Abstract
Background
Transmitted drug resistance (TDR) compromises clinical management and outcomes. Transmitted drug resistance surveillance and identification of growing transmission clusters are needed in the Southeast, the epicenter of the US HIV epidemic. Our study investigated prevalence and transmission dynamics in North Carolina.
Methods
We analyzed surveillance drug resistance mutations (SDRMs) using partial pol sequences from patients presenting to 2 large HIV outpatient clinics from 1997 to 2014. Transmitted drug resistance prevalence was defined as ≥1 SDRMs among antiretroviral therapy (ART)–naïve patients. Binomial regression was used to characterize prevalence by calendar year, drug class, and demographic and clinical factors. We assessed the transmission networks of patients with TDR with maximum likelihood trees and Bayesian methods including background pol sequences (n = 15 246).
Results
Among 1658 patients with pretherapy resistance testing, ≥1 SDRMs was identified in 199 patients, with an aggregate TDR prevalence of 12% (95% confidence interval, 10% to 14%) increasing over time (P = .02). Resistance to non-nucleoside reverse transcriptase inhibitors (NNRTIs; 8%) was common, followed by nucleoside reverse transcriptase inhibitors (4%) and protease inhibitors (2%). Factors associated with TDR were being a man reporting sex with men, white race, young age, higher CD4 cell count, and being a member of a transmission cluster. Transmitted drug resistance was identified in 106 clusters ranging from 2 to 26 members. Cluster resistance was primarily NNRTI and dominated by ART-naïve patients or those with unknown ART initiation.
Conclusions
Moderate TDR prevalence persists in North Carolina, predominantly driven by NNRTI resistance. Most TDR cases were identified in transmission clusters, signifying multiple local transmission networks and TDR circulation among ART-naïve persons. Transmitted drug resistance surveillance can detect transmission networks and identify patients for enhanced services to promote early treatment.
Keywords: antiretroviral therapy, drug resistance, HIV-1, molecular epidemiology, Southeastern United States
The widespread use of antiretroviral therapy (ART) has a profound impact on reducing morbidity and mortality of people living with HIV and lowering the risk of onward HIV transmission [1, 2]. However, the emergence of drug resistance mutations can compromise the effectiveness of ART and, for some regimens, result in a longer time to viral suppression and jeopardize treatment success [3]. Most drug resistance mutations are transmissible, resulting in treatment-naïve persons with drug-resistant virus [4]. Globally, transmitted drug resistance (TDR) limits effective options of the treatment of HIV among treatment-naïve persons, a critical challenge to the consistent gains of effective ART [5]. In recognition of this emerging problem, since 2007 the US Department of Health and Human Services (DHHS) has recommended HIV genotyping for all treatment-naïve patients before initiation of ART [6]. The presence of a surveillance drug resistance mutation (SDRM) in pretreatment HIV genotyping is indicative of TDR [7].
National and global estimates of TDR vary widely [4, 8]. In a meta-analysis of studies from 2000 to 2013, TDR prevalence ranged from 2.8% in sub-Saharan Africa to 11.5% in North America [9]. From 2007 to 2010, however, up to 16% of persons with new HIV diagnoses among 10 US surveillance sites had TDR [10]. An increasing trend in TDR was specifically observed in mutations to non-nucleoside reverse transcriptase inhibitors (NNRTIs). Similarly, TDR prevalence has increased in San Diego [11] and is as high as 20% in Washington, DC [12, 13]. Conversely, several European countries recently reported stabilized TDR prevalence of around 8% [8, 14, 15] or evidence that TDR may be decreasing [16–19]. Phylogenetic clustering studies have revealed that persistence of TDR, particularly for mutations to NNRTIs, is largely due to transmission from treatment-naïve persons [15, 18–20]. Thus, TDR may persist within transmission networks despite decreasing use of NNRTIs in resource-rich settings. Given prior findings of TDR circulation among treatment-naïve persons, TDR may serve as a marker of recent transmission and possible cluster growth. It could be studied as a potential indication for enhanced testing, linkage, and prevention services.
We investigated the prevalence and transmission dynamics of TDR in North Carolina, drawing from a large cohort with genetic sequencing and clinical data available since 1997 and using phylogenetic reconstruction of HIV transmission networks. Our objectives were to characterize temporal trends in TDR by ART drug class, identify the demographic and clinical factors associated with TDR, and assess genetic clustering of TDR within statewide transmission networks. A better understanding of TDR trends and transmission dynamics can help identify where surveillance and prevention efforts could be intensified, such as for implementation of rapid ART and pre-exposure prophylaxis.
METHODS
Study Population
Clinical Cohort
We assessed TDR prevalence among HIV patients who were enrolled in the University of North Carolina Center for AIDS Research HIV Clinical Cohort (UCHCC) or who received HIV care at the Duke University Infectious Diseases Clinic. The UNC and Duke clinics provide HIV care for most HIV patients in central NC. Patients were included if they had ≥1 pretherapy HIV-1 pol sequences available for analysis. HIV-1 pol sequences were derived from a statewide data set of 15 246 adult patients (aged 18 years or older) with routine drug resistance testing performed by the largest reference laboratory in North Carolina (Laboratory Corporation of America) from 1997 through June 2014. Sequences were matched to clinical data by specimen identifiers and medical record numbers.
At UNC, the UCHCC collects comprehensive data from institutionally available electronic health and administrative records and performs expansive medical record reviews. The data cover demographic factors, clinical diagnoses, laboratory findings, and medication provision. At Duke, clinical data were retrieved by query of the Duke Enterprise Data Unified Content Explorer and manual abstraction of electronic medical records. Patients who received care at both clinics were identified, and data associated with the earliest visit were retained. Following final data collection, all phylogenetic and statistical analyses were performed on de-identified data sets to protect participants’ anonymity. The study was approved by the UNC Institutional Review Board (No. 16-0228).
Statewide HIV Sequence Data
To characterize transmission clusters involving TDR, the study population was expanded beyond the clinical cohorts of UNC and Duke to include all sequences in the statewide data set (n = 15 246). This data set is estimated to represent more than 50% of people living with HIV in North Carolina [21]. Within the statewide data set, we identified transmission clusters that included ≥1 sequences from a clinical cohort patient with TDR. We analyzed these clusters to understand the connectivity of patients with TDR to other sequences that had SDRMs but did not come from patients in the clinical cohorts.
Sequence and Drug Resistance Analysis
Most genotypic resistance tests were GenoSure MG assay. Sequences spanned protease nucleotide positions 1–297 and reverse transcriptase positions 1–1200 and were aligned using MUSCLE [22] and edited manually in Bioedit [23]. Gapped positions were stripped, and the final sequence length was 1497 bases. We determined HIV subtypes using the Context-based Modeling for Expeditious Typing (COMET) tool [24]. We identified mutations by ART drug class (non-nucleoside reverse transcriptase inhibitor, nucleoside reverse transcriptase inhibitor [NRTI], protease inhibitor [PI]) using the Stanford University HIV Drug Resistance Database genotypic resistance interpretation algorithm (Sierra, v. 1.1) [25]. Major mutations were selected using the 2009 standardized list of SDRMs from the World Health Organization [7]. We defined TDR as the presence of ≥1 SDRMs in a pretreatment genotype.
Phylogenetic Analyses
Using sequence data from all patients (n = 15 246), a maximum likelihood (ML) phylogenetic tree was constructed in FastTree, v. 2.1.4 [26], with the general time-reversible model of nucleotide substitution using the earliest available sequence from each person. Statistical support of clades was assessed with local branch support values (Shimodaira-Hasegawa-like test [SH test]) in FastTree [27]. Putative transmission clusters were identified using the automated tool ClusterPicker, v. 1.3 [28]. We defined clusters as clades with high branch support (probability ≥0.90, SH test) and a maximum pairwise genetic distance <3.5% difference between all sequences (ie, no 2 sequences in the cluster with a pairwise genetic distance ≥0.035 substitutions per site) [29, 30].
Putative clusters of subtype B sequences in the ML tree were confirmed using Bayesian Markov Chain Monte Carlo (MCMC) inference in BEAST, v. 1.8.2 [31]. Non-B subtypes were investigated in a separate analysis [32]. Sequences clustered in the initial ML tree were split into 40 alignments along cluster lines with <200 sequences per file to decrease computational time in BEAST. Care was taken not to divide potential clusters. All analyses were conducted using the SRD06 nucleotide substitution model, a log-normal relaxed molecular clock model, and the Bayesian Skyline model as coalescent tree prior. The MCMC chain was run for 50–100 million generations with 1–5 runs performed for each file. Convergence of the estimates was considered satisfactory when the effective sample size was >200 in all parameters, as calculated in Tracer, v. 1.6.0 [31]. BEAST log and tree files obtained for data sets with multiple runs were combined using LogCombiner, v. 1.8.2; 10% of the generations were discarded as burn-in. Maximum clade credibility trees (MCCTs) were summarized using TreeAnnotator, v. 1.8.2, keeping the median height over the posterior distribution of trees [31]. Clades with a posterior probability ≥0.95 were considered highly supported.
We analyzed clusters that included ≥1 sequences with TDR from the clinical cohort to determine the degree of connectivity to other sequences with SDRMs. The time of the most recent common ancestor (tMRCA) was estimated for each cluster as the difference in the sampling date of the most recent sequence in the tree from the median height of the basal node of the cluster identified in the MCCT. Cluster time span was calculated as the difference between each cluster’s tMRCA and the sampling date of the most recent sequence in the cluster.
Statistical Analyses
We estimated the prevalence of TDR over the study period and by year among the patients whose first genotype occurred before ART initiation. We calculated binomial exact 95% confidence intervals for prevalence estimates. Binomial regression models with an identity link were used to assess trends in prevalence by sequencing year and to compare TDR by demographic and clinical factors. Our regression models estimated prevalence differences (PDs), the absolute difference in TDR prevalence (in percentage points) when comparing 1 group with another. All models were unadjusted for other covariates, given that our focus was to identify general associations rather than infer causal relationships. Statistical significance was defined at the P <.05 level. All analyses were conducted using R, v. 3.4.3 (R Foundation for Statistical Computing, Vienna, Austria).
RESULTS
Study Population
Between 1997 and mid-2014, HIV-1 pol sequences for 4477 patients were sent from UNC or Duke to the reference laboratory, and 3711 of those patients were successfully matched to UCHCC or Duke clinical data. For 1658 of 3711 (45%) patients, ≥1 sequences were available before ART exposure, and these 1658 patients were the focus of our analysis (Table 1). Most participants were male (73%) and black (62%). The average age of patients at the time of genotyping (interquartile range [IQR]) was 37 (27–46) years. The most commonly reported risk factor for transmission was being a man who has sex with men (MSM; 50%), with few patients reporting injection drug use (7%). At the time of genotyping (within 90 days), the median HIV viral load (IQR) was 4.8 (4.1–5.3) log10 copies/mL, and the median CD4 cell count (IQR) was 283 (75–465) cells/mm3. Nearly all patients were infected with HIV-1 subtype B (98%).
Table 1.
Characteristics of 1658 ART-Naïve Patients in Central NC at First Genotype Sequencing During the Study Period, 1997–2014
| Characteristic | No. (%) |
|---|---|
| Sex | |
| Male | 1211 (73) |
| Female | 447 (27) |
| Race/ethnicity | |
| Black | 1024 (62) |
| White | 454 (27) |
| Other race or Hispanic/Latino | 180 (11) |
| Age, y | |
| <20 | 56 (3) |
| 20–29 | 473 (29) |
| 30–39 | 456 (28) |
| 40–49 | 439 (26) |
| ≥50 | 234 (14) |
| HIV transmission risk factora | |
| MSM | 763 (50) |
| IDU | 69 (7) |
| MSM + IDU | 16 (2) |
| HIV-1 subtype B | 1620 (98) |
| CD4 cell count, cells/mm3 | |
| <200 | 544 (40) |
| 200–349 | 272 (20) |
| 350–499 | 264 (19) |
| ≥500 | 295 (21) |
| Median CD4 (IQR) | 283 (75–465) |
| No CD4 within 90 d of sequencing | 283 |
| Viral load, copies/mL | |
| Median viral load | 64 381 |
| Median log10 viral load (IQR) | 4.8 (4.2–5.3) |
| No viral load within 90 d of sequencing | 280 |
| Transmission cluster | |
| Sequence identified in cluster | 1084 (65) |
| Sequence not in cluster | 574 (35) |
Abbreviations: ART, antiretroviral therapy; IDU, injection drug user; IQR, interquartile range; MSM, men who have sex with men.
aFor MSM, there were 135 missing values, and the percentage is based on the number of patients with nonmissing data (n = 1523). For IDU, data were only available for UNC clinical cohort patients, and the percentage is based on the number of UNC patients with nonmissing data (n = 975). The percentage for MSM + IDU is based on the total number of UNC patients with nonmissing data on both MSM and IDU (n = 956).
Overall Prevalence of TDR
Among 1658 ART-naïve patients, the prevalence of TDR was 12% (95% confidence interval [CI], 10% to 14%); 199 patients had ≥1 SDRMs for NNRTI, NRTI, or PI resistance (Table 2). Among patients with TDR, most had 1 drug class mutation (174/1658, 10.5%), whereas dual- and triple-class TDR were uncommon, at 1.2% (20/1658) and 0.3% (5/1658), respectively. Most patients with TDR harbored resistance to NNRTIs (132/1658, 8%), followed by NRTIs (70/1658, 4%) and PIs (27/1658, 2%). The most frequent SDRM was NNRTI-associated K103N, which was observed for 5.9% of patients (97/1658). For NRTI resistance, thymidine analogue mutations (TAMs) were most commonly observed, with T215Y/F/I/S/D/E/C/V mutations seen in 2.2% of patients (37/1658), followed by M41L in 1.2% (20/1658). M184V was seen in 0.5% (9/1658). Of the PI-associated mutations, L90M was seen in 0.8% of patients (13/1658).
Table 2.
Prevalence of TDR From 1997 to 2014 Among 1658 Patients in Central NC
| TDR Category | No. (%) | Common SDRMs | No. (%) |
|---|---|---|---|
| Any TDR | 199 (12.0) | K103N | 97 (5.9) |
| NNRTI | 113 (6.8) | T215Y/F/S/C/D/E/I/V | 37 (2.2) |
| NRTI | 48 (2.9) | M41L | 20 (1.2) |
| PI | 13 (0.8) | G190A | 19 (1.1) |
| NNRTI + NRTI | 11 (0.7) | Y181C | 13 (0.8) |
| NRTI + PI | 6 (0.4) | L90M | 13 (0.8) |
| NNRTI + NRTI + PI | 5 (0.3) | M46I/L | 11 (0.7) |
| NNRTI + PI | 3 (0.2) | L210W | 10 (0.6) |
Abbreviations: NNRTI, non-nucleoside reverse transcriptase inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; PI, protease inhibitor; SDRM, surveillance drug resistance mutation; TDR, transmitted drug resistance.
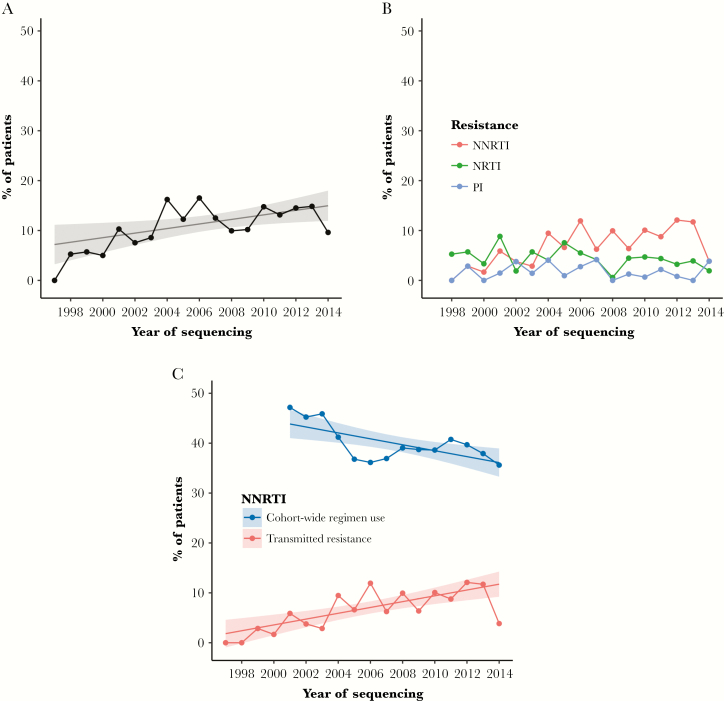
The yearly prevalence of TDR varied from 1997 to mid-2014 but, in general, increased over time (Figure 1A). We found an absolute increase in TDR prevalence of 0.5% (95% CI, 0.1% to 0.8%; P = .02) per calendar year. Although TDR prevalence decreased in the first half of 2014 (data only available through June 2014), we found no statistical evidence that the temporal relationship was parabolic rather than linear (quadratic term P = .12, no change in Akaike information criterion (AIC) AIC [linear model AIC, 1216; quadratic model AIC, 1216]). The prevalence of TDR over time varied by drug class; NNRTI-associated SDRMs increased across the study period (P < .01), whereas NRTI and PI resistance did not show a significant trend (Figure 1B).
Figure 1.
A, Overall transmitted drug resistance (TDR) prevalence by year among 1658 antiretroviral therapy (ART)–naïve patients from 1997 to mid-2014. Fitted linear model indicated an increasing trend in the percentage of ART-naïve patients with ≥1 surveillance drug resistance mutations. B, Prevalence of TDR broken down by drug class resistance (non-nucleoside reverse transcriptase inhibitor [NNRTI], nucleoside reverse transcriptase inhibitor [NRTI], protease inhibitor [PI]). C, Increasing prevalence of NNRTI-associated TDR among 1658 ART-naïve patients despite decreasing prevalence of NNRTI-containing regimen use among UNC and Duke patients on ART. ART regimen data were abstracted starting in 2001.
Increasing NNRTI resistance over the study period is in contrast to the declining use of NNRTI regimens among UNC and Duke cohort patients (Figure 1C). Cohort-wide use of ART regimens that included an NNRTI decreased by 6% per year from 2001 to 2014 (95% CI, –10% to –2%; P < .01). Despite this declining use, the prevalence of NNRTI-associated TDR increased by 0.6% each year over the same period (95% CI, 0.3% to 0.9%; P < .01), driving the overall rise in TDR prevalence. Consistent with this overall trend in TDR, we observed a decrease in NNRTI resistance in the first half of 2014; however, again, we found no statistical evidence that the temporal relationship was parabolic rather than linear (quadratic term P = .21, no change in AIC [linear AIC, 913; quadratic AIC, 913]).
Factors Associated With TDR
We explored the association between TDR and the demographic and clinical characteristics of patients using absolute differences in TDR prevalence (PDs) (Table 3). Compared with MSM, TDR was less common among men who did not report sex with men (PD, –4.4%; 95% CI, –8.6% to 0.3%; P = .03) and women (PD, –4.0%; 95% CI, –7.7% to –0.3%; P = .03). TDR prevalence was highest among patients younger than age 20 years; for patients aged 19–29, 30–39, 40–49, and ≥50 years, the PDs were –12.2%, –9.8%, –12.5%, and –12.1%, respectively (all P < .10). Compared with white patients, TDR prevalence was lower among black patients (PD, –3.9%; 95% CI, –7.7% to –0.1%; P = .04) and patients of Hispanic ethnicity or other race (PD, –5.0%; 95% CI, –10.5% to 0.5%; P = .07). At pretherapy sequencing, patients with TDR had higher CD4 cell counts (median, 331 vs 277; P = .04) but similar HIV viral loads (median, 4.79 vs 4.81; P = .18). TDR prevalence was higher among patients identified in a transmission cluster than among patients whose sequence did not cluster (PD, 5.0%; 95% CI, 1.9% to 8.1%; P = .001).
Table 3.
Prevalence of TDR Associated With Patient Characteristics, With Absolute Differences in TDR Prevalence Estimated With Binomial Regression Using an Identity Link
| Patient Characteristic | TDR, No. (%) | No TDR, No. (%) | Prevalence Difference (95% CI), % |
|---|---|---|---|
| Sex | |||
| Male, MSM | 108 (14) | 671 (86) | Ref |
| Male, not MSM | 28 (9) | 269 (91) | –4.4 (–8.6 to –0.3)a |
| Female | 44 (10) | 403 (90) | –4.0 (–7.7 to –0.3)a |
| Age, y | |||
| <20 | 13 (23) | 43 (77) | Ref |
| 20–29 | 52 (11) | 421 (89) | –12.2 (–23.6 to –0.8)a |
| 30–39 | 61 (13) | 395 (87) | –9.8 (–21.3 to 1.7) |
| 40–49 | 47 (11) | 392 (89) | –12.5 (–23.9 to –1.1)a |
| ≥50 | 26 (11) | 208 (89) | –12.1 (–23.9 to –0.3)a |
| Race | |||
| White | 68 (15) | 386 (85) | Ref |
| Black | 113 (11) | 911 (89) | –3.9 (–7.7 to –0.1)a |
| Hispanic or other | 18 (10) | 162 (90) | –5.0 (–10.5 to 0.5) |
| Transmission cluster | |||
| In cluster | 149 (14) | 935 (86) | 5.0 (1.9 to 8.1)a |
| Not in cluster | 50 (9) | 524 (91) | Ref |
| Median CD4 cell count (IQR) | 331 (147–505) | 277 (72–459) | 0.7 (0.04 to 1.4)a per 100 cells/mm3 |
| Median log10 viral load (IQR) | 4.79 (4.13–5.22) | 4.81 (4.16–5.32) | –1.3 (–3.2 to 0.6) per log10 copies/mL |
Abbreviations: CI, confidence interval; IQR, interquartile range; MSM, men who have sex with men; TDR, transmitted drug resistance.
a P < .05.
Phylogenetic Clusters With TDR
Using sequences from all 15 246 patients with routine genotyping in NC from 1997 to 2014, 7591 sequences (50%) were identified in 2297 transmission clusters. Approximately half of the clusters (1111/2297, 48%) included ≥1 patients from the UNC or Duke clinical cohorts (data available on ART exposure and presence of TDR). The prevalence of TDR was strongly associated with transmission clusters; 75% of patients with TDR (149/199) were identified in a transmission cluster, whereas 64% of patients without TDR (935/1459) were in a cluster (P = .001). Therefore, our phylogenetic analysis focused on clusters containing ≥1 patients with TDR. We identified TDR in 106 transmission clusters that ranged from 2 to 26 members (35% pairs, 54% 3–9, 11% 10–26) and involved a total of 546 persons (296/546 from clinical cohorts, 54%). Most sequences in TDR clusters had ≥1 SDRMs (348/546, 64%), and only a minority of patients with an SDRM were known to be ART-exposed (41/348, 12%). Most patients in clusters with an SDRM were either known to be ART-naïve (149/348, 43%) or had missing data on ART initiation if they were not in the UNC or Duke clinical cohorts (158/348, 45%).
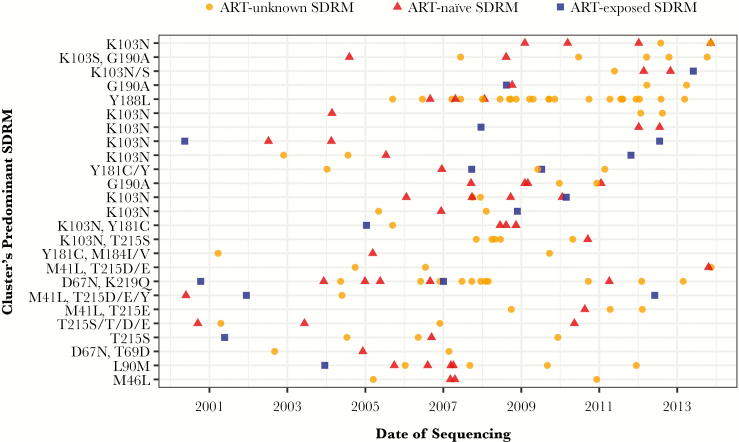
Of 69 TDR clusters with 3 or more members, 25 (36%) were composed of sequences that all had ≥1 SDRMs (Figure 2; phylogenetic trees in Supplementary Figure 1). In 20 of 25 clusters, all members shared an identical SDRM. NNRTI drug resistance was common, and overall resistance was dominated by patients known to be ART-naïve or with unknown ART initiation (89% of patients in 25 clusters). Among these 25 clusters, most had time of the most recent common ancestor before 2005 (median year of tMRCA, 1997), and the time span of clusters ranged from 3.6 to 20.3 years (median, 12.5 years). In comparison, among all 2297 clusters identified in the statewide data set, the median year of tMRCA was slightly more recent (2000), and the median time span was slightly shorter (11.0 years). Most of the 25 clusters were male-dominated; more than half of the members in 20 of 25 clusters were male, and of these clusters, 75% (15/20) consisted only of men. Most members of these 25 clusters were aged 30 years or older, although 7 of 25 clusters were dominated by persons younger than age 30 years.
Figure 2.
Twenty-five transmission clusters illustrating resistance circulation among antiretroviral therapy (ART)–naïve and ART-unknown members. Each horizontal line of dots represents 1 cluster, and each dot indicates a single patient in the cluster by date of first sequencing. All clusters have at least 3 members, all with a surveillance drug resistance mutation (SDRM) and ≥1 members with transmitted drug resistance. Clusters are ordered by drug class resistance (non-nucleoside reverse transcriptase inhibitor [NNRTI], NNRTI/nucleoside reverse transcriptase inhibitor [NRTI], NRTI, protease inhibitor) based on the SDRM shared by the majority of sequences. Within each drug class, clusters are ordered by the most recent member’s sequencing date.
DISCUSSION
Transmitted drug resistance persists among treatment-naïve persons and can impact successful HIV therapy, particularly in resource-limited settings. We found a moderate TDR prevalence estimate of 12% overall, but TDR has steadily increased over the 15 years of observation. The TDR rise was driven largely by NNRTI resistance; few cases of dual and triple class resistance were noted. By incorporating >15 000 HIV-1 pol sequences collected statewide, we traced genetic networks involving TDR, revealing significant clustering of NNRTI mutations among ART-naïve persons, including several clusters of young MSM. Altogether, our study provides important insight into TDR trends and characteristics of drug resistance clustering in the Southeast, which has the greatest burden of HIV infection compared with other US regions [33, 34].
Although our prevalence estimate of 12% is consistent with other US-based studies [10, 11], recent trends in TDR are higher compared with European studies reporting stable [8, 14, 15] or decreasing [16–19] TDR prevalence. The persistence of TDR in North Carolina is predominantly driven by transmitted resistance to NNRTI-containing regimens. Despite declining use of these regimens in our clinical cohort, most transmitted resistance mutations were NNRTI-associated, which increased over time. The prevalence of NNRTI-transmitted resistance in our study (8.0%) mirrors a recent study in San Diego [11]; over the same time period, they observed that prevalence was 8.5% and that it increased in more recent years. This continued propagation of NNRTI SDRMs could be driven by the high transmissibility of the K103N mutation, corroborated by prior work [35]. Despite the absence of drug pressure, the persistence of K103N and other NNRTI-associated SDRMs may be a result of low fitness costs (ie, limited effect of the mutation on pathogen replication) or compensation through additional mutations [36]. In contrast, transmitted resistance to NRTIs and PIs was less common and remained at low levels throughout the study period. Our study could not assess transmitted resistance to integrase inhibitors (INSTIs) given that pre-ART integrase genotyping was not yet routine in the UNC or Duke clinics. However, recent work indicates that major INSTI-transmitted resistance is rare in North Carolina [37].
We found that the prevalence of TDR was highest among young white MSM. Globally, the highest rates of TDR have been reported for MSM [36], whereas research is more limited on other demographic or clinical factors associated with transmission of drug resistance. The disproportionately high risk of TDR among MSM could result from unprotected sexual intercourse in the early stages of infection, when HIV is undiagnosed and more likely to have SDRMs (before possible reversion) [15, 38]. The factors found to be associated with TDR—higher CD4 count and transmission cluster member, in addition to young white MSM—represent markers of earlier diagnosis and potentially more recent transmission. The enhanced ability to detect TDR in earlier infection may then have resulted in the observed associations.
In addition to examining individual risk factors, we used phylogenetic reconstruction of HIV transmission networks to understand the degree of connectivity of patients with TDR. Most TDR cases were identified in clusters, signifying multiple local transmission networks. Resistance in clusters was dominated by patients known to be ART-naïve or with unknown ART initiation, with few known to be ART-exposed. This suggests that TDR circulation is driven by ART-naïve persons, which is consistent with previous reports from Europe that treatment-naïve persons are the major source of TDR [4, 9, 15, 18–20]. Compared with prior work in the United States, in our study, we identified a higher proportion of TDR cases in local transmission clusters (75%) than previously found in San Diego (24%) and Washington, DC (6%) [11, 12]. Phylogenetic analyses are limited by the inability to infer direction of transmission and the fact that genetic linkage does not indicate direct transmission as unsampled persons may be involved in the transmission chain. Nonetheless, transmission networks appear to be an important driver of propagation of SDRM in North Carolina (as opposed to de novo acquisition of resistance), and their continued identification and surveillance will be critical for future intervention efforts and to promote early treatment. In fact, the consolidation of TDR cases in clusters suggests that intervention based on phylogenetic analyses may be particularly effective in identifying and abrogating onward transmission of SDRMs in our region. Similar studies are needed in other locales to better define whether this consolidation of TDR is a unique feature of transmission networks in the Southeast.
Although our data stem from a large HIV patient population in North Carolina, the cohort may not be representative of all people living with HIV in the state, as only persons diagnosed and engaged in care were included. The statewide data set included more than 15 000 patients with sequences, but we could only measure TDR prevalence for patients with ≥1 sequences before known ART exposure, resulting in a sample of 1658 patients. However, our phylogenetic analysis involved all 15 246 patients with genotyping and provides an indication of potential TDR cases missed by our study; of the 546 sequences identified in TDR clusters, 158 contained ≥1 SDRMs but were from patients with missing data on ART initiation. In addition, the demographic and clinical data on our sample of 1658 patients are derived from observational clinical cohorts with potential for measurement error, and in particular, the data on transmission risk categories (MSM, injection drug user [IDU]) were not available for all patients (8% missing MSM, 41% missing IDU). We did not have estimates of recency of infection, which would have been informative in understanding the persistence of NNRTI drug resistance. Despite these limitations, this study still provides an important indication of the propagation of TDR in the Southeastern United States, where little work has been done previously.
In conclusion, TDR has persisted in North Carolina and is predominantly driven by NNRTI resistance, despite declining use of NNRTI-containing regimens. Most TDR cases were identified in transmission clusters, where resistance was dominated by patients known to be ART-naïve or with unknown ART initiation; few were known to be ART-exposed. Consistent with previous findings in Europe, TDR persistence appears to be due to circulation among ART-naïve persons, rather than those failing therapy. Continued TDR surveillance may help detect transmission networks and identify local clusters at high risk of propagating TDR to inform future intervention.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgment
We thank the patients and staff of the UNC Center for AIDS Research Clinical Cohort and the Duke Infectious Diseases Clinic.
Disclaimer. This content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Financial support. This work was supported by the National Center for Advancing Translational Sciences (Grant Award Number UL1TR001111), the National Institute of Allergy and Infectious Diseases (Grant Award Number K08AI112432-02), and the UNC Center for AIDS Research (Grant Award Number P30AI50410). Traineeship support for S.N.L. was provided by the National Institute of Allergy and Infectious Diseases (Grant Award Number T32AI070114-10).
Prior presentation. Portions of this study were presented at the 2017 International AIDS Society Conference on HIV Science.
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Bradley H, Mattson CL, Beer L, et al. ; Medical Monitoring Project Increased antiretroviral therapy prescription and HIV viral suppression among persons receiving clinical care for HIV infection. AIDS 2016; 30:2117–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cohen MS, Chen YQ, McCauley M, et al. ; HPTN 052 Study Team Prevention of HIV-1 infection with early antiretroviral therapy. N Engl J Med 2011; 365:493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wittkop L, Günthard HF, de Wolf F, et al. ; EuroCoord-CHAIN Study Group Effect of transmitted drug resistance on virological and immunological response to initial combination antiretroviral therapy for HIV (EuroCoord-CHAIN joint project): a European multicohort study. Lancet Infect Dis 2011; 11:363–71. [DOI] [PubMed] [Google Scholar]
- 4. Yang WL, Kouyos R, Scherrer AU, et al. ; Swiss HIV Cohort Study Assessing the paradox between transmitted and acquired HIV type 1 drug resistance mutations in the Swiss HIV Cohort Study from 1998 to 2012. J Infect Dis 2015; 212:28–38. [DOI] [PubMed] [Google Scholar]
- 5. Bertagnolio S, Perno CF, Vella S, Pillay D. The impact of HIV drug resistance on the selection of first- and second-line ART in resource-limited settings. J Infect Dis 2013; 207(Suppl 2):S45–8. [DOI] [PubMed] [Google Scholar]
- 6. DHHS Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the Use of Antiretroviral Agents in HIV-1-Infected Adults and Adolescents. United States Department of Health and Human Services: Washington DC; 2007. Available at: https://aidsinfo.nih.gov/contentfiles/AdultandAdolescentGL000721.pdf. Accessed 30 April 2018. [Google Scholar]
- 7. Bennett DE, Camacho RJ, Otelea D, et al. . Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS One 2009; 4:e4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schmidt D, Kollan C, Fätkenheuer G, et al. ; ClinSurv-HIV Drug Resistance Study Group in CHAIN Estimating trends in the proportion of transmitted and acquired HIV drug resistance in a long term observational cohort in Germany. PLoS One 2014; 9:e104474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rhee SY, Blanco JL, Jordan MR, et al. . Geographic and temporal trends in the molecular epidemiology and genetic mechanisms of transmitted HIV-1 drug resistance: an individual-patient- and sequence-level meta-analysis. PLoS Med 2015; 12:e1001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim D, Ziebell R, Saduvala N, et al. . Trend in transmitted HIV-1 antiretroviral drug resistance-associated mutations, 10 HIV surveillance areas, United States, 2007–2010. Paper presented at: CROI 3-6 March 2013; Atlanta, GA. Abstract 149. [Google Scholar]
- 11. Panichsillapakit T, Smith DM, Wertheim JO, et al. . Prevalence of transmitted HIV drug resistance among recently infected persons in San Diego, CA 1996-2013. J Acquir Immune Defic Syndr 2016; 71:228–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kassaye SG, Grossman Z, Balamane M, et al. . Transmitted HIV drug resistance is high and longstanding in metropolitan Washington, DC. Clin Infect Dis 2016; 63:836–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aldous AM, Castel AD, Parenti DM; DC Cohort Executive Committee Prevalence and trends in transmitted and acquired antiretroviral drug resistance, Washington, DC, 1999-2014. BMC Res Notes 2017; 10:474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hofstra LM, Sauvageot N, Albert J, et al. ; SPREAD Program Transmission of HIV drug resistance and the predicted effect on current first-line regimens in Europe. Clin Infect Dis 2016; 62:655–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Drescher SM, von Wyl V, Yang WL, et al. ; Swiss HIV Cohort Study Treatment-naive individuals are the major source of transmitted HIV-1 drug resistance in men who have sex with men in the Swiss HIV Cohort Study. Clin Infect Dis 2014; 58:285–94. [DOI] [PubMed] [Google Scholar]
- 16. Tostevin A, White E, Dunn D, et al. ; UK HIV Drug Resistance Database Recent trends and patterns in HIV-1 transmitted drug resistance in the United Kingdom. HIV Med 2017; 18:204–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pineda-Peña AC, Schrooten Y, Vinken L, et al. . Trends and predictors of transmitted drug resistance (TDR) and clusters with TDR in a local Belgian HIV-1 epidemic. PLoS One 2014; 9:e101738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vega Y, Delgado E, Fernández-García A, et al. ; Spanish Group for the Study of New HIV-1 Diagnoses in Galicia and Basque Country Epidemiological surveillance of HIV-1 transmitted drug resistance in Spain in 2004-2012: relevance of transmission clusters in the propagation of resistance mutations. PLoS One 2015; 10:e0125699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Paraskevis D, Kostaki E, Magiorkinis G, et al. . Prevalence of drug resistance among HIV-1 treatment-naive patients in Greece during 2003-2015: transmitted drug resistance is due to onward transmissions. Infect Genet Evol 2017; 54:183–91. [DOI] [PubMed] [Google Scholar]
- 20. Mbisa JL, Fearnhill E, Dunn DT, et al. ; UK HIV Drug Resistance Database Evidence of self-sustaining drug resistant HIV-1 lineages among untreated patients in the United Kingdom. Clin Infect Dis 2015; 61:829–36. [DOI] [PubMed] [Google Scholar]
- 21. North Carolina HIV/STD Surveillance Unit. North Carolina HIV/STD Epidemiologic Profile (2013). Raleigh, NC: NC Department of Health and Human Services; 2015; Available at: http://epi.publichealth.nc.gov/cd/stds/figures/Epi_Profile_2013.pdf. Accessed 3 July 2018. [Google Scholar]
- 22. Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 2004; 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 98/98/ NT. Nucleic Acids Symp Ser 1999; 41:95–8. Accessed 30 April 2018. [Google Scholar]
- 24. Struck D, Lawyer G, Ternes AM, et al. . COMET: adaptive context-based modeling for ultrafast HIV-1 subtype identification. Nucleic Acids Res 2014; 42:e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu TF, Shafer RW. Web resources for HIV type 1 genotypic-resistance test interpretation. Clin Infect Dis 2006; 42:1608–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 2009; 26:1641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guindon S, Dufayard JF, Lefort V, et al. . New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 2010; 59:307–21. [DOI] [PubMed] [Google Scholar]
- 28. Ragonnet-Cronin M, Hodcroft E, Hué S, et al. ; UK HIV Drug Resistance Database Automated analysis of phylogenetic clusters. BMC Bioinformatics 2013; 14:317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hassan AS, Pybus OG, Sanders EJ, et al. . Defining HIV-1 transmission clusters based on sequence data. AIDS 2017; 31:1211–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pasquale DK, Doherty IA, Sampson LA, et al. . Leveraging phylogenetics to understand HIV transmission and partner notification networks. J Acquir Immune Defic Syndr 2018; 78:367–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rambaut A, Drummond A.. Tracer v1.4. 2007. Available at: http://beast.bio.ed.ac.uk/Tracer. Accessed 30 April 2018. [Google Scholar]
- 32. Dennis AM, Hué S, Learner E, et al. . Rising prevalence of non-B HIV-1 subtypes in North Carolina and evidence for local onward transmission. Virus Evol. 2017; 3(1): vex013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Centers for Disease Control and Prevention. HIV in the United States by Geography. Division of HIV/AIDS Prevention, National Center for HIV/AIDS, Viral Hepatitis, STD, andTB Prevention, Centers for Disease Control and Prevention;2017. Available at: https://www.cdc.gov/hiv/statistics/overview/geographicdistribution.html. Accessed 30 April 2018. [Google Scholar]
- 34.Centers for Disease Control and Prevention. HIV Surveillance Report, 2016; vol. 28. Available at: http://www.cdc.gov/hiv/library/reports/hiv-surveillance.html. Published November 2017. Accessed 30 April 2018. [Google Scholar]
- 35. Wertheim JO, Oster AM, Johnson JA, et al. . Transmission fitness of drug-resistant HIV revealed in a surveillance system transmission network. Virus Evol. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang WL, Kouyos RD, Böni J, et al. ; Swiss HIV Cohort Study Persistence of transmitted HIV-1 drug resistance mutations associated with fitness costs and viral genetic backgrounds. PLoS Pathog 2015; 11:e1004722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Menza TW, Billock R, Samoff E, et al. . Pre-treatment integrase strand transfer inhibitor resistance in North Carolina from 2010–2016. AIDS 2017; 31:2235–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pham QD, Wilson DP, Law MG, et al. . Global burden of transmitted HIV drug resistance and HIV-exposure categories: a systematic review and meta-analysis. AIDS 2014; 28:2751–62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.