15].
Normal colonic microbiota convert the resulting unconjugated primary bile acids (cholic and chenodeoxycholic acids) to secondary bile salts (deoxycholic and lithocholic acids). This occurs by dehydroxylation and epimerization of primary bile acid 3-, 7- and 12-hydroxyl groups by bacterial bile acid hydroxysteroid dehydrogenases (HSDHs). The most important process in humans is 7α-dehydroxylation under anaerobic conditions [16]. Epimerization requires the action of both α- and β-HSDHs, which can be present in a single bacterial species or shared by two different species. Many colonic bacteria dehydrogenate unconjugated primary bile acids, but very few species perform 7α-dehydroxylation of primary bile acids, including anaerobic Clostridium and Eubacterium spp. from the Firmicutes phylum [15]. Faecal Clostridial species also epimerize chenodeoxycholic acid to ursodeoxycholic acid (UDCA). UDCA makes up 3% of the total human bile acid pool [16].
Secondary bile acids effectively inhibit spore germination and vegetation of C. difficile cells [14, 15]. Broad-spectrum antibiotics suppress normal gut flora and prevent the deconjugation of primary bile acids, changing cholate/chenodeoxycholate ratios and decreasing secondary bile acid formation [17]. This permits C. difficile proliferation and provides a survival advantage to antibiotic-resistant C. difficile strains. Use of clindamycin, fluoroquinolones, third-generation cephalosporins and penicillins predispose patients to CDI [18, 19]. CDI can occur after single-dose antibiotic prophylaxis in surgical patients [20, 21]. CDI usually starts during or shortly after antibiotic administration, but can occur up to 3 months later. Impairment of colonization resistance can still occur after narrow-spectrum antibiotic treatment for CDI, including oral vancomycin, metronidazole or fidaxomicin, contributing to persistent intestinal dysbiota, altered bile acid metabolism and recurrent CDI. This is because non-toxigenic C. difficile spp. and other beneficial anaerobes are suppressed by CDI antibiotic treatment [22]. Treatment with subinhibitory concentrations of antibiotics, particularly clindamycin and ampicillin, increase colonization factors of C. difficile. Such factors include three adhesins, Cwp66, the S-layer protein P47 and Fbp68 (2- to 10-fold increase), and a cysteine protease, Cwp84 (2- to 41-fold increase). Co-amoxiclav induces spore germination and toxin production in RT027 CD strains. Cefotaxime use is associated with increased C. difficile toxin production, as compared to piperacillin/tazobactam [23].
Different Clostridium difficile toxins affect virulence of CDI
Clostridium difficile causes diarrhoea and pseudomembranous colitis via enterotoxin A (TcdA) and cytotoxin B (TcdB) production by vegetative cells. The genes that encode for toxin A (tcdA) and toxin B (tcdB) are part of the pathogenicity locus (PaLoc), which also includes tcdR (positive regulator) and tcdC (negative regulator) genes [24]. Binding of TcdA and TcdB toxins to enterocyte receptors leads to glucosylation and inactivation of the Rho family GTPases Rho, Rac and Cdc42 [25]. This results in disruption of colonic mucosal integrity, secretory diarrhoea and acute colitis. More virulent strains of C. difficile include UK PCR ribotypes (RT) 001, 018, 027, 078 and 126 [26]. The hypervirulent RT027 strain (North American pulsed-field-type NAP1) was implicated in a severe outbreak in Quebec in 2003 during which incidence and mortality increased 5- and 3-fold, respectively [27]. The most important risk factor in the RT027 CDI epidemics in North American hospitals in 2002–06 was administration of fluoroquinolones in hospital patients. RT027 strain has higher sporulation rates, fluoroquinolone resistance, increased secretion of toxins A and B (via loss of tcdC regulator gene) and produces binary toxin (C. difficile transferase, CDT). Binary toxin production in RT027 strains is controlled by the orphan response regulator CdtR, which also up-regulates TcdA and TcdB production [28].
Other virulence factors, which are shared with epidemic or outbreak ribotype strains, allow C. difficile to adhere to host enterocytes, germinate in the presence of primary bile acids, sporulate when stressed, burrow under intestinal mucus, form biofilms, survive and adapt to host defences and adverse environmental conditions. Important virulence factors include adhesion molecules (S-layer subunits, Cwp66 protein, fibronectin-binding protein Fbp68, collagen-binding protein CbpA, lipoprotein CD0873), spore-germinant CspC, flagellar proteins (fliC, fliD), heat-shock protein GroEL, type IV fimbriae, sporulation initiator spo0A and proteases such as Cwp84 or Zmp1 [25, 29–32]. Heat-shock protein GroEL is part of the heat-shock protein (HSP) 60 family and enhances C. difficile adhesion to enterocytes in response to heat shock, acidic pH or low iron levels [33].
CDT belongs to the binary ADP-ribosylating toxin family, including C. botulinum C2 toxin, C. perfringens iota toxin, C. spiroforme toxin and the B. cereus/thuringiensis vegetative insecticidal proteins. CDT is an iota-like toxin with two components: CDTb binds to the LSR cell surface receptor on enterocytes and interacts with the enzyme component CDTa. The CDT-LSR complex is then endocytosed and the CDTa component induces depolymerization of actin tubules and destruction of the enterocyte actin cytoskeleton [34]. Translocation of CDTa into the enterocyte is dependent on intracellular helper proteins, including HSP90 [34].
Clostridium difficile subtypes have varying pathogenicity
C. difficile bacteria are divided into five clades and each is prevalent in specific continents. These include clade 1 (Europe), clade 2 (North America), clade 3 (Africa), clade 4 (Asia) and clade 5 (Australia). Human CDI mortality is closely related to clade type and binary toxin production. In a large study of human CDI from Oxfordshire, UK, the 14-day mortality was 25% in clade 5 (PCR RT078), 20% in clade 2 (PCR RT027) and 12% in clade 1 CDI (P < 0.001) [35]. Several techniques have been developed to identify different strains of C. difficile in order to study its epidemiology [36]. These include multilocus sequence typing (MLST), multilocus variable number tandem repeat analysis (MLVA), restriction endonuclease analysis (REA), pulsed-field gel electrophoresis (PFGE), repetitive-element PCR typing, toxinotyping and UK PCR ribotyping [21].
PCR ribotyping identifies ribosomal RNA genes using primers complimentary to the 16 s and 23s RNA regions. There have been 116 distinct ribotypes of C. difficile identified. This technique has been used to track CDI in the UK since 1995. PCR detection of CDT genotypes (cdtA+ and cdtB+) allows identification of binary toxin producing C. difficile strains [34]. These include UK PCR RT027, 078, 244, 126/127, 033 and 251 [37]. In contrast, PFGE analyses the whole genome using specific restriction enzymes, but is more expensive and time-consuming [36]. Toxinotyping analyses changes in the pathogenicity locus (PaLoc), with 11 toxinotypes identified (0, I–X) [38].
The epidemiology of non-RT027 binary toxin producing strains and CDI has changed dramatically since 1990. Binary toxin producing isolates were not identified before 1990 in an Italian study of human CDI, but comprised 24% (1991–99) and 45% (2000–01) in later analyses [39]. Similarly, in 2005, CDT positive strains made up 17.2% of all toxinogenic strains in 14 EU countries and, in 2008, 23% of all strains from 34 EU countries [40]. Only 5% of the isolates were RT027. Binary toxin production was associated with an increased 30-day all-cause mortality compared to patients infected with C. difficile isolates without binary toxin genes (31% vs 14%, P = 0.02), in a study from a London NHS trust in 2011 [41]. In this study, only 8% of the isolates were RT027, but binary toxin genes were detected in 28% of isolates. A 2011 Danish study found the 30-day CDI mortality was 28% for RT027 isolates, 27.8% for binary toxin non-RT027 strains and 17% for TcdA+, TcdB+, CDT-strains [42]. Non-RT027 binary toxin positive isolates included RT 078 (33%), RT 066 (36%), RT 023 and nine other ribotypes (31%) [34].
Clostridium difficile subtype strains in Australia differ
Australia has different distributions of CDI ribotypes compared to the USA and Europe. Australian hospital surveillance of CDI was mandated in 2010, which improved source tracing and epidemiology. In 2012, the ribotype proportions of CDI from Australian hospitals and private laboratories were RT014/020 (25.5%), RT002 (10.5%), RT056 (5.9%), RT070 (4.2%), RT244 (2.4%), RT027 (1.6%) and RT078 (0.9%) [37]. Ribotype 244 shares clade 2 with RT027 and produces CDT, but is fluoroquinolone-sensitive. The Australasian outbreak of RT244 in 2011–12 was predominantly community-acquired and associated with severe disease, with a 42% 30-day mortality [43, 44]. Ribotype 078 has not been found in Australian livestock, but similar CDT-producing clade-5 ribotypes 126/127, 237 and 033 have been isolated. For example, a study of Australian neonatal veal carcass contamination by C. difficile identified binary toxin positive RTs in 70.3% (71/101) of isolates; 127 (A+, B+, CDT+, 32.7%), 288 (A–, B–, CDT+, 28.7%), 033 (A–, B–, CDT+, 6.9%) and 126 (A+, B+, CDT+, 2.0%). Degree of C. difficile contamination included 66.7% (10/15) of faecal subset samples (range 2.0 × 103 to 2.3 × 106 CFU/mL, median count 2.5 × 104 CFU/mL) and in 16.7% (25/150) of carcass samples (range 3–33 CFU/cm2, median count 7 CFU/cm2) [45]. Together with clade 1 RT014 and RT056, these livestock-associated Clostridia were also isolated in Australian human CDI [37, 46, 47].
CDI is increasingly recognized in the community
Up to 75% of CDI begins in patients who are not hospitalized, including recently discharged patients, outpatients and nursing-home residents [48]. CACDI is defined as CDI in persons with no overnight stay in an inpatient healthcare facility in the 12 weeks prior to symptom onset [48]. Hospital-acquired CDI (HACDI) in contrast is defined as a positive stool CD culture result >72 hours after admission or earlier with hospital contact in the previous 4 weeks [5].
The reported relative incidence of CACDI/overall CDI varies between countries, including Singapore (13.6%), Australia (26%), Canada (27%) and the USA (20–32%) [24]. Variability may be related to underdiagnosis or differences in public health reporting. The reported 30-day mortality of CACDI is 1.3% (USA), 3.2% (Finland) and 4% (Sweden). This is lower than the observed HACDI 30-day mortality of 6.9% in Quebec before 2003 [49], 13.8% in the Quebec outbreak in 2003 [27], 9.3% in the USA in 2003 [50] and 12.7% in Finland in 2013 [51, 52]. This compares to 11% for CACDI and 8% for HACDI in a retrospective 2014 Australian study [5].
Additional risk factors for CDI and recurrent CDI susceptibility include advanced age (>65 years), severe comorbidity, immunosuppression, chemotherapy, inflammatory bowel disease or renal failure [53].
Hospital contamination with spores, symptomatic inpatient ‘super shedders’, host susceptibility and antibiotic use are recognized risk factors for HACDI [54, 55]. The majority of community-onset CDI is also related to nosocomial acquisition (onset after discharge or through frequent hospital visits/contacts) or antibiotic treatment. Up to 25% of CDI patients have no traditionally recognized risk factors [56, 57]. Patients with HACDI are usually elderly (median age 72), immunosuppressed or recently received antibiotics. When compared to HACDI, CACDI patients are more often younger (median age 50 years), likely to be female (72 vs 60%) and 27% have had no exposure to antibiotics 180 days prior to CDI diagnosis [37, 56, 58]. Antibiotic treatment is still, however, seven times more likely to produce CACDI than no prior antibiotic treatment. Clindamycin (OR 20.43), fluoroquinolones (OR 5.65), cephalosporins (OR 4.47), penicillins (OR 3.25), macrolides (OR 2.55) and sulphonamides/trimethoprim (OR 1.84) were the most-implicated antibiotic agents in a 2013 meta-analysis of eight studies and 30 184 patients with CACDI [19]. Gastric acid suppression is associated with 18% of CACDI cases, more so with proton pump inhibitors (PPIs) than H2 antagonists [24, 56]. Antibiotics and PPIs may be synergistic risk factors for CACDI, particularly with widespread community use of PPIs [59, 60]. PPIs change the gut microbiome by decreasing Bacteroidetes and increasing Firmicutes species, leading to favourable conditions for C. difficile spores to germinate and vegetate [61–63]. PPIs may also interfere with intestinal neutrophil phagocytosis and lysosome killing of C. difficile [31, 64].
Clostridium difficile spore exposure from asymptomatic carriers, food sources or the environment may be important community sources of CDI. Both symptomatic patients and asymptomatic carriers excrete C. difficile spores in high numbers. Carriage is common in neonates and infants, with up to 70% colonized with C. difficile [65] and 13% harbouring toxigenic strains [66]. Neonatal acquisition appears to be from environmental rather than maternal sources. Human infants rarely develop pseudomembranous colitis. This may be related to cellular membrane toxin receptor expression, protective factors in colostrum or neonatal gut flora [67]. Females are twice as likely to develop CACDI than males, particularly aged 15–44 years [51]. CACDI is increasingly described in peripartum women, who now comprise 1% of all cases [68]. Human infants may be an important reservoir for C. difficile excretion [66]. Contact with infants younger than 2 years of age is a significant risk factor for CACDI, which may explain the female preponderance [24, 57].
Clostridium difficile is prevalent in the environment and the food chain
The number of C. difficile spores required to cause CDI in susceptible humans is unknown, but estimated to be low (100–1000 spores) [61]. Several animal species have been used to assess C. difficile susceptibility, including rabbit, rat, guinea pig, Syrian Golden hamster, conventional and germ-free mouse and germ-free piglet models [69]. Hamsters are extremely susceptible to CDI after antibiotic administration [70, 71] with as few as 1–2 CFUs sufficient to cause enterocolitis and death [72]. Mouse and piglet models have been developed to more closely match the disease course in humans, with susceptibility to CD spores induced by antibiotic treatment or by a lack of intestinal microbiome in gnotobiotic animals. The severity of the disease in conventional mouse models is related to the size of the spore inoculum. Using RT 027 spores in mice pretreated with a single dose of clindamycin, Sun et al. showed a dose of 104 CFUs of spores caused 0% mortality and 30% diarrhoea, 105 CFUs a 30% mortality and 70% diarrhoea, and 106 CFUs a 50% mortality and 100% diarrhoea [73]. Mice which were immunosuppressed with dexamethasone were more susceptible to severe and fulminant CDI. Using a conventional mouse model with 5 days of cefoperazone pretreatment, it was shown that only 100 C. difficile spores were sufficient to consistently cause CDI in mice. The spores were heat-treated for 20 minutes at 65°C prior to oral gavage, which is designed to kill any vegetative cells but also stimulates spore germination [74].
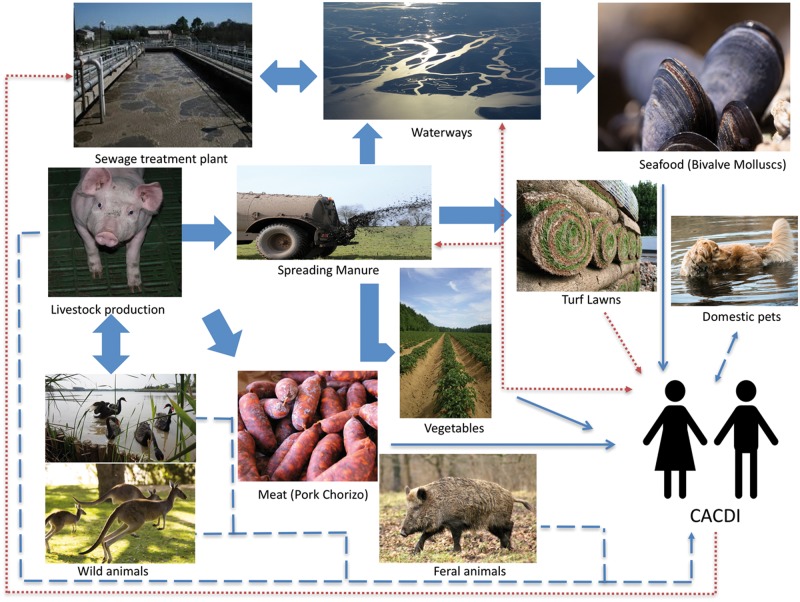
Transmission of spores between healthy asymptomatic carriers in CACDI was confirmed by PCR ribotyping and pulse field gel electrophoresis [75]. Potential modes of long-range environmental C. difficile spore dissemination include treated piggery waste water, bioeffluent, interstate stock transportation, reclaimed irrigation water, composting, biosolids, manure, turf lawns, estuarine sludge, river sediments and slaughtering of colonized pigs [61] (Figure 1). Clostridium difficile spores have been identified in soil samples from Swedish horse farms, rural parks and gardens [76], Zimbabwean farmers’ markets [77], suburban soil in South Wales [78] and Australian municipal lawns [79]. Clostridium difficile spores were identified in 59% of lawn soils in Perth, Western Australia, 39% of which were RT014. The highest viable count was 1200 CFU/g. Contamination with RT014 spores was thought to be due to turf lawns being grown with pig manure [79]. The frequency of toxigenic C. difficile spore contamination of community environs in Houston, Texas, was highest in parks (24.6%), followed by homes (17.1%), commercial shops (8.1%) and fast-food restaurants (6.5%), as compared to 16.5% positive isolates from hospitals. Spores isolated from community environmental sources were more frequently ribotypes 014/020 (21%), 002 (12%) and 078/126 (7%), as compared to RT027 (4.5%). A similar distribution of ribotypes between environmental isolates and clinical cases was found, with the exception of RT027. Hospital wards (patient bathrooms and tables) were more likely to be colonized with RT027 (32% of C. difficile-positive hospital-environment isolates) and clinical CDI cases were more likely to be caused by RT027 (24.1%). Clinical cases were patients who were hospitalized with CDI, but no data on CACDI versus HACDI was provided [80].
Figure 1.
The cycling and recycling of C. difficile from zoonotic (- - - - -), environmental (........) or food-borne (_______) sources implicated in community-associated C. difficile infection (CACDI). Adapted from Warriner et al [61].
Clostridium difficile spores are highly resistant to extreme physical or biochemical environments. They exist on hard surfaces for up to 6 months and even longer on concrete, wooden or dirt surfaces [81]. Freezing (to –80°C), heating (to 85°C), drying, ultraviolet radiation, alcohol gel and most disinfectants have proved ineffective in eradication [82–84]. Vaporized hydrogen peroxide or chlorine-based disinfectants are sporicidal. Quaternary ammonium/surfactant-based detergents are not sporicidal and may actually increase sporulation in virulent outbreak strains such as RT027 and RT001 [85]. Spores are easily spread by the faecal–oral route, contaminated hands or airborne dispersal in hospitals and nursing homes. These facilities are rapidly and extensively colonized, with spores being deposited on most surfaces and fomites [85–87].
Few studies have analysed the prevalence of C. difficile in household or community environments. Toxigenic C. difficile spores were detected in 32% of samples collected from 25/30 households in Houston, Texas. The most frequently contaminated surfaces were soles of shoes (39.7%), floor dust (33.3%) and bathroom/toilet surfaces (33.3%). Spore transfer on the soles of shoes was considered important in domestic contamination [88]. Clostridium difficile ribotypes from HACDI and CACDI were compared at two tertiary hospitals in Australia and 79% of hospital isolates had matching ribotypes in the community, suggesting transmission between the two reservoirs [89]. Whole-genome sequencing of 1250 CDI cases between 2007 and 2011 in healthcare settings and the community in Oxfordshire, UK, found 45% of cases were not due to contact exposure with symptomatic patients. Acquisition was more likely from asymptomatic carriers or non-hospital environmental sources [90]. In Europe and North America, HACDI is more likely to be associated with C. difficile RT027 and RT001, and CACDI more diverse ribotypes, including livestock-associated RT078 strain [51, 54, 91, 92]. It has been postulated that, in Australia, the emergence of livestock-associated C. difficile CDT producing ribotypes 127, 126 and 033 may parallel that of Northern Hemisphere RT078 strains in the pathogenesis of human CACDI [93].
In Australia, 20% of vegetables grown in enriched soils are contaminated with C. difficile spores. Vegetable spore contamination rates include carrots (5%), onions (6%), beetroots (22%) and potatoes (50%) [94]. Prevalence was lower in the USA (0% root vegetables, 2.4% other vegetables) [95] and Canada (4.5%) [96]. Clostridium difficile has also been identified in 7.5% of ready-to-eat salads in Scotland [97]. Of C. difficile spores isolated from river sediment in Ontario, Canada, 92% were toxigenic [98]. RT078 spores survived municipal water treatment for domestic housing in Ontario [99]. River and estuarine sediments containing C. difficile spores have been implicated in seafood contamination, including bottom-dwelling molluscs harbouring RT078/126 (22.2%), 010 (19.4%) and 001 (8.3%) in Italy. Filter feeders such as oysters, mussels and clams bioaccumulate pathogens of animal and human origin. However, in the Troiano and Montazeri studies, C. difficile contamination of molluscs was not correlated with indicators of human faecal pollution [100, 101] (Table 1).
Table 1.
Incidence and ribotypes of environmental C. difficile
| Source | Location | Proportion contaminated (%) | Main ribotypes (%) | Reference |
|---|---|---|---|---|
| Ground beef (uncooked) | Arizona, USA | 13/26 (50) | 027 (11.5)078 (30.8) | [102] |
| Beef Sausage (cooked) | Arizona, USA | 1/7 (14.3) | 027 (14.3) | [102] |
| Ground beef | Ontario/Quebec, Canada | 11/53 (20.8) | 077, M31, 014, M26 | [103] |
| Ground beef | Canada | 10/149 (6.7) | M26, 077, J, 014, C, F, H | [104] |
| Ground beef | Canada | 14/115 (12.2) | 027, 078, C | [105] |
| Ground pork (uncooked) | Arizona, USA | 3/7 (42.9) | 027 (14.3)078 (28.6) | [80] |
| Braunschweiger (cooked) | Arizona, USA | 10/16 (62.5) | 027 (18.8)078 (43.8) | [102] |
| Ground pork | Canada | 14/115 (12.2) | 027, 078, C, E, Y | [105] |
| Pork sausage (uncooked) | Arizona, USA | 3/13 (23.1) | 027 (7.7) 078 (15.4) | [102] |
| Chorizo (uncooked) | Arizona, USA | 3/10 (30) | 027 (10)078 (20) | [102] |
| Chicken | Ontario, Canada | 26/203 (12.8) | 078 (12.8) | [106] |
| Ground Turkey (uncooked) | Arizona, USA | 4/9 (44.4) | 078 (44.4) | [102] |
| Molluscs | Italy | 36/925 (3.9) | 078/126 (22.2, 8/36)010 (19.4, 7/36)001 (8.3, 3/36) | [100] |
| Oysters | Louisiana, USA | 9/19 (47.4) | tcdB positive (100, 9/9) | [101] |
| Raw vegetables | Canada | 5/111 (4.5) | 078 | [96] |
| Raw vegetables | Australia | 14/71 (19.7) | – | [94] |
| Salads | Scotland | 3/40 (7.5) | 017, 001 | [97] |
| Shoe soles | Texas, USA | 25/63 (39.7) | 001, 002, UM-8 | [88] |
| Lawns | Australia, Perth | 182/311 (58.5) | 014/020 (39) | [79] |
The contribution of C. difficile spore contamination of food in the development of human CDI remains to be established. This is because, after ingestion, spore germination and vegetation are normally inhibited by an intact gut microbiome, with CDI occurring only in susceptible individuals. Clostridium difficile spores have been identified in North American retail meat products, including chicken (12.5%), turkey (44.4%), ground beef (50%), ground pork (43%) and Braunschweiger pork sausage (63%) [61, 106, 107] (Table 1). In the USA, these were predominantly RT078 (75%), but some were RT027 strain. Viable spores were found in both ready-to-eat, cooked meats and uncooked meats, although absolute spore counts were low. Twenty to 60 spores per gram were identified by direct culture of ground pork, 20–240 spores per gram from ground beef [105] and <100 CFU/g in chicken meat [106]—counts sufficient to cause disease in susceptible hosts [61, 74, 108].
The growth of C. difficile bacteria (as opposed to spores) is rare in foods because germination and vegetation of spores require primary bile salts and neutral to alkaline conditions (pH = 5.5–9.0) [12]. No germination of RT027 or RT078 spores occurred in beef or fish extracts without the addition of sodium taurocholate [61]. Clostridium difficile bacteria are heterotrophic obligate anaerobes, and require intestinal fermentation of organic substrates such as amino acids to produce ATP. The presence of amino acids such as glycine or histidine enhance cholate-induced germination after spore ingestion [11, 12, 109]. This differs from microaerophilic Campylobacter jejuni or facultative anaerobic Salmonella enterica bacteria, which can grow in contaminated food, making source tracing of CDI challenging.
Clostridium difficile spores are resilient
The persistence of C. difficile spores in cooked meat demonstrates their survival ability in adverse environments [82, 110]. Clostridium difficile spores have several lamellations of the spore coat contributing to resilience [109]. RT078 is particularly heat-resistant compared to other ribotypes, including RT027 [111]. Clostridium difficile spores can survive cooking to a core temperature of 74°C and RT078 up to 96°C. Heat selection may explain the recent emergence of RT078 strain CDI in humans in the USA and Europe. Heating meat to 63 and 71°C (minimal recommended temperatures for cooking seafood and hamburgers, respectively) eliminates all vegetative microbiota but increases subsequent C. difficile spore germination by 30%. Sublethal cooking temperatures in modern food preparation instead of traditional methods such as pressure cooking or boiling may induce heat-shock protein expression in C. difficile RT078 and thence antibiotic resistance and virulence pathogenicity genes [111].
Clostridium difficile carriage and antibiotic resistance in domestic farm animals
Domestic farm animals have varying levels of symptomatic and asymptomatic C. difficile carriage. Neonatal animals are much more likely to be affected than adult animals. Carriage rates in a Texas, USA, piggery were 50.0% (61/122) in suckling pigs, followed by 23.8% (34/143) in lactating sows [112]. All isolates were positive for binary toxin gene and 93% were tcdA+ and tcdB+. In a 2009 Spanish study, 26% (140/541) of newborn piglets were found to have C. difficile on rectal swabs and 94% (132/140) were toxigenic strains (tcdA+, tcdB+) [113]. This included animals with and without diarrhoea, and animals from control farms without diarrhoea. Older piglets (1–2 months old) did not show any carriage of C. difficile. Seemingly healthy piglets that tested positive for C. difficile still had classic features of acute colitis on histology, suggesting a subclinical course in some animals [114]. In a Dutch study of C. difficile acquisition, all caesarean-section-derived piglets were C. difficile-negative, but were rapidly colonized with RT078 strain within 48 hours [115].
Transmission was thought to be from lactating sows or from the farm environment. In a Belgian study of C. difficile prevalence in beef cattle farms, there was a higher colonization rate of calves less than 6 months of age versus older calves >11 months old [116]. Some studies have suggested that intensive farming of pigs and cattle increases the carriage of toxigenic C. difficile, particularly with antibiotic use in lactating animals with mastitis or in suckling neonatal calves and piglets [61, 113, 114, 117]. However, other studies of C. difficile in pig herds have found no differences in overall carriage rates in pigs from conventional or organic farms [118]. Farm system, size or the presence of other animal species on the farm did not result in statistically significant differences in the C. difficile carriage rate.
Agricultural antibiotic use may influence Clostridium difficile strains and pathogenicity in humans
The emergence of CACDI may be related to zoonotic transmission. Clostridium difficile colonizes many domestic and wild animals, including cats, dogs, horses, pigs, calves, poultry, goats, rats, rabbits, raccoons, kangaroos, feral swine, birds, elephants, ostriches and Kodiak bears [61, 119–121] (Figure 1). Juvenile animals are most commonly affected, including 20% of beef calves and 90% of piglets [122]. Ribotype 027 has been isolated in livestock in the USA, but RT078 appears to predominate, particularly in poultry, calves and piglets [123]. Ribotype 078 is the most common cause of CACDI in the Northern Hemisphere [120].
Antibiotics are widely used in the agricultural industry as ‘growth promoters’. In the USA, 80% of all antibiotics are used in agriculture, 70% of which are considered ‘medically important’, i.e. also used in human medical therapy. In 2011, over 26 tons of cephalosporins were used in the US agricultural industry [124]. Because of the risk of antibiotic-selection pressure leading to emergence of multiresistant organisms and vertical transmission, attempts have been made to improve antibiotic stewardship in the agricultural industry. In 2012, the US Food and Drug Administration (FDA) issued an order to prohibit ‘extra-label’ use of cephalosporins. However, ‘approved’ indications allow continued widespread use. The 2006 ban on antibiotic use in EU countries for livestock growth promotion did not lead to a decrease in antibiotic consumption [125].
Toxigenic C. difficile have developed high levels of resistance to beta lactams, macrolides, tetracyclines and aminoglycosides. This is due to antibiotic selection pressure, gene mutations and acquisition from other gut bacteria via conjugative and mobilizable transposons and bacteriophages [31]. The off-label use of ceftiofur, a third-generation cephalosporin, for the treatment and prevention of post-weaning diarrhoea in pigs caused by enterotoxigenic Escherichia coli, represented the biggest risk to public health faced by the Australian pork industry in 2014 [126]. This is because ceftiofur is analogous to ceftriaxone and cefotaxime, which are used in human medicine. Ceftiofur use can drive the amplification and transmission of multiresistant organisms such as C. difficile in intensive animal farming. Up to 25% of large piggeries in Australia use off-label ceftiofur in their herds. Ceftiofur is also registered for use for respiratory infections in cattle in Australia [127]. High carriage levels of binary toxin producing C. difficile ribotypes have been isolated in neonatal piglets and veal calves in Australia, which may provide a reservoir for vertical transmission to humans. In veal calves, CD was isolated in 53% and, of these CDI strains, 76% were CDT producing, including ribotypes 127, 126 and 033 [93].
The One Health Commission began in 2007 as a collaboration between the American Medical and Veterinary Associations. It aims to achieve optimal health outcomes, recognizing the interconnection between people, animals, plants and their shared environment. This has led to further initiatives to improve antibiotic stewardship [128].
Many human enteric infections have a zoonotic origin linked to agricultural antibiotic use. The emergence of fluoroquinolone resistance in human Campylobacter jejuni and Salmonella typhimurium-definitive phage type104 gastroenteritis was associated with the use of fluoroquinolones (enrofloxacin) in the poultry industry since 1993. The US FDA withdrew fluoroquinolones in poultry production in 2005. Their use in poultry production is banned in Nordic European countries and Australia. The UK and most EU countries continue to allow their use in turkey, duck, geese and chicken meat production, with ciprofloxacin resistance rates in human Campylobacter infections as high as 97.9% in Portugal and 84.7% in Spain and Lithuania in 2014 [129]. The average rate of ciprofloxacin resistance in human Campylobacter infections in EU countries was 60% (2014 data), compared to 22.3% in the USA (2013 data), 14% in Sweden (2014 data), 11.6% in Finland (2012 data) and 2% in Australia (2001–02 data) [130]. Treatment failures, higher hospitalization rates and a 2-fold increase in mortality were observed in patients with fluoroquinolone-resistant S. typhimurium DT104 infections. Fluoroquinolones have never been approved in Australia for use in the meat and livestock industry [129–131]. The conservative use of fluoroquinolones in agriculture and clinical medicine may explain the lack of domestic emergence of fluoroquinolone-resistant C. difficile RT027 in Australia [37].
Antibiotic selection of resistant organisms and their transfer from livestock to farm workers have been widely demonstrated. A 1997 Netherlands study reported higher carriage of vancomycin resistant enterocooci in turkeys (50%) treated with avoparcin (a vancomycin-like growth promoter) and their farmers (39%) compared to local residents (14%) [131, 132]. Avoparcin was banned as a growth promoter in animal feeds in Europe in 1999. Tetracycline-resistant E. coli was identified in both chickens and chicken farmers in 1975, when tetracyclines were used for prophylaxis and growth promotion [133]. Tetracycline was banned as a growth promoter in food animal production in EU countries in 2006, due to the emergence of tetracycline-resistant S. typhimurium and E. coli. Whole-genome analysis of C. difficile RT078 strains in the Netherlands from 2002 to 2011 found identical strains were shared between pigs and pig farmers, indicating transmission between the two groups [91]. These included identical streptomycin- and tetracycline-resistance determinants. Other non-clonal strains suggested alternative reservoirs for the community spread of RT078, including wild animals and environmental sources [91]. There is genomic evidence that RT078 and 027 porcine strains are similar to strains isolated from human CDI, indicating interspecies transmission [120].
Whole-genome and proteome analysis by Knight et al. demonstrated substantial similarities between human and porcine strains of C. difficile (RT014/NAP4) in Australia [94]. Specific CDI strains can also be traced through pan-genome analysis and characterization of antibiotic resistance, prophage content and in silico virulence potential [134]. This enables source origin, vector patterns, CDI risk factors and modes of environmental contamination to be established [120]. RT014/020 is the most prevalent ribotype in Australia, accounting for 24% of human CDI cases. It is also the most common ribotype in neonatal pigs, found in 23% of isolates collected thousands of kilometres and many months apart in Australia. Interspecies transmission was substantiated in 42% of human strains and 37.5% of porcine strains based on single nucleotide variant analysis. Of these interspecies clonal groups, 50% of human strains were classified as CACDI. This suggests that, in Australia, porcine C. difficile can be transferred to the human population and cause CACDI.
Conclusion
There is accumulating evidence of a persistent community reservoir of C. difficile. This involves spore contamination of soil, water, food, households, shoes, lawns and public spaces. The mechanism of this reservoir leading to disease in susceptible hosts requires further exploration. Clostridium difficile genome analysis and differing demographic patterns between CACDI and HACDI suggest a zoonotic origin in Australian CACDI, particularly porcine-derived RT014/020. This may be driven by antibiotic use in the agricultural industry. The importance of One Health initiatives, antibiotic stewardship and source control in human and veterinarian medicine as well as the agricultural industry is emphasized.
Conflict of interest statement: none declared.
References
- 1. McDonald LC, Owings M, Jernigan DB.. Clostridium difficile infection in patients discharged from US short-stay hospitals, 1996–2003. Emerg Infect Dis 2006;12:409–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lessa FC, Mu Y, Bamberg WM. et al. Burden of Clostridium difficile infection in the United States. N Engl J Med 2015;372:825–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Centers for Disease Control and Prevention (CDC). Severe Clostridium difficile-associated disease in populations previously at low risk--four states, 2005. Morb Mortal Wkly Rep 2005;54:1201–5. [PubMed] [Google Scholar]
- 4. Nanwa N, Sander B, Krahn M. et al. A population-based matched cohort study examining the mortality and costs of patients with community-onset Clostridium difficile infection identified using emergency department visits and hospital admissions. PLoS One 2017;12:e0172410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clohessy P, Merif J, Post JJ.. Severity and frequency of community-onset Clostridium difficile infection on an Australian tertiary referral hospital campus. Int J Infect Dis 2014;29:152–5. [DOI] [PubMed] [Google Scholar]
- 6. Anderson DJ, Rojas LF, Watson S. et al. Identification of novel risk factors for community-acquired Clostridium difficile infection using spatial statistics and geographic information system analyses. PLoS One 2017;12:e0176285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hensgens MPM, Keessen EC, Squire MM. et al. Clostridium difficile infection in the community: a zoonotic disease? Clin Microbiol Infect 2012;18:635–45. [DOI] [PubMed] [Google Scholar]
- 8. Holý O, Chmelař D.. Oxygen tolerance in anaerobic pathogenic bacteria. Folia Microbiol 2012;57:443–6. [DOI] [PubMed] [Google Scholar]
- 9. Jump RL, Pultz MJ, Donskey CJ.. Vegetative Clostridium difficile survives in room air on moist surfaces and in gastric contents with reduced acidity: a potential mechanism to explain the association between proton pump inhibitors and C. difficile-associated diarrhea? Antimicrob Agents Chemother 2007;51:2883–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rao A, Jump RLP, Pultz NJ. et al. In vitro killing of nosocomial pathogens by acid and acidified nitrite. Antimicrob Agents Chemother 2006;50:3901–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wheeldon LJ, Worthington T, Lambert PA.. Histidine acts as a co-germinant with glycine and taurocholate for Clostridium difficile spores. J Appl Microbiol 2011;110:987–94. [DOI] [PubMed] [Google Scholar]
- 12. Paredes-Sabja D, Bond C, Carman RJ. et al. Germination of spores of Clostridium difficile strains, including isolates from a hospital outbreak of Clostridium difficile-associated disease (CDAD). Microbiology (Reading, Engl) 2008;154:2241–50. [DOI] [PubMed] [Google Scholar]
- 13. Weingarden AR, Dosa PI, DeWinter E. et al. Changes in colonic bile acid composition following fecal microbiota transplantation are sufficient to control Clostridium difficile germination and growth. PLoS One 2016;11:e0147210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barrett KE, Barman SM, Boitano S. et al. Ganong’s Review of Medical Physiology. New York: McGraw-Hill Medical, 2010. [Google Scholar]
- 15. Winston JA, Theriot CM.. Impact of microbial derived secondary bile acids on colonization resistance against Clostridium difficile in the gastrointestinal tract. Anaerobe 2016;41:44–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gérard P. Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens 2013;3:14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sun X, Hirota SA.. The roles of host and pathogen factors and the innate immune response in the pathogenesis of Clostridium difficile infection. Mol Immunol 2015;63:193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leffler DA, Lamont JT.. Clostridium difficile infection. N Engl J Med 2015;372:1539–48. [DOI] [PubMed] [Google Scholar]
- 19. Deshpande A, Pasupuleti V, Thota P. et al. Community-associated Clostridium difficile infection and antibiotics: a meta-analysis. J Antimicrob Chemother 2013;68:1951–61. [DOI] [PubMed] [Google Scholar]
- 20. Privitera G, Scarpellini P, Ortisi G. et al. Prospective study of Clostridium difficile intestinal colonization and disease following single-dose antibiotic prophylaxis in surgery. Antimicrob Agents Chemother 1991;35:208–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sartelli M, Malangoni MA, Abu-Zidan FM. et al. WSES guidelines for management of Clostridium difficile infection in surgical patients. World J Emerg Surg 2015;10:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Almeida R, Gerbaba T, Petrof EO.. Recurrent Clostridium difficile infection and the microbiome. J Gastroenterol 2016;51:1–10. [DOI] [PubMed] [Google Scholar]
- 23. Deneve C, Delomenie C, Barc MC. et al. Antibiotics involved in Clostridium difficile-associated disease increase colonization factor gene expression. J Med Microbiol 2008;57:732–8. [DOI] [PubMed] [Google Scholar]
- 24. Bloomfield LE, Riley TV.. Epidemiology and risk Factors for community-associated Clostridium difficile infection: a narrative review. Infect Dis Ther 2016;5:231–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Janoir C. Virulence factors of Clostridium difficile and their role during infection. Anaerobe 2016;37:13–24. [DOI] [PubMed] [Google Scholar]
- 26. Vindigni SM, Surawicz CM.. C. difficile infection: changing epidemiology and management paradigms. Clin Trans Gastroenterol 2015;6:e99.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pépin J, Valiquette L, Alary ME. et al. Clostridium difficile-associated diarrhea in a region of Quebec from 1991 to 2003: a changing pattern of disease severity. CMAJ 2004;171:466–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lyon SA, Hutton ML, Rood JI. et al. CdtR regulates TcdA and TcdB production in Clostridium difficile. PLoS Pathog 2016;12:e1005758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kansau I, Barketi-Klai A, Monot M. et al. Deciphering adaptation strategies of the epidemic Clostridium difficile 027 strain during Infection through in vivo transcriptional analysis. PLoS One 2016;11:e0158204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vohra P, Poxton IR.. Comparison of toxin and spore production in clinically relevant strains of Clostridium difficile. Microbiology 2011;157:1343–53. [DOI] [PubMed] [Google Scholar]
- 31. Johanesen PA, Mackin KE, Hutton ML. et al. Disruption of the gut microbiome: Clostridium difficile infection and the threat of antibiotic resistance. Genes 2015;6:1347–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Barketi-Klai A, Monot M, Hoys S. et al. The Flagellin FliC of Clostridium difficile is responsible for pleiotropic gene regulation during in vivo infection. PLoS One 2014;9:e96876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kirk JA, Banerji O, Fagan RP.. Characteristics of the Clostridium difficile cell envelope and its importance in therapeutics. Microb Biotechnol 2017;10:76–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gerding DN, Johnson S, Rupnik M. et al. Clostridium difficile binary toxin CDT. Gut Microbes 2014;5:15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Walker AS, Eyre DW, Wyllie DH. et al. Relationship between bacterial strain type, host biomarkers, and mortality in Clostridium difficile infection. Clin Infect Dis 2013;56:1589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brazier JS. Typing of Clostridium difficile. Clin Microbiol Infect 2001;7:428–31. [DOI] [PubMed] [Google Scholar]
- 37. Collins DA, Putsathit P, Elliott B. et al. Laboratory-based surveillance of Clostridium difficile strains circulating in the Australian healthcare setting in 2012. Pathology 2017;49:309–13. [DOI] [PubMed] [Google Scholar]
- 38. Rupnik M, Avesani V, Janc M. et al. A novel toxinotyping scheme and correlation of toxinotypes with serogroups of Clostridium difficile isolates. J Clin Microbiol 1998;36:2240–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Spigaglia P, Mastrantonio P.. Comparative analysis of Clostridium difficile clinical isolates belonging to different genetic lineages and time periods. J Med Microbiol 2004;53:1129–36. [DOI] [PubMed] [Google Scholar]
- 40. Barbut F, Mastrantonio P, Delmée M. et al. Prospective study of Clostridium difficile infections in Europe with phenotypic and genotypic characterisation of the isolates. Clin Microbiol Infect 2007;13:1048–57. [DOI] [PubMed] [Google Scholar]
- 41. Goldenberg SD, French GL.. Lack of association of tcdC type and binary toxin status with disease severity and outcome in toxigenic Clostridium difficile. J Infect 2011;62:355–62. [DOI] [PubMed] [Google Scholar]
- 42. Bacci S, Mølbak K, Kjeldsen MK. et al. Binary toxin and death after Clostridium difficile infection. Emerg Infect Dis 2011;17:976–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Eyre DW, Tracey L, Elliott B. et al. Emergence and spread of predominantly community-onset Clostridium difficile PCR ribotype 244 infection in Australia, 2010 to 2012. Euro Surveill 2015;20:21059. [DOI] [PubMed] [Google Scholar]
- 44. Huber CA, Hall L, Foster NF. et al. Surveillance snapshot of Clostridium difficile infection in hospitals across Queensland detects binary toxin producing ribotype UK 244. Commun Dis Intell Q Rep 2014;38:E279–84. [PubMed] [Google Scholar]
- 45. Knight DR, Putsathit P, Elliott B. et al. Contamination of Australian newborn calf carcasses at slaughter with Clostridium difficile. Clin Microbiol Infect 2016;22:266.e1–7. [DOI] [PubMed] [Google Scholar]
- 46. Mc Govern AM, Foster NF, Pereira LA. et al. Human Clostridium difficile infection caused by a livestock-associated PCR ribotype 237 strain in Western Australia. JMM Case Rep 2016;3:e005062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Elliott B, Dingle KE, Didelot X. et al. The complexity and diversity of the pathogenicity locus in Clostridium difficile clade 5. Genome Biol Evol 2014;6:3159–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cohen SH, Gerding D, Johnson S. et al. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA). Infect Control Hosp Epidemiol 2010;31:431–55. [DOI] [PubMed] [Google Scholar]
- 49. Loo VG, Poirier L, Miller MA. et al. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N Engl J Med 2005;353:2442–9. [DOI] [PubMed] [Google Scholar]
- 50. Ricciardi R, Rothenberger DA, Madoff RD. et al. Increasing prevalence and severity of Clostridium difficile colitis in hospitalized patients in the United States. Arch Surg 2007;142:624–31. [DOI] [PubMed] [Google Scholar]
- 51. Kotila SM, Mentula S, Ollgren J. et al. Community- and healthcare-associated Clostridium difficile infections, Finland, 2008 − 2013. Emerg Infect Dis 2016;22:1747–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mitchell BG, Gardner A.. Mortality and Clostridium difficile infection: a review. Antimicrob Resist Infect Control 2012;1:20.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bignardi GE. Risk factors for Clostridium difficile infection. J Hosp Infect 1998;40:1–15. [DOI] [PubMed] [Google Scholar]
- 54. Kumar N, Miyajima F, He M. et al. Genome-based infection tracking reveals dynamics of Clostridium difficile transmission and disease recurrence. Clin Infect Dis 2016;62:746–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lawley TD, Clare S, Walker AW. et al. Antibiotic treatment of clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect Immun 2009;77:3661–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kuntz JL, Chrischilles EA, Pendergast JF. et al. Incidence of and risk factors for community-associated Clostridium difficile infection: a nested case-control study. BMC Infect Dis 2011;11:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gupta A, Khanna S.. Community-acquired Clostridium difficile infection: an increasing public health threat. Infect Drug Resist 2014;7:63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. DePestel DD, Aronoff DM.. Epidemiology of Clostridium difficile infection. J Pharm Pract 2013;26:464–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Freeman J, Bauer MP, Baines SD. et al. The changing epidemiology of Clostridium difficile infections. Clin Microbiol Rev 2010;23:529–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mezoff EA, Cohen MB.. Acid suppression and the risk of Clostridium difficile infection. J Pediatr 2013;163:627–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Warriner K, Xu C, Habash M. et al. Dissemination of Clostridium difficile in food and the environment: significant sources of C. difficile community-acquired infection? J Appl Microbiol 2017;122:542–53. [DOI] [PubMed] [Google Scholar]
- 62. Arriola V, Tischendorf J, Musuuza J. et al. Assessing the risk of hospital-acquired Clostridium difficile infection with proton pump inhibitor use: a meta-analysis. Infect Control Hosp Epidemiol 2016;37:1408–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tariq R, Singh S, Gupta A. et al. Association of gastric acid suppression with recurrent Clostridium difficile infection: a systematic review and meta-analysis. JAMA Intern Med 2017;177:784–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Paredes-Sabja D, Cofre-Araneda G, Brito-Silva C. et al. Clostridium difficile spore-macrophage interactions: spore survival. PLoS One 2012;7:e43635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yamamoto-Osaki T, Kamiya S, Sawamura S. et al. Growth inhibition of Clostridium difficile by intestinal flora of infant faeces in continuous flow culture. J Med Microbiol 1994;40:179–87. [DOI] [PubMed] [Google Scholar]
- 66. Rousseau C, Poilane I, De Pontual L. et al. Clostridium difficile carriage in healthy infants in the community: a potential reservoir for pathogenic strains. Clin Infect Dis 2012;55:1209–15. [DOI] [PubMed] [Google Scholar]
- 67. Jangi S, Lamont JT.. Asymptomatic colonization by Clostridium difficile in infants: implications for disease in later life. J Pediatr Gastroenterol Nutr 2010;51:2–7. [DOI] [PubMed] [Google Scholar]
- 68. Lessa FC. Community-associated Clostridium difficile infection: how real is it? Anaerobe 2013;24:121–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chen X, Katchar K, Goldsmith JD. et al. A mouse model of Clostridium difficile-associated disease. Gastroenterology 2008;135:1984–92. [DOI] [PubMed] [Google Scholar]
- 70. Price AB, Larson HE, Crow J.. Morphology of experimental antibiotic-associated enterocolitis in the hamster: a model for human pseudomembranous colitis and antibiotic-associated diarrhoea. Gut 1979;20:467–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sambol SP, Tang JK, Merrigan MM. et al. Infection of hamsters with epidemiologically important strains of Clostridium difficile. J Infect Dis 2001;183:1760–6. [DOI] [PubMed] [Google Scholar]
- 72. Larson HE, Borriello SP.. Quantitative study of antibiotic-induced susceptibility to Clostridium difficile enterocecitis in hamsters. Antimicrob Agents Chemother 1990;34:1348–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sun X, Wang H, Zhang Y. et al. Mouse relapse model of Clostridium difficile infection. Infect Immun 2011;79:2856–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Koenigsknecht MJ, Theriot CM, Bergin IL. et al. Dynamics and establishment of Clostridium difficile infection in the murine gastrointestinal tract. Infect Immun 2015;83:934–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kato H, Kita H, Karasawa T. et al. Colonisation and transmission of Clostridium difficile in healthy individuals examined by PCR ribotyping and pulsed-field gel electrophoresis. J Med Microbiol 2001;50:720–7. [DOI] [PubMed] [Google Scholar]
- 76. Båverud V, Gustafsson A, Franklin A. et al. Clostridium difficile: prevalence in horses and environment, and antimicrobial susceptibility. Equine Vet J 2010;35:465–71. [DOI] [PubMed] [Google Scholar]
- 77. Simango C, Mwakurudza S.. Clostridium difficile in broiler chickens sold at market places in Zimbabwe and their antimicrobial susceptibility. Int J Food Microbiol 2008;124:268–70. [DOI] [PubMed] [Google Scholar]
- 78. Al Saif N, Brazier JS.. The distribution of Clostridium difficile in the environment of South Wales. J Med Microbiol 1996;45:133–7. [DOI] [PubMed] [Google Scholar]
- 79. Moono P, Lim SC, Riley TV.. High prevalence of toxigenic Clostridium difficile in public space lawns in Western Australia. Sci Rep 2017;7:41196.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Alam MJ, Walk ST, Endres BT. et al. Community environmental contamination of toxigenic Clostridium difficile. Open Forum Infect Dis 2017;4:ofx018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kaatz GW, Gitlin SD, Schaberg DR. et al. Acquisition of Clostridium difficile from the hospital environment. Am J Epidemiol 1988;127:1289–94. [DOI] [PubMed] [Google Scholar]
- 82. Deng K, Plaza-Garrido A, Torres JA. et al. Survival of Clostridium difficile spores at low temperatures. Food Microbiol 2015;46:218–21. [DOI] [PubMed] [Google Scholar]
- 83. Edwards AN, Karim ST, Pascual RA. et al. Chemical and stress resistances of Clostridium difficile spores and vegetative cells. Front Microbiol 2016;7:97–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Connor M, Flynn PB, Fairley DJ. et al. Evolutionary clade affects resistance of Clostridium difficile spores to cold atmospheric plasma. Sci Rep 2017;7:41814–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Gerding DN, Muto CA, Owens RC.. Measures to control and prevent Clostridium difficile infection. Clin Infect Dis 2008;46:S43–9. [DOI] [PubMed] [Google Scholar]
- 86. Best EL, Fawley WN, Parnell P. et al. The potential for airborne dispersal of Clostridium difficile from symptomatic patients. Clin Infect Dis 2010;50:1450–7. [DOI] [PubMed] [Google Scholar]
- 87. Donskey CJ. Preventing transmission of Clostridium difficile: is the answer blowing in the wind? Clin Infect Dis 2010;50:1458–61. [DOI] [PubMed] [Google Scholar]
- 88. Alam MJ, Anu A, Walk ST. et al. Investigation of potentially pathogenic Clostridium difficile contamination in household environs. Anaerobe 2014;27:31–3. [DOI] [PubMed] [Google Scholar]
- 89. Furuya-Kanamori L, Riley TV, Paterson DL. et al. Comparison of Clostridium difficile ribotypes circulating in Australian hospitals and communities. J Clin Microbiol 2017;55:216–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Eyre DW, Cule ML, Wilson DJ. et al. Diverse sources of C. difficile infection identified on whole-genome sequencing. N Engl J Med 2013;369:1195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Knetsch CW, Connor TR, Mutreja A. et al. Whole genome sequencing reveals potential spread of Clostridium difficile between humans and farm animals in the Netherlands, 2002 to 2011. Euro Surveill 2014;19:20954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Davies KA, Ashwin H, Longshaw CM. et al. Diversity of Clostridium difficile PCR ribotypes in Europe: results from the European, multicentre, prospective, biannual, point-prevalence study of Clostridium difficile infection in hospitalised patients with diarrhoea (EUCLID), 2012 and 2013. Euro Surveill 2016;21:30294. [DOI] [PubMed] [Google Scholar]
- 93. Knight DR, Thean S, Putsathit P. et al. Cross-sectional study reveals high prevalence of Clostridium difficile non-PCR ribotype 078 strains in Australian veal calves at slaughter. Appl Environ Microbiol 2013;79:2630–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Knight DR, Squire MM, Collins DA. et al. Genome analysis of Clostridium difficile PCR ribotype 014 lineage in Australian pigs and humans reveals a diverse genetic repertoire and signatures of long-range interspecies transmission. Front Microbiol 2017;7:2716–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Rodriguez-Palacios A, Ilic S, LeJeune JT.. Clostridium difficile with moxifloxacin/clindamycin resistance in vegetables in Ohio, USA, and prevalence meta-analysis. J Pathog 2014;2014:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Metcalf DS, Costa MC, Dew WM. et al. Clostridium difficile in vegetables, Canada. Lett Appl Microbiol 2010;51:600–2. [DOI] [PubMed] [Google Scholar]
- 97. Bakri MM, Brown DJ, Butcher JP. et al. Clostridium difficile in ready-to-eat salads, Scotland. Emerg Infect Dis 2009;15:817–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Xu C, Weese JS, Flemming C. et al. Fate of Clostridium difficile during waste water treatment and incidence in Southern Ontario watersheds. J Appl Microbiol 2014;117:891–904. [DOI] [PubMed] [Google Scholar]
- 99. Bazaid F. Distribution and sources of Clostridium difficile present in water sources. MSc thesis, University of Guelph 2013.
- 100. Troiano T, Harmanus C, Sanders IM. et al. Toxigenic Clostridium difficile PCR ribotypes in edible marine bivalve molluscs in Italy. Int J Food Microbiol 2015;208:30–4. [DOI] [PubMed] [Google Scholar]
- 101. Montazeri N, Liu D, Janes ME.. Occurrence of toxigenic Clostridium difficile in Louisiana Oysters (Crassostrea virginica) and environmental waters. Fns 2015;06:6: 1065–70. [Google Scholar]
- 102. Songer JG, Trinh HT, Killgore GE. et al. Clostridium difficile in retail meat products, USA, 2007. Emerg Infect Dis 2009;15:819–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Rodriguez-Palacios A, Staempfli HR, Duffield T. et al. Clostridium difficile in retail ground meat, Canada. Emerg Infect Dis 2007;13:485–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Rodriguez-Palacios A, Reid-Smith RJ, Staempfli HR. et al. Possible seasonality of Clostridium difficile in retail meat, Canada. Emerg Infect Dis 2009;15:802–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Weese JS, Avery BP, Rousseau J. et al. Detection and enumeration of Clostridium difficile spores in retail beef and pork. Appl Environ Microbiol 2009;75:5009–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Weese JS, Reid-Smith RJ, Avery BP. et al. Detection and characterization of Clostridium difficile in retail chicken. Lett Appl Microbiol 2010;50:362–5. [DOI] [PubMed] [Google Scholar]
- 107. Gould LH, Limbago B.. Clostridium difficile in food and domestic animals: a new foodborne pathogen? Clin Infect Dis 2010;51:577–82. [DOI] [PubMed] [Google Scholar]
- 108. Lawley TD, Croucher NJ, Yu L. et al. Proteomic and genomic characterization of highly infectious Clostridium difficile 630 spores. J Bacteriol 2009;191:5377–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Paredes-Sabja D, Shen A, Sorg JA.. Clostridium difficile spore biology: sporulation, germination, and spore structural proteins. Trends Microbiol 2014;22:406–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Rodriguez-Palacios A, LeJeune JT.. Moist-heat resistance, spore aging, and superdormancy in Clostridium difficile. Appl Environ Microbiol 2011;77:3085–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Rodriguez-Palacios A, Ilic S, LeJeune JT.. Subboiling moist heat favors the selection of enteric pathogen Clostridium difficile PCR ribotype 078 spores in food. Can J Infect Dis Med Microbiol 2016;2016:1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Norman KN, Harvey RB, Scott HM. et al. Varied prevalence of Clostridium difficile in an integrated swine operation. Anaerobe 2009;15:256–60. [DOI] [PubMed] [Google Scholar]
- 113. Alvarez-Perez S, Blanco JL, Bouza E. et al. Prevalence of Clostridium difficile in diarrhoeic and non-diarrhoeic piglets. Vet Microbiol 2009;137:302–5. [DOI] [PubMed] [Google Scholar]
- 114. Silva RO, Guedes RM, Lobato FCF.. Clostridium difficile infection: main features and occurrence in domestic species in Brazil. Cienc Rural 2012;43:73–80. [Google Scholar]
- 115. Hopman NEM, Keessen EC, Harmanus C. et al. Acquisition of Clostridium difficile by piglets. Vet Microbiol 2011;149:186–92. [DOI] [PubMed] [Google Scholar]
- 116. Rodriguez C, Hakimi DE, Vanleyssem R. et al. Clostridium difficile in beef cattle farms, farmers and their environment: assessing the spread of the bacterium. Vet Microbiol 2017;210:183–7. [DOI] [PubMed] [Google Scholar]
- 117. Bandelj P, Blagus R, Briski F. et al. Identification of risk factors influencing Clostridium difficile prevalence in middle-size dairy farms. Vet Res 2016;47:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Keessen EC, van den Berkt AJ, Haasjes NH. et al. The relation between farm specific factors and prevalence of Clostridium difficile in slaughter pigs. Vet Microbiol 2011;154:130–4. [DOI] [PubMed] [Google Scholar]
- 119. Bauer MP, Kuijper EJ.. Potential sources of Clostridium difficile in human infection. Infect Dis Clin North Am 2015;29:29–35. [DOI] [PubMed] [Google Scholar]
- 120. Knight DR, Elliott B, Chang BJ. et al. Diversity and evolution in the genome of Clostridium difficile. Clin Microbiol Rev 2015;28:721–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Stone NE, Sidak-Loftis LC, Sahl JW. et al. More than 50% of Clostridium difficile isolates from pet dogs in Flagstaff, USA, carry toxigenic genotypes. PLoS One 2016;11:e0164504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Songer JG, Post KW, Larson DJ. et al. Infection of neonatal swine with Clostridium difficile. Swine Health Prod 2000;8:185–9. [Google Scholar]
- 123. Goorhuis A, Debast SB, van Leengoed LA. et al. Clostridium difficile PCR ribotype 078: an emerging strain in humans and in pigs? J Clin Microbiol 2008;46:1157–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. US Food and Drug Administration. Summary Report on Antimicrobials Sold or Distributed for Use in Food-Producing Animals 2013. US Department of Health and Human Services.
- 125. Woolhouse M, Ward M, van Bunnik B. et al. Antimicrobial resistance in humans, livestock and the wider environment. Philos Trans R Soc Lond B Biol Sci 2015;370:20140083.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Shaban RZ, Simon GI, Trott DJ.. Surveillance and Reporting of Antimicrobial Resistance and Antibiotic Usage in Animals and Agriculture in Australia CC BY 30, 2014. Australian Department of Agriculture.
- 127. Jordan D, Chin JC, Fahy VA. et al. Antimicrobial use in the Australian pig industry: results of a national survey. Aust Vet J 2009;87:222–9. [DOI] [PubMed] [Google Scholar]
- 128. D'Angeli MA, Baker JB, Call DR. et al. Antimicrobial stewardship through a one health lens: observations from Washington state. Int J Health 2016;21:114–30. [Google Scholar]
- 129. Nelson JM, Chiller TM, Powers JH. et al. Fluoroquinolone-resistant campylobacter species and the withdrawal of fluoroquinolones from use in poultry: a public health success story. Clin Infect Dis 2007;44:977–80. [DOI] [PubMed] [Google Scholar]
- 130. European Food Safety Authority, European Centre for Disease Prevention and Control. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2014. EFSA J 2016;14:1–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. World Health Organization. Tackling Antibiotic Resistance from a Food Safety Perspective in Europe 2011. World Health Organization Regional Office for Europe.
- 132. Van den Bogaard AE, Jensen LB, Stobberingh EE.. Vancomycin-resistant enterococci in turkeys and farmers. N Engl J Med 1997;337:1558–9. [DOI] [PubMed] [Google Scholar]
- 133. Levy SB, FitzGerald GB, Macone AB.. Changes in intestinal flora of farm personnel after introduction of a tetracycline-supplemented feed on a farm. N Engl J Med 1976;295:583–8. [DOI] [PubMed] [Google Scholar]
- 134. Tian TT, Zhao JH, Yang J. et al. Molecular characterization of Clostridium difficile isolates from human subjects and the environment. PLoS One 2016;11:e0151964. [DOI] [PMC free article] [PubMed] [Google Scholar]