Abstract
Rim1 is the mitochondrial single-stranded DNA binding protein in Saccharomyces cerevisiae and functions to coordinate replication and maintenance of mtDNA. Rim1 can form homo-tetramers in solution and this species has been assumed to be solely responsible for ssDNA binding. We solved structures of tetrameric Rim1 in two crystals forms which differ in the relative orientation of the dimers within the tetramer. In testing whether the different arrangement of the dimers was due to formation of unstable tetramers, we discovered that while Rim1 forms tetramers at high protein concentration, it dissociates into a smaller oligomeric species at low protein concentrations. A single point mutation at the dimer–dimer interface generates stable dimers and provides support for a dimer–tetramer oligomerization model. The presence of Rim1 dimers in solution becomes evident in DNA binding studies using short ssDNA substrates. However, binding of the first Rim1 dimer is followed by binding of a second dimer, whose affinity depends on the length of the ssDNA. We propose a model where binding of DNA to a dimer of Rim1 induces tetramerization, modulated by the ability of the second dimer to interact with ssDNA.
INTRODUCTION
Single-stranded DNA binding proteins (SSBs) are ubiquitous and are involved in all DNA processing steps that generate ssDNA. In eukaryotes, the hetero-trimeric RPA protein provides the major non-specific single-stranded DNA binding activity in the nucleus (1–3). In mitochondria, single-stranded DNA binding activity is provided by a homo-tetrameric SSB that bears strong similarity to the SSB from Escherichia coli (EcSSB). Human mitochondrial SSB (HsmtSSB) forms stable homo-tetramers that have DNA binding activities similar to EcSSB (4–7), as does the mitochondrial SSB from Saccharomyces cerevisiae (Rim1) (8,9). While formation of homo-tetramers appears to be a characteristic of most eubacterial SSBs, phage-encoded SSBs display a wider range of oligomeric states (10–15). Interestingly, Mtp1/mtTBP, a Rim1 homologue in Candida parapsilosis with dual functions as a general SSB and as a sequence specific SSB for the telomeric ends of its mitochondrial DNA, does not appear to form stable tetramers in solution, forming a significant fraction of dimers (16). These observations indicate that single-stranded DNA binding activity can be carried out by more than one oligomeric species of SSB and raise the question of whether formation of unstable tetramers as observed for Mtp1/mtTBP is unique or shared by other mitochondrial SSBs in fungi.
Rim1 harbors a natural tyrosine instead of a histidine at a highly conserved position in EcSSB, HsmtSSB and mouse mtSSB (6,9,17,18). Mutation of the conserved histidine to tyrosine (H55Y) in EcSSB leads to formation of unstable tetramers, that at low protein concentrations cooperatively dissociate into monomers (19–21). Similarly, mutation of H69Y of mouse mtSSB leads to destabilization of tetramers (18). The presence in Rim1 of a tyrosine at this position does not appear to affect its ability to form a homo-tetramer. Recent studies clearly confirmed the original observation that Rim1 forms tetramers in solution, at least at the high concentrations tested (8). However, at the protein concentration at which Rim1 forms tetramers, EcSSB harboring the H55Y mutation forms tetramers as well (19–21), albeit it dissociates into monomers at lower protein concentration. Rim1 has been assumed to form stable tetramers across all regimes of protein concentrations and the possibility that, at low protein concentration, Rim1 may dissociate to form smaller oligomeric species has not been considered. In this regard, we note that some of the reported data (8) suggest that Rim1 may have a behavior more complex than expected for a pre-formed stable homo-tetramer.
Interaction of SSBs with ssDNA not only provides protection of the DNA from nucleolytic digestion but it also affords a platform for binding and recruitment of various SSB interacting proteins (SIPs) (22). EcSSB interacts via its C-terminal disordered tails with at least fourteen SIPs (22). Interestingly, HsmtSSB and Rim1 do not possess long C-terminal tails and lack the conserved residues at the ‘tip’ that mediate interactions with the SIPs in EcSSB. Yet, HsmtSSB directly interacts with p53 (5) and specifically stimulates the unwinding activity of the mitochondrial Twinkle helicase (23). Of note, it has recently been reported that Rim1physically interacts with the Pif1 helicase and stimulates the unwinding activity of the helicase (8). In an effort to further understand the function of this interaction we discovered that, in solution, the behavior of Rim1 is more complex than previously assumed. Using a combination of X-ray crystallography, analytical sedimentation and DNA binding studies, in this work we show that while at high protein concentrations Rim1 forms tetramers, consistent with previous reports (8,9), at low protein concentrations Rim1 dissociates into smaller oligomeric species. At low protein concentrations DNA binding is dominated by the high affinity interaction of Rim1 dimers, with binding of additional dimers being modulated by the length of the ssDNA. We propose a model where binding of DNA induces tetramerization of Rim1 dimers. The ability of S. cerevisiae Rim1 to dissociate into dimers suggests the presence of species-specific changes in the oligomeric state of mitochondrial SSBs.
MATERIALS AND METHODS
Protein expression and purification
The coding sequence for S. cerevisiae Rim1 (Rim1) was synthesized and provided in pUC57 (GenScript), while the one for human mitochondrial SSB (HsmtSSB) was synthetized as a gBlocks® (IDT). Both coding sequences were optimized for expression in E. coli. The mitochondrial forms, comprising amino acids 17–135 for Rim1 and 17–148 for HsmtSSB, were first sub-cloned in a pET28a vector (Novagen) between NcoI and XhoI restriction sites, followed by removal by site-directed mutagenesis of the extra glycine from the NcoI site. Rim1 harboring the mutation Y85H (Rim1Y85H) was generated by site directed mutagenesis. The proteins were over-expressed in E. coli BL21 (DE3) cells by induction with 0.4 mM IPTG and grown overnight at 16°C, in LB medium for Rim1s and 2xYT for HsmtSSB. Seleno-l-methionine labeling of Rim1 was produced with a protocol for non-auxotroph E. coli BL21 (DE3) cells (24). Detailed purification protocols are presented in Supplementary material. Before use, the purified Rim1s and HsmtSSB (Supplementary Figure S1) were dialyzed in the indicated buffer and quantified spectrophotometrically using extinction coefficients ϵ280 = 14 800 M−1 cm−1 for Rim1 (25), ϵ280 = 12 950 M−1 cm−1 for Rim1Y85H (26) and ϵ280 = 19 940 M−1 cm−1 for HsmtSSB (5). The experiments were performed in Buffer HKxM5, where H is 20 mM HEPES pH 7.4, 1% (v/v) glycerol; Kx is KCl and the subscript x is its concentration in mM; M5 is 5 mM MgCl2 when present.
Crystallization and structure determination of Rim1
The native and seleno-l-methionine labeled Rim1 proteins were dialyzed in Buffer F (20 mM HEPES pH 8.2, 100 mM NaCl, 1 mM EDTA, 4% (v/v) glycerol) and concentrated to ∼8 mg/ml. Crystallization of Rim1 was performed using the hanging-drop vapor-diffusion method at 22°C. Crystals were obtained by mixing equal volumes of protein (∼8 mg/ml) and the precipitant solution present in the well (0.2 M MgCl2 and 20% (w/v) PEG 3350). Crystals grew to final dimensions of about 100 μm × 40 μm × 40 μm within a week. The crystals were flash frozen in liquid nitrogen after a brief transfer into mother liquor with added 10% PEG 3350. Diffraction data for the native and seleno-l-methionine labeled Rim1 crystals were collected under cryogenic conditions (100 K) at the Beamline ALS 4.2.2. Native Rim1 crystallized in two crystal forms with space groups C2221 (Form 1, PDB: 6CQK) and P212121 (Form 2, PDB: 6CQM), while seleno-l-methionine labeled Rim1 crystallized with space group P212121 (Form 2, PDB: 6CQO). The structure of the seleno-l-methionine labeled Form2 was determined first, using phases obtained by the anomalous diffraction of selenium measured at single wavelength (Se-SAD). This structure was further refined with native data to obtain the native structure in Form2. The native structure of Form1 was determined by molecular replacement (MR) by using monomer of seleno-l-methionine Form2. Detailed information on diffraction data processing and models building are provided in Supplementary material, and data collection, phasing and refinement statistics are summarized in Supplementary Table S1.
DNA substrates
Poly (dT) was purchased from Midland Certified Reagents and it had an average size larger than 250 nt. Fluorescently modified oligo-(dT)n of lengths up to 38 nt were purchased from Integrated DNA Technology. Fluorescently modified oligo-(dT)n longer than 38 nt and the ones modified with a donor–acceptor couple were a kind gift from Dr Lohman (Washington University School of Medicine, St. Louis, MO, USA). DNA concentrations were determined spectrophotometrically using an extinction coefficient ϵ260 = 8100 M−1 cm−1 for dT, corrected for the contribution at 260 nm of the fluorophores. The average degree of labeling was higher than 90%.
Analytical ultracentrifugation
All sedimentation experiments were collected on an Optima XL-A analytical ultracentrifuge using a An60Ti rotor (Beckman Coulter), as previously described (27–29). Sedimentation velocity experiments were performed using Epon charcoal-filled double-sector centerpieces at 50 000 rpm, monitoring absorbance at either 280 or 230 nm. Sedimentation equilibrium experiments were performed using Epon charcoal-filled six-sector centerpieces at 12 000–27 000 rpm, monitoring absorbance at either 280 or 230 nm for protein alone, or at 550 nm for the complexes of protein with Cy3-labeled ssDNA. Sedimentation equilibrium and velocity data were processed and analyzed with SedFit/SedPhat (NIBIB, NIH). A single-species model was sufficient to fit most of the equilibrium absorbance profiles, unless otherwise indicated. The standard error of the fits was smaller than the size of the symbols used in the figures, and it was therefore omitted. The apparent molecular weights were determined using the partial specific volume of 0.74 ml/g for Rim1 and a partial specific volume of 0.52 ml/g for DNA.
Equilibrium fluorescence titrations
All fluorescence titrations were performed with an L-format PC1 spectrofluorimeter (ISS, Champaign, IL, USA) equipped with Glan-Thompson polarizers, as previously described (27,28,30). Measurements were recorded using excitation and emission wavelengths as follows: λex = 520 nm and λem = 565 nm for Cy3 labeled DNA; λex = 520 nm and λem = 662 nm for Cy5–Cy3 labeled DNA; λex = 520 nm and λem = 706 nm for Cy5.5–Cy3 labeled DNA; λex = 490 nm and λem = 530 nm for FAM- and FL-labeled DNA. All titrations were carried out at 20°C. Comparison of two independent Rim1 preparations using a reference DNA substrate showed that the preparation to preparation variability is <5%.
RESULTS
Crystal structure of Rim1
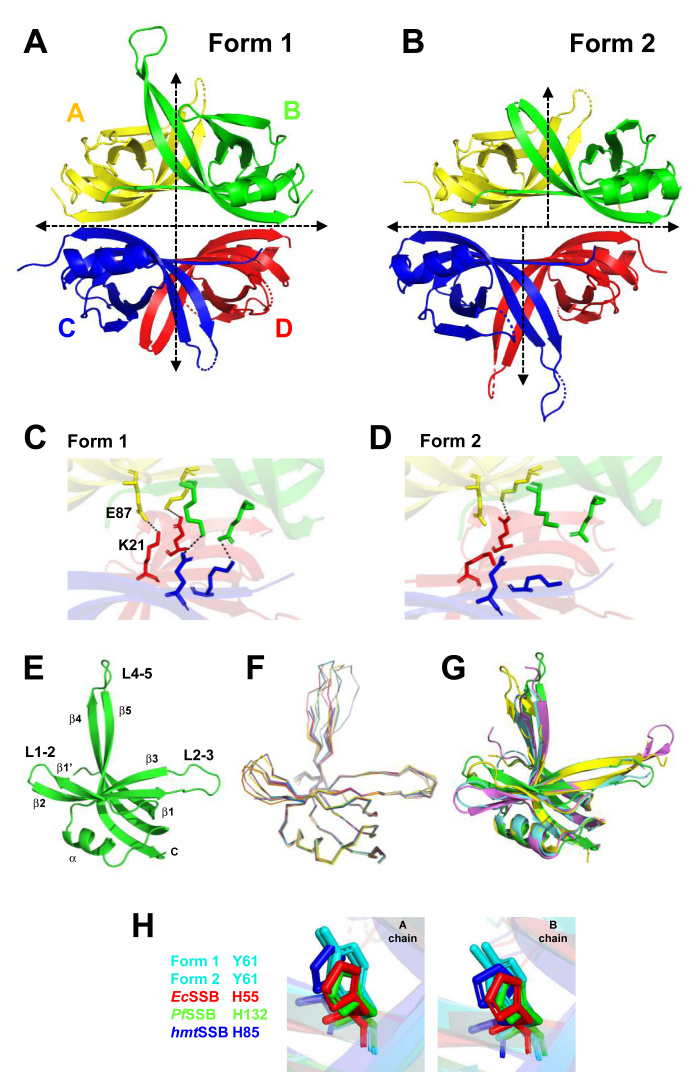
We determined crystal structures of S. cerevisiae Rim1 at 2.8–3.0 Å resolution in two crystal forms from the same crystal growth conditions, with space groups C2221 (Form 1, PDB: 6CQK) and P212121 (Form 2, PDB: 6CQM and 6CQO). The electron density of 16 aa at the C terminus was missing in both forms, suggesting a high degree of flexibility of this region as for other SSBs (6,17,31,32). The asymmetric unit of crystal Form 1 contains one tetramer with two dimers AB and CD in a head to head arrangement with perfect 222 symmetry (Figure 1A), similar to other known SSBs (6,17,31). The asymmetric unit of crystal Form 2 contains three dimers and two monomers, which after symmetry operations generate two tetramers that do not have a perfect 222 symmetry (Figure 1B). In crystal Form 2 the two dimers AB and CD are slightly tilted and translated relative to each other. In both crystal forms tetramer assembly is mediated by interactions of a six beta-stranded sheet, which includes interfaces from monomers A and C, A and D, B and C, and B and D. The total tetramer contact surface area at the interface of the AB and CD dimers is approximately 1938 and 2135 Å2 for crystal Form 1 and Form 2, respectively. In addition to hydrophobic interactions, the tetramer in crystal Form 1 is stabilized by four symmetric pairs of potential salt bridges (3.2–4.4 Å) between K21 and E87 of monomers A and D, and B and C (Figure 1C), which is characteristic of other SSBs (6,17,31). However, due to the asymmetric tetramer conformation, in crystal Form 2 only one potential salt bridge is present, suggesting weaker tetramer assembly (Figure 1D).
Figure 1.
Crystal structures of Rim1. (A, B) Models of Rim1 tetramers determined from crystal Form 1 in A and 2 in B. (C, D) Zoom-in of the dimer–dimer interfaces highlighting the potential salt-bridges (dash lines) in crystal Form 1 in C and Form 2 in D. (E) Model of a Rim1 monomer. (F) Superimposition of all the Rim1 monomers from crystal Form 1 and 2. (G) Superimposition of the monomer in E with monomers from EcSSB (PDB: 1sru, cyan), Plasmodium falciparum SSB (PDB: 3ulp, pink), and HsmtSSB (PDB: 3ull, yellow). (H) Position of Y61 of Rim1 in crystal Form 1 and 2 relative to the histidine of EcSSB, PfSSB and HsmtSSB.
The difference in conformations of the tetramers of crystal Form 1 and 2 does not seem to originate from either a change in the conformation of the monomers or the AB (or CD) dimer interfaces, but rather from a rigid body offset upon formation of the tetramer. The structure of the Rim1 monomers from residues 17 to 119 contains a characteristic OB-fold composed of a six-stranded closed β-barrel domain (β1–β1′–β2–β3–β4–β5) with a α-helix between the β3 and β4 strands and three long connecting loops (Figure 1E, one subunit of crystal Form 1). Loops L1–2 (aa 35–43), L2–3 (aa 50–59), and L4–5 (aa 95–103) exhibit considerable structure variation and their electron density is not well defined in some subunits in either crystal form. All monomers from crystal Form 1 and Form 2 have a RMSD of 0.43–0.8 Å over 89–94 equivalent Cα atoms, except for the loop regions that show higher conformational heterogeneity (Figure 1F). Also, the monomer structures show an RMSD between 1.12 and 1.25 Å over 56–66 equivalent Cα atoms when compared to monomers from other known SSBs (Figure 1G). Finally, two adjacent monomers (A and B or C and D) form a stable dimer with a 2-fold non-crystallographic symmetry axis in both crystal forms. Each dimer is stabilized by main chain hydrogen bonds involving two intermolecular antiparallel β1 strands (aa 18–25) of two adjacent units, forming a flat beta pleated sheet composed of six antiparallel strands (β1, β4 and β5 of each subunit). The dimer is further stabilized by two potential salt bridges between D18 and R27 and hydrophobic interactions between F19, V25 and L106 of adjacent subunits. The total buried surface area for the dimer interface is ∼2100 Å2 which is close to the one found in other SSBs (6,17,31). A H55Y mutation in EcSSB leads to an unstable tetramer that cooperatively dissociates into monomers (19,20). At the corresponding position Rim1 harbors a natural tyrosine (Y61). Interestingly, within the A or B monomer of Rim1 Y61 occupies a similar position as the corresponding histidine in the A or B monomers of other SSBs (Figure 1H), suggesting that Y61 in Rim1 may not be as destabilizing to the AB (or CD) dimer interface.
Rim1 is not a stable tetramer in solution
To test whether the two conformations of Rim1 observed in the crystal structures possibly reflect formation of unstable tetramers, we examined its oligomeric state in solution, using analytical sedimentation approaches. Previous analyses of the oligomeric state of Rim1 indicated formation of a stable tetramer (8,9), at high protein concentrations.
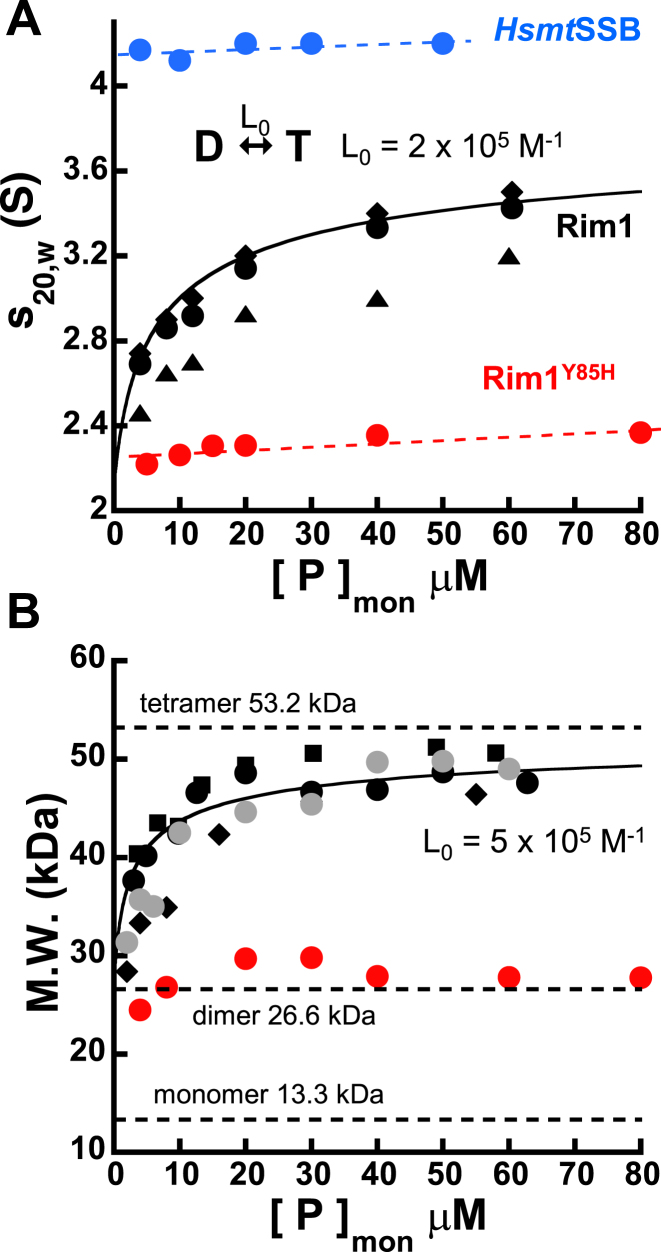
Sedimentation velocity analysis of Rim1 showed the presence of a single distribution of sedimentation coefficients, at any protein concentration tested (Supplementary Figure S2A). However, the calculated s20,w decreases as the Rim1 concentration decreases (Figure 2A), suggesting that Rim1 may not form a single stable species in solution, even in the presence of 1 M KCl. This is in stark contrast to the behavior of HsmtSSB (Figure 2A), that forms stable tetramers in solution (5). The presence in solution of an oligomeric species of Rim1 smaller than a tetramer was confirmed by the molecular weight (M.W.) determined from equilibrium analytical sedimentation experiments performed at different Rim1 concentrations. At each Rim1 concentration, a single species model is sufficient to fit the data (Supplementary Figure S2B). At high protein concentrations the calculated M.W. is consistent with formation of a tetramer of Rim1, as reported (8,9). However, below ∼20 μM of Rim1 monomer the calculated M.W. decreases, independent of the solution conditions (Figure 2B). A decreasing M.W. obtained from fitting the data with a single species suggests that the observed M.W. is a weight average of at least two species. The loss of the monitored absorbance signal limits the range of low protein concentrations that is accessible experimentally and, therefore, the minimum species formed by Rim1 in solution remains uncertain. As a result, the currently available experimental data prevent us from clearly determining the mechanism of oligomerization; that is to say, whether the Rim1 tetramer dissociates into a stable dimer (dimer–tetramer) or monomer (monomer-tetramer or monomer-dimer–tetramer). The solid lines in Figure 2A and B are tentative analyses of the data assuming a simple dimer–tetramer equilibrium (Supplementary material), with an intrinsic oligomerization constant L0 of 2 × 105 to 5 × 105 M−1. Independent of a precise model for oligomerization, the observed decrease in both the sedimentation coefficient and apparent molecular weight indicates that Rim1 does not form stable tetramers in solution, which is different from previous reports (8,9).
Figure 2.
Rim1 does not form stable tetramers in solution. (A) The sedimentation coefficient as a function of Rim1 concentration (monomers) in Buffer HK150, for two preparations of the protein (• and ▴) and in Buffer HK1000 (♦). The solid black line is the fit of the data collected in Buffer HK150 to a dimer–tetramer oligomerization model (equations s1 and s4 in Supplementary material) with L0 = (2 ± 0.3) x 105 M−1 and sT = (3.9 ± 0.1) S, keeping sD fixed at 2.1 S. The protein concentration dependences for HsmtSSB and Rim1Y85H are shown in blue and red, respectively. The dashed lines are linear fits meant to indicate little protein concentration dependence of the s20,w. (B) Protein concentration dependence of the molecular weight of Rim1 in Buffers HK150 (•), HK150M5 ( ), HK20 (▪) and HK1000 (♦). The data in red are for Rim1Y85H. The solid black line is a simulation with a dimer–tetramer with L0 = 5 × 105 M−1, meant to capture the trend of the data.
), HK20 (▪) and HK1000 (♦). The data in red are for Rim1Y85H. The solid black line is a simulation with a dimer–tetramer with L0 = 5 × 105 M−1, meant to capture the trend of the data.
To probe further the oligomeric state of Rim1, we reasoned that mutations that would alter the stability of the Rim1 tetramer may provide a means to test the origin and nature of the smaller oligomeric states. The simplest explanation for an unstable tetramer of Rim1 would be the presence of a natural tyrosine at position 61, rather than a histidine (Supplementary Figure S1B). A H55Y mutation in EcSSB leads to an unstable tetramer that at low protein concentration cooperatively dissociates into monomers (19,20), and one would predict that changing Y61 back to a histidine would lead to stable tetramers of Rim1. However, mutation Y61H in Rim1 appears to change the properties of protein and we were not able to purify this protein construct to homogeneity. Thus, we chose to test mutations at the dimer–dimer interface predicted to destabilize the Rim1 tetramer. For EcSSB it has been shown that mutation Y78R at the dimer–dimer interface leads to dissociation of the tetramer into dimers (33). Thus, we generated a Rim1 construct with a tyrosine to histidine mutation at position 85 (Rim1Y85H, Supplementary Figure S1B). We chose histidine, rather than arginine as for EcSSB, because sequence analysis suggests that other mitochondrial SSBs at this position have a leucine, isoleucine, phenylalanine or histidine. At difference with wild-type Rim1, the sedimentation coefficient of Rim1Y85H shows little change over the protein concentration range tested (Figure 2A), indicating the presence in solution of a single, stable species. Sedimentation equilibrium experiments indicate that Rim1Y85H forms a stable dimer, over the same protein concentration range (Figure 2B). These data argue that in solution either Rim1 forms a stable dimer or, if the dimer dissociates into monomers, the M-D equilibrium dissociation constant is at least one order of magnitude lower than the lowest concentration of protein tested. Also, the similar values of the s20,w extrapolated at low protein concentration for Rim1 and Rim1Y85H suggest that the tetramer may dissociate into a AB/CD dimer rather than a AC/BD dimer.
Rim1 binds ssDNA with a site size of ∼14 nt per monomer
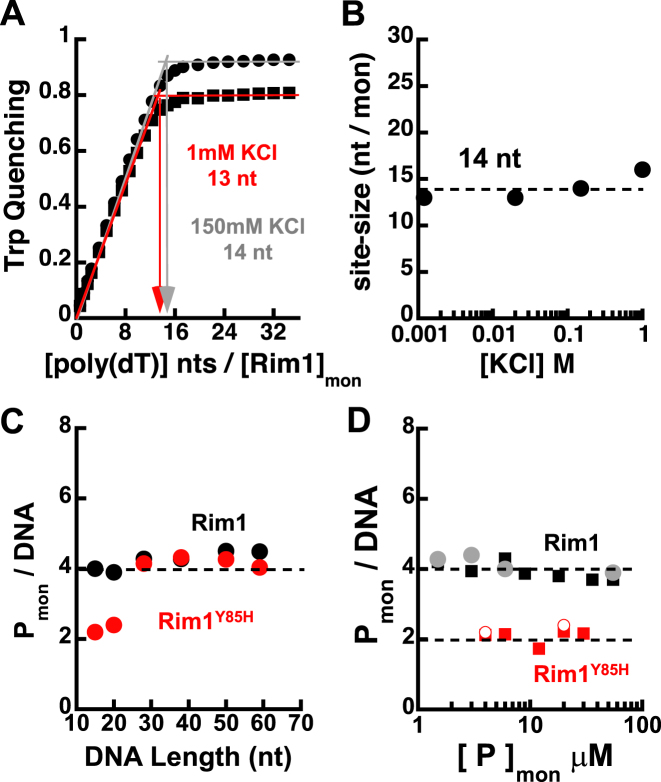
To determine which species of Rim1 is competent for DNA binding we performed studies at different Rim1 and DNA concentrations. First, we determined the occluded ssDNA binding site-size, a parameter not known for Rim1 and needed to interpret binding to ssDNA of different lengths. To this end, we performed equilibrium fluorescence titrations monitoring the large quenching of Rim1 intrinsic tryptophan fluorescence induced by binding of poly(dT) to the protein (Figure 3A). The calculated site-size indicates that ∼14 nt are occluded by a Rim1 monomer (Figure 3B), independent of the KCl concentration.
Figure 3.
Rim1 binds ssDNA with an occluded site-size of ∼14 nt per monomer. (A) Quenching of tryptophan fluorescence as a function of the ratio of the concentration of poly (dT) (nt) to the concentration of Rim1 (monomers), in Buffers HK150 (•) and HK1 (▪). (B) Occluded site-size in nucleotides per Rim1 monomer as a function of KCl concentration. (C) Stoichiometry of Rim1-DNA complexes (•) formed with different lengths of ssDNA in Buffer HK150, under conditions where Rim1 is predominantly a tetramer. In red are the data for Rim1Y85H. (D) Stoichiometry of Rim1-DNA complexes formed in Buffer HK150 with dT15-Cy3-T (▪) and dT20-Cy3-T ( ), as a function of Rim1 concentration. In red (solid and open symbol) are the corresponding data for Rim1Y85H.
), as a function of Rim1 concentration. In red (solid and open symbol) are the corresponding data for Rim1Y85H.
Next, we examined Rim1 binding to DNA under conditions where Rim1 forms a stable tetramer in solution. For this, we performed equilibrium analytical ultracentrifugation experiments monitoring the absorbance of Cy3-labeled ssDNAs (3 μM) of different lengths, in the presence of a high loading concentration of Rim1 (55 μM), in Buffer HK150. For each DNA length, a single species model was sufficient to describe the data. The stoichiometry (Rim1mon/DNA) calculated for complexes assembled at high Rim1 concentration with different lengths of ssDNA (Figure 3C) indicates that a single pre-formed Rim1 tetramer binds to these lengths of ssDNA. The observation that a single tetramer binds to a ssDNA 59 nt long suggests a site-size of ∼ 15 nt per monomer, consistent with the one independently determined in Figure 3B.
To test whether a species of Rim1 smaller than a tetramer would bind to DNA we performed sedimentation equilibrium experiments, in Buffer HK150, monitoring the absorbance of 3 μM Cy3-labeled dT15 and dT20 (short enough to bind a single Rim1 monomer) in the presence of different loading concentrations of Rim1. For concentrations of Rim1 lower than 6 μM a two-species model (free DNA and DNA-Rim1 complex) was used to fit the data. The calculated stoichiometry of the DNA-Rim1 complexes (Figure 3D) indicates that a Rim1 tetramer forms on DNA, even for Rim1 concentrations which are lower than the concentration of DNA. In light of the observation in Figure 2 that Rim1 alone does not form a stable tetramer, we would have expected smaller oligomeric species (e.g. a dimer) bound to these lengths of DNA. To clarify this discrepancy, we performed the same experiment using Rim1Y85H, which forms stable dimers in solution (Figure 2). Independent of the protein concentration, Rim1Y85H forms a dimer with Cy3-labeled dT15 and dT20 (Figure 3D). However, at high protein concentration Rim1Y85H forms a tetramer on ssDNA longer than 20 nt (Figure 3C). Despite being a stable dimer on its own, Rim1Y85H can form a tetramer even on a DNA that is long enough to engage with no more than two monomers (e.g. 28 nt), suggesting that at high protein concentration Rim1Y85H forms tetramers upon DNA binding. A similar scenario may explain the discrepancy between the data for wild-type Rim1 in Figures 2 and 3D.
High affinity binding of a first Rim1 dimer to ssDNAs short enough to engage less than two monomers is followed by lower affinity binding of a second dimer
The data in the previous section suggested the possibility that rather than a pre-formed tetramer binding to DNA, DNA binding may induce dimerization of Rim1 dimers to form tetramers on DNA. To test this possibility, we performed equilibrium DNA binding experiments at concentrations of DNA and Rim1 lower than the ones used in the previous experiments.
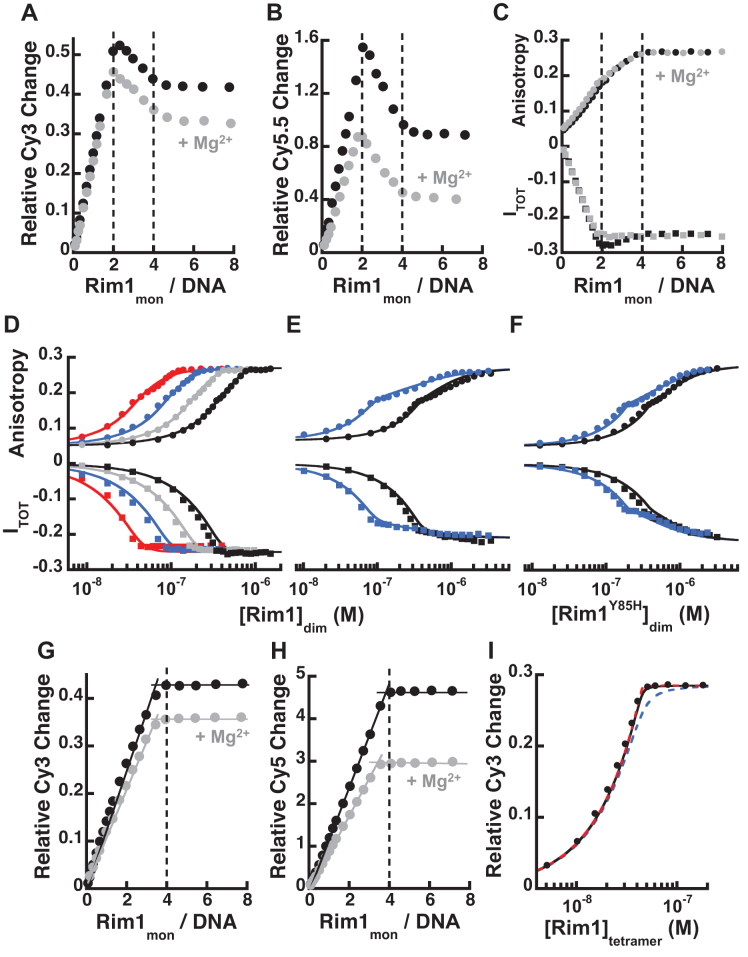
Binding of Rim1 to Cy3-labeled ssDNAs induces a sufficient increase in the fluorescence of Cy3 to monitor the interaction. For ssDNA shorter than 28 nt and a Cy3 label at the 3′-end, the Cy3 fluorescence change is dominated by binding of a single dimer of Rim1 (Supplementary Figure S3A), providing direct evidence that this is the dominant species in solution. The same behavior is observed independent of MgCl2 being present in solution, indicating that the divalent cation does not alter the oligomeric state of the protein, as also observed in Figure 2B. However, based on these data one would erroneously conclude that Rim1 binds to DNA exclusively as a dimer. This is an example of a limiting case where high affinity binding of the first ligand dominates the monitored signal and binding of a second ligand occurs with much lower affinity and with insignificant contribution to the signal (Supplementary material).
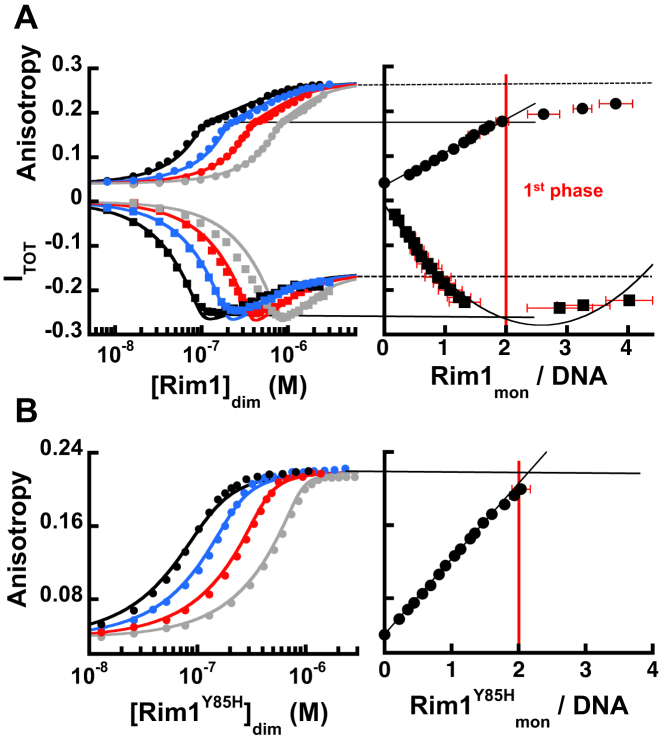
This conclusion is supported by DNA experiments using a carboxy-fluorescein labeled ssDNA (FAM-dT20), and monitoring changes in its fluorescence anisotropy and total intensity as a function of Rim1 concentration. The changes in both anisotropy and total fluorescence of FAM-dT20 upon binding of Rim1 are bi-phasic (Figure 4A) in Buffer HK150M5. This behavior is not consistent with Rim1 binding as a pre-formed tetramer (8,9) and is indicative of multiple Rim1 molecules binding. The stoichiometry calculated via the general method (34–38) using titrations performed at multiple DNA concentrations indicates that the first phase in the signal change is dominated by binding of a Rim1 dimer to FAM-dT20 (Figure 4A, right panel), consistent with the data obtained with Cy3-labeled DNAs. However, increasing the Rim1 concentration leads to a further change in both monitored signals and suggests lower affinity binding of an additional Rim1 dimer, consistent with formation of a tetramer as observed in the sedimentation equilibrium experiments. The solid lines in the left panel of Figure 4A show that a 2:1 binding model captures the behavior of the isotherms, with the DNA binding constant of the first Rim1 dimer K20 = (1.6 ± 0.7) x 109 M−1 and the binding constant for the second dimer L20 is 1.5 × 106 M−1 to 3 × 106 M−1 (see Supplementary material for details).
Figure 4.
On short ssDNA high-affinity binding of one Rim1 dimer is accompanied by lower affinity binding of a second one. (A) The left panel shows the change in fluorescence anisotropy (circles) and total intensity (squares) of FAM-dT20 at different concentrations (50 nM black, 100 nM blue, 200 nM red, 400 nM grey) as a function of Rim1 concentration (dimer), in Buffer HK150M5. The solid lines are the fits with a 2:1 binding model (equations s6 and s10 in Supplementary material). The right panel shows the dependence of the monitored signals as a function of the degree of binding calculated from the data in the left panel. The lines indicate the estimated values for different signals corresponding to one and two ligand-bound states and their associated stoichiometry. (B) Same as in A but for Rim1Y85H and the change in fluorescence anisotropy.
We also examined the binding of Rim1Y85H to FAM-dT20. Figure 4B shows the change in fluorescence anisotropy of FAM-dT20 as a function of Rim1Y85H concentration in Buffer HK150M5 (quenching of fluorescence intensity is less then ∼5%). Consistent with the sedimentation equilibrium data in Figure 3D showing that a single dimer of Rim1Y85H binds to this length of DNA, analysis of the stoichiometry with the general method (34–38) indicates that a dimer of Rim1Y85H binds to FAM-dT20 (Figure 4B, right panel). The solid lines in the left panel of Figure 4B are the fits of the data with a simple 1:1 binding model (i.e. one dimer/DNA) with a binding constant K20Y85H = (5.5 ± 0.5) x 107 M−1. The mutation Y85H not only dramatically alters the oligomerization state of Rim1 but it also affects its DNA binding properties. Considering the position of this mutation, at the interface of the AB/CD dimer (Supplementary Figure S1B), the effect on the DNA binding affinity is surprising. We currently do not know the reason for this change; however, it is possible that it may reflect the mode in which the dimer binds to this ssDNA, short enough to bind mainly to a single monomer within the dimer. For a longer ssDNA the effect of the Y85H mutation on the DNA binding affinity of the dimer appears to be less drastic (see below).
Binding of Rim1 to DNA long enough to engage with more than two monomers indicates that binding of a first dimer is followed by binding of a second one, which is modulated by DNA
The data in the previous section indicate that at low protein concentrations a dimer of Rim1 is present in solution and DNA binding favors dimerization of Rim1 dimers (‘tetramerization’) even on DNA lengths that are too short to accommodate more than one dimer. Next, we asked how Rim1 would bind to DNAs that can accommodate more than one dimer.
Figure 5A shows the Cy3 fluorescence increase in Buffer HK150 (in the absence or presence of MgCl2) of dT38-Cy3-T as a function of the ratio of the total concentration of Rim1 monomers to the total concentration of DNA (200 nM). For this longer DNA substrate, the Cy3 fluorescence as function of Rim1 concentration initially increases and peaks at a ratio of two monomers of Rim1, i.e. one dimer. However, further increase in the Rim1 concentration leads to a quenching in the Cy3 fluorescence followed by a plateau at four monomers per DNA, i.e. two Rim1 dimers. Similar results were observed when monitoring FRET changes of Cy5.5-dT40-Cy3-T (Figure 5B) or Cy5.5-dT45-Cy3-T (Supplementary Figure S4A) as a function of Rim1 concentration. Even though the change in acceptor intensity is a complex convolution of the protein induced fluorescence enhancement of the donor (PIFE, Figure 5A) and FRET due to conformational changes of DNA (e.g. wrapping), the data strongly argue that binding of a second Rim1 dimer is accompanied by a large conformational change of the complex. The observation of a similar behavior in the presence and absence of MgCl2 indicates that the divalent cation does not alter the oligomeric state of the protein, albeit it affects the monitored signals, suggesting differences in the conformation of the formed Rim1-DNA complexes. Also, the very observation of a switch in the monitored signals as a function of protein concentration provides further strong evidence that Rim1 does not bind as a pre-formed tetramer (8,9).
Figure 5.
The affinity of the second Rim1 dimer is modulated by the length of ssDNA available for interaction. (A) Change in fluorescence intensity of dT38-Cy3-T as a function of the ratio of Rim1 concentration to the concentration of the DNA (200 nM), in Buffer HK150 (•) and HK150M5 ( ). (B) Same as in A but for Cy5.5-dT40-Cy3-T, monitoring the change in Cy5.5 fluorescence intensity upon excitation of Cy3. (C) The change in fluorescence anisotropy (circles) and total intensity (squares) of dT38-FL as a function of Rim1 concentration (monomer) to the concentration of the DNA (200 nM), in Buffer HK150 (•) and HK150M5 (
). (B) Same as in A but for Cy5.5-dT40-Cy3-T, monitoring the change in Cy5.5 fluorescence intensity upon excitation of Cy3. (C) The change in fluorescence anisotropy (circles) and total intensity (squares) of dT38-FL as a function of Rim1 concentration (monomer) to the concentration of the DNA (200 nM), in Buffer HK150 (•) and HK150M5 ( ). (D) The change in fluorescence anisotropy (circles) and total intensity (squares) of dT38-FL at different concentrations (50 nM red, 100 nM blue, 200 nM gray, 400 nM black) as a function of Rim1 concentration (dimer), in Buffer HK150M5. The solid lines are the fits with a 2:1 binding model (equations s6 and s10 in Supplementary material). (E) Same as in D but in Buffer HK1000M5. (F) Same as in D but for Rim1Y85H. (G, H) Same as in A and B but for dT59-Cy3-T in G and Cy5-dT68-Cy3-T in H. (I) Analysis with a 1:1 binding model (Supplementary material) with different binding constants (1 × 109 M−1 blue, 1 × 1010 M−1 black, and 1 × 1011 M−1 red) and assuming that in Buffer HK1000M5 Rim1 binds to dT59-Cy3-T (50 nM) as a preformed tetramer.
). (D) The change in fluorescence anisotropy (circles) and total intensity (squares) of dT38-FL at different concentrations (50 nM red, 100 nM blue, 200 nM gray, 400 nM black) as a function of Rim1 concentration (dimer), in Buffer HK150M5. The solid lines are the fits with a 2:1 binding model (equations s6 and s10 in Supplementary material). (E) Same as in D but in Buffer HK1000M5. (F) Same as in D but for Rim1Y85H. (G, H) Same as in A and B but for dT59-Cy3-T in G and Cy5-dT68-Cy3-T in H. (I) Analysis with a 1:1 binding model (Supplementary material) with different binding constants (1 × 109 M−1 blue, 1 × 1010 M−1 black, and 1 × 1011 M−1 red) and assuming that in Buffer HK1000M5 Rim1 binds to dT59-Cy3-T (50 nM) as a preformed tetramer.
Because of the potential caveats associated with monitoring changes of Cy3 fluorescence at this position on the DNA (see above), we also examined binding of Rim1 to a fluorescein labeled ssDNA (dT38-FL), monitoring both fluorescence anisotropy and total intensity of as function of Rim1 concentration (Figure 5C), in Buffer HK150 in the absence or presence of 5 mM MgCl2. The change in fluorescence anisotropy is dominated by binding of the first Rim1 dimer, followed by additional contributions to the signal from binding of a second dimer. Interestingly, in the absence of Mg2+ the change in total intensity is bi-phasic as observed for FAM-dT20, indicative of two binding events. However, this is not the case in the presence of Mg2+, where the change in total intensity reaches a plateau at ∼ one dimer of Rim1 per DNA, despite the change in anisotropy indicates binding of two dimers. As for the data in Figure 4A, this discrepancy can be explained by a lower affinity binding of a second dimer that does not significantly contribute to the signal. Indeed, the DNA binding isotherms in Buffer HK150M5 in Figure 5D can be fitted with a 2:1 binding model, assuming that binding of the second dimer does not have additional contribution to the total intensity. The binding constant for the first dimer is estimated to be K38 ∼ 3 × 109 M−1 and the binding constant for the second dimer to be L38 ∼ 7 × 107 M−1 (see Supplementary material for details). We interpret the effect of MgCl2 on the fluorescence intensity as originating from differences in the conformation of the formed Rim1–DNA complexes.
Binding of a second Rim1 dimer becomes evident when the salt concentration is raised to 1 M (Figure 5E), and K38 and L38 are reduced to ∼6 × 108 M−1 and ∼3 × 106 M−1, respectively. Moreover, for Rim1Y85H, which forms stable dimers in solution, a second dimer can bind to dT38-FL (Figure 5F), while only one dimer bound to FAM-dT20 (Figure 4B). Binding of two dimers of Rim1Y85H is consistent with the sedimentation equilibrium data in Figures 3C and suggests that for this longer ssDNA binding to the first dimer favors formation of tetramers (see Discussion). For this ssDNA, long enough to bind both monomers within the dimer, the DNA binding affinity of the first Rim1Y85H dimer is, within error, the same as wild-type Rim1 (Supplementary Material). However, the L38 ∼ 3 × 106 M−1 for Rim1Y85H is much lower than wild-type Rim1 and indicates a strongly reduced propensity to tetramerize.
Finally, we examined binding of Rim1 to ssDNAs sufficiently long to allow interaction with at least four Rim1 monomers. Figures 5G shows the change of Cy3 intensity of dT59-Cy3-T (Buffer HK150 in the absence and presence of 5 mM MgCl2) as a function of the ratio of Rim1 concentration to DNA concentration (200nM). Figure 5H shows the change in Cy5 intensity of Cy5-dT68-Cy3-T. Independent of the signal monitored and the presence of Mg2+, the data indicate binding of a tetramer of Rim1. Indeed, assuming that Rim1 is a tetramer the change in Cy3 intensity of dT59-Cy3-T can be analyzed with a simple 1:1 binding model and with an apparent binding constant that can only be estimated to be at least 1010 M−1, even in Buffer HK1000M5 (Figure 5I). On the sole basis of these data one would conclude that Rim1 binds to these lengths of ssDNA as a pre-formed tetramer; however, this is not consistent with the clear presence of two binding events with shorter DNAs (see Discussion).
DISCUSSION
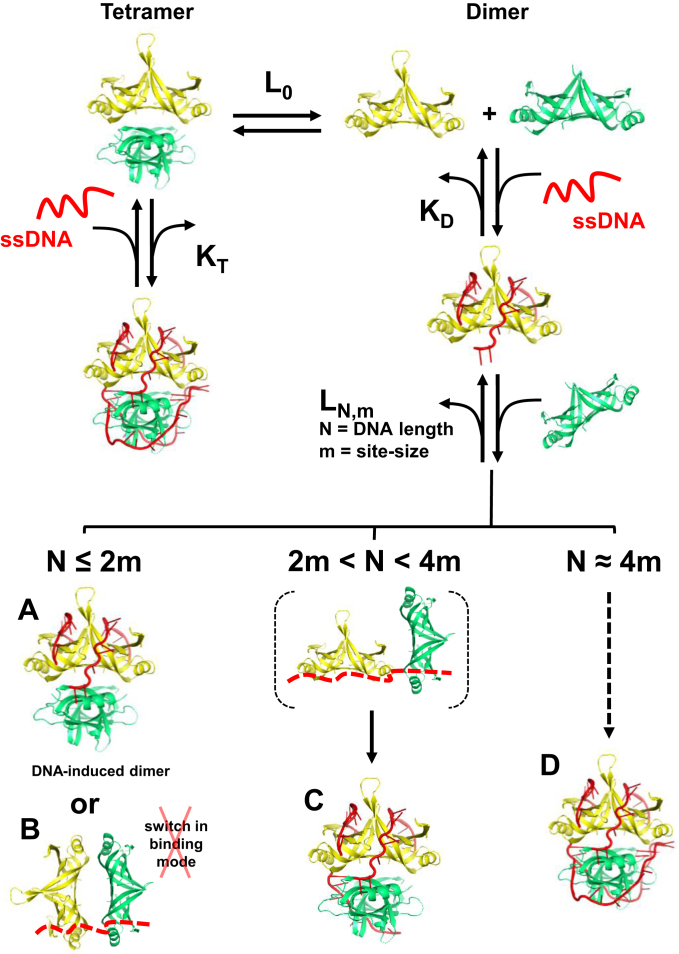
The mitochondrial single-stranded DNA binding proteins from humans and S. cerevisiae have been shown to form homo-tetramers in solution (5,8,9), as does the prototypical SSB from E. coli (20,21,39). In this work, we showed that Rim1 crystallizes as a tetramer in two different space groups. We note that Zybailov et al. (40) reported having solved the structure for Rim1. However, no information has been made available yet and it remains to be determined whether the two crystal forms we detected are the only two forms that Rim1 can adopt. In crystal Form 1 the structure of the Rim1 tetramer is barely distinguishable from the structure of other known SSBs (6,17,31). However, within the tetramer in crystal Form 2 the AB and CD dimers of Rim1 are offset and tilted relative to each other, suggesting that this conformation may originate from an alternative oligomerization of stable dimers. Analytical sedimentation experiments showed that Rim1 forms a tetramer at high protein concentrations, consistent with previous reports (8,9). Under conditions where Rim1 forms a tetramer in solution (e.g. 55 μM monomer), the tetramer is the species that binds to DNA with an occluded site-size of 14–15 nt per monomer, consistent with the one independently determined at lower protein concentrations. Surprisingly, the occluded site-size is constant over the range of KCl concentrations examined, suggesting that at difference with other SSBs (4,31,41,42) Rim1 does not efficiently access different binding modes on DNA, in the presence of these monovalent ions. Because of the high Rim1 concentrations needed to populate the tetramer species, the binding constant of a pre-formed Rim1 tetramer remains undetermined (KT in Figure 6).
Figure 6.
Working model showing how DNA binding is coupled to the oligomeric state of Rim1. Rim1 exists in solution in a dimer–tetramer equilibrium with L0 intrinsic oligomerization constant. In the left branch of the model, Rim1 binds ssDNA as a pre-formed tetramer with an equilibrium binding constant KT that is currently undetermined. The models for ssDNA bound to Rim1 were based on the DNA in the crystal structure of EcSSB (PDB:1eyg). The right branch of the model shows binding of two dimers of Rim1 and possible protein–DNA complexes (A-D) that can be formed depending of the length of the ssDNA. Independent of the ssDNA length, the binding constant of the second Rim1 dimer, LN,m, is larger than L0, indicating that DNA binding to the first Rim1 dimer favors tetramerization. Also, for N ≈ 4 m LN,m increases to the point that it cannot be distinguished from KD, and Rim1 appears to bind as a pre-formed tetramer.
At low protein concentrations Rim1 dissociates into a smaller species that we interpret to be stable dimers. Although we cannot exclude that Rim1 dissociates into monomers, multiple lines of evidence suggest that monomers are not significantly populated at the protein concentrations tested. A Y85H mutation at the AB/CD interface of Rim1 leads to the formation of stable dimers, with little evidence of monomers. This is surprising because at the interface of the AB (or CD) dimer Rim1 harbors a natural tyrosine (Y61). At the corresponding position in EcSSB a H55Y mutation leads to an unstable tetramer that at low protein concentrations cooperatively dissociates into monomers (20). Thus, we would have expected that the combination of Y85H and the presence of Y61 would have given rise to Rim1 monomers in solution. We note that in the structures of Rim1 Y61 occupies a similar position to the corresponding histidine in SSBs that form stable homo-tetramers (Figure 1H), suggesting that Y61 in Rim1 may not be as destabilizing to the AB dimer interface. In EcSSB second-site compensatory mutations Q76L and Q110L of a H55K variant lead to stabilization of the tetramer (43). Interestingly, the amino acid corresponding to EcSSB Q76 in Rim1 is L83. Whether Rim1 L83 has a second-site compensatory role as in EcSSB remains to be tested. However, we note that the role in homo-tetramerization of the conserved histidine in bacterial SSBs appears to be more complex as M. tuberculosis SSB, which harbors a leucine and has no second-site compensatory mutations, forms tetramers, at least at high concentrations (44).
Formation of Rim1 dimers rather than monomers as a major species in solution is also supported by the equilibrium DNA binding experiments using short ssDNAs. Binding of a dimer of Rim1 dominates the observed signals and this is especially true for the Cy3-labeled ssDNAs in Figure 4A, where the signal is exclusively sensitive to binding of a dimer of Rim1. Analysis of equilibrium fluorescence titrations of short Cy3-labeled DNAs show that, assuming Rim1 is a dimer in solution, a simple 1:1 binding model is sufficient to fit the data (Supplementary Figure S5). The ability to analyze these DNA binding isotherms with a simple 1:1 binding model that assumes that Rim1 is a stable dimer suggests that if a monomer exists in solution it has to be a minor populated species, supporting our conclusion that a dimer of Rim1 is the major species formed in solution. However, Rim1 does not bind to DNA just as a single dimer. The DNA binding studies indicate that while tight binding of a dimer of Rim1 dominates the observed signal, it is followed by the lower affinity binding of a second Rim1 dimer. The 2:1 binding model used to analyze the data in Figures 4A and 5D–F is sufficient to recapitulate the behavior of the binding isotherms; however, the model simply says that a second dimer of Rim1 binds to the DNA, but it does not provide information on how it does it.
Because of a site-size of 14 nt per Rim1 monomer, it is unlikely that on the short FAM-dT20 in Figure 4A the second Rim1 dimer interacts directly with DNA. Thus, we interpret the binding of a second Rim1 dimer as originating from DNA-induced tetramerization of Rim1 (Figure 6, complex A) rather than binding of the second dimer due to a change in binding mode of the first one (Figure 6, complex B). If complex B in Figure 6 were to form, one would expect a site-size <10 nt per monomer and three to four dimers would bind to the longer DNAs; however, this is not the case. The ∼10-fold increase in L20 (Figure 4A) relative to L0, the intrinsic oligomerization constant (Figure 2), suggests that binding of DNA to the first Rim1 dimer favors formation of tetramers. This interpretation is further supported by experiments performed in 1 M KCl (Supplementary Figure S4B). The increase in salt concentration mainly affects the binding of the first dimer, while it has comparatively less effect on tetramerization, consistent with sedimentation equilibrium data indicating that oligomerization is not very sensitive to salt concentration.
For the longer dT38-FL (Figure 5D) the binding constant of the second Rim1 dimer, L38, is ∼10-fold higher than L20. If binding of DNA to the first dimer were to favor only tetramerization one would expect L to be independent of DNA length. We interpret the increase in L38 relative to L20 as originating from tetramerization favored by the additional interaction of the second dimer with available DNA (Figure 6, complex C), a possibility that, because of the site-size of 14 nt per monomer, would be precluded on a 20 nt long ssDNA. The conclusion that interaction of the second dimer with DNA stabilizes formation of a tetramer is further supported by two observations. First, increasing the salt concentration to 1 M lowers L38 (Figure 5E) to a value that is similar to L20. Second, for Rim1Y85H that forms stable dimers in solution, one dimer binds to a 20 nt ssDNA (Figure 4B) while two dimers bind to dT38-FL (Figure 5F).
Finally, for ssDNA lengths that can accommodate two dimers, the binding isotherms might be mistakenly interpreted as representing binding of a stable pre-formed Rim1 tetramer. In light of the data with shorter ssDNAs, the apparent binding of Rim1 as a pre-formed tetramer can still be explained with a DNA-induced tetramerization model in which binding of the second dimer, because of its interaction with DNA, becomes so tight that the L and K equilibrium constants can no longer be distinguished (Figure 6, complex D).
Previous studies showed that Rim1 forms a tetramer at relatively high concentrations and assumed that this is the species that binds DNA (8), even at the much lower protein concentrations used for DNA binding studies. Interestingly, titration of a short 20 nt ssDNA with Rim1 (tetramer) was analyzed with a Hill equation with a coefficient of 2.5 (8). By definition, the Hill equation implies binding of more than one Rim1 tetramer to the 20 nt ssDNA with a site-size of no more than 5 nt per monomer, shorter than the one we determined in Figure 4 and the one determined for other SSBs (4,31,45). Rather than assuming that Rim1 is a tetramer, the data by Ramanagoudr-Bhojappa et al. (8) may be explained with our observation that indeed Rim1 is not a tetramer at low protein concentration and DNA binding is coupled to protein oligomerization. Moreover, Ramanagoudr-Bhojappa et al. (8) observed an increase in anisotropy of labeled Rim1 in the presence of unlabeled protein and interpreted it as originating from subunit exchange. Alternatively, this may be explained by a change in the oligomeric state of Rim1.
In addition to DNA binding, a major function of SSBs is their interaction with other proteins to either recruit them to their site of function and/or to modulate their activity (22). Rim1 interacts with the Pif1 helicase and stimulates its unwinding activity (8). Interestingly, interaction of Rim1 with Pif1 was detected at a concentration of Rim1 at which our analytical sedimentation studies indicate that Rim1 is not a stable tetramer. Therefore, it remains to be determined whether interaction of Pif1 occurs with a tetramer or dimer of Rim1. Of note, the Rim1–Pif1 interaction was shown to be lost upon Rim1 binding to a 70 nt ssDNA (40), making it difficult to rationalize the stimulatory effect of Rim1 on the unwinding activity of Pif1 (8). If Pif1 were to bind to a dimer of Rim1, loss of Rim1–Pif1 interaction upon binding of DNA to Rim1 may be explained by the highly cooperative tetramerization of Rim1 on such a long ssDNA (Figure 6, complex D).
Our findings that Rim1 in solution does not form a stable tetramer and binds to DNA predominantly as a dimer raises the question of whether this is an exclusive property of Rim1 or a more general property, shared among mitochondrial SSBs. Analytical ultracentrifugation (5) and DNA binding studies (4,5) indicate that human mtSSB forms stable tetramers, over the same protein concentration range where Rim1 dissociates into dimers. However, Mtp1/mtTBP, the mitochondrial Rim1 homologue in Candida parapsilosis, can be cross-linked as tetramers but it predominantly forms dimers (16), suggesting that smaller oligomeric species other than a tetramer may be formed by other fungi mitochondrial SSBs. We note that C. parapsilosis Mtp1/mtTBP harbors an isoleucine at the position corresponding to Y85 in Rim1, that when mutated to histidine leads to formation of a stable dimer (Rim1Y85H in Figure 2). Whether this single amino acid change in C. parapsilosis is a major contributor to the instability of the tetramer remains to be determined. Preliminary sequence analysis of mitochondrial SSBs from different species suggests that other mitochondrial SSBs, rather than a tyrosine, harbor a leucine, isoleucine, phenylalanine or histidine at this position. We speculate that in mitochondrial SSBs from different species these changes and the amino acid sequence context at the dimer–dimer interface can give rise to oligomeric species other than a tetramer.
Finally, what the functional oligomeric species of Rim1 is in mitochondria remains an open question. A Y78R mutation that in EcSSB leads to stable dimers does not yield viable strains, indicating that dimers are not functional in E. coli. Whether Rim1Y85H, which forms stable dimers, would sustain mitochondrial functions is currently unknown. Also, C. parapsilosis Mtp1/mtTBP, that appears to form a distribution of oligomeric species dominated by dimers, cannot functionally replace Rim1 (16). However, in this case the situation may be complicated by species-specific interaction with protein partners or from sequence specific DNA binding properties acquired by Mtp1/mtTBP (16,46). The discovery that in vitro Rim1 can form oligomeric species smaller than a tetramer (e.g. a dimer), suggest the possibility that modulation of its oligomeric state, either via changes in protein concentration, or specific cation and/or anion effects, or DNA binding, could be a means to regulate its function in vivo. In this regard, we note that, at least at the level of mRNA, RIM1 expression appears to be regulated by the type of carbon source used for growth (47). Also, a recent single-molecule study showed that DNA synthesis can modulate the DNA binding-mode with which HsmtSSB binds to the newly generated ssDNA (48), favoring a low site-size mode. It is tempting to speculate that binding to newly synthetized ssDNA by dimers of Rim1 may mimic the low site-size mode of HsmtSSB, and that the additional DNA-dependent tetramerization may play a regulatory function. Discovery of other mitochondrial SSBs that either display a dimer–tetramer equilibrium or that exclusively form stable dimers will provide further support for the potential functional role in mitochondria of oligomeric species other than a tetramer.
DATA AVAILABILITY
Atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession numbers: 6CQK (native Rim1 in space group C2221), 6CQM (native Rim1 in space group P212121), and 6CQO (seleno-l-methionine labeled Rim1 in space group P212121).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Kozlov and Prof. Lohman for sharing DNA substrates, suggestions and countless discussions. We thank Dr Ellenberger for help with the initial stages of crystallization of Rim1 and suggestions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [2R01GM098509 to R.G., 7R15GM110671 to E.A.]. Funding for open access charge: National Institute of General Medical Sciences [2R01GM098509].
Conflict of interest statement. None declared.
REFERENCES
- 1. Wold M.S. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997; 66:61–92. [DOI] [PubMed] [Google Scholar]
- 2. Iftode C., Daniely Y., Borowiec J.A.. Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 1999; 34:141–180. [DOI] [PubMed] [Google Scholar]
- 3. Zou Y., Liu Y., Wu X., Shell S.M.. Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses. J. Cell. Physiol. 2006; 208:267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Qian Y., Johnson K.A.. The human mitochondrial single-stranded DNA-binding protein displays distinct kinetics and thermodynamics of DNA binding and exchange. J. Biol. Chem. 2017; 292:13068–13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wong T.S., Rajagopalan S., Townsley F.M., Freund S.M., Petrovich M., Loakes D., Fersht A.R.. Physical and functional interactions between human mitochondrial single-stranded DNA-binding protein and tumour suppressor p53. Nucleic Acids Res. 2009; 37:568–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang C., Curth U., Urbanke C., Kang C.. Crystal structure of human mitochondrial single-stranded DNA binding protein at 2.4 A resolution. Nat. Struct. Biol. 1997; 4:153–157. [DOI] [PubMed] [Google Scholar]
- 7. Webster G., Genschel J., Curth U., Urbanke C., Kang C., Hilgenfeld R.. A common core for binding single-stranded DNA: structural comparison of the single-stranded DNA-binding proteins (SSB) from E. coli and human mitochondria. FEBS Lett. 1997; 411:313–316. [DOI] [PubMed] [Google Scholar]
- 8. Ramanagoudr-Bhojappa R., Blair L.P., Tackett A.J., Raney K.D.. Physical and functional interaction between yeast Pif1 helicase and Rim1 single-stranded DNA binding protein. Nucleic Acids Res. 2013; 41:1029–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Van Dyck E., Foury F., Stillman B., Brill S.J.. A single-stranded DNA binding protein required for mitochondrial DNA replication in S. cerevisiae is homologous to E. coli SSB. EMBO J. 1992; 11:3421–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim Y.T., Richardson C.C.. Acidic carboxyl-terminal domain of gene 2.5 protein of bacteriophage T7 is essential for protein-protein interactions. J. Biol. Chem. 1994; 269:5270–5278. [PubMed] [Google Scholar]
- 11. Kim Y.T., Tabor S., Bortner C., Griffith J.D., Richardson C.C.. Purification and characterization of the bacteriophage T7 gene 2.5 protein. A single-stranded DNA-binding protein. J. Biol. Chem. 1992; 267:15022–15031. [PubMed] [Google Scholar]
- 12. Alberts B., Frey L., Delius H.. Isolation and characterization of gene 5 protein of filamentous bacterial viruses. J. Mol. Biol. 1972; 68:139–152. [DOI] [PubMed] [Google Scholar]
- 13. Folmer R.H., Folkers P.J., Kaan A., Jonker A.J., Aelen J.M., Konings R.N., Hilbers C.W.. Secondary structure of the single-stranded DNA binding protein encoded by filamentous phage Pf3 as determined by NMR. Eur. J. Biochem. 1994; 224:663–676. [DOI] [PubMed] [Google Scholar]
- 14. Gascon I., Gutierrez C., Salas M.. Structural and functional comparative study of the complexes formed by viral o29, Nf and GA-1 SSB proteins with DNA. J. Mol. Biol. 2000; 296:989–999. [DOI] [PubMed] [Google Scholar]
- 15. Carroll R.B., Neet K., Goldthwait D.A.. Studies of the self-association of bacteriophage T4 gene 32 protein by equilibrium sedimentation. J. Mol. Biol. 1975; 91:275–291. [DOI] [PubMed] [Google Scholar]
- 16. Nosek J., Tomaska L., Pagacova B., Fukuhara H.. Mitochondrial telomere-binding protein from Candida parapsilosis suggests an evolutionary adaptation of a nonspecific single-stranded DNA-binding protein. J. Biol. Chem. 1999; 274:8850–8857. [DOI] [PubMed] [Google Scholar]
- 17. Raghunathan S., Ricard C.S., Lohman T.M., Waksman G.. Crystal structure of the homo-tetrameric DNA binding domain of Escherichia coli single-stranded DNA-binding protein determined by multiwavelength x-ray diffraction on the selenomethionyl protein at 2.9-A resolution. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:6652–6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li K., Williams R.S.. Tetramerization and single-stranded DNA binding properties of native and mutated forms of murine mitochondrial single-stranded DNA-binding proteins. J. Biol. Chem. 1997; 272:8686–8694. [DOI] [PubMed] [Google Scholar]
- 19. Bujalowski W., Lohman T.M.. Monomers of the Escherichia coli SSB-1 mutant protein bind single-stranded DNA. J. Mol. Biol. 1991; 217:63–74. [DOI] [PubMed] [Google Scholar]
- 20. Bujalowski W., Lohman T.M.. Monomer-tetramer equilibrium of the Escherichia coli ssb-1 mutant single strand binding protein. J. Biol. Chem. 1991; 266:1616–1626. [PubMed] [Google Scholar]
- 21. Williams K.R., Murphy J.B., Chase J.W.. Characterization of the structural and functional defect in the Escherichia coli single-stranded DNA binding protein encoded by the ssb-1 mutant gene. Expression of the ssb-1 gene under lambda pL regulation. J. Biol. Chem. 1984; 259:11804–11811. [PubMed] [Google Scholar]
- 22. Shereda R.D., Kozlov A.G., Lohman T.M., Cox M.M., Keck J.L.. SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 2008; 43:289–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Korhonen J.A., Gaspari M., Falkenberg M.. TWINKLE Has 5′ → 3′ DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. J. Biol. Chem. 2003; 278:48627–48632. [DOI] [PubMed] [Google Scholar]
- 24. Doublie S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 1997; 276:523–530. [PubMed] [Google Scholar]
- 25. Gill S.C., von Hippel P.H.. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989; 182:319–326. [DOI] [PubMed] [Google Scholar]
- 26. Pace C.N., Vajdos F., Fee L., Grimsley G., Gray T.. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995; 4:2411–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barranco-Medina S., Galletto R.. DNA binding induces dimerization of Saccharomyces cerevisiae Pif1. Biochemistry. 2010; 49:8445–8454. [DOI] [PubMed] [Google Scholar]
- 28. Feldmann E.A., Galletto R.. The DNA-binding domain of yeast Rap1 interacts with double-stranded DNA in multiple binding modes. Biochemistry. 2014; 53:7471–7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kozlov A.G., Galletto R., Lohman T.M.. SSB-DNA binding monitored by fluorescence intensity and anisotropy. Methods Mol. Biol. 2012; 922:55–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Feldmann E.A., Koc K.N., Galletto R.. Alternative arrangements of telomeric recognition sites regulate the binding mode of the DNA-binding domain of yeast Rap1. Biophys. Chem. 2015; 198:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Antony E., Weiland E.A., Korolev S., Lohman T.M.. Plasmodium falciparum SSB tetramer wraps single-stranded DNA with similar topology but opposite polarity to E. coli SSB. J. Mol. Biol. 2012; 420:269–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bernstein D.A., Eggington J.M., Killoran M.P., Misic A.M., Cox M.M., Keck J.L.. Crystal structure of the Deinococcus radiodurans single-stranded DNA-binding protein suggests a mechanism for coping with DNA damage. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:8575–8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Landwehr M., Curth U., Urbanke C.. A dimeric mutant of the homotetrameric single-stranded DNA binding protein from Escherichia coli. Biol. Chem. 2002; 383:1325–1333. [DOI] [PubMed] [Google Scholar]
- 34. Bujalowski W. Thermodynamic and kinetic methods of analyses of protein-nucleic acid interactions. From simpler to more complex systems. Chem. Rev. 2006; 106:556–606. [DOI] [PubMed] [Google Scholar]
- 35. Bujalowski W., Lohman T.M.. A general method of analysis of ligand-macromolecule equilibria using a spectroscopic signal from the ligand to monitor binding. Application to Escherichia coli single-strand binding protein-nucleic acid interactions. Biochemistry. 1987; 26:3099–3106. [DOI] [PubMed] [Google Scholar]
- 36. Lohman T.M., Bujalowski W.. Thermodynamic methods for model-independent determination of equilibrium binding isotherms for protein-DNA interactions: spectroscopic approaches to monitor binding. Methods Enzymol. 1991; 208:258–290. [DOI] [PubMed] [Google Scholar]
- 37. Galletto R., Jezewska M.J., Bujalowski W.. Interactions of the Escherichia coli DnaB helicase hexamer with the replication factor the DnaC protein. Effect of nucleotide cofactors and the ssDNA on protein-protein interactions and the topology of the complex. J. Mol. Biol. 2003; 329:441–465. [DOI] [PubMed] [Google Scholar]
- 38. Galletto R., Rajendran S., Bujalowski W.. Interactions of nucleotide cofactors with the Escherichia coli replication factor DnaC protein. Biochemistry. 2000; 39:12959–12969. [DOI] [PubMed] [Google Scholar]
- 39. Weiner J.H., Bertsch L.L., Kornberg A.. The deoxyribonucleic acid unwinding protein of Escherichia coli. Properties and functions in replication. J. Biol. Chem. 1975; 250:1972–1980. [PubMed] [Google Scholar]
- 40. Zybailov B., Gokulan K., Wiese J., Ramanagoudr-Bhojappa R., Byrd A.K., Glazko G., Jaiswal M., Mackintosh S., Varughese K.I., Raney K.D.. Analysis of protein-protein interaction interface between yeast mitochondrial proteins Rim1 and Pif1 using chemical cross-linking mass spectrometry. J. Proteomics Bioinform. 2015; 8:243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bujalowski W., Overman L.B., Lohman T.M.. Binding mode transitions of Escherichia coli single strand binding protein-single-stranded DNA complexes. Cation, anion, pH, and binding density effects. J. Biol. Chem. 1988; 263:4629–4640. [PubMed] [Google Scholar]
- 42. Lohman T.M., Overman L.B.. Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes. Modulation by NaCl concentration. J. Biol. Chem. 1985; 260:3594–3603. [PubMed] [Google Scholar]
- 43. Carlini L., Curth U., Kindler B., Urbanke C., Porter R.D.. Identification of amino acids stabilizing the tetramerization of the single stranded DNA binding protein from Escherichia coli. FEBS Lett. 1998; 430:197–200. [DOI] [PubMed] [Google Scholar]
- 44. Purnapatre K., Varshney U.. Cloning, over-expression and biochemical characterization of the single-stranded DNA binding protein from Mycobacterium tuberculosis. Eur. J. Biochem. 1999; 264:591–598. [DOI] [PubMed] [Google Scholar]
- 45. Bujalowski W., Lohman T.M.. Escherichia coli single-strand binding protein forms multiple, distinct complexes with single-stranded DNA. Biochemistry. 1986; 25:7799–7802. [DOI] [PubMed] [Google Scholar]
- 46. Tomaska L., Makhov A.M., Nosek J., Kucejova B., Griffith J.D.. Electron microscopic analysis supports a dual role for the mitochondrial telomere-binding protein of Candida parapsilosis. J. Mol. Biol. 2001; 305:61–69. [DOI] [PubMed] [Google Scholar]
- 47. Li Z., Ling F., Shibata T.. Glucose repression on RIM1, a gene encoding a mitochondrial single-stranded DNA-binding protein, in Saccharomyces cerevisiae: a possible regulation at pre-mRNA splicing. Curr. Genet. 1998; 34:351–359. [DOI] [PubMed] [Google Scholar]
- 48. Morin J.A., Cerron F., Jarillo J., Beltran-Heredia E., Ciesielski G.L., Arias-Gonzalez J.R., Kaguni L.S., Cao F.J., Ibarra B.. DNA synthesis determines the binding mode of the human mitochondrial single-stranded DNA-binding protein. Nucleic Acids Res. 2017; 45:7237–7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession numbers: 6CQK (native Rim1 in space group C2221), 6CQM (native Rim1 in space group P212121), and 6CQO (seleno-l-methionine labeled Rim1 in space group P212121).