Abstract
Self-cleaving ribozymes are biologically relevant RNA molecules which catalyze site-specific cleavage of the phosphodiester backbone. Gathering knowledge of their three-dimensional structures is critical toward an in-depth understanding of their function and chemical mechanism. Equally important is collecting information on the folding process and the inherent dynamics of a ribozyme fold. Over the past years, Selective-2′-Hydroxyl Acylation analyzed by Primer Extension (SHAPE) turned out to be a significant tool to probe secondary and tertiary interactions of diverse RNA species at the single nucleotide level under varying environmental conditions. Small self-cleaving ribozymes, however, have not been investigated by this method so far. Here, we describe SHAPE probing of pre-catalytic folds of the recently discovered ribozyme classes twister, twister-sister (TS), pistol and hatchet. The study has implications on Mg2+-dependent folding and reveals potentially dynamic residues of these ribozymes that are otherwise difficult to identify. For twister, TS and pistol ribozymes the new findings are discussed in the light of their crystal structures, and in case of twister also with respect to a smFRET folding analysis. For the hatchet ribozyme where an atomic resolution structure is not yet available, the SHAPE data challenge the proposed secondary structure model and point at selected residues and putative long-distance interactions that appear crucial for structure formation and cleavage activity.
INTRODUCTION
Nine classes of small self-cleaving ribozymes have been validated to date (1), including the recently discovered twister (2), twister-sister (TS) (3), pistol (3) and hatchet (3) motifs. Except for the latter, crystal structures have been solved to shed light on the three-dimensional (3D) folds (4–10), with all of the currently available structures representing pre-catalytic conformations. So far, no structures of transition-state analogs or post-catalytic states of these ribozymes have been published. Furthermore, information about their folding process and the underlying structural dynamics are rare. Only with respect to the twister ribozyme class, folding and dynamics have been investigated in detail, based on single-molecule fluorescence-resonance-energy-transfer (smFRET) imaging (11,12). Since smFRET and other high-resolution techniques to study structural dynamics, such as nuclear magnetic resonance (NMR) spectroscopy (13,14) and recently emerging mix-and-inject XFEL serial X-ray crystallography (15) are time consuming and challenging to implement, more straightforward biochemical probing methods take an important role to shed light on ribozyme folding and folding-induced ribozyme catalysis.
A wide variety of chemical probing approaches have proven to be powerful tools for validating existing structure models, testing RNA structure-function hypotheses as well as generating new models. Of these, Selective 2′-Hydroxyl Acylation analyzed by Primer Extension (SHAPE) has emerged as particularly useful (16,17), since it uses small hydroxyl-selective electrophilic reagents to probe the reactivity of the 2′-hydroxyl group in the ribose ring, a chemical functionality that is available for all nucleosides of the target RNA independent of nucleobase identities. The reactivity of this group is gated by local nucleotide flexibility, meaning that the 2′-OH is reactive at single-stranded and conformationally flexible positions, whereas nucleotides constrained by base pairing are in general unreactive.
SHAPE has already been used to investigate diverse RNA species at single-nucleotide resolution, ranging from small ncRNAs such as RNA thermometers (18), to snRNAs and rRNAs (17), up to entire viral RNA genomes (HIV-1) (19,20). Furthermore, SHAPE provided insights into the effects of ligand binding and of the cellular environment on riboswitch structure and folding (21,22), allowed selecting appropriate sites for 2-aminopurine nucleobase substitutions (23) and fluorophore labeling (22), and has recently also been implemented into in vivo studies (24–26).
Here, we established a protocol that enables SHAPE probing on non-cleavable derivatives of small self-cleaving ribozymes containing a 2′-deoxyribose modification next to the scissile phosphate. The approach allows to investigate the effect of distinct environmental parameters, such as divalent metal ions and temperature on their pre-catalytic RNA folds. Our results complement insights from existing chemical, biochemical and biophysical studies and suggest several novel aspects on Mg2+-dependent folding and structural dynamics of the four recently discovered ribozyme classes.
MATERIALS AND METHODS
Solid-phase synthesis of oligonucleotides
For each of the ribozyme SHAPE constructs, the following two separate RNA strands (half molecules, H1 and H2) were synthesized by solid-phase RNA synthesis (depicted in the 5′-3′ direction): Twister H1 GUGCUUUU(dU)AAUGAAGCCACAUUCGUGUGAGGGUCCUA, Twister H2 p-AGCCCCUAAUUCAGAAGGGAAAAACAGAUGACAGAACUAACGAUUCG, Pistol H1 GUGCCGUGGUUAGGGCCACGUUAAAUAGUUGCUUAAGCCCUAAGCGUUGAUA, Pistol H2 p-UUAUCAG(dG)UGCAAAACAGAUGACAGAACUAACGAUUCG, Twister-sister H1 GUGCACCCGCAAGGCCGACGGCUUCGGCCGCCGCUGGUGCAAGUCCAGCCACGCGA, Twister-sister H2 p-AAGCGUGGGCGCU(dC)AUGGGUAACAGAUGACAGAACUAACGAUUCG, Hatchet H1 GUGC(dC)UCAGAAAAUGACAAACCUGUGGGGCGUAAGCACCAGA, Hatchet H2 p-AAUGGUGUGAUCGUGCAGACGUUAAAAUCAGGUAACAGAUGACAGAACUAACGAUUCG.
Standard phosphoramidite chemistry using 2′-O-TOM or 2′-O-tBDMS ribonucleoside phosphoramidite building blocks (ChemGenes, Sigma Aldrich) and controlled pore glass (ChemGenes, 1000 Å, 36.8 μmol/g) was applied (27,28). 2′-Deoxyguanosine, 2′-deoxycytidine and 2′-deoxyuridine phosphoramidites were purchased from ChemGenes. All oligonucleotides were synthesized on ABI 392 or 394 nucleic acid synthesizers following standard methods: detritylation (80 s) with dichloroacetic acid/1,2-dichloroethane (4/96); coupling (2.0 min) with phosphoramidites/acetonitrile (0.1 M × 130 μl) and benzylthiotetrazole/acetonitrile (0.3 M × 360 μl); capping (3 × 0.4 min, Cap A/Cap B = 1/1) with Cap A: THF/phenoxyacetic anhydride (95/5) and Cap B: 10% 1-Methylimidazole in THF/sym-collidine (8/1); oxidation (1.0 min) with I2 (20 mM) in THF/pyridine/H2O (35/10/5). The solutions of amidites, tetrazole, and acetonitrile were dried over activated molecular sieves (4 Å) overnight.
Deprotection of oligonucleotides
The solid support was treated each with ammonium hydroxide (28–30%, 0.5 ml) and methylamine in water (40%, 0.5 ml) for 5 h at room temperature. The supernatant was removed and the solid support was washed 3 × with ethanol/water (1/1, v/v). The supernatant and washings were combined and evaporated to dryness. To remove the 2′-silyl protecting groups the resulting residue was treated with tetrabutylammonium fluoride trihydrate (TBAF·3H2O) in THF (1 M, 2 ml) at 37°C overnight. The reaction was quenched by the addition of triethylammonium acetate (1 M, pH 7.4, 2 ml). The volume of the solution was reduced and the solution was desalted with a size exclusion column (GE Healthcare, HiPrep 26/10 Desalting; 2.6 × 10 cm; Sephadex G25) eluting with H2O, the collected fraction was evaporated to dryness and dissolved in 1 ml H2O. Analysis of the crude RNA after deprotection was performed by anion-exchange chromatography on a Dionex DNAPac PA-100 column (4 mm × 250 mm) at 80°C. Flow rate: 1 ml/min, eluant A: 25 mM Tris·HCl (pH 8.0), 6 M urea; eluant B: 25 mM Tris·HCl (pH 8.0), 0.5 M NaClO4, 6 M urea; gradient: 0–60% B in A within 60 min, UV detection at 260 nm.
Purification of RNA
Crude RNA products were purified on a semipreparative Dionex DNAPac PA-100 column (9 × 250 mm) at 80°C with flow rate 2 ml/min. Fractions containing RNA were loaded on a C18 SepPak Plus cartridge (Waters/Millipore), washed with 0.1 M (Et3NH)+HCO3−, H2O and eluted with H2O/CH3CN (1/1). RNA containing fractions were lyophilized. Analysis of the quality of purified RNA was performed by anion-exchange chromatography with same conditions as for crude RNA. The molecular weight was confirmed by LC-ESI mass spectrometry. Yield determination was performed by UV photometrical analysis of oligonucleotide solutions.
Mass spectroscopy of RNA
All experiments were performed on a Finnigan LCQ Advantage MAX ion trap instrumentation connected to an Amersham Ettan micro LC system. RNA sequences were analyzed in the negative-ion mode with a potential of −4 kV applied to the spray needle. LC: Sample (200 pmol RNA dissolved in 30 μl of 20 mM ethylenediaminetetraacetic acid (EDTA) solution; average injection volume: 30 μl); column (Waters XTerraMS, C18 2.5 μm; 1.0 × 50 mm) at 21°C; flow rate: 30 μl/min; eluant A: 8.6 mM triethylamine (TEA), 100 mM 1,1,1,3,3,3-hexafluoroisopropanol in H2O (pH 8.0); eluant B: methanol; gradient: 0–100% B in A within 30 min; UV detection at 254 nm.
Enzymatic RNA ligation
The ribozyme constructs for SHAPE analysis were prepared by splinted enzymatic ligation of two chemically synthesized RNA fragments using T4 DNA ligase (Thermo Fisher Scientific), in analogy to previously published protocols (29–32). Briefly, 10 μM of each RNA strand, 10 μM of a DNA splint oligonucleotide (IDT), and a final ligase concentration of 0.5 U/μl in a total volume of 2 ml, were incubated for 3 h at 37°C. Following splint sequences were used (depicted in the 5′-3′ orientation): Twister TTAGGGGCTTAGGACCCTCACACG, Pistol TTGCACCTGATAATAATCAAGCTTA, Twister-sister GCGCCCACGCTTTCGCGTGGCTGG, Hatchet GATCACACCATTTCTGGTGCTTAC. Analysis of the ligation reaction efficiency and purification of the ligated RNA constructs were performed by anion exchange chromatography, as already described above.
RNA 2′-hydroxyl acylation by benzoyl cyanide
Reaction mixtures containing 10 pmol ligated and purified RNA, 50 mM of MOPS at pH of 7.5 and 100 mM of KCl in the presence or absence of 5–20 mM of MgCl2 were heated at 65°C for 2 min, cooled to 4°C for 5 min, and incubated at 7, 37 or 57°C (as indicated) for 25 min in an Eppendorf Mastercylcer personal (VWR). Following incubation, the control background reaction was treated with DMSO, while the probing reagent benzoyl cyanide (BzCN), dissolved in anhydrous dimethyl sulfoxide (DMSO), was added to the probing reaction mixtures for a final concentration of 55 mM. The reaction with BzCN was complete within 2 s (33,34). The RNA was recovered by ethanol precipitation with sodium acetate and glycogen. After centrifugation, the RNA samples were resuspended in 8 μl water.
Primer extension assay and data analysis
A total of 8 μl of RNA from BzCN 2′-hydroxyl acylation were combined with 4 μl of an Alexa Fluor 647 5′-end labeled DNA primer (2 pmol/μl; IDT; 5′-/5Alexa647N/CGAATCGTTAGTTCTGTC-3′) and allowed to anneal at 65°C for 5 min, followed by an incubation at 35°C for 5 min and cooling to 4°C for 1 min (performed in an Eppendorf Mastercycler personal). A total of 8 μl of a mix containing 4 μl of 5× first strand buffer (250 mM Tris·HCl pH 8.3, 375 mM KCl, 15 mM MgCl2), 1 μl of 0.1 M DTT, 1 μl of 5 mM dNTPs mixture and 2 μl of DMSO were then added to the reactions, followed by incubation at 61°C for 1 min, addition of 0.4 μl of SuperScript III reverse transcriptase (200 U/μl; Invitrogen), and further incubation at 61°C for 10 min. Reactions were then stopped by addition of 1 μl 4 M NaOH and incubation at 95°C for 5 min and cooling to 4°C. Alexa Fluor 647 labeled cDNA strands were recovered by ethanol precipitation with sodium acetate and glycogen. After centrifugation, the samples were resuspended in 8 μl of gel loading buffer (97% formamide, 10 mM EDTA). Sequencing ladders were produced by adding 1 μl of 10 mM ddNTPs in addition to the 8 μl of reaction mixture to unmodified RNA samples, prior to incubation at 61°C. Samples were loaded next to a migration control dye (0.1% xylene cyanol, 95% formamide, 10 mM EDTA) on 10% polyacrylamide gels with 7 M urea and run for approximately 100 min at 45 W. The primer extension labeling was revealed by scanning the gel at 635 nm with a Typhoon FLA 9500 instrument (GE Healthcare). Band intensities visualized by gel electrophoresis were quantified using SAFA v.1.1 (Semi-Automated Footprinting Analysis) (35). The nucleotide identity of each band was identified from dideoxy sequencing lanes. It has to be kept in mind that the intensity of a band on the gel represents the degree of 2′-hydroxyl acylation of the last nucleotide upstream or the 5′-adjacent nucleotide of the cDNA and thus of the primary RNA sequence. Datasets were normalized for loading variations and reverse transcription (RT) efficiency by dividing all intensities by the intensity of the last base of primer extension. Final results for graphical representation were obtained by subtracting the DMSO control background from the BzCN-probed reaction intensities. The relative SHAPE reactivity was then calculated by normalizing individual datasets to a scale in which 0 indicates an unreactive site and 1.0 a highly reactive site. The normalization factor for each dataset was generated based on the 2/8% rule (25,36,37), and determined by first excluding the most reactive 2% of peak intensities and then calculating the average for the next 8% of peak intensities. All reactivities were then divided by this average value. In the scale, reactivities ≤ 0.25 are considered as not reactive, 0.26–0.59 as moderately reactive and ≥ 0.6 as very reactive. For all four ribozymes investigated, the normalized reactivity data of each nucleotide with the mean of at least three independent experiments are shown in the Supplementary Data.
Statistical analysis
For assessing statistical significance of differences in mean SHAPE reactivities, a two-sided paired student’s t-test was applied using the R software package ‘stats’, where P-values ≤ 0.1 were considered statistically significant. The corresponding plots are depicted in the Supplementary Data for all four ribozymes. Based on this analysis, meaningful individual nucleotides and/or nucleotide regions with the corresponding SHAPE reactivities are highlighted in graphs of the main figures.
RESULTS AND DISCUSSION
Strategy to obtain ribozyme probes for SHAPE probing
Self-cleaving ribozymes are frequently designed in a bimolecular fashion for their use in biochemical assays and crystallographic studies. These RNA constructs consist of two annealed RNA strands, one assigned to the ‘enzyme’ and the other one to the ‘substrate’. In general, the enzyme strand includes the conserved nucleotide core, whereas the substrate strand contains the site of cleavage. For the intended SHAPE studies, we needed unimolecular non-cleavable RNA constructs comprising a primer binding site in addition (32). Therefore, we designed RNA probes containing (i) a 2′-deoxy modification at the active site so as to prevent cleavage, (ii) an extra loop connecting ‘enzyme’ and ‘substrate’ strands, (iii) a primer annealing segment enabling RT of the full-length ribozyme, as well as (iv) 5′ and 3′ spacers (Supplementary Figure S1). The reason for the 5′ spacer is, that at the very 5′ end of the RNA, nucleotides often cannot be quantified after RT due to the intensity of the band corresponding to the full-length extension product. Additionally, 10–20 nt adjacent to the primer binding site cannot be quantified due to the presence of fragments that reflect pausing by the reverse transcriptase during the initiation phase of RT, sustaining the need for the 3′ spacer. To smoothly access the required lengths of RNA with a single 2′-deoxyribose modification, we decided for a combination of RNA solid-phase synthesis and enzymatic ligation (29) (Supplementary Figure S1). Hence, we synthesized the corresponding RNA half-molecules first. After deprotection, the RNA strands were purified via high pressure liquid chromatography (HPLC) and enzymatically ligated using a DNA splint and T4 DNA ligase to obtain the full-length RNAs. For the SHAPE experiments, they were incubated at different Mg2+ concentrations before 2′-O benzoylations were introduced by treatment with benzoyl cyanide (BzCN). To read out the probing results, RT with a fluorescently labeled primer was applied, followed by fractionation of the resulting cDNAs via polyacrylamide gel electrophoresis. Gel electrophoresis band analysis and data processing was performed as described in the ‘Materials and Methods’ section.
Mg2+ concentrations
Divalent magnesium ions represent a major determinant to achieve the physiologically active conformation of a ribozyme. With the goal to reveal Mg2+ induced structural changes at the single nucleotide level, we probed the four ribozyme classes in the absence and presence of increasing concentrations of Mg2+ ions. We decided for concentrations of 5, 10 and 20 mM Mg2+ at physiological pH to be comparable with previous studies. Twister and pistol ribozymes are fully folded in vitro at concentrations above 2 mM Mg2+ (2–5,8,9,38,39). For TS, the activity profile showed that the maximal cleavage rates were obtained at a Mg2+ concentration of 1 mM and therefore this concentration should be sufficient to reach the folded state (3,6); however, other studies indicated that the required concentrations might be higher for some of the four-way junctional systems of this ribozyme class (7). For hatchet ribozymes, the Mg2+ concentration for significant folding is assumed to be around 10 mM according to the cleavage activity profile (3,40).
The twister ribozyme
We started our comprehensive SHAPE investigation to explore folding of self-cleaving RNA scaffolds with the twister ribozyme which is the best characterized out of the four new classes. Its secondary as well as tertiary structure and folding have been described based on crystallographic (4,5,38,41), NMR (38,42) and FRET (11,12) investigations. To enable direct comparison, we designed an env22 twister construct capable for SHAPE probing (Figure 1A and Supplementary Figure S1a) which contained the sequence that we previously used for crystallography (5,38).
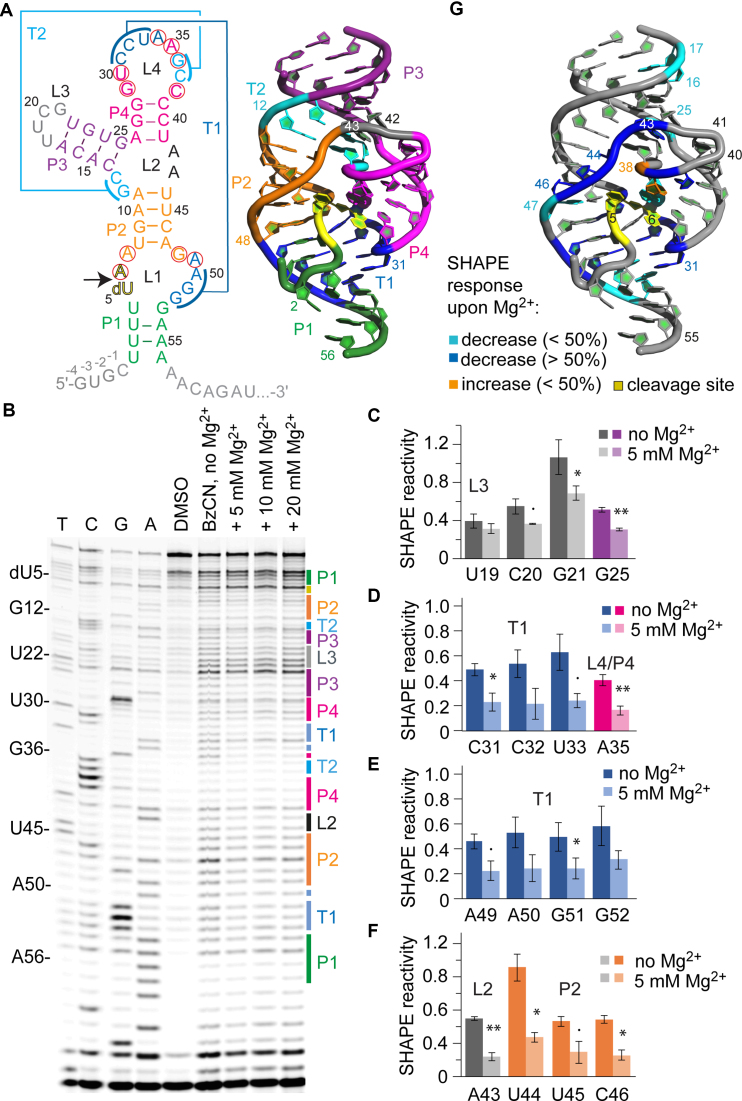
Figure 1.
SHAPE probing of the env22 twister ribozyme. (A) Secondary structure representation of the twister RNA with 5′ and 3′ spacer sequences in gray (left). Red circles indicate highly conserved nucleosides. 3D structure (PDB accession number: 4RGE) using the same color code (right). (B) Typical gel for the probing of the twister RNA structure with BzCN at 37°C. Lanes from left to right: T, C, G and A ladders, control in the absence of probing reagent (DMSO), probing with BzCN, probing with BzCN and in the presence of either 5, 10 or 20 mM MgCl2. Note that the fragment in the sequencing ladder that matches the size of the extension product defines the precise nucleotide that corresponds to +1 (therefore the color-coded stem-loop assignments at the right side of the gel is shifted +1 to nucleoside numbering on the left side). (C–F) Relative 2′-OH reactivity for selected bases obtained from quantification and normalization of the SHAPE probing results (mean of at least three independent experiments, error bars show standard deviation; two-sided paired t-test with **P ≤ 0.01, *P ≤ 0.05, • P ≤ 0.1). For quantification of the complete env22 twister RNA sequence see Supplementary Figure S3. (G) Projection of SHAPE reactivities of individual nucleotides on the 3D structure of the twister ribozyme (PDB accession number: 4RGE) following the color code as indicated in the legend.
Representative examples of twister ribozyme probing after urea-acrylamide gel electrophoresis are shown in Figure 1B and Supplementary Figure S2. First of all, we observed that the loop L3 that was inserted in order to obtain a unimolecular ribozyme construct (U18-U19-C20-G21; Figure 1A) was reactive under each of the tested conditions, with decreases in reactivity for C20 and G21 in response to Mg2+ ions (Figure 1C), consistent with the formation of an exposed and structured loop (UNCG motif). Furthermore, we could not only confirm the main stems (P2-P4) according to the secondary structure model (Figure 1A, left), but also the long distance interactions of both pseudoknots (PKs) (T1 and T2; Figure 1A and B). Here, a strong protection of the involved nucleotides was observed in the presence of Mg2+ ions, as shown in Figure 1D and E for PK T1 at 5 mM final concentration. Whereas C31-C32-U33 and A50-G51-G52 are known to form Watson–Crick base pairs, the highly conserved A34 and A49 form a non-canonical trans-Watson–Crick base pair that stacks onto U33-A50 (5). Notably, SHAPE probing reflects the formation of this interaction in response to Mg2+ by a decrease in reactivity of A49 (Figure 1E). Likewise, extension of helix P4 by the non-canonical U30•A35 base pair is reflected by a decrease in reactivity of A35 (Figure 1D). The U30•A35 pair is a crucial part of the catalytic pocket since A6 of the cleavage site stacks on top of it. Also, the A34•A49 pair molds the pocket and is crucial for positioning (stacking) of A7•G48 that directly interacts with the scissile phosphate.
Furthermore, the data nicely picture the Mg2+-induced structuring of the three-way junction (reactivity decrease of G25, Figure 1C) including L2 (reactivity decrease of A43, Figure 1F) which fixates the 180°–backbone turn (upstream between C40 and U41) and which continues in full formation of stem P2 (reactivity decreases of U44 to C46, Figure 1F) oriented antiparallel to P4 (Figure 1G). Like T1, also P2 is extended by a non-Watson–Crick basepair, namely a sheared trans-Hoogsteen sugar edge pair between A7 and G48, located coaxially between T1 and P2 (Figure 1G, right). Again, SHAPE monitors the formation of this interaction at the catalytic pocket by reduced reactivity of one of the nucleotides (A7, Supplementary Figure S3) in response to Mg2+.
Altogether, the data obtained from the SHAPE experiments on the twister ribozyme suggested the rather rigid formation of the helical core segments P2, P3, P4 and of PKs T1 and T2, at a concentration of 5 mM Mg2+ (no further significant changes in SHAPE reactivities were observed for the higher Mg2+ concentrations investigated; see also comparative discussion below and Supplementary Figures S10–12). This is coinciding with other published biochemical and spectroscopic investigations. Importantly, the nucleosides assigned to the weak A-U rich stem P1 (U1–U4 and G53–A56) showed no significant SHAPE response to increasing concentrations of Mg2+ (not even at 20 mM concentration), suggesting that P1 is breathing and not stably formed for this particular RNA (env22) at 37°C. This observation is consistent with the finding that P1 is not essential for cleavage activity (38) and the observation that the A-U rich stem P1 adopted a different topology (with only two Watson–Crick base pairs formed (5)) compared to the C-G rich stem P1 (fully base-paired) in the corresponding X-ray structure of an Oryza sativa derived sequence (4). This finding is also consistent with a recent smFRET study that demonstrated that T1 and P1 breathing is required to enable free tumbling of the 5′-tail of the ribozyme; only when this tail becomes properly pre-orientated, T1 and P1 can form stably and thereby lock the cleavage-competent conformation (11).
The pistol ribozyme
Two crystal structures of the pistol ribozyme from environmental probes (env25 and env27) have been published to date; their overall folds as well as nucleobase arrangements at their cleavage sites are in good agreement (8,9). Additionally, a recent chemical study shed light on the mechanism and the important role of an innersphere coordinated, hydrated Mg2+ ion in general acid-base catalysis (43).
For the SHAPE experiments, we focused on the env25 pistol sequence (8) (Figure 2A and Supplementary Figure S1b). This RNA consists of three stems, a hairpin loop and an internal loop, with a PK being formed between complementary segments of the two loops. First, we probed the structure under increasing concentrations of MgCl2 as described above for the twister ribozyme. Compared to the Mg2+-free conditions, we observed clear distinctions in band intensities that indicated a fold compaction already at the lowest Mg2+ concentration investigated (5 mM). Thereby, the additional loop (L3) that joins the ribozyme and the substrate RNA to a monomolecular system, served as internal control for SHAPE reactivity. Loop L3 is structurally exposed and not interacting with the rest of the molecule. As expected, all nucleotides of this loop, U47a–U47d (Figure 2B and C; Supplementary Figure S4), showed pronounced reactivity with BzCN, rather independent from the absence or presence of Mg2+. In contrast, folding of stem P3 (which represents one of the two substrate recognition arms) was accompanied by decreases in SHAPE reactivities upon Mg2+ addition (U26, A57, Supplementary Figure S5).
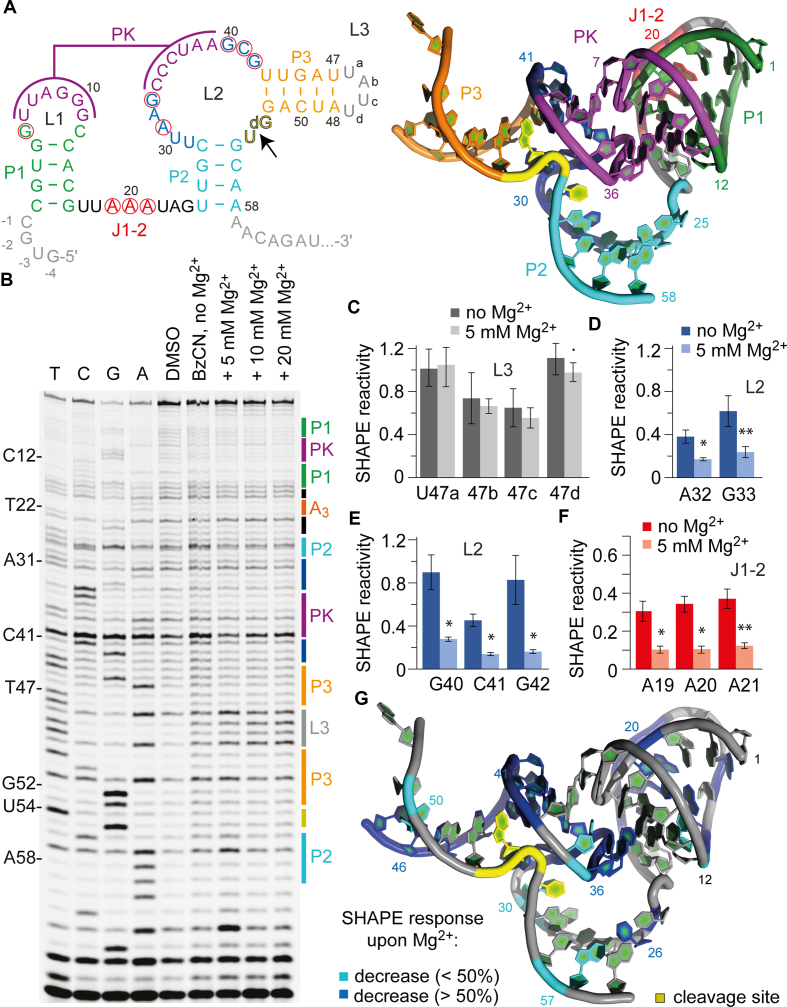
Figure 2.
SHAPE probing of the env25 pistol ribozyme. (A) Secondary structure representations of the pistol RNA motif with 5′ and 3′ spacer sequences in gray. Red circles indicate highly conserved nucleosides. 3D structure (PDB accession number: 5K7C) using the same color code (right). (B) Typical gel for the probing of the pistol RNA structure with BzCN at 37°C. Lanes from left to right: T, C, G and A ladders, control in the absence of probing reagent, probing with BzCN, probing with BzCN and in the presence of 5, 10 and 20 mM MgCl2. Note that the fragment in the sequencing ladder that matches the size of the extension product defines the precise nucleotide that corresponds to +1 (therefore the color-coded stem-loop assignments at the right side of the gel is shifted +1 to nucleoside numbering on the left side). (C–F) Relative 2′-OH reactivity for selected bases obtained from quantification and normalization of the SHAPE probing results (mean of at least three independent experiments, error bars show standard deviation; two-sided paired t-test with ** P ≤ 0.01, * P ≤ 0.05, • P ≤ 0.1). For quantification of the SHAPE probing data for the complete env25 pistol RNA sequence see Supplementary Figure S5. (G) Projection of SHAPE reactivities of individual nucleotides on the 3D structure of the pistol ribozyme (PDB accession number: 5K7C) following the color code as indicated in the legend.
Interestingly, for nucleosides 1–16 of stem-loop P1-L1, SHAPE reactivities were rather poor suggesting that this part of the sequence may represent a pre-organized compact unit (Supplementary Figure S5). Annealing of the six consecutive nucleotides 34–39 of L2 with L1 to form the coaxially stacked PK is however well reflected by clear decreases in their SHAPE reactivities (except for U37 and A38) (Supplementary Figure S5). Likewise, the majority of the remaining eight nucleosides of L2 that are intensively involved in active site formation and fixation of the substrate moiety by employing complex tertiary interactions (8), displayed significant decreases in reactivities. The formation of these interactions critically depends on the presence of Mg2+ ions. For instance, while in the absence of Mg2+, U30, A32-G33 and G40-C41-G42 well reacted with BzCN (Figure 2 and Supplementary Figure S5), their reactivities were decreased about 2- to 3-fold in the presence of Mg2+ (Figure 2D and E; Supplementary Figure S5). This is consistent with the compaction of the structure by defined triplet alignment of G40•(G33-C41), and second, with A32 being essential for proper positioning (via its 2′-OH) of G42 and, in turn, of the catalytically active G40 (8). Functional evidence for these hydrogen bond networks was independently derived from atomic mutagenesis experiments (43).
The importance of Mg2+ for pistol ribozyme folding was further reflected in the SHAPE reactivity decreases observed for A19-A20-A21 (Figure 2F). This A3 sequence is highly conserved in pistol ribozymes, as part of a 7 nt single-stranded segment J1–2 that connects stems P1 and P2 (7). Crystal structures showed that A19-A20-A21 interact with the minor groove of stem P1 (Figure 2A), where each adenine forms an A-minor triple with adjacent base pairs of the stem (8).
We mention that we had to face relatively high background rates in the DMSO control lane as well as high false-positive rates in the BzCN probing lanes due to spurious RT stops. We hypothesized that these stops might be structurally induced and thus decided to test the effect of different temperatures on the SHAPE probing pattern. Whereas low temperatures should stabilize the secondary structure, higher temperatures are expected to disrupt base pairing and thus to unfold the structure (44). By comparing the different lanes on the gel depicted in Supplementary Figure S4, we did not find pronounced differences between the BzCN probing pattern for the folding at 7 and 37°C, both in the presence of 10 mM Mg2+ concentration. However, we observed distinctions in relative band intensities that indicated the unfolding of this RNA when the temperature was raised to 57°C, in particular for the P3-L3 stem-loop (Supplementary Figure S4) and the PK regions (see also comparative discussion below and Supplementary Figures S10–12). Interestingly, the P1-L1 stem-loop of the pistol RNA showed hardly any differences in SHAPE reactivity compared to the low temperature folding conditions, suggesting that these secondary structure segment is rather rigid and largely pre-organized also at elevated temperatures.
The twister-sister ribozyme
The TS and the twister ribozyme classes share similarities in their secondary structure models (3). For instance, several TS RNAs have P1 through P5 stems in an alike arrangement to twister and a comparable nucleotide composition of the P4 loop. However, most obvious is that the two ribozyme classes cleave at different sites of their internal loop L1 (compare Figures 1A and 3A) (3), thus having made their 3D structure determinations a critical factor to shed light on the question whether they are indeed closely related. So far, two groups have been successful in solving crystal structures of these ribozymes with good resolutions (4–7). Whereas Ren and coworkers reported on four-way junctional TS (7) and P3-comprising twister RNAs (5), Lilley and coworkers solved three-way junctional TS (6) and P3-lacking twister (4) RNAs. These structures reveal that although twister and TS form a comparable PK involving L4 and L1, nucleobase alignments of their active sites are distinct (10).
Figure 3.
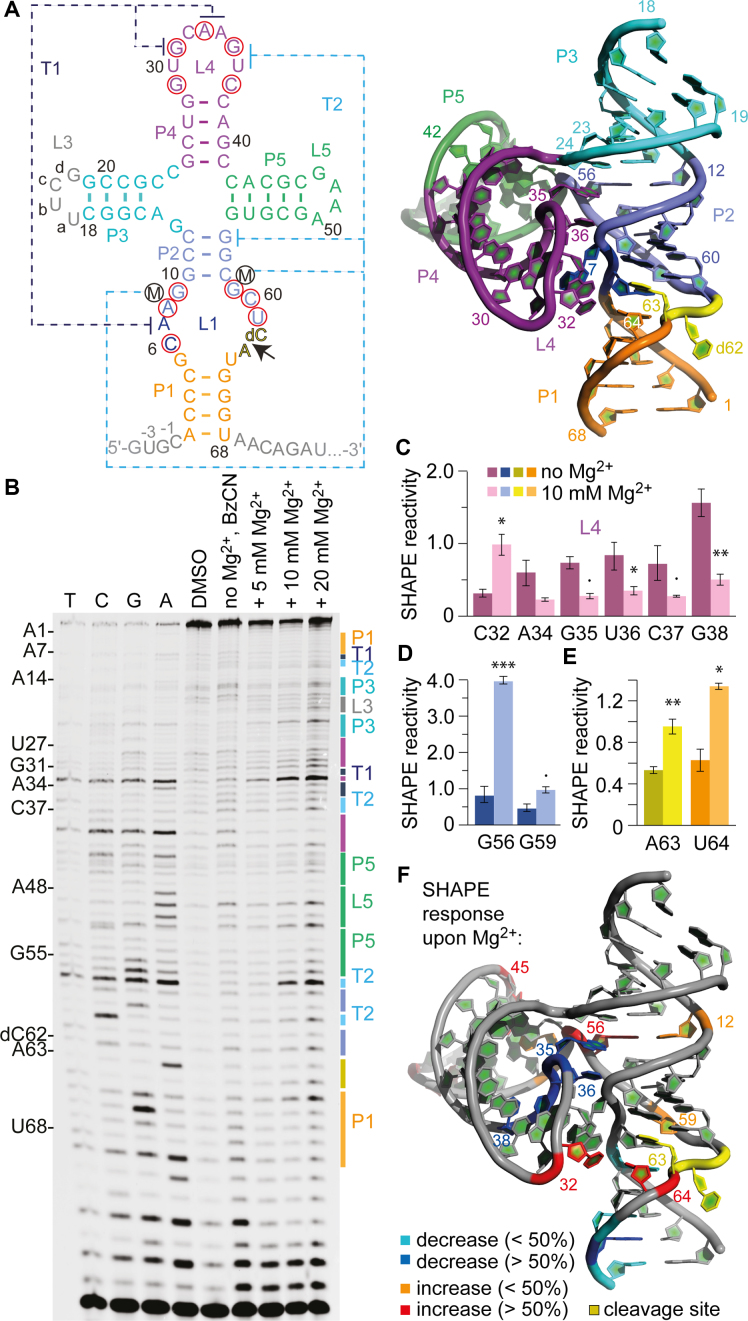
SHAPE probing of the TS ribozyme. (A) Secondary structure representations of the TS RNA motif with 5′ and 3′ spacer sequences in gray. Red circles indicate highly conserved nucleosides. 3D structure (PDB accession number: 5Y87) using the same color code (right). (B) Typical gel for the probing of the pistol RNA structure with BzCN at 37°C. Lanes from left to right: T, C, G and A ladders, control in the absence of probing reagent, probing with BzCN, probing with BzCN and in the presence of 5, 10 and 20 mM MgCl2. Note that the fragment in the sequencing ladder that matches the size of the extension product defines the precise nucleotide that corresponds to +1 (therefore the color-coded stem-loop assignments at the right side of the gel is shifted +1 to nucleoside numbering on the left side). (C–E) Relative 2′-OH reactivity for selected bases of PK T1 obtained from quantification and normalization of the SHAPE probing results (mean of at least three independent experiments, error bars show standard deviation; two-sided paired t-test with *** P ≤ 0.001, ** P ≤ 0.01, * P ≤ 0.05, • P ≤ 0.1). For quantification of the SHAPE probing data for the complete TS RNA sequence see Supplementary Figure S7. (F) Projection of SHAPE reactivities of individual nucleotides on the 3D structure of the TS ribozyme (PDB accession number: 5Y87) following the color code as indicated in the legend.
For the SHAPE experiments (Figure 3B and Supplementary Figure S6), we focused on a construct similar to the one we have used for crystallization (7) (Figure 3A). The only differences concerned the introduction of a loop (L3), that was needed to join the two strands, and the replacement of the UNCG to a GNRA L5 loop motif (U47-U48-C49-G50 to G47-A48-A49-A50) to improve enzymatic ligation yields (Figure 3A and Supplementary Figure S1c).
We were particularly interested in the long-range interactions between L4 and L1, since these contain all of the highly conserved residues (marked with red circles in Figure 3A). The L4-L1 interaction can be broken down to two PKs T1 and T2, each involving only few nucleosides. PK T1 employs A33-A34 of L4 in order to sandwich the extruded A7 from the zippered-up loop L1 (Figure 3A) (7). Additionally, A7 is anchored in place by formation of a non-canonical A7•G31 base pair. For PK T2, the Hoogsteen interaction of G56 and G35 constitutes the major long-range interaction, with G56 being additionally base-paired to C12 forming a base triple (Figure 3A). Furthermore, U36 finds itself in an extruded conformation like G56 and forms two interactions mediated by hydrated Mg2+ ions, one with G59 (via O2) and another one with the non-bridging phosphate oxygens of A8 and G9 (via O4) (Figure 3D, lower panel) (7).
Upon formation of T1 and T2, C32 becomes significantly exposed with its ribose unit, representing the turn of the loop with its 2′-OH directed toward outside. Consistently, our SHAPE data revealed that C32 in between the T1 forming nucleosides G31 and A33–A34 reacted significantly with BzCN in the presence of Mg2+, reaching maximal modification at a concentrations of 10 mM (Figure 3C and F; Supplementary Figure S7). Also for the long-range interaction T2, the SHAPE probing efficiencies for the corresponding nucleosides became clearly distinct in the absence versus presence of 10 mM Mg2+. In particular, G56 but also G59 were accompanied by pronounced increases in the corresponding band intensities (Figure 3D), consistent with their 2′-OH groups becoming exposed and/or dynamic. For the nucleosides A34-G35-U36-C37-G38 we observed SHAPE reactivity decreases upon Mg2+ addition, consistent with their shielded position upon compaction of the structure (Figure 3C and F).
We point out that G56, which participates in the G35•G56-C12 base triple, is part of the highly structured four-way junction with an innersphere coordinated Mg2+ ion that bridges G55-G56 and C23-C24 on opposite sides of the junction as seen in the X-ray structure (Figure 3A, right panel) (7). The formation of this crucial structural unit is accompanied by the strongest reactivity enhancement (4-fold) in response to Mg2+ of a nucleoside (G56) in comparison to all other nucleosides in the sequence (Figure 3D and Supplementary Figure S7; see also comparative discussion below and Supplementary Figures S10–12).
At the cleavage site, the nucleosides dC62 and A63 adopt a splayed apart conformation, with dC62 directed outward and A63 directed inward into the ribozyme core (Figure 3A) (7). Since dC62 lacks the 2′-OH (to prevent cleavage), the corresponding band intensities reflect spontaneous or structure-based loss of RT that is not due to BzCN modification; indeed, dC62 lanes were rather weak and not varying in intensities under all conditions tested. However, for A63 and U64 the reactivity to BzCN was increased significantly in the presence of 10 mM Mg2+ when compared to the Mg2+-free conditions (Figure 3E and F). This is consistent with the A63 nucleobase directed inward (7), and leaving its ribose 2′-OH accessible for probing. The high SHAPE reactivity of A63 implies flexibility also in the folded ribozyme. This flexibility seems even more pronounced for the nucleoside that follows A63. Uridine-64 is stacked on A63 and forms a single hydrogen bond to G5 (O2 of U64 to H2N-C2 of G5) but it is not involved in further interactions and well accessible from the solvent side (7). Our SHAPE experiments demonstrated that the ribose 2′-OH of U64 became significantly modified upon Mg2+ addition (Figure 3E and F). Both observations suggest that the conformational flexibility that is required to accommodate the cleavage site and to achieve the transition state likely originates from the nucleosides downstream of the scissile phosphate (A63 and U64). Of note, the three-way junctional TS ribozyme which contained an additional U residue between stem P1 and the dC-A cleavage site compared to the four-way junctional counterpart, displayed no electron density for one of these uridines in the X-ray density map (6). This also indicates increased flexibility of the downstream nucleosides. The authors thus speculated that this uridine is probably extra-helical and mobile (6) which is now strongly supported by the chemical probing results.
At last, we point out that for TS ribozymes, the formation of closing stem P1 is strongly Mg2+-dependent and well reflected by decreased SHAPE reactivites of the corresponding nucleotides (Supplementary Figure S7). This was not the case for the twister ribozyme where closing stem P1 is weakly formed and breathing as described above (see also Supplementary Figure S3).
The hatchet ribozyme
Of all newly discovered small self-cleaving ribozymes, the hatchet RNA (3) has been rather neglected thus far. There are as yet only biochemical data (40) but no 3D structures available. The proposed secondary structure consists of four stems (labeled P1 through P4 in Figure 4A), two loops (L1 and L4) and two internal bulges (L2 and L3) (7). The 13 highly conserved residues (marked with red circles in Figure 4A) are positioned in the linker J1-2 between P1 and P2, and in the asymmetric internal bulge that bridges P3 and P4. Several of these nucleotides likely participate in the organization of the active site for RNA cleavage, which occurs at the base of stem P1. The location of the cleavage site at the very 5′-end of the hatchet consensus motif is distinct from the other three ribozyme classes described in this study and is reminiscent of the glmS riboswitch-ribozyme (45,46).
Figure 4.
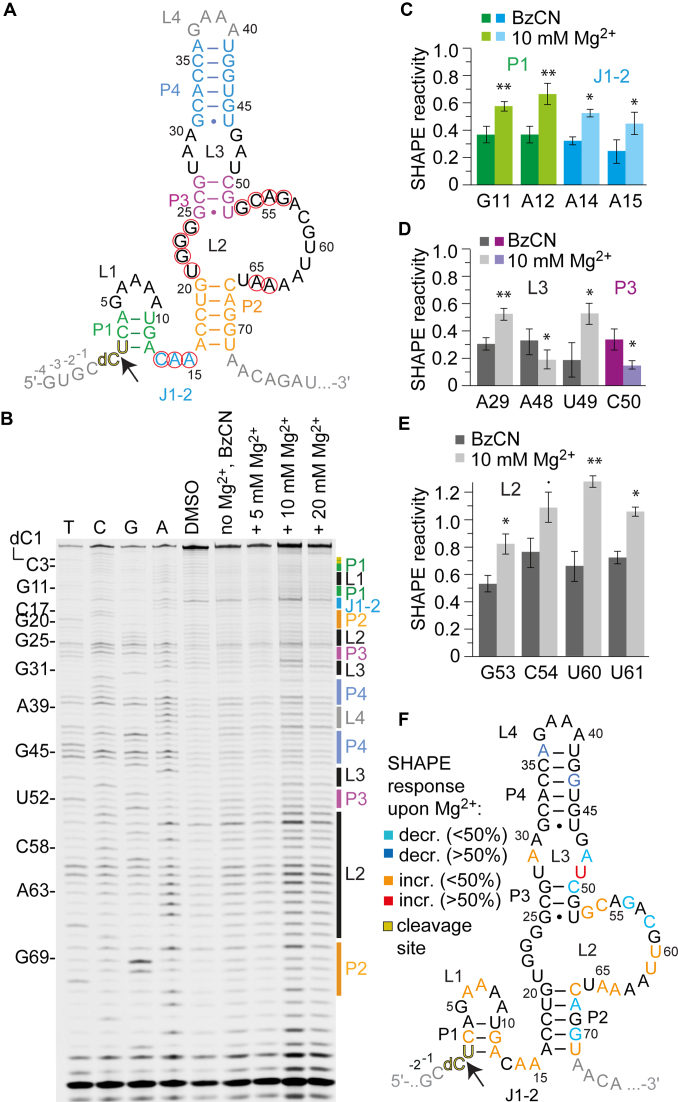
SHAPE probing of the hatchet ribozyme. (A) Secondary structure representations of the hatchet RNA motif with 5′ and 3′ spacer sequences in gray. (B) Typical gel for the probing of the pistol RNA structure with BzCN at 37°C. Lanes from left to right: T, C, G and A ladders, control in the absence of probing reagent, probing with BzCN, probing with BzCN and in the presence of 5, 10 and 20 mM MgCl2. Note that the fragment in the sequencing ladder that matches the size of the extension product defines the precise nucleotide that corresponds to +1 (therefore the color-coded stem-loop assignments at the right side of the gel is shifted +1 to nucleoside numbering on the left side). (C–E) Relative 2′-OH reactivity for selected bases obtained from quantification and normalization of the SHAPE probing results (mean of at least three independent experiments, error bars show standard deviation; two-sided paired t-test with ** P ≤ 0.01, * P ≤ 0.05, • P ≤ 0.1). For quantification of the SHAPE probing data for the complete hatchet RNA sequence see Supplementary Figure S9. (F) Projection of SHAPE reactivities of individual nucleotides on the secondary structure model of the hatchet ribozyme following the color code as indicated in the legend.
For our SHAPE experiments, we chose a metagenomic hatchet sequence (Figure 4A) that was previously investigated by Breaker and coworkers (40). Again, we prepared a monomolecular construct with a 2′-deoxyribose modification at the cleavage site (dC1-U2) to prevent cleavage and the corresponding sequence additions required for reliable SHAPE probing (Supplementary Figure S1d).
Representative gels are depicted in Figure 4B and Supplementary Figure S8; like for the three preceding ribozymes, the SHAPE probing patterns of hatchet RNA showed several significant reactivity changes between absence versus presence of Mg2+ ions, however distinct to the previous ribozymes, more increases rather than decreases in SHAPE reactivities were observed (see also comparative discussion below and Supplementary Figures S10–12). In particular, the 5′ part of the sequence that comprises the predicted short stem P1 showed nucleotides with slight increases although decreases were expected for stable stem formation at high (≥10 mM) Mg2+ concentrations. The J1-2 linker with a conserved CAA motif also displayed an enhancement of SHAPE reactivities that appeared strongest for A14 (≥10 Mg2+ mM) (Figure 4B and C; Supplementary Figure S9). In general, concentrations of 10 mM Mg2+ (or higher) were needed for the hatchet RNA to observe significant SHAPE reactivity changes. This corresponds with the Mg2+ concentration that is needed for maximal cleavage rates (40); it is significantly higher compared to the three other ribozymes.
Formation of the predicted stems P2, P3 and P4 was consistent with the general observation of weak SHAPE reactivities for the majority of the nucleosides involved. Only few significant decreases upon Mg2+ addition were observed (A36, G43, C50, A68, A70; Figure 4D and F;Figure S9). Potential inconsistencies with the prediction of stem P2 concerned both terminal base pairs G20-C67 and A16-U71, with C67 and U71 showing small but significant increases rather than the expected decreases (Figure 4F and Supplementary Figure S9). This might be a hint at their involvement in more complex tertiary interactions.
Of particular interest was the analysis of the internal bulge L2 because it contains most of the highly conserved nucleotides. Surprisingly, only very little distinctions in intensities can be made out for the highly conserved G22, G23 and G24, however, significantly increased reactivities in the presence of Mg2+ were observed for G53 and C54 (conserved), U60 and U61 (not conserved), and for A64 and A65 (conserved) (Figure 4F and Supplementary Figure S9).
For the smaller internal loop L3, nucleotide identities are not highly conserved however the loop motif itself is conserved in the hatchet ribozyme (3,40). Indeed, significant increases in SHAPE reactivities were observed for two out of the six nucleotides, namely A29 and U49 (Figure 4D).
Although strong efforts to solve the 3D structure at atomic resolution of the hatchet ribozyme are needed to create a profound basis for mechanistic studies and to elucidate the catalytic mechanism, the SHAPE probing experiments allow for the following insights: our data support a folding model for this ribozyme, in which stems P4 and P3 seem rather static, separated by an internal loop L3 that shows significant restructuring upon Mg2+ addition. The large internal loop L2 also becomes restructured in its 3′ single-stranded half. Interestingly, its 5′ half (the short 4 nt connecting sequence U21-G22-G23-G24 between P2 and P3) did hardly respond to an increase in Mg2+ concentration and we speculate that these guanosines might be engaged in a rigid structural scaffold (e.g. tetrad formation). Such a scaffold could be needed to bring the conserved nucleosides of L2 into vicinity of the cleavage site and the conserved J1-2 element. In this context, stem P2 seems to play a pivotal role, with both terminal base pairs becoming exposed and/or dynamic, likely because of their involvement in a structural scaffold that is crucial for the active conformation.
Temperature and Mg2+ dependencies of folding of the four ribozymes
We point out that the SHAPE data of twister, pistol and TS ribozymes display clear temperature dependencies with respect to their PK formations. At 10 mM Mg2+ concentration, defolding of twister PK T1 at elevated temperature (57°C) was most pronounced and reflected in significantly higher SHAPE reactivities of the involved nucleotides compared to almost similar reactivities at 7 and 37°C (Supplementary Figures S2 and 10). For pistol and TS, SHAPE reactivities of the nucleosides forming their PKs (PK and T1–T2) were gradually increasing from 7 to 37 to 57°C (Supplementary Figures S4, 6 and 10). In this context, we point out that smFRET imaging revealed structural dynamics of PK T1 to significantly contribute to twister cleavage rate kinetics (11). For TS and pistol ribozymes, quantitative smFRET analyses of PK dynamics remain to be performed. Based on the generally low SHAPE reactivities observed for pistol, breathing of its PK seems less likely (compared to twister and TS), rather it is the nucleation determinant together with stem-loop P1-L1 that molds the ribozyme core and catalytic pocket.
With respect to the influence of divalent metal ions, twister and pistol ribozymes showed comparable SHAPE reactivities at 5, 10 and 20 mM Mg2+ concentrations, consistent with observation that these two ribozyme classes become fully folded at concentrations higher than 2 mM (Supplementary Figure S11). For the four-way junctional TS ribozyme, we however observed clear increases of SHAPE reactivity with increasing Mg2+ concentrations. While for the PK-sensitive residue C32, a 2-fold increase in SHAPE reactivity for 10 and 20 mM Mg2+ compared to 5 mM Mg2+ was observed, the corresponding increase for G56 was nearly 20-fold. We underline that the phosphate backbone of this nucleoside is innersphere coordinated to a Mg2+ ion that bridges to the phosphate of G25, again through innersphere coordination: this most critical interaction of the four-way junction seems to be fully folded at 10 mM Mg2+ with no further increase observed at 20 mM Mg2+. In contrast, for U64 we observed a stepwise increase in reactivity from 5 to 10 to 20 mM Mg2+ (4-fold and another 2-fold). We recall that the conformational flexibility that is required to accommodate the cleavage site and to achieve the transition state likely originates from the two nucleosides A63 and U64 downstream of the scissile phosphate; these conformational adaptions are strongly Mg2+ dependent as observed here.
Like TS, also the hatchet ribozyme requires Mg2+ concentrations above 5 mM to become fully folded. This is reflected by the SHAPE behavior of several of its residues, three of them (G53, U61 and U71) are depicted in Supplementary Figure S11.
CONCLUSION
In this study, we have been able to show that SHAPE probing can be successfully applied on small ribozymes, exemplified for pre-catalytic folds of twister, pistol, TS and hatchet ribozymes. The obtained data not only assist in the evaluation of proposed structure models but are especially useful to reveal Mg2+ induced folding of molecular architectures at single nucleotide resolution which concern PKs, junctions, bulges, internal loops and even single loop-closing base pairs at the termini of helices. SHAPE reactivity changes of single nucleotides in RNA originate from their enhanced (or reduced) conformational flexibility to sample conformations that are more (or less) reactive toward SHAPE reagents. The interpretation of SHAPE data with respect to structural dynamics has to be carefully considered because nucleosides that become locked in a specific conformation can turn reactive although their inherent dynamics are decreased. Taking the specific knowledge from X-ray structures of three of the four ribozymes into account, we could for instance show that PK formation of twister, pistol and TS ribozymes are strictly Mg2+ dependent, although the required concentrations for their formation differ. Moreover, the pistol ribozyme seems to fold most stably with structural segments that remain well pre-organized even at elevated temperatures; its 5′-stem-loop and PK likely represent the nucleation core of the pistol ribozyme. Another important finding concerns the twister ribozyme for which phylogenetics implies that the closing stem P1 is a mandatory factor for activity (2,3). For the twister ribozyme investigated here, SHAPE probing however does not indicate stable formation of this stem, rather it remains modified by the reagent at equal levels, even at high concentrations of Mg2+ which implies a significant degree of P1 breathing. This is consistent with the finding that this stem is negligible for cleavage activity (38). Concerning the TS ribozyme, an interesting result is that structuring of the four-way junction which has a specific innersphere coordination site for Mg2+, requires much higher Mg2+ (10 mM) compared to the concentration of about 1 mM that has been reported to reach maximal cleavage rates. Finally, for the hatchet ribozyme, where a 3D structure is not yet available, the secondary structure model is in agreement with the reactivity patterns of stems P3 and P4 but can be challenged with respect to stem P1. Unexpected features with respect to structural integration of the ends of stem P2 can also be expected according to the SHAPE data and make structure elucidation of the hatchet ribozyme eagerly awaited.
Moreover, the information from SHAPE probing is invaluable for the experimental design of quantitative RNA folding and RNA–small molecule binding studies that involve fluorescence spectroscopic techniques as we have shown earlier for another RNA species, namely for riboswitches (22,23,47). In this context, we note that the SHAPE data presented here have already successfully directed the selection of positions for fluorophore attachment that were needed for a smFRET study of the twister ribozyme (11) and the data will be further used in our laboratory for comprehensive fluorescence spectroscopic studies on TS and hatchet ribozyme folding.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Elisabeth Fuchs for RNA mass spectrometric analysis, and Nikola Vusurovic, Christoph Falschlunger and Elisabeth Mairhofer for discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Austrian Science Fund FWF [I1040, P27947 to R.M.]; Austrian Research Promotion Agency FFG [West-Austrian BioNMR 858017]; Swiss National Foundation SNF [Early Postdoc.Mobility to J.G.]. Funding for open access charge: Austrian Science Fund FWF [I1040, P27947].
Conflict of interest statement. None declared.
REFERENCES
- 1. Jimenez R.M., Polanco J.A., Lupták A.. Chemistry and biology of self-cleaving ribozymes. Trends Biochem. Sci. 2015; 40:648–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Roth A., Weinberg Z., Chen A.G., Kim P.B., Ames T.D., Breaker R.R.. A widespread self-cleaving ribozyme class is revealed by bioinformatics. Nat. Chem. Biol. 2014; 10:56–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Weinberg Z., Kim P.B., Chen T.H., Li S., Harris K.A., Lünse C.E., Breaker R.R.. New classes of self-cleaving ribozymes revealed by comparative genomics analysis. Nat. Chem. Biol. 2015; 11:606–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu Y., Wilson T.J., McPhee S.A., Lilley D.M.. Crystal structure and mechanistic investigation of the twister ribozyme. Nat. Chem. Biol. 2014; 10:739–744. [DOI] [PubMed] [Google Scholar]
- 5. Ren A., Košutić M., Rajashankar K.R., Frener M., Santner T., Westhof E., Micura R., Patel D.J.. In-line alignment and Mg2+ coordination at the cleavage site of the env22 twister ribozyme. Nat. Commun. 2014; 5:5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu Y., Wilson T.J., Lilley D.M.J.. The structure of a nucleolytic ribozyme that employs a catalytic metal ion. Nat. Chem. Biol. 2017; 13:508–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zheng L., Mairhofer E., Teplova M., Zhang Y., Ma J., Patel D.J., Micura R., Ren A.. Structure-based insights into self-cleavage by a four-way junctional twister-sister ribozyme. Nat. Commun. 2017; 8:1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ren A., Vušurović N., Gebetsberger J., Gao P., Juen M., Kreutz C., Micura R., Patel D.J.. Pistol ribozyme adopts a pseudoknot fold facilitating site-specific in-line cleavage. Nat. Chem. Biol. 2016; 12:702–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nguyen L.A., Wang J., Steitz T.A.. Crystal structure of Pistol, a class of self-cleaving ribozyme. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:1021–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ren A., Micura R., Patel D.J.. Structure-based mechanistic insights into catalysis by small self-cleaving ribozymes. Curr. Opin. Chem. Biol. 2017; 41:71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vušurović N., Altman R.B., Terry D.S., Micura R., Blanchard S.C.. Pseudoknot formation seeds the twister ribozyme cleavage reaction coordinate. J. Am. Chem. Soc. 2017; 139:8186–8193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Panja S., Hua B., Zegarra D., Ha T., Woodson S.A.. Metals induce transient folding and activation of the twister ribozyme. Nat. Chem. Biol. 2017; 13:1109–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Juen M.A., Wunderlich C.H., Nußbaumer F., Tollinger M., Kontaxis G., Konrat R., Hansen D.F., Kreutz C.. Excited states of nucleic acids probed by proton relaxation dispersion NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 2016; 55:12008–12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nielsen G., Jonker H.R., Vajpai N., Grzesiek S., Schwalbe H.. Kinase in motion: insights into the dynamic nature of p38α by high-pressure NMR spectroscopic studies. Chembiochem. 2013; 14:1799–1806. [DOI] [PubMed] [Google Scholar]
- 15. Stagno J.R., Liu Y., Bhandari Y.R., Conrad C.E., Panja S., Swain M., Fan L., Nelson G., Li C., Wendel D.R. et al. Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography. Nature. 2017; 541:242–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Deigan K.E., Li T.W., Mathews D.H., Weeks K.M.. Accurate SHAPE-directed RNA structure determination. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Spitale R.C., Flynn R.A., Torre E.A., Kool E.T., Chang H.Y.. RNA structural analysis by evolving SHAPE chemistry. Wiley Interdiscip. Rev. RNA. 2014; 5:867–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Choi E.K., Ulanowicz K.A., Nguyen Y.A.H., Frandsen J.K., Mitton-Fry R.M.. SHAPE analysis of the htrA RNA thermometer from Salmonella enterica. RNA. 2017; 23:1569–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Watts J.M., Dang K.K., Gorelick R.J., Leonard C.W., Bess J.W., Swanstrom R., Burch C.L., Weeks K.M.. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009; 460:711–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Weeks K.M. Toward all RNA structures, concisely. Biopolymers. 2015; 103:438–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rice G.M., Busan S., Karabiber F., Favorov O.V., Weeks K.M.. SHAPE analysis of small RNAs and riboswitches. Methods Enzymol. 2014; 549:165–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Soulière M.F., Altman R.B., Schwarz V., Haller A., Blanchard S.C., Micura R.. Tuning a riboswitch response through structural extension of a pseudoknot. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E3256–E3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Soulière M.F., Haller A., Rieder R., Micura R.. A powerful approach for the selection of 2-aminopurine substitution sites to investigate RNA folding. J. Am. Chem. Soc. 2011; 133:16161–16167. [DOI] [PubMed] [Google Scholar]
- 24. Spitale R.C., Crisalli P., Flynn R.A., Torre E.A., Kool E.T., Chang H.Y.. RNA SHAPE analysis in living cells. Nat. Chem. Biol. 2013; 9:18–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kwok C.K., Ding Y., Tang Y., Assmann S.M., Bevilacqua P.C.. Determination of in vivo RNA structure in low-abundance transcripts. Nat. Commun. 2013; 4:2971. [DOI] [PubMed] [Google Scholar]
- 26. Watters K. E., Abbott T. R., Lucks J. B.. Simultaneous characterization of cellular RNA structure and function with in-cell SHAPE-Seq. Nucleic Acids Res. 2015; 44:e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Micura R. Small interfering RNAs and their chemical synthesis. Angew. Chem. Int. Ed. 2002; 41:2265–2269. [DOI] [PubMed] [Google Scholar]
- 28. Pitsch S., Weiss P. A., Jenny L., Stutz A., Wu X.. Reliable chemical synthesis of oligoribonucleotides (RNA) with 2′-O-[(Triisopropylsilyl) oxy] methyl (2′-O-tom)-protected phosphoramidites. Helv. Chim. Acta. 2001; 84:3773–3795. [Google Scholar]
- 29. Lang K., Micura R.. The preparation of site-specifically modified riboswitch domains as an example for enzymatic ligation of chemically synthesized RNA fragments. Nat. Protoc. 2008; 3:1457–1466. [DOI] [PubMed] [Google Scholar]
- 30. Höbartner C., Micura R.. Chemical synthesis of selenium-modified oligoribonucleotides and their enzymatic ligation leading to an U6 SnRNA stem-loop segment. J. Am. Chem. Soc. 2004; 126:1141–1149. [DOI] [PubMed] [Google Scholar]
- 31. Rieder R., Höbartner C., Micura R.. Enzymatic ligation strategies for the preparation of purine riboswitches with site-specific chemical modifications. Methods Mol. Biol. 2009; 540:15–24. [DOI] [PubMed] [Google Scholar]
- 32. Soulière M.F., Micura R.. Use of SHAPE to select 2AP substitution sites for RNA-ligand interactions and dynamics studies. Methods Mol. Biol. 2014; 1103:227–239. [DOI] [PubMed] [Google Scholar]
- 33. Mortimer S.A., Weeks K.M.. Time-resolved RNA SHAPE chemistry. J. Am. Chem. Soc. 2008; 130:16178–16180. [DOI] [PubMed] [Google Scholar]
- 34. Mortimer S.A., Weeks K.M.. Time-resolved RNA SHAPE chemistry: quantitative RNA structure analysis in one-second snapshots and at single-nucleotide resolution. Nat. Protoc. 2009; 4:1413–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Laederach A., Das R., Vicens Q., Pearlman S.M., Brenowitz M., Herschlag D., Altman R.B.. Semiautomated and rapid quantification of nucleic acid footprinting and structure mapping experiments. Nat. Protoc. 2008; 3:1395–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Low J.T., Weeks K.M.. SHAPE-directed RNA secondary structure prediction. Methods. 2010; 52:150–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cordero P., Kladwang W., VanLang C.C., Das R.. Quantitative dimethyl sulfate mapping for automated RNA secondary structure inference. Biochemistry. 2012; 51:7037–7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Košutić M., Neuner S., Ren A., Flür S., Wunderlich C., Mairhofer E., Vušurović N., Seikowski J., Breuker K., Höbartner C. et al. A mini-twister variant and impact of residues/cations on the phosphodiester cleavage of this ribozyme class. Angew. Chem. Int. Ed. 2015; 54:15128–15133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Harris K.A., Lünse C.E., Li S., Brewer K. I., Breaker R. R.. Biochemical analysis of pistol self-cleaving ribozymes. RNA. 2015; 21:1852–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li S., Lünse C.E., Harris K.A., Breaker R.R.. Biochemical analysis of hatchet self-cleaving ribozymes. RNA. 2015; 21:1845–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gebetsberger J., Micura R.. Unwinding the twister ribozyme: from structure to mechanism. Wiley Interdiscip. Rev. RNA. 2017; 8:e1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Neuner S., Kreutz C., Micura R.. The synthesis of 15N(7)-hoogsteen face-labeled adenosine phosphoramidite for solid-phase RNA synthesis. Monatsh. Chem. 2017; 148:149–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Neuner S., Falschlunger C., Fuchs E., Himmelstoss M., Ren A., Patel D.J., Micura R.. Atom-Specific mutagenesis reveals structural and catalytic roles for an active-site adenosine and hydrated Mg2+ in pistol ribozymes. Angew. Chem. Int. Ed. Engl. 2017; 56:15954–15958. [DOI] [PubMed] [Google Scholar]
- 44. Cruz J.A., Westhof E.. The dynamic landscapes of RNA architecture. Cell. 2009; 136:604–609. [DOI] [PubMed] [Google Scholar]
- 45. Barrick J.E., Corbino K.A., Winkler W.C., Nahvi A., Mandal M., Collins J., Lee M., Roth A., Sudarsan N., Jona I. et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:6421–6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Klein D.J., Ferré-D’Amaré A.R.. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006; 313:1752–1756. [DOI] [PubMed] [Google Scholar]
- 47. Lang K., Rieder R., Micura R.. Ligand-induced folding of the thiM TPP riboswitch investigated by a structure-based fluorescence spectroscopic approach. Nucleic Acids Res. 2007; 35:5370–5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.