Abstract
We report new crystallographic structures of Thermus thermophilus ribosomes complexed with long mRNAs and native Escherichia coli tRNAs. They complete the full set of combinations of Watson–Crick G•C and miscoding G•U pairs at the first two positions of the codon–anticodon duplex in ribosome functional complexes. Within the tight decoding center, miscoding G•U pairs occur, in all combinations, with a non-wobble geometry structurally indistinguishable from classical coding Watson–Crick pairs at the same first two positions. The contacts with the ribosomal grip surrounding the decoding center are all quasi-identical, except in the crowded environment of the amino group of a guanosine at the second position; in which case a G in the codons may be preferred. In vivo experimental data show that the translational errors due to miscoding by G•U pairs at the first two positions are the most frequently encountered ones, especially at the second position and with a G on the codon. Such preferred miscodings involve a switch from an A-U to a G•U pair in the tRNA/mRNA complex and very rarely from a G = C to a G•U pair. It is concluded that the frequencies of such occurrences are only weakly affected by the codon/anticodon structures but depend mainly on the stability and lifetime of the complex, the modifications present in the anticodon loop, especially those at positions 34 and 37, in addition to the relative concentration of cognate/near-cognate tRNA species present in the cellular tRNA pool.
INTRODUCTION
The process of translation is the last stage in the genetic information transfer and it depends on the correct matching between mRNA codons and corresponding tRNA anticodons within the ribosomal complexes. Among all involved steps, translation is estimated to be the least accurate one with an error rate in the range 6 × 10−4 − 5 × 10−3 (1–3). On the whole, the estimated frequency of translation errors (∼10−4 − 10−3) converts into at least one misincorporated amino acid in 18% of expressed protein molecules (assuming an average gene length of 400 codons) (4). Although such random substitutions could be beneficial for withstanding evolutionary pressure, subtle regulation of protein expression or promoting cell fitness and homeostasis, specific single substitutions can be deleterious to protein folding and function.
There are several sources for such errors, but the main source is attributed to the erroneous matching between mRNA codons and tRNA anticodons (1,5–7). At the molecular level, the discrimination between correct (cognate) and incorrect (near-cognate) tRNAs primarily hinges on complementary base pairing between codon and anticodon triplets. Here, we follow the definition of near-cognate tRNA as those tRNAs able to form a mini-helix with A-site codons in the ribosomal decoding center (8). However, this interaction alone cannot provide the required accuracy. The nature of tRNA–mRNA interactions on the 70S ribosome has been structurally investigated over the course of more than a decade (9–11). In particular, during the last five years a large amount of evidence was collected regarding the chemistry of translation errors and their molecular recognition basis (12–18). The experimental data indicate that discrimination between near-cognate and cognate tRNAs is based primarily on steric fit and shape complementarity rather than on the number of hydrogen bonds between the rigid decoding center and the codon–anticodon duplex (12,15,19,20). Such observations further strengthen the view that spatial mimicry, due either to base tautomerism or ionization, dominates the translation infidelity mechanism. A systematic understanding of the mechanisms underlying translational errors, together with the possible influence of these errors on the proteome and cell homeostasis, will further improve our knowledge of evolutionary mechanics behind the genetic code.
MATERIALS AND METHODS
Materials
The 70S ribosomes from Thermus thermophilus strain H8 were purified according to the published protocol (13). Both native tRNAfMet and tRNAArg from Escherichia coli were purchased from ‘Chemical Block’ (Russia). Native tRNAThr from E. coli was purchased from ‘Plenum Company’ (USA). The mRNA constructs whose sequences are specified below were purchased from Thermo Scientific (USA) and deprotected following the supplier procedure.
Nomenclature
Since this study is dedicated to the A-site codon–anticodon interactions, for simplicity we will use the following notation for the A-site codon nucleotides: B1B2B3; instead of more universally accepted numbering, starting from the first nucleotide of the P-codon. The tRNA anticodon numbering is as usual with nucleotide B34 pairing to B3, nucleotide B35–B2 and nucleotide B36–B1. Throughout, we will use the following nomenclature: A-U/U-A or G = C/C = G for Watson–Crick pairs, GoU/UoG for standard wobble pairs (geometry distinct from the ‘classic’ Watson–Crick pair because the U moves in the major deep groove of the RNA helix), G•U/U•G for Watson–Crick-like tautomeric pairs (such pairs are isosteric and occupy the same volumes than Watson–Crick pairs). In all those notations the slash character means ‘or’; for example G•U/U•G stands for a G•U pair or a U•G pair.
Complex formation and crystallization
The ribosomal complexes were formed in 10 mM Tris-acetate, 40 mM KCl, 7.5 mM Mg (CH3COO)2, 0.5 mM Dithiothreitol at pH7.0 at 37°C. For all complexes the 70S ribosomes (3 μM) were incubated with 5-fold stoichiometric excess of mRNA and 3- to 5-fold excess of tRNAs. For both the cognate and near-cognate 70S ribosome complexes the mRNA sequences contained GGC.AAG.GAG.GCA.AAA (Z) at the 5′-end, the Shine-Dalgarno sequence is underlined (21). The cognate Thr complex mRNA sequence contained threonine A codon ACC following Met codon AUG (underlined) and was as follows: mRNA-1 = ZAUGACCA8. Here, C2 base of the Thr codon (in bold) will pair with G35 in the middle of anticodon of tRNAThr. The near-cognate Thr complex contained isoleucine AUC codon that modeled the U2•G35 mismatch at the second codon–anticodon was as follows: mRNA-2 = ZAUGAUCA8. The cognate Arg complex mRNA sequence contained arginine A codon CGA allowing a G2 codon base to pair with C35 of tRNAArg was as follows: mRNA-3 = ZAUGCGAA8 (the start codon and the Shine-Dalgarno sequence are underlined). The 70S ribosomes (3 μM) were pre-incubated with mRNA-1 or mRNA-2 or mRNA-3 and tRNAfMet for 15 min to fill the P-site. The complexes modeling the above cognate or near-cognate codon–anticodon at the A-site were obtained by subsequent incubation with tRNAThr or tRNAArg, or tRNAArg respectively. Crystals were grown at 24°C via vapor diffusion in sitting-drop plates (CrysChem, Hampton Research) as described before (13).
Data collection, processing and structure determination
Data for all complexes were collected at the X06SA beamline of Swiss Light Source, Switzerland, at 100K. A low dose very fine slicing mode was used to collect high-redundancy data (22,23). The data were indexed, integrated and scaled using XDS (24). All crystals belong to space group P212121 and contain two ribosomes per asymmetric unit. One of the previously published structures (12), with tRNA, mRNA and metal ions removed, was used for refinement with Phenix (25). The initial model was placed within each dataset by rigid body refinement with each biopolymer chain as a rigid body. The resulting electron density maps were inspected in Coot (26) and the tRNA and mRNA chains were built in. During several cycles of manual rebuilding followed by coordinate and isotropic B-factor refinement, magnesium ions were added and the final refinement round took place. The data collection and refinement as well as model geometry statistics are presented in the Table 1.
Table 1.
Data collection and refinement statistics
| Cognate Thra | UG mismatchb | Cognate Argc | |
|---|---|---|---|
| PDB ID | 6gsj | 6gsk | 6gsl |
| Data collection | |||
| Space group | P212121 | P212121 | P212121 |
| Cell dimensions | |||
| a, b, c (Å) | 209.41 449.24 618.40 | 209.82, 449.75, 618.71 | 210.05, 449.39, 618.66 |
| α, β, γ (°) | 90.0 90.0 90.0 | 90.0 90.0 90.0 | 90.0 90.0 90.0 |
| Resolution (Å) | 300–2.96 (3.04–2.96)* | 300–3.36 (3.45–3.36) | 300–3.16 (3.24–3.16) |
| R meas | 29.2 (470.5) | 36.4 (382.6) | 38.6 (806.0) |
| I / σI | 20.60 (1.03) | 11.53 (0.99) | 14.66 (1.00) |
| CC(1/2) (42) | 99.9 (24.8) | 99.8 (31.6) | 99.7 (45.2) |
| Completeness (%) | 100.0 (100.0) | 99.9 (100) | 100 (99.9) |
| Redundancy | 87.13 (41.67) | 35.99 (23.56) | 97.11 (84.61) |
| Refinement | |||
| Resolution (Å) | 224.620–2.96 | 224.875–3.36 | 153.44–3.16 |
| No. reflections | 2 348 293 | 1 609 958 | 1 936 360 |
| R work/Rfree | 19.60/25.19 | 20.03/26.42 | 19.21/24.01 |
| No. atoms | |||
| RNA | 201 697 | 200 857 | 205 176 |
| Protein | 88 484 | 88 467 | 94 344 |
| Ligand/ion/water | 4077 | 3321 | 6532 |
| B-factors | |||
| RNA | 95.73 | 118.23 | 110.39 |
| Protein | 103.45 | 127.03 | 118.59 |
| Ligand/ion/water | 83.10 | 96.33 | 89.83 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.009 | 0.009 | 0.010 |
| Bond angles (°) | 1.536 | 1.490 | 1.649 |
*Values in parentheses are for highest-resolution shell.
Number of crystals used for data collection: a8; b2; c5.
RESULTS
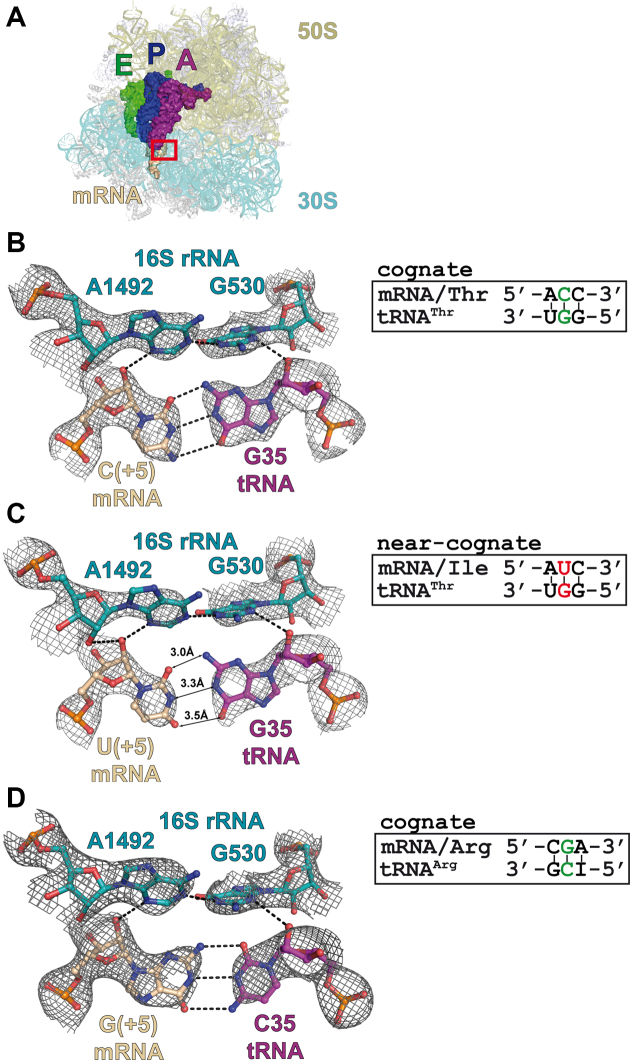
In this communication, we present three novel X-ray structures of the 70S ribosomes complexed with a long mRNA and three tRNAs at the A-, P- and E-sites (Figure 1A and Table 1). In one of the structures, tRNAThr (anticodon GGU) pairs with the cognate threonine codon (ACC) in the A-site (Figure 1B); in the second one, the same tRNAThr pairs with the near-cognate isoleucine codon AUC in the A-site and thus involves a U2•G35 base opposition (Figure 1C). The third structure corresponds to a cognate interaction between tRNAArg (anticodon ICG) and one of its cognate codon CGA (Figure 1D). In this latter case, it is the first time that a G = C pair with a G on the codon at the second position of the codon, a G2 = C35 pair, is shown. Taken together with the previously published structures (12,16), this new set of structures completes the ensemble of the four possible combinations of Watson–Crick pairs and Watson–Crick-like G•U/U•G pairs at the first and second positions of the codon–anticodon duplex on 70S ribosomes with long mRNAs and three tRNAs bound.
Figure 1.
Complexes of the Thermus thermophilus 70S ribosome with mRNA and tRNA in the A-site. (A) Overview of the complex structure. The red frame marks the decoding center. 16S rRNA is shown in teal, 23S rRNA—in olive, A-site tRNA—in purple, P-site tRNA—in blue, E-site tRNA—in green and mRNA—in wheat. Views of the second codon–anticodon base pair in the cognate (B) and near-cognate (C) tRNAThrGGU complexes. (D) Second codon–anticodon base pair in the cognate tRNAArgICG complex. Putative hydrogen bonds (distances ≤3.3 Å) are shown as dashed lines and interatomic distances for the mismatched pair are indicated. 2Fo–Fc electron density maps are contoured at 1.2 σ.
The G•U/U•G pairs, usually termed ‘wobble’ base pair, are of special interest for at least two reasons. First, they are widespread naturally in a majority of cellular RNAs. Second, several in vivo studies of translational errors (e.g. see refs. (3,7,27,28)) single out these mismatches as strongly predominant. As observed in previous structures (12,16) with near-cognate complexes containing a G•U mismatch in the second position of the codon–anticodon duplex, we confirm here that the G•U pair adopts geometry structurally identical to that of the cognate G•C Watson–Crick pair and not the classical wobble geometry (Figure 1B and C). Additionally, the conformational changes of the ribosomal decoding center involving the crucial and invariant nucleotides A1492, A1493 and G530 are identical in response to binding of either cognate or near-cognate tRNA in the A site (29). The recently proposed mechanism of translation infidelity (12,15) definitively puts forward spatial Watson–Crick base pair mimicry as the way for mismatched pairs to bypass ribosomal discrimination. As discussed elsewhere (12,19,30–32), such mimicry can be achieved via rare tautomeric states of the bases involved in mismatched base pair and G•U/U•G pairs are more likely to be in such states compared to other base combinations.
DISCUSSION
In order to compare the effects of various U•G/G•U substitutions we use the recently published (33) representation of the codon table as a wheel. This representation, based on the Turner energy (34) of RNA–RNA triplets (from ref. (35)), displays the 64 codons as Strong (north quadrant), Intermediate (east and west quadrants) and Weak (south quadrant) (see Supplementary Figure S1a). In the complete system, additional structural elements of the tRNA molecules, especially of the anticodon loops and stems also contribute to the global stability of codon–anticodon association (as discussed in ref. (33)). Such structural elements can be mapped on the wheels and such mappings illustrate how these elements vary with the codon positions on the wheel (see Supplementary Figure S1 and Supplementary Materials of ref. (33)), attesting that the sequence identities of each of the individual tRNAs of the cellular pool extend beyond the anticodon triplet and there are strong correlations between the anticodon triplet and the neighboring nucleotides. In particular, tRNA modifications at positions 34 and 37 are critical for stabilizing and tuning the efficiency and accuracy of codon–anticodon duplexes (Supplementary Figures S1b–d and 2).
Overall, the tRNA identities and modifications tend to decrease the binding affinities of the codon–anticodon duplex in the northern semicircle of the wheel (mostly to restrict miscoding). On the other hand, in the southern semicircle of the wheel, both specific tRNA identities and modifications are needed, besides restricting miscoding, to increase the binding strengths of the codon–anticodon duplex and fine tuning of translation so that each native aminoacyl-tRNA binds to the A site of the ribosome with the about the same average optimal binding energy, whatever the base composition of the codon/anticodon mini-helix (Supplementary Figure S1b–d).
Structural consequences of a G•U/ U•G pair at the first or second position in bacterial systems
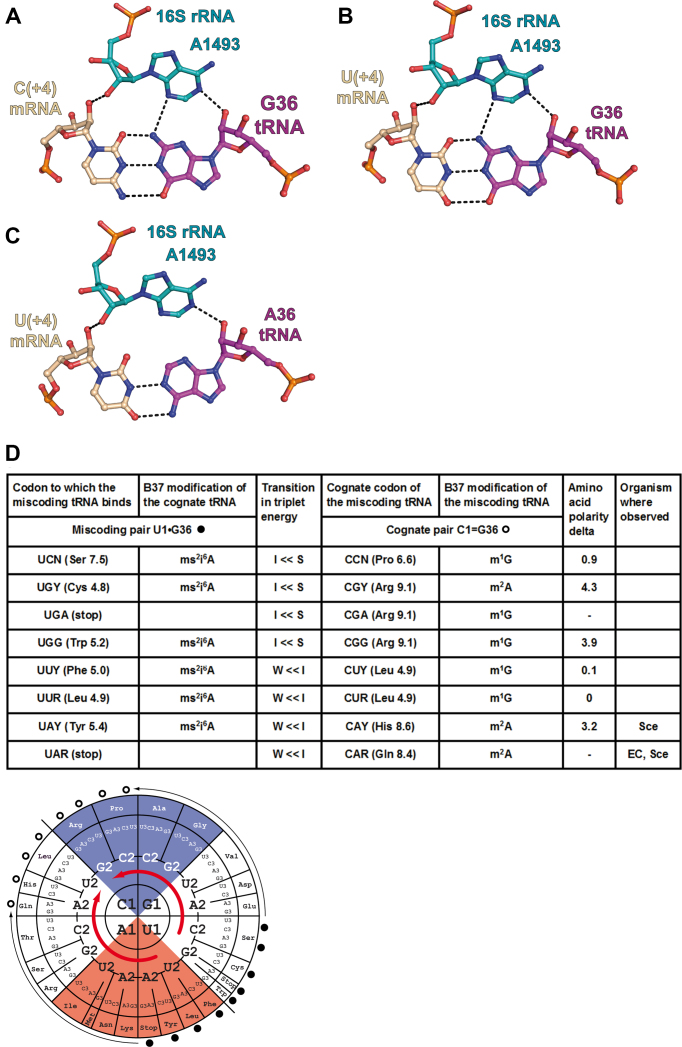
We now have crystallographic examples of every combination of Watson–Crick geometries for G = C/U-A and G•U/U•G pairs at the first two positions within a bacterial ribosome environment (see Figures 1B–D, 2A–C, 3A–C, 4A–C and 5A–C). A full comparison between the interactions of the first two decoding pairs with the key elements of the 30S ribosomal decoding center can be made. These elements are namely the 16S rRNA nucleotides A1492, A1493 and G530 that interact with the minor groove of the codon–anticodon duplex via specific interactions: in particular A-minor interactions between A1493 and the first pair of the mini-helix (nucleotides mRNA B1 and tRNA B36) and A-minor interactions between A1492, G530 and the second pair of the mini-helix (nucleotides mRNA B2 and tRNA B35). Structural comparisons lead to important observations on bacterial ribosomal constraints.
With a G at the first position (either G1 in the codon (Figure 2A) or G36 in the anticodon (Figure 3A)), a favorable additional H-bond occurs between the exocyclic amino group of G and the N3 of A1493, compared to the situation with an A, because the exocyclic amino group N2 falls at close-by positions in a G = C or a C = G pair. In short, a G on either side at the first position presents an additional H-bond to the ribosomal grip.
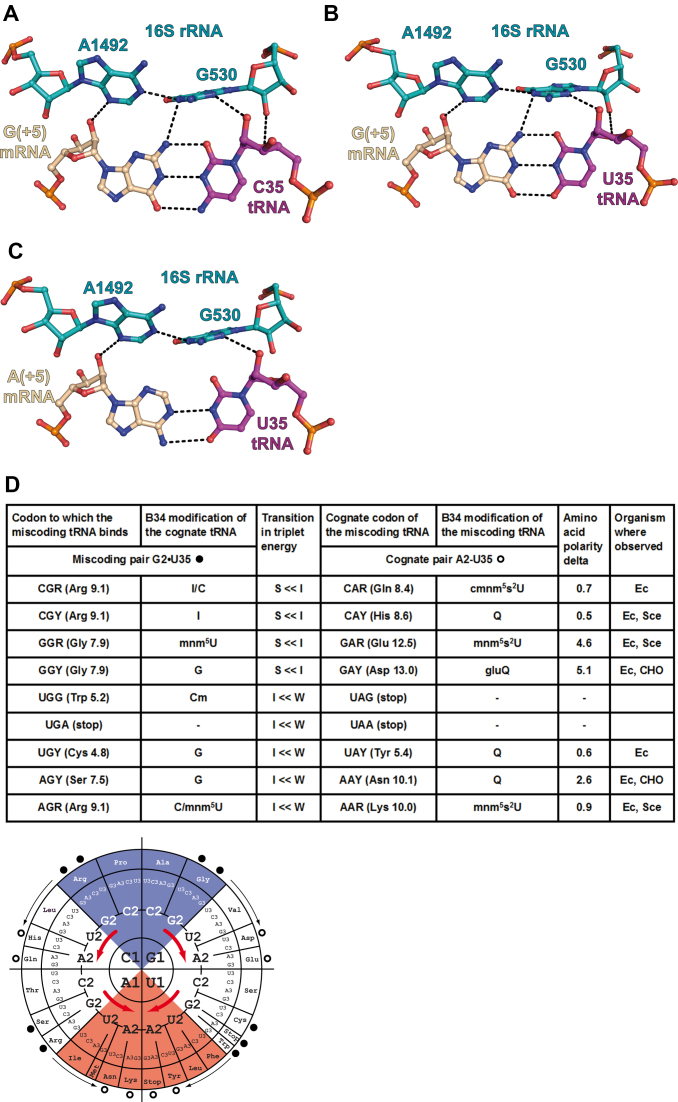
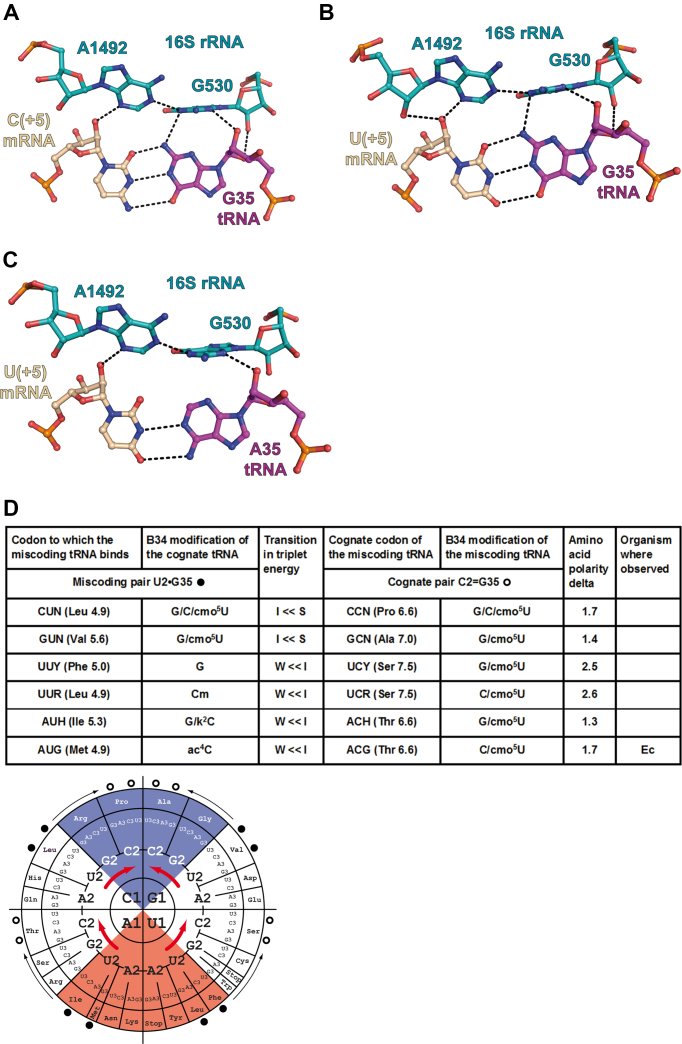
With a G at the second position (either G2 in the codon (Figure 4A) or G35 in the anticodon (Figure 5A)), the exocyclic amino group N2 is in a tight or crowded environment, lodged between A1492 and G530. In both cases, the C2-H of A1492 points to the exocyclic amino group N2 of either G2 or G35. Here, the cases where G is present in the codon or in the anticodon appear not identical. However, on the basis of the contacts alone, it is very difficult to evaluate which of the base pairs C2 = G35 or G2 = C35 is in a more favorable environment. Also, the interactions between G530 and the anticodon base B35 are unusual; one may surmise that the O2(C35) (slightly negative) pointing to the center of G530 (positively charged) might be slightly more favorable than the N3(G35). Overall, the environment of the second pair appears more crowded with possibly a slight preference for a G2 = C35 pair compared to a C2 = G35 pair.
Since the miscoding G•U/U•G pairs at either of the first two positions are Watson–Crick-like, the same considerations apply to the interactions with near-cognate tRNAs. Consequently, the energy and structural consequences of the introduction of a miscoding G•U/U•G pair are not identical for the first two base pair positions. The additional H-bond between the G in either the G•U or U•G pair occurs for the first position (Figures 2B and 3B). Further, with a miscoding G•U/U•G pairs at the second position (Figures 4B and 5B), a slight energetic preference for a G in the B2 codon position may be present.
Figure 2.
Substitution of a G1 = C36 pair or a A1-U36 pair by a G1•U36 base pair in the first position of the codon–anticodon duplex. (A) Cognate G1 = C36 pair from the complex of 70S ribosome with mRNA and tRNAVal (PDB ID: 5ibb); (B) near-cognate G1•U36 pair from the complex of 70S ribosome with mRNA and tRNALys (PDB ID: 5ib8); (C) cognate A1-U36 pair from the complex of 70S ribosome with mRNA and tRNALys (PDB ID: 5e7k). The nucleotides of the 16S rRNA are depicted in teal, tRNA in magenta and mRNA in wheat. The dashed lines indicate the putative hydrogen bonds (interatomic distance ≤3.3Å). (D, top) Summary table of all amino acid substitutions resulting from the incorporation of a miscoding pair with comparisons of amino acid polarity, transition in triplet energy between the two cognate tRNAs (S = Strong, I = Intermediate, W = Weak), the experimentally observed G•U pairing miscoding available in the literature (Escherichia coli (EC), Saccharomyces cerevisiae (Sce) and CHO cells). The in vivo data for E. coli and S. cerevisiae are from (6,7) and (28); those for the CHO cells are from (27). The transition in triplet energy between cognate tRNAs and the base modification at nucleotide 37 in the respective cognate tRNAs (for E. coli) are both indicated to point to the competition between the tRNAs for the A site. The cognate codon of the miscoding tRNA is at the right and, at the left, the miscoding pair formed by the miscoding tRNA. (D, bottom) The corresponding tRNA modifications and relative positions of cognate and near-cognate codons on the energy wheel. The miscoding pairs (dark circles) are indicated with the thin arrows pointing to the miscoding tRNA (open circles) (for example, a tRNAThr miscodes an Ala codon or a tRNALys miscodes a Glu codon). The thick red arrows indicate the energetic transitions between Strong (blue) and Intermediate (white) codon–anticodon triplets as well as between Intermediate and Weak (red) ones. Because the substitutions are at the first position, the transitions start in the top right quadrant and end in the bottom left quadrant. The miscoding tRNAs are highly modified and replace stronger, but less modified, tRNA binders. Many instances have been reported in the literature.
Figure 3.
Substitution of a C1 = G36 pair or a U1-A36 pair by a U1•G36 base pair in the first position of the codon–anticodon duplex. (A) Cognate C1 = G36 pair from the complex of 70S ribosome with mRNA and tRNALeu (PDB ID: 4v87); (B) near-cognate U1•G36 pair from the complex of 70S ribosome with mRNA and tRNALeu (PDB ID: 4v8b); (C) cognate U1-A36 pair from the complex of 70S ribosome with mRNA and tRNATyr (PDB ID: 4v8d). Same comments as for Figure 2. (D, top) The in vivo data for Escherichia coli and Saccharomyces cerevisiae are from (6,7) and (28); those for the CHO cells are from (27). (D, bottom) The miscoding pairs (dark circles) are indicated with the thin arrows pointing to the miscoding tRNA (open circles) (e.g. a tRNAHis miscodes a Tyr codon). The thick red arrows indicate the energetic transitions between Intermediate (white) and Strong (blue) codon–anticodon triplets as well as between Weak (red) and Intermediate ones. Because the substitutions are at the first position, the transitions start in the bottom right quadrant and end in the top left quadrant. The miscoding tRNAs are good binders and replace weaker, but more modified, tRNA binders. Only two instances could be found in the literature.
Figure 4.
Substitution of a G2 = C35 pair or a A2-U35 pair by a G2•U35 base pair in the second position of the codon–anticodon duplex. (A) Cognate G2 = C35 pair from the complex of 70S ribosome with mRNA and tRNAArg (this study, PDB ID: 6gsl); (B) near-cognate G2•U35 pair from the complex of 70S ribosome with mRNA and tRNATyr (PDB ID: 4v8e); (C) A2-U35 pair from the complex of 70S ribosome with mRNA and tRNALys (PDB ID: 5e7k). The nucleotides of the 16S rRNA are depicted in teal, tRNA in magenta and mRNA in wheat. The dashed lines indicate the putative hydrogen bonds (interatomic distance ≤3.3Å). (D, top) Summary table of all amino acid substitutions resulting from the incorporation of the mismatched pair with comparisons of amino acid polarity, transition in triplet energy between the two cognate tRNAs (S = Strong, I = Intermediate, W = Weak), the experimentally observed G•U pairing miscoding available in the literature (Escherichia coli (EC), Saccharomyces cerevisiae (Sce) and CHO cells). The in vivo data for E. coli and S. cerevisiae are from (6,7) and (28); those for the CHO cells are from (27). The transition in triplet energy between cognate tRNAs and the base modification at nucleotide 37 in the respective cognate tRNAs (for E. coli) are both indicated to point to the competition between the tRNAs for the A site. The cognate codon of the miscoding tRNA is at the right and at the left the miscoding pair formed by the miscoding tRNA. (D, bottom) The corresponding tRNA modifications and relative positions of cognate and near-cognate codons on the energy wheel. The miscoding pairs (dark circles) are indicated with the thin arrows pointing to the miscoding tRNA (open circles) (e.g. a tRNAHis miscodes an Arg codon or a tRNAAsp miscodes a Gly codon). The thick red arrows indicate the energetic transitions between Strong (blue) and Intermediate (white) codon–anticodon triplets as well as between Intermediate and Weak (red) ones. Because the substitutions are at the second position, the transitions occur within each of the four quadrants. The miscoding tRNAs are highly modified and replace stronger, but less modified, tRNA binders. Many instances have been reported in the literature.
Figure 5.
Substitution of a C2 = G35 pair or a U2-A35 pair by a U2•G35 base pair in the second position of the codon–anticodon duplex. (A) Cognate C2 = G35 pair from the complex of 70S ribosome with mRNA and tRNAThr (this study, PDB ID 6gsj); (B) near-cognate U2•G35 pair from the complex of 70S ribosome with mRNA and tRNAThr (this study, PDB ID 6gsk); (C) U2-A35 pair from the complex of 70S ribosome with mRNA and tRNAVal (PDB ID: 5ibb). Same comments as in Figure 4. (D, top) The in vivo data for Escherichia coli and Saccharomyces cerevisiae are from (6,7) and (28); those for the CHO cells are from (27). (D, bottom) The miscoding pairs (dark circles) are indicated with the thin arrows pointing to the miscoding tRNA (open circles) (e.g. a tRNAThr miscodes a Met codon). The thick red arrows indicate the energetic transitions between Intermediate (white) and Strong (blue) codon–anticodon triplets as well as between Weak (red) and Intermediate ones. Because the substitutions are at the second position, the transitions occur within each of the four quadrants. The miscoding tRNAs replace weaker tRNAs. Concerning the modifications, the situation is more difficult to evaluate because of the tRNA isoacceptors. Only one instance could be found in the literature.
Translation consequences of a G•U/U•G pair at the first or second position
Here, we focus on the situation where one given tRNA reads a near-cognate codon instead of its cognate codon (see Supplementary Materials, legend to Supplementary Figure S1). Similar considerations would apply to the case where a given codon is read by either its cognate or a near-cognate tRNA. In both cases, the final outcome is strongly influenced by kinetics factors such as optimal lifetimes of the codon–anticodon associations that should be sufficient for the ribosome to grip the complex in the A-decoding site and, last but not least, the competition with the isoacceptor and isodecoder tRNAs present in the tRNA pool (correlated with the distribution of codon usage that also varies between organisms) (see e.g. (36,37). The introduction of a G•U/U•G mismatch during translation has two major consequences: destabilization of the codon–anticodon interaction and insertion of an amino acid different from that expected by the genetic code.
- Although miscoding G•U pairs are structurally equivalent to Watson–Crick pairs, the introduction of a miscoding G•U pair requires the formation of a tautomeric state in one of the two bases with a concomitant unpredictable decrease in base pair energy (19). We lack experimental data regarding the relative stability of the tautomeric Watson–Crick-like G•U base pair and energy considerations are not feasible at the present time. However, in terms of the cognate duplex strength and the wheel code, one can notice the following.
- The introduction of a single miscoding G•U/U•G pair at the first position of the codon–anticodon duplex results in a move from one quadrant to another across the diagonal of the codon wheel (Figures 2D and 3D). Importantly, the northern and southern quadrants are correlated with tRNAs harboring modified purines at position 37 displaying different stabilizing power (stacking) on the adjacent base pair (Supplementary Figure S2b). Therefore the presence of the hypermodified purine-37, like ct6A or m6t6A adjacent to a U36 (minitable in Figure 2D) should help formation of an adjacent U36•G1 base pair; while the presence of a simpler modified purine-37, like m2A or m1G adjacent to G36 (minitable in Figure 3D), should not have the same stabilizing effect, thus limiting formation of a G36•U1 miscoded RNA–RNA duplex (see also Supplementary Figure S2b).
- The introduction of a single miscoding G•U pair at the second position of the codon–anticodon duplex results in a move within a given quadrant (Figures 4D and 5D). The important modifications to consider are the frequently modified nucleotide located at position 34. They also vary with the quadrant and the nature of the base at the middle position of the RNA–RNA duplexes (minitables in Figures 4D and 5D). The main role of base-34 modification is to allow decoding of a subset of near-cognate codons varying at position 3; but also, for a few of them, like the 2-thiolated uridine, to enhance stacking with the adjacent base pair at the middle position of the RNA–RNA duplex, as for a G2•U35 miscoding pair (14,38).
The end result of a G•U/U•G substitution in the first or second position during translation is an amino acid mutation in the protein chain. To gauge the severity of the amino acid substitution, we evaluated the corresponding changes in amino acid polarity (39,40). Results for a G1•U36 (or U1•G36) pair in the first position and a G2•U35 (or U2•G35) pair in the second position of the codon–anticodon duplex are given in minitables in Figures 2D, 3D, 4D and 5D. See Supplementary Materials for analysis.
Comparisons with experimental data on in vivo translation miscodings
All possible mispairs of the type G•U/U•G that could occur at the first or second position of a codon in mRNA and the corresponding bases in the anticodon of a fully matured cellular tRNA are listed in the minitables of Figures 2D, 3D, 4D and 5D, some of them having been successfully crystallized on the ribosome (see Supplementary Material for detailed descriptions of the experimental protocols and results).
Only some of them have been described as a recurrent source of missense errors during in vivo translation in various wild-type organisms (see Supplementary Figure S3 for E. coli and Supplementary Figure S4 for Saccharomyces cerevisiae). Among them are those bearing a G2 in the mRNA codon with U35 in the tRNA anticodon (five cases in E. coli out of a total seven theoretical ones—cf. Figure 4D and Supplementary Figure S3). The two missing ones result from the absence in the E. coli tRNA pool of the corresponding potential near-cognate tRNAs. About those codons with U2 pairing with G35 on the tRNA, there is only one case (out of six potential ones, cf. Figure 5D and Supplementary Figure S3). From these experimental data, it appears that the probability of mismatching a G in the middle position of the codon–anticodon duplex is easier when G is in the mRNA rather than in the tRNA. Likewise, codons bearing G1 on the mRNA with U36 on the tRNA occur five cases over eight potential ones (cf. Figure 3D and Supplementary Figure S3). For the reverse situation of U1 on the mRNA and G36 in the tRNA, there are only two cases over a total of eight potential ones (compare Figure 2D and Supplementary Figure S3), and they read each a distinct stop codon UAA or UAG. The situation in S. cerevisiae (Supplementary Figure S4) looks the same as in E. coli (Supplementary Figure S3), in the sense that what has been observed in yeast is valid also in E. coli but not necessarily the reverse situation. Thus, in yeast, misreading of a given codon appears in general less prone to errors than in E. coli (7).
These observations point to the following regularities.
A more frequent occurrence of mismatches at the second position than at the first and, in both cases, with a strong bias for G on the codon.
These miscoding bias result from a transition between an A1-U36 (A2-U35) and a G1•U36 (G2•U35) pair in the near-cognate mRNA/tRNA complex with the G1•U36 (G2•U35) pair replacing the cognate G1 = C36 (G2 = C35) pair in the cognate mRNA/tRNA complex.
Most of the experimentally observed miscoding tRNAs belong to 2-codon boxes.
For the non-observed miscoding tRNAs, the cognate triplets are stronger than those with which they compete.
The frequent tRNAs forming a G1•U36 contain heavily modified A37 (ct6A or m6t6A) and the first nucleotide G1 forms an additional H-bond compared to the cognate A1-U36.
Among the frequent tRNAs forming a G2•U35 are those bearing the stabilizing nucleotides 2-thioU or Q at position 34.
The other miscoding tRNAs have less differences between their modifications of the anticodon loops.
There is no correlation between the variations in amino acid polarities resulting from the G•U/U•G miscodings and the experimentally observed miscoded amino acids.
In sum, the decoding properties of the tRNAs from the southern part of the genetic wheel (like tRNALys or tRNATyr), which depend very much on an ensemble of specific post-transcriptional modifications to stabilize their duplexes with cognate codons, are also the same modified tRNAs that miscode, hopefully to a small but detectable level, a limited number of near-cognate codons. Any minor change in the subtle modification status of these tRNAs, due to misfunction of the corresponding modification enzymes or the lack of necessary cofactors due to unbalanced cellular nutrients (41), is expected to lead to a substantial decrease in miscoding without necessarily affecting to the same degree the reading of cognate codons. This effect will be further modulated by the nature of the third base pair because of the strong dissymmetry (19) between G34oU3, where modifications do occur but are not structurally necessary and U34*•G3, where modifications are required for forming long-lived binding.
CONCLUSION
The crystallographic studies describe the structural aspects of a codon–anticodon duplex involving U•G or G•U at either of the first or second position of a codon duplex within the decoding A-site of a bacterial ribosome. Extrapolation of the occurrence of a given miscoding event during in vivo mRNA translation to the different organisms is risky because of the large differences in tRNA library and the modes of post-transcriptional modifications in the different organisms. Under crystallization conditions, the problem of competition between the various aminoacyl tRNAs at the A-site of the ribosome is irrelevant and only the need of a sufficiently stable complex involving a G•U/U•G mismatch at the first or second position of the codon–anticodon duplex within the subsequent ribosomal grip have to be taken into account.
The crystal structures show that structurally Watson–Crick pairs and Watson–Crick-like pairs at the first and second positions are indistinguishable, with the environment of a G = C/C = G or G•U/U•G pair at the second position more crowded and possibly presenting a weak energetic preference for a G in the codon. However, experimentally, G•U pairs do occur more frequently in the second position with a clear preference for the G on the codon. Thus, other factors are controlling the occurrence of miscodings with near-cognate tRNAs. No correlation exists between the occurrence of G•U pair and polarity changes (see Minitables and supplementary materials).
But, the tRNAs involved in the introduction of a miscoding G•U pair are heavily modified in their anticodon loops (necessary for stabilizing their interactions with A-U rich triplets on the ribosome). As a result, they become prone to miscoding through Watson–Crick-like G•U pairs. Among the experimentally observed miscoding events are found mainly tRNAs that are highly modified and that replace stronger cognate triplets formed between codons and less modified tRNAs. It is therefore mainly stability considerations (most frequently when a tRNA instead of binding to its cognate A1 or A2 in the mRNA binds to G1 or G2, respectively), coupled with the modifications on the miscoding tRNA, together with kinetic factors and the relative amounts of cognate and near-cognate tRNAs in the cellular pool, that have to be considered for understanding the initial step of miscoding of a codon involving a G•U or U•G before complex formation and proceeding by the ribosomal decoding center.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
French National Research Agency [ANR-15-CE11-0021-01 to G.Y., E.W.]; LABEX [ANR-10-LABX-0036_NETRNA to E.W.]; ‘La Fondation pour la Recherche Médicale’, France, [DBF20160635745 to A.R., G.Y.]; European Research Council Advanced Grant [294312 to M.Y.]; Russian Government Program of Competitive Growth of Kazan Federal University (to M.Y.). Funding for open access charge: ANR [ANR-15-CE11-0021-01].
Conflict of interest statement. None declared.
REFERENCES
- 1. Drummond D.A., Wilke C.O.. The evolutionary consequences of erroneous protein synthesis. Nat. Rev. Genet. 2009; 10:715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wohlgemuth I., Pohl C., Mittelstaet J., Konevega A.L., Rodnina M.V.. Evolutionary optimization of speed and accuracy of decoding on the ribosome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011; 366:2979–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Manickam N., Nag N., Abbasi A., Patel K., Farabaugh P.J.. Studies of translational misreading in vivo show that the ribosome very efficiently discriminates against most potential errors. RNA. 2014; 20:9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Drummond D.A., Wilke C.O.. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008; 134:341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Parker J. Errors and alternatives in reading the universal genetic code. Microbiol. Rev. 1989; 53:273–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kramer E.B., Vallabhaneni H., Mayer L.M., Farabaugh P.J.. A comprehensive analysis of translational missense errors in the yeast Saccharomyces cerevisiae. RNA. 2010; 16:1797–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kramer E.B., Farabaugh P.J.. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA. 2007; 13:87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Plant E.P., Nguyen P., Russ J.R., Pittman Y.R., Nguyen T., Quesinberry J.T., Kinzy T.G., Dinman J.D.. Differentiating between near- and non-cognate codons in Saccharomyces cerevisiae. PLoS One. 2007; 2:e517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ogle J.M., Ramakrishnan V.. Structural insights into translational fidelity. Annu. Rev. Biochem. 2005; 74:129–177. [DOI] [PubMed] [Google Scholar]
- 10. Demeshkina N., Jenner L., Yusupova G., Yusupov M.. Interactions of the ribosome with mRNA and tRNA. Curr. Opin. Struct. Biol. 2010; 20:325–332. [DOI] [PubMed] [Google Scholar]
- 11. Voorhees R.M., Ramakrishnan V.. Structural basis of the translational elongation cycle. Annu. Rev. Biochem. 2013; 82:203–236. [DOI] [PubMed] [Google Scholar]
- 12. Demeshkina N., Jenner L., Westhof E., Yusupov M., Yusupova G.. A new understanding of the decoding principle on the ribosome. Nature. 2012; 484:256–259. [DOI] [PubMed] [Google Scholar]
- 13. Rozov A., Demeshkina N., Westhof E., Yusupov M., Yusupova G.. Structural insights into the translational infidelity mechanism. Nat. Commun. 2015; 6:7251–7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rozov A., Demeshkina N., Khusainov I., Westhof E., Yusupov M., Yusupova G.. Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code. Nat. Commun. 2016; 7:10457–10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rozov A., Demeshkina N., Westhof E., Yusupov M., Yusupova G.. New structural insights into translational miscoding. Trends Biochem. Sci. 2016; 41:798–814. [DOI] [PubMed] [Google Scholar]
- 16. Rozov A., Westhof E., Yusupov M., Yusupova G.. The ribosome prohibits the G*U wobble geometry at the first position of the codon-anticodon helix. Nucleic Acids Res. 2016; 44:6434–6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schmeing T.M., Voorhees R.M., Kelley A.C., Ramakrishnan V.. How mutations in tRNA distant from the anticodon affect the fidelity of decoding. Nat. Struct. Mol. Biol. 2011; 18:432–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weixlbaumer A., Murphy F.V.T., Dziergowska A., Malkiewicz A., Vendeix F.A., Agris P.F., Ramakrishnan V.. Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines. Nat. Struct. Mol. Biol. 2007; 14:498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Westhof E., Yusupov M., Yusupova G.. Recognition of Watson-Crick base pairs: constraints and limits due to geometric selection and tautomerism. F1000Prime Rep. 2014; 6:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Khade P.K., Shi X., Joseph S.. Steric complementarity in the decoding center is important for tRNA selection by the ribosome. J. Mol. Biol. 2013; 425:3778–3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yusupova G.Z., Yusupov M.M., Cate J.H., Noller H.F.. The path of messenger RNA through the ribosome. Cell. 2001; 106:233–241. [DOI] [PubMed] [Google Scholar]
- 22. Mueller M., Wang M., Schulze-Briese C.. Optimal fine phi-slicing for single-photon-counting pixel detectors. Acta Crystallogr. D Biol. Crystallogr. 2012; 68:42–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Casanas A., Warshamanage R., Finke A.D., Panepucci E., Olieric V., Noll A., Tampe R., Brandstetter S., Forster A., Mueller M. et al. . EIGER detector: application in macromolecular crystallography. Acta Crystallogr. D Struct. Biol. 2016; 72:1036–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. . PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang Z., Shah B., Bondarenko P.V.. G/U and certain wobble position mismatches as possible main causes of amino acid misincorporations. Biochemistry. 2013; 52:8165–8176. [DOI] [PubMed] [Google Scholar]
- 28. Manickam N., Joshi K., Bhatt M.J., Farabaugh P.J.. Effects of tRNA modification on translational accuracy depend on intrinsic codon-anticodon strength. Nucleic Acids Res. 2016; 44:1871–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ogle J.M., Brodersen D.E., Clemons W.M. Jr, Tarry M.J., Carter A.P., Ramakrishnan V.. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001; 292:897–902. [DOI] [PubMed] [Google Scholar]
- 30. Vendeix F.A., Murphy F.V.t., Cantara W.A., Leszczynska G., Gustilo E.M., Sproat B., Malkiewicz A., Agris P.F.. Human tRNA(Lys3)(UUU) is pre-structured by natural modifications for cognate and wobble codon binding through keto-enol tautomerism. J. Mol. Biol. 2012; 416:467–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kimsey I.J., Petzold K., Sathyamoorthy B., Stein Z.W., Al-Hashimi H.M.. Visualizing transient Watson-Crick-like mispairs in DNA and RNA duplexes. Nature. 2015; 519:315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Szymanski E.S., Kimsey I.J., Al-Hashimi H.M.. Direct NMR Evidence that transient tautomeric and anionic states in dG.dT form Watson-Crick-like base pairs. J. Am. Chem. Soc. 2017; 139:4326–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grosjean H., Westhof E.. An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res. 2016; 44:8020–8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Turner D.H., Sugimoto N., Kierzek R., Dreiker S.D.. Free energy increments for hydrogen bonds in nucleic acid base pairs. J. Am. Chem. Soc. 1987; 109:3783–3785. [Google Scholar]
- 35. Chen J.L., Dishler A.L., Kennedy S.D., Yildirim I., Liu B., Turner D.H., Serra M.J.. Testing the nearest neighbor model for canonical RNA base pairs: revision of GU parameters. Biochemistry. 2012; 51:3508–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Czech A., Fedyunin I., Zhang G., Ignatova Z.. Silent mutations in sight: co-variations in tRNA abundance as a key to unravel consequences of silent mutations. Mol. Biosyst. 2010; 6:1767–1772. [DOI] [PubMed] [Google Scholar]
- 37. Brule C.E., Grayhack E.J.. Synonymous Codons: choose wisely for expression. Trends Genet. 2017; 33:283–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sochacka E., Lodyga-Chruscinska E., Pawlak J., Cypryk M., Bartos P., Ebenryter-Olbinska K., Leszczynska G., Nawrot B.. C5-substituents of uridines and 2-thiouridines present at the wobble position of tRNA determine the formation of their keto-enol or zwitterionic forms - a factor important for accuracy of reading of guanosine at the 3-end of the mRNA codons. Nucleic Acids Res. 2017; 45:4825–4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Woese C.R., Dugre D.H., Dugre S.A., Kondo M., Saxinger W.C.. On the fundamental nature and evolution of the genetic code. Cold Spring Harb. Symp. Quant. Biol. 1966; 31:723–736. [DOI] [PubMed] [Google Scholar]
- 40. Mathew D.C., Luthey-Schulten Z.. On the physical basis of the amino acid polar requirement. J. Mol.Evol. 2008; 66:519–528. [DOI] [PubMed] [Google Scholar]
- 41. Müller M., Hartmann M., Schuster I., Bender S., Thüring K.L., Helm M., Katze J.R., Nellen W., Lyko F., Ehrenhofer-Murray A.E.. Dynamic modulation of Dnmt2-dependent tRNA methylation by the micronutrient queuine. Nucleic Acids Res. 2015; 43:10952–10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Karplus P.A., Diederichs K.. Linking crystallographic model and data quality. Science. 2012; 336:1030–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.