Abstract
Past work on relatively small, single-site studies using regional volumetry, and more recently machine learning methods, has shown that widespread structural brain abnormalities are prominent in schizophrenia. However, to be clinically useful, structural imaging biomarkers must integrate high-dimensional data and provide reproducible results across clinical populations and on an individual person basis. Using advanced multi-variate analysis tools and pooled data from case–control imaging studies conducted at 5 sites (941 adult participants, including 440 patients with schizophrenia), a neuroanatomical signature of patients with schizophrenia was found, and its robustness and reproducibility across sites, populations, and scanners, was established for single-patient classification. Analyses were conducted at multiple scales, including regional volumes, voxelwise measures, and complex distributed patterns. Single-subject classification was tested for single-site, pooled-site, and leave-site-out generalizability. Regional and voxelwise analyses revealed a pattern of widespread reduced regional gray matter volume, particularly in the medial prefrontal, temporolimbic and peri-Sylvian cortex, along with ventricular and pallidum enlargement. Multivariate classification using pooled data achieved a cross-validated prediction accuracy of 76% (AUC = 0.84). Critically, the leave-site-out validation of the detected schizophrenia signature showed accuracy/AUC range of 72–77%/0.73–0.91, suggesting a robust generalizability across sites and patient cohorts. Finally, individualized patient classifications displayed significant correlations with clinical measures of negative, but not positive, symptoms. Taken together, these results emphasize the potential for structural neuroimaging data to provide a robust and reproducible imaging signature of schizophrenia. A web-accessible portal is offered to allow the community to obtain individualized classifications of magnetic resonance imaging scans using the methods described herein.
Keywords: psychosis, neuroimaging, MRI, brain, biomarker
Introduction
Schizophrenia is an often devastating illness that results in substantial morbidity and mortality worldwide.1 Structural brain abnormalities were noted by early anatomists,2,3 and results from in vivo magnetic resonance imaging (MRI) have supported the notion that schizophrenia is a brain disorder.4,5 Despite such advances, neuroimaging is typically not used as part of standard clinical care of psychotic disorders, in part due to heterogeneous results. Advances in clinical practice require robust imaging biomarkers of disease that can be used for diagnosis, determination of prognosis, and integration within trials of novel therapeutics.
As imaging technology has advanced and costs have declined, both the sample sizes and number of studies examining structural brain abnormalities in schizophrenia have expanded dramatically. Results from small single-site studies have often been heterogeneous, leading investigators to integrate findings across studies using meta-analyses of published data from hundreds of studies and thousands of patients.6–12 These studies provide increasingly consistent evidence of widespread patterns of structural deficits in schizophrenia, including gray matter loss that is prominent in the insular, cingulate, prefrontal, and temporal cortex.
However, such retrospective, literature-based meta-analyses are inherently limited by methodological variation. Scanner effects, processing choices, and analytic strategy can have a dramatic impact on results of individual studies, adding noise to results. The ENIGMA consortium was created to overcome such obstacles through the use of prospective meta-analyses, where all sites perform the same data processing, quality assurance, and group-level analyses.13,14 Using this approach, the ENIGMA-schizophrenia working group recently examined subcortical volumetric abnormalities in a sample of over 2000 patients and a similar number of controls, and reported volume reductions of small-moderate effect size (Cohen’s d: 0.2–0.4). These findings have subsequently been replicated by the COCORO consortium, which used a similar approach.15 Despite the enormous sample size provided by ENIGMA, such regional analyses ultimately provide a coarse-grained account of structural brain abnormalities associated with schizophrenia, and at present are limited to the subcortex. In contrast, one large recent study directly pooled data in a voxel-based, whole-brain mega-analysis, and delineated widespread loss of gray matter density that was maximal in insular cortex.16
Importantly, despite accelerating efforts to pool structural imaging data to study psychosis, such meta- and mega-analyses have focused on mass-univariate approaches applied at the group level. Alternatively, brain-wide multivariate neuroanatomical patterns can be used to classify individuals as cases or controls using machine learning methods.17–19 By concisely summarizing high-dimensional data, multivariate classification has clear advantages for clinical translation.20,21 Single-site studies over the past decade have demonstrated that multivariate analyses can accurately discriminate patients and controls, as well as predict progression.18,19,22–26 One recent meta-analysis of single-site multivariate classification studies reported pooled classification sensitivity of 76%, with 79% specificity.27 However, no study to date has demonstrated single-subject classification in a large, multi-site, and multi-ethnic setting. In particular, it remains unknown whether classifiers trained on data acquired at one site will perform similarly on data acquired from a new site that was not included in the training set. Any practical clinical application would require an imaging biomarker that is robust to site differences, so that algorithms developed at academic centers could be applied in the community.
Here, we report results from a large-scale analysis of structural MRI data from 941 participants across 5 sites. Importantly, by pooling images instead of derived measures, and uniformly processing all data via identical pipelines, we were able to harmonize structural measurements and attenuate intersite differences. Building on prior efforts, our first hypothesis was that widespread structural brain abnormalities in adult patients with schizophrenia would be detectable across all cohorts. Our second hypothesis was that a reproducible neuroanatomical signature of schizophrenia would be present across sites and diverse populations, and be detectable at the individual patient level. Our approach included mass-univariate analyses of regional volumes defined using state-of-the-art multi-atlas segmentation,28 optimally discriminative voxel based analyses (ODVBA)29,30 for local-multivariate pattern analysis, and individual level classification using consensus-based machine learning.
Methods
Participants and Image Acquisition
Data from 5 MRI studies were included in the current multi-site mega-analysis. All subjects were part of previous studies overseen by local institutional policies.31–39 Sample demographics of these data sets are given in table 1. Detailed MRI acquisition protocol information for each site is given in supplementary table 1. Anonymized data provided by the 5 sites were provided to and analyzed by the University of Pennsylvania’s Center for Biomedical Image Computing and Analytics. Raw MR scans were initially examined for motion, image artifacts, or restricted field-of-view. Scans were also checked for lesions, but this population of patients being relatively young, there were few subjects with lesions, and the total lesion load of these subjects was low. Further details of quality control procedures are given in supplementary methods.
Table 1.
Sample Demographics
| Harmonized (n = 835) | SET1 (n = 325) | SET2 (n = 180) | SET3 (n = 330) | SET4 (n = 42) | SET5 (n = 64) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group, No. (%) | Controls | Patients | Controls | Patients | Controls | Patients | Controls | Patients | Controls | Patients | Controls | Patients |
| 448 (53.7) | 387 (46.3) | 170 (52.3) | 155 (47.7) | 105 (58.3) | 75 (41.7) | 173 (52.4) | 157 (47.6) | 24 (57.1) | 18 (42.9) | 29 (45.3) | 35 (54.7) | |
| Female sex, no. (%) | 215 (48.0) | 148 (38.2) | 100 (58.8) | 70 (45.2) | 61 (58.1) | 39 (52.0) | 54 (31.2) | 39 (24.8) | 11 (45.8) | 12 (66.7) | 13 (44.8) | 15 (42.9) |
| Age, mean (SD) | 33.9 (11.6) | 35.3 (11.0) | 35.7 (13.5) | 39.5 (12.0) | 35.0 (11.3) | 35.5 (8.7) | 31.5 (9.2) | 31.0 (9.1) | 28.2 (4.9) | 24.0 (4.8) | 23.1 (3.7) | 23.5 (4.7) |
| ICV, mean (SD), cm3 | 1415.8 (148.5) | 1398.2 (145.4) | 1410.8 (166.5) | 1378.8 (149.3) | 1382.3 (122.9) | 1361.6 (153.1) | 1441.0 (139.0) | 1434.9 (128.7) | 1405.1 (93.3) | 1409.1 (161.3) | 1470.8 (108.9) | 1438.5 (148.8) |
| CPZ dosage, mean (min–max)a | N/A | 436.0 (0.0–2766.7) | N/A | 506.8 (25.0–2766.7) | N/A | 478.0 (0.0–1775.0) | N/A | 375.9 (10.0– 1850.0) | Not available | Not available | Not available | Not available |
| Duration of illness, mean (min–max), yearsb | N/A | 10.78 (0.00–43.90) | N/A | 18.21 (0.60–43.90) | N/A | 9.58 (0.00–27.00) | N/A | 4.44 (0.04–39.41) | Not available | Not available | Not available | Not available |
| Clinical symptom scores, no. (%), type | N/A | 357 (92.2), Various | N/A | 143 (92.3), SAPS/SANS | N/A | 59 (78.7), PANSS | N/A | 155 (98.7), PANSS | Not available | Not available | Not available | Not available |
| Acquisition protocol | Various | MPRAGE | BRAVO | MPRAGE | MPRAGE | MPRAGE | ||||||
| Field strength | Various | 3 T | 3 T | 1.5 T | 3 T | 3 T | ||||||
| Location | Various | University of Pennsylvania, Philadelphia, PA | Tianji Anning Hospita, Tianjin, China | Ludwig-Maximilian-University, Munich, Germany | Xijing Hospital, Xi’an, China | Peking University, Beijing, China | ||||||
Note: N/A = not applicable.
aCPZ dosage equivalents available for 71 of SET1 patients, 59 of SET2 patients, and 125 of SET3 patients.
bDuration of illness available for 130 of SET1 patients, 59 of SET2 patients, and 153 of SET3 patients.
Image Processing
Following magnetic field inhomogeneity correction,40 MR images were first segmented into a hierarchically organized set of 259 anatomical regions, ranging from total brain volume down to individual cortical gyri and deep structures. To parcellate the brain into anatomically defined regions, we used a highly accurate multi-atlas consensus labeling procedure,28 which was recently the top-ranked method at an independent international competition.41 Besides regional volumes, we also calculated a set of rich regional descriptors based on intensity, shape, size, and texture properties of each region, to be used as an additional set of input features in subsequent multivariate classification. See supplementary methods for more details.
To perform high-resolution voxel-based analysis, each image’s gray matter segmentation was obtained using a previously reported and validated method, MICO.42 Segmented maps were warped into a common template space using a highly accurate deformable registration (DRAMMS),43–45 yielding regional volumetric maps (RAVENS) that quantify gray matter volume at each voxel.46 We masked out subcortical gray matter structures on RAVENS maps, because available structural T1-weighted images do not provide sufficient contrast for reliable intensity-based segmentation;47 analysis of subcortical structures was limited to the more robust atlas-based regional parcellation described above.
Intersite Image Harmonization
Scanner and demographic differences across MRI samples offer challenges for combining data. Accordingly, we harmonized data across sites by estimating intracranial volume (ICV), site, age, and sex effects on each imaging measure within a pooled sample of controls using a linear model. The coefficients estimated from this model were then applied to the whole sample including patient data. Effectively, this removed the influence of site and demographic effects on the difference between patients and controls. Importantly, this control-based harmonization model was always cross-validated, ie, it was only derived from the training set, and subsequently applied to the test set. Due to the small sample-sizes of the SET4 and SET5 sites, these sites were not included in the harmonization procedure and were set aside for use as validation cohorts in the classification experiments described later. The harmonization procedure was applied independently on each input image feature, including regional features and voxelwise RAVENS maps.
Group-Level Statistical Analyses
All group-level analyses were conducted within the harmonized cohort (n = 835). Regional case-control comparisons were conducted using 2 sample t-tests. Results are reported as effect sizes (Cohen’s d-statistic). Effect of medication and disease duration were each evaluated separately using linear regression. Voxelwise analyses of RAVENS maps used ODVBA,29,30 a local-multivariate pattern analysis method that has been shown in prior studies to have greater sensitivity than both standard voxel-based morphometry and searchlight-based local-multivariate analyses. Type I error was controlled using the false discovery rate (FDR) correction (Q < 0.05). Voxelwise effect sizes were reported in regions that were found to be significant by ODVBA.
Multivariate Classification
The above analyses provide a description of structural brain differences between schizophrenia and control groups. As a final step, we used machine learning techniques to create a multivariate classifier that provides individual-level prediction of group status. We used a linear support vector machine (SVM) classifier,48 a widely used method with extensive validation in various similar high dimensional pattern classification problems. The input features included voxelwise values from RAVENS maps, regional volumes, and an extended set of shape, intensity and texture features computed for each region. A classifier was run separately for each feature type using 10-fold cross-validation. A consensus prediction was then obtained by combining the predictions of each classifier through a weighted averaging based on their cross-validated individual performances. This procedure was performed using an external 10-fold cross-validation (supplementary figure 4), except in experiments that use independent training and testing sets.
To maximize generalizability, we applied the SVM classification using default parameters (C = 1), without feature selection. (Optimization of such parameters would likely further improve accuracy, albeit at the risk of overfitting the data.) See supplementary methods for further details.
Multivariate models were trained under 3 different conditions to evaluate the impact of pooling data across sites and training/testing on different sites. First, models were trained and tested separately within each site using 10-fold cross-validation. Second, a single 10-fold cross-validated SVM classification analysis was conducted using the complete sample of harmonized pooled data. Third, we performed training/testing on different sites through leave-one-site-out cross-validation. This procedure included applying a model trained on the complete pooled data to the 2 independent validation cohorts. This procedure aimed to evaluate the generalizability of the classifier to a totally new data set sampled from an entirely different population and scanner. Importantly, in this procedure harmonization was done only within the training set, and the control-based regression model was applied to the test set. Before training the classifier, the training and testing sets were z-scored separately, with the assumption that the test set has a similar patient to control ratio as the training set. For all analyses, individual-subject classification performance was evaluated using receiver operating characteristic (ROC) curves. Classification performance was summarized using both the area under the curve (AUC) and the classification accuracy metrics.
Finally, to assess the clinical relevance of the MRI-based classification, we investigated correlations between the classifier output and clinical symptom scores of individual subjects. The symptom scores for different sites were obtained using common symptom rating scales in schizophrenia research: the Scale for the Assessment of Positive Symptoms,49 the Scale for the Assessment of Negative Symptoms,50 and the Positive And Negative Syndrome Scale.51,52Table 1 details the available type of scores for each site. The scores were normalized by z-scoring them within each site and pooled together. For this analysis, individualized pseudo-probabilities of having the neuroanatomical signature of schizophrenia were estimated from the classifier’s output using sigmoid fits. These pseudo-probabilities correspond to the degree to which a given individual’s brain appears to match the schizophrenia pattern.
Results
Regional and Voxelwise Analyses Reveal Distributed Structural Brain Abnormalities in Schizophrenia
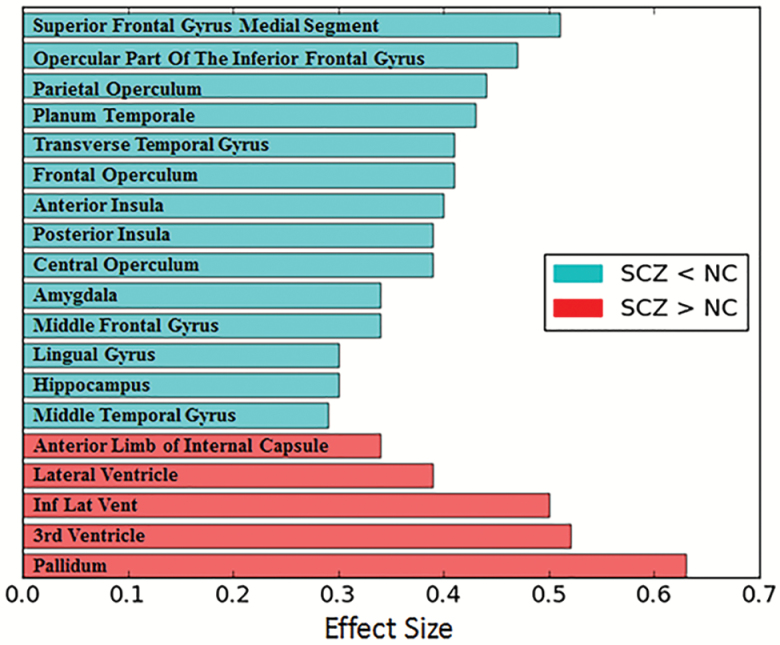
Mass-univariate regional analyses of pooled data delineated robust abnormalities of brain structure that survived FDR correction. The structures that show the most significant group differences, both in positive and negative directions, are shown in figure 1. Patients with schizophrenia had marked ventricular expansion, as well as larger pallidum volumes. Cortical gray matter volume loss was also quite evident, with maximal effects in the prefrontal cortex (superior frontal gyrus; d = 0.51) as well as temporal cortex, parietal cortex, insula, and amygdala. The effect sizes of volumetric changes were strongly correlated across sites (pairwise correlation coefficient range: 0.71–0.81), indicating strong consistency despite protocol, scanner, and demographic differences across sites. A complete list of all regions with significant group differences (q < 0.05) is provided in supplementary table 2. Site-specific analysis results are provided in supplementary table 3.
Fig. 1.
Key regional volume differences between patients (SCZ) and controls (NC). ROIs with the highest effect sizes (absolute effect size ≥ 0.29) between controls (N = 448) and patients (N = 387), calculated using pooled, harmonized data. For a complete list of all regions with significant group differences (q < 0.05), see supplementary table 1.
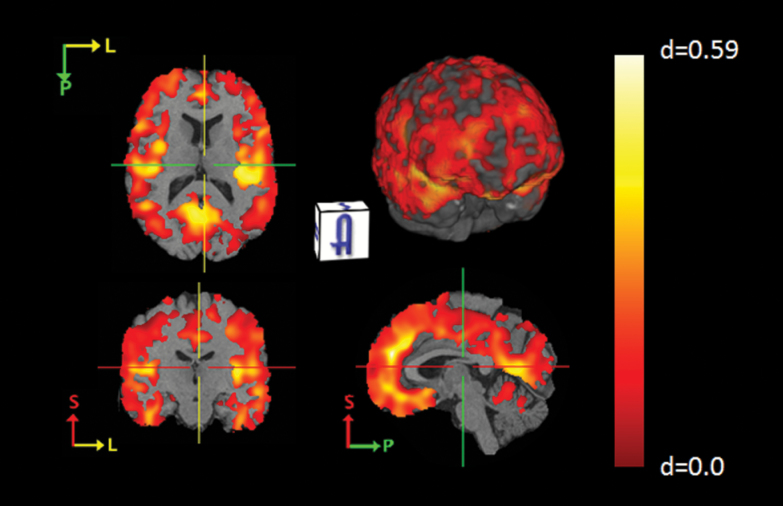
High-resolution voxelwise analyses of harmonized RAVENS maps in the pooled sample using ODVBA delineated a distributed pattern of volume loss (figure 2), and showed good agreement with the results of the ROI-based analysis (supplementary figure 1). The results of the ODVBA analyses performed using each data set separately are shown in supplementary figure 2. The effects found for individual data sets were consistent with each other and with the pooled analysis, but less significant than the pooled analysis results, emphasizing the enhanced power of the pooled analysis compared to the smaller single-site analyses.
Fig. 2.
Voxelwise gray matter volume differences between patients (SCZ) and controls (NC). Effect-size maps between controls (N = 448) and patients (N = 387) calculated using pooled, harmonized data. The highlighted regions that show significant group differences were calculated using the output of ODVBA thresholded at the FDR-corrected significance value of q < 0.05. Note: The location of the crosshairs is unchanged between each of the cross-sections. A color figure is available online at Schizophrenia Bulletin.
We also evaluated the impact of disease duration and antipsychotic load (chlorpromazine equivalents53) on the regional data (supplementary table 4). In contrast to the very robust and distributed group differences, medication effects were only seen in the frontal operculum, where higher medication dose was associated with lower volume. Duration of illness was associated with larger lateral ventricles and pallidum, as well as reduced volume of regions including the middle frontal gyrus, parahippocampal gyrus, and hippocampus.
Multivariate Classification is Accurate Using Data Pooled Across Sites
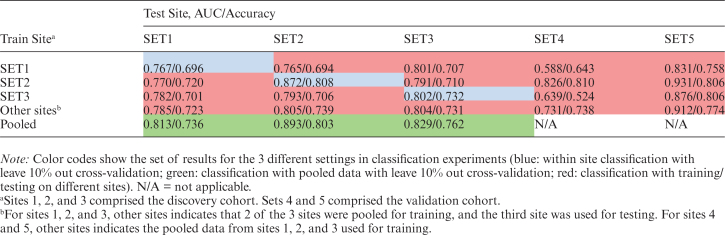
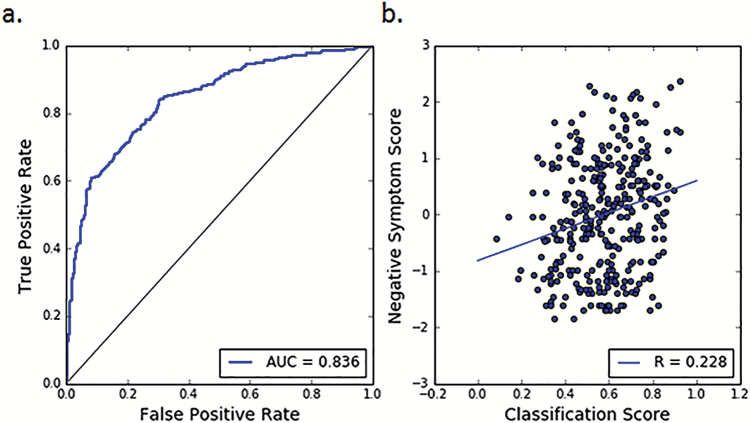
Classification results are detailed in table 2 for the within-site, pooled, and between-site experiments. Compared to individual within-site classification, pooled classification resulted in higher classification accuracy for each of SET1, SET2, and SET3, with 0.02 to 0.045 increase in AUC. The ROC curve for the cross-validated classification within the pooled-data is shown in figure 3a. These results indicate that pooling data across sites is not only viable, but it actually improves the classification results. The leave-site-out cross-validation procedure on SET1, SET2 and SET3, ie, using models trained exclusively on data collected at different sites, achieved AUC values between 0.78 and 0.80, which were comparable to pooled or within site cross-validation results. Besides, combination of training data from 2 separate external sites consistently outperformed training on each external site individually, suggesting that pooling a larger number of data sets for training helps to increase accuracy and robustness of the classifier. Classification results for the independent validation cohorts, SET4 and SET5 were also high (accuracies 0.738 and 0.774), further emphasizing the feasibility of using models trained exclusively on data collected at a different site. A comparison of the relative performance of different types of input feature sets used in the classification experiments is shown in supplementary figure 3.
Table 2.
Classification Results
Fig. 3.
Classification results. (a) ROC curve for patient vs control classification. The classification was performed using the pooled data and with leave-10%-out cross-validation through consensus-voting of all input feature sets, ie, RAVENS maps, regional volumes, and region-based descriptors. (b) Correlation between classifier output and negative clinical symptom scores (SANS for SET1, PANSS Negative for sets 2 and 3). Classification scores were obtained by converting the distances of the test samples from the discrimination hyperplane to pseudo-probabilities using sigmoid fits. Negative symptom scores were z-scored within data sets and then pooled together. The correlation for classifier score with negative symptom score was equal to r = .228 (P = .00003). No correlations were found between positive symptom scores and the classifier output (r = −.019, P = .73).
Classification Probability Is Related to Negative Symptoms
We found a significant positive correlation (r = .228, P = .00003) between the individual pseuoprobabilities of having neuroanatomical pattern and the negative symptom scores of the subjects (figure 3b). No significant correlations were found between the positive symptom scores and the classification pseudoprobabilities, nor was there a significant relationship to the duration of illness.
Discussion
In a large-scale analysis of data pooled across sites, we demonstrated that the widespread structural brain abnormalities associated with schizophrenia can be used for accurate single-subject classification using machine learning techniques. Application of state-of-the-art consensus-based classifiers on different types of imaging features yielded high performance, and indicated that multivariate neuroanatomical signatures have the potential to become a quantitative imaging biomarker for schizophrenia. Importantly, these procedures remained accurate when trained and tested on data from completely different sites, populations, and scanners, highlighting the reproducibility of this imaging signature and, therefore, its translational potential.
Structural Brain Abnormalities in Schizophrenia Are Widespread and Visible at Multiple Scales
Capitalizing on a sample of nearly 1000 participants amassed via a multi-site, mega-analytic design, we provide robust evidence of widespread structural brain abnormalities in schizophrenia. These were present on multiple scales, and were evident in both analyses of regional volumes as well as high-resolution voxelwise analyses. Brain abnormalities in schizophrenia have been studied using brain imaging for 3 decades.2,4 While abnormalities have been consistently documented by single-site studies, the relative degree to which different brain regions were impacted has varied considerably by sample. Large-scale meta-analyses have pooled data,8,10–12,54 but they have been limited by important differences in data processing across studies. The ENIGMA and COROCO consortia, as well the study of Gupta et al.16 have overcome this limitation with rigorous meta-analyses of data processed in consistent fashion (albeit not always with identical pipelines harmonized on raw data), and recently provided evidence of subcortical volume loss of moderate effect size.14,15 Our analysis of subcortical structures provided convergent results to those provided by ENIGMA and other studies, including loss of hippocampus, thalamus, and amygdala volume; expansion of the pallidum; and ventricular enlargement. However, we build upon such findings from the subcortex and demonstrate cortical abnormalities of often large effect size,11,16 with a maximal deficit in the prefrontal cortex. In line with previous studies, duration of illness was associated with increasing expansion of the pallidum and ventricles.9,11,14,55 In contrast to the lack of medication effects found in ENIGMA, but in line with a previous large-scale retrospective meta-analysis, higher dose of antipsychotics was associated with larger pallidum and reduced gray matter volume in several frontal regions.9,11,18
Multivariate Classification of Individuals Is Accurate Even Using Data From Different Scanners and Sites
While mass-univariate analyses provide a valuable description of the structural brain abnormalities associated with schizophrenia at the group level, they cannot function as biomarkers in individual patients. Accordingly, our primary focus was to use machine learning techniques to create a classifier that leveraged the complex multivariate pattern of structural deficits.21 The classifier we described here could differentiate patients and controls with a high degree of accuracy, with levels comparable to those previously reported in single-site studies.18,24,25,27,56
A highly significant positive correlation was found between the classification output and negative symptom scores. Thus, the probability to which an individual’s multivariate pattern of structural anatomy was classified as consistent with schizophrenia was correlated with the burden of negative symptoms. In contrast, there was not a significant correlation with positive symptoms. This result is consistent with extensive prior research linking negative symptoms to structural brain abnormalities in schizophrenia.57 To the degree to which both structural brain abnormalities and negative symptoms portend a poor response to standard pharmacotherapies,58 these results may assist in stratification within clinical trials of new therapies which integrate structural neuroimaging and machine-learning tools.
Critically, classification results were obtained with established algorithms and methods, to ensure that the resultant biomarker would be accessible for widespread use. Nonetheless, it is likely that better classification could potentially be achieved via parameter tuning, feature selection, and use of nonlinear kernels or deep-leaning approaches. However, such gains might also carry lead to greater model complexity, higher risk of over-fitting to training data, and reduced generalizability to new data sets. Such generalizability is of paramount importance for clinical translation.
Indeed, the single most important finding from this approach was that multivariate classifiers could retain accurate prediction even when trained and tested on data from separate scanners. Machine learning can be quite robust to intersite variations, by virtue of depending on the relative weights, ie, contrasts, of a complex pattern rather than on absolute volume-based thresholds, which can be impacted by slight variations of image contrast across scanners. These results suggest that models trained on data collected at specialized academic centers have the potential to be applied to data acquired in the community in order to calculate an imaging biomarker of schizophrenia. Importantly, leave-one-site-out was tested without the need for harmonization of the testing data with the training data set, but only via z-scoring within the new test site data. The classification pipeline is available in our image processing portal as a web-accessible application (CBICA Image Processing Portal: https://ipp.cbica.upenn.edu/). The web application allows users to submit single or multiple T1-weighted MRI images and outputs individualized scores, computed using automatically extracted imaging features and the presaved classification model from the harmonized training data. Thus, for any new data set, this web portal provides the scientific and clinical community with a freely accessible quantitative index of the imaging-based biomarker described here.
Limitations and Future Directions
Although this study benefited from a large sample size and advanced analytics, certain limitations should be noted. While developing a distinctive imaging signature of schizophrenia is an important endeavor, its value in predicting disease progression, treatment response, and in assisting differential diagnosis needs to be investigated in future studies. We note that the proposed imaging index, which can be calculated at an individual level using the provided online tool, cannot be used as a direct diagnostic index, as it has not been validated in heterogeneous clinical populations. Applications to samples that are not enriched for schizophrenia may result in a high false-positive rate. Like most other psychiatric conditions, schizophrenia is associated with substantial co-morbidity of other psychiatric conditions (such as mood disorders). Unfortunately, co-morbid conditions were not consistently recorded across the participating sites, and thus could not be evaluated in the present analysis. Future studies should evaluate co-morbidity of other psychiatric conditions on classification performance. Expanded data sets that seek to parse heterogeneity within diagnostic groups will be critical.39,59–61 Such work is particularly important for studies of youth with psychosis-spectrum or prodromal symptoms, where there is substantial diversity of clinical outcomes.55,62–66 Recent work67 using heterogeneity analyses offers hope that semi-supervised machine learning methods can provide a better understanding of the heterogeneity of neuroanatomical signatures of schizophrenia. Finally, future work that combines large-scale imaging data sets in youth with well-characterized longitudinal clinical follow up with advanced multivariate analytics will be critical areas of focus moving forward.
Conclusions
Using a pooled mega-analytic strategy, the present data provide among the most robust evidence to date of structural brain abnormalities in adults with schizophrenia. Furthermore, these results emphasize that such signals can be used to derive highly accurate multivariate models that allow for discrimination at the level of individual patients, thereby providing a robust neuroanatomical signature of schizophrenia. Critically, this signature remains accurate even when the classifier is trained using data from different sites and scanners. Taken together, our findings highlight the accelerating promise of imaging-based biomarkers in major neuropsychiatric illnesses such as schizophrenia. The classifier used in this paper for classification is publicly available online at the CBICA Image Processing Portal.
Supplementary Material
Supplementary data are available at Schizophrenia Bulletin online.
Funding
The research at the University of Pennsylvania site was supported in part by the National Institute of Health (P50MH096891 to R.E.G., R01MH107703 to T.D.S., R01MH101111 to D.H.W.) and the Dowshen Program for Neuroscience.
Supplementary Material
Acknowledgments
At the University of Pennsylvania site, thanks to Monica Calkins, PhD for assistance with clinical phenotyping.
Disclosures: R.E.G. participated in an advisory board for Otsuka Pharmaceuticals and RCG receives royalties from the Brain Resource Center and serves without compensation on an advisory board for Lumosity. All other authors have no disclosures.
References
- 1. Freedman R. Schizophrenia. N Engl J Med. 2003;349:1738–1749. [DOI] [PubMed] [Google Scholar]
- 2. Stevens JR. An anatomy of schizophrenia?Arch Gen Psychiatry. 1973;29:177–189. [DOI] [PubMed] [Google Scholar]
- 3. Gross G, Huber G, Schüttler R. Computerized tomography studies on schizophrenic diseases. Arch Psychiatr Nervenkr (1970). 1982;231:519–526. [DOI] [PubMed] [Google Scholar]
- 4. Shenton ME, Dickey CC, Frumin M, McCarley RW. A review of MRI findings in schizophrenia. Schizophr Res. 2001;49:1–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Insel TR. Rethinking schizophrenia. Nature. 2010;468:187–193. [DOI] [PubMed] [Google Scholar]
- 6. Bora E, Fornito A, Radua J et al. . Neuroanatomical abnormalities in schizophrenia: a multimodal voxelwise meta-analysis and meta-regression analysis. Schizophr Res. 2011;127:46–57. [DOI] [PubMed] [Google Scholar]
- 7. Cooper D, Barker V, Radua J, Fusar-Poli P, Lawrie SM. Multimodal voxel-based meta-analysis of structural and functional magnetic resonance imaging studies in those at elevated genetic risk of developing schizophrenia. Psychiatry Res. 2014;221:69–77. [DOI] [PubMed] [Google Scholar]
- 8. Ellison-Wright I, Glahn DC, Laird AR, Thelen SM, Bullmore E. The anatomy of first-episode and chronic schizophrenia: an anatomical likelihood estimation meta-analysis. Am J Psychiatry. 2008;165:1015–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fusar-Poli P, Smieskova R, Kempton MJ, Ho BC, Andreasen NC, Borgwardt S. Progressive brain changes in schizophrenia related to antipsychotic treatment? A meta-analysis of longitudinal MRI studies. Neurosci Biobehav Rev. 2013;37:1680–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Glahn DC, Laird AR, Ellison-Wright I et al. . Meta-analysis of gray matter anomalies in schizophrenia: application of anatomic likelihood estimation and network analysis. Biol Psychiatry. 2008;64:774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Haijma SV, Van Haren N, Cahn W, Koolschijn PC, Hulshoff Pol HE, Kahn RS. Brain volumes in schizophrenia: a meta-analysis in over 18 000 subjects. Schizophr Bull. 2013;39:1129–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Honea R, Crow TJ, Passingham D, Mackay CE. Regional deficits in brain volume in schizophrenia: a meta-analysis of voxel-based morphometry studies. Am J Psychiatry. 2005;162:2233–2245. [DOI] [PubMed] [Google Scholar]
- 13. Thompson PM, Stein JL, Medland SE et al. ; Alzheimer’s Disease Neuroimaging Initiative, EPIGEN Consortium, IMAGEN Consortium, Saguenay Youth Study (SYS) Group The ENIGMA Consortium: large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging Behav. 2014;8:153–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Erp TG, Hibar DP, Rasmussen JM et al. . Subcortical brain volume abnormalities in 2028 individuals with schizophrenia and 2540 healthy controls via the ENIGMA consortium. Mol Psychiatry. 2016;21:585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Okada N, Fukunaga M, Yamashita F et al. . Abnormal asymmetries in subcortical brain volume in schizophrenia. Mol Psychiatry. 2016;21:1460–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gupta CN, Calhoun VD, Rachakonda S et al. . Patterns of gray matter abnormalities in schizophrenia based on an international mega-analysis. Schizophr Bull. 2015;41:1133–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bansal R, Staib LH, Laine AF et al. . Anatomical brain images alone can accurately diagnose chronic neuropsychiatric illnesses. PLoS One. 2012;7:e50698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Davatzikos C, Shen D, Gur RC et al. . Whole-brain morphometric study of schizophrenia revealing a spatially complex set of focal abnormalities. Arch Gen Psychiatry. 2005;62:1218–1227. [DOI] [PubMed] [Google Scholar]
- 19. Koutsouleris N, Meisenzahl EM, Davatzikos C et al. . Use of neuroanatomical pattern classification to identify subjects in at-risk mental states of psychosis and predict disease transition. Arch Gen Psychiatry. 2009;66:700–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fan Y, Shen D, Gur RC, Gur RE, Davatzikos C. COMPARE: classification of morphological patterns using adaptive regional elements. IEEE Trans Med Imaging. 2007;26:93–105. [DOI] [PubMed] [Google Scholar]
- 21. Veronese E, Castellani U, Peruzzo D, Bellani M, Brambilla P. Machine learning approaches: from theory to application in schizophrenia. Comput Math Methods Med. 2013;2013:867924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nakamura K, Kawasaki Y, Suzuki M et al. . Multiple structural brain measures obtained by three-dimensional magnetic resonance imaging to distinguish between schizophrenia patients and normal subjects. Schizophr Bull. 2004;30:393–404. [DOI] [PubMed] [Google Scholar]
- 23. Kawasaki Y, Suzuki M, Kherif F et al. . Multivariate voxel-based morphometry successfully differentiates schizophrenia patients from healthy controls. Neuroimage. 2007;34:235–242. [DOI] [PubMed] [Google Scholar]
- 24. Koutsouleris N, Gaser C, Bottlender R et al. . Use of neuroanatomical pattern regression to predict the structural brain dynamics of vulnerability and transition to psychosis. Schizophr Res. 2010;123:175–187. [DOI] [PubMed] [Google Scholar]
- 25. Nieuwenhuis M, van Haren NE, Hulshoff Pol HE, Cahn W, Kahn RS, Schnack HG. Classification of schizophrenia patients and healthy controls from structural MRI scans in two large independent samples. Neuroimage. 2012;61:606–612. [DOI] [PubMed] [Google Scholar]
- 26. Schnack HG, Nieuwenhuis M, van Haren NE et al. . Can structural MRI aid in clinical classification? A machine learning study in two independent samples of patients with schizophrenia, bipolar disorder and healthy subjects. Neuroimage. 2014;84:299–306. [DOI] [PubMed] [Google Scholar]
- 27. Kambeitz J, Kambeitz-Ilankovic L, Leucht S et al. . Detecting neuroimaging biomarkers for schizophrenia: a meta-analysis of multivariate pattern recognition studies. Neuropsychopharmacology. 2015;40:1742–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Doshi J, Erus G, Ou Y et al. ; Alzheimer’s Neuroimaging Initiative MUSE: MUlti-atlas region Segmentation utilizing Ensembles of registration algorithms and parameters, and locally optimal atlas selection. Neuroimage. 2016;127:186–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang T, Davatzikos C. ODVBA: optimally-discriminative voxel-based analysis. IEEE Trans Med Imaging. 2011;30:1441–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang T, Davatzikos C. Optimally-discriminative voxel-based morphometry significantly increases the ability to detect group differences in schizophrenia, mild cognitive impairment, and Alzheimer’s disease. Neuroimage. 2013;79:94–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Satterthwaite TD, Wolf DH, Loughead J et al. . Association of enhanced limbic response to threat with decreased cortical facial recognition memory response in schizophrenia. Am J Psychiatry. 2010;167:418–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wolf DH, Satterthwaite TD, Kantrowitz JJ et al. . Amotivation in schizophrenia: integrated assessment with behavioral, clinical, and imaging measures. Schizophr Bull. 2014;40:1328–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang H, Zeng LL, Chen Y, Yin H, Tan Q, Hu D. Evidence of a dissociation pattern in default mode subnetwork functional connectivity in schizophrenia. Sci Rep. 2015;5:14655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhu J, Zhuo C, Liu F, Xu L, Yu C. Neural substrates underlying delusions in schizophrenia. Sci Rep. 2016;6:33857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhuo C, Ma X, Qu H, Wang L, Jia F, Wang C. Schizophrenia patients demonstrate both inter-voxel level and intra-voxel level white matter alterations. PLoS One. 2016;11:e0162656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yan H, Tian L, Wang Q et al. . Compromised small-world efficiency of structural brain networks in schizophrenic patients and their unaffected parents. Neurosci Bull. 2015;31:275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yan H, Tian L, Yan J et al. . Functional and anatomical connectivity abnormalities in cognitive division of anterior cingulate cortex in schizophrenia. PLoS One. 2012;7:e45659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang P, Xi Y, Lu ZL et al. . Decreased bilateral thalamic gray matter volume in first-episode schizophrenia with prominent hallucinatory symptoms: A volumetric MRI study. Sci Rep. 2015;5:14505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang T, Koutsouleris N, Meisenzahl E, Davatzikos C. Heterogeneity of structural brain changes in subtypes of schizophrenia revealed using magnetic resonance imaging pattern analysis. Schizophr Bull. 2015;41:74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tustison NJ, Avants BB, Cook PA et al. . N4ITK: improved N3 bias correction. IEEE Trans Med Imaging. 2010;29:1310–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Doshi JJ EG, Ou Y, Davatzikos C. Ensemble-based medical image labeling via sampling morphological appearance manifold. In: MICCAI Challenge Workshop on Segmentation: Algorithms, Theory and Applications.Nagoya, Japan; 2013. [Google Scholar]
- 42. Li C, Gore JC, Davatzikos C. Multiplicative intrinsic component optimization (MICO) for MRI bias field estimation and tissue segmentation. Magn Reson Imaging. 2014;32:913–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yangming Ou AS, Aristeidis Sotiras DRAMMS Distribution General Website; 2009. http://www.med.upenn.edu/sbia/dramms.html
- 44. Ou Y, Sotiras A, Paragios N, Davatzikos C. DRAMMS: deformable registration via attribute matching and mutual-saliency weighting. Med Image Anal. 2011;15:622–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ou Y, Akbari H, Bilello M, Da X, Davatzikos C. Comparative evaluation of registration algorithms in different brain databases with varying difficulty: results and insights. IEEE Trans Med Imaging. 2014;33:2039–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Davatzikos C, Genc A, Xu D, Resnick SM. Voxel-based morphometry using the RAVENS maps: methods and validation using simulated longitudinal atrophy. Neuroimage. 2001;14:1361–1369. [DOI] [PubMed] [Google Scholar]
- 47. Helms G, Draganski B, Frackowiak R, Ashburner J, Weiskopf N. Improved segmentation of deep brain grey matter structures using magnetization transfer (MT) parameter maps. Neuroimage. 2009;47:194–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vapnik VN. An overview of statistical learning theory. IEEE Trans Neural Netw. 1999;10:988–999. [DOI] [PubMed] [Google Scholar]
- 49. Andreasen N. The Scale for the Assessment of Positive Symptoms (SAPS). Iowa City: University of Iowa; 1984. [Google Scholar]
- 50. Andreasen N. The scale for the assessment of negative symptoms (SANS). Iowa City: University of Iowa; 1983. [Google Scholar]
- 51. Kay SR, Fiszbein A, Opler LA. The positive and negative syndrome scale (PANSS) for schizophrenia. Schizophr Bull. 1987;13:261–276. [DOI] [PubMed] [Google Scholar]
- 52. Kay SR, Opler LA, Lindenmayer JP. The Positive and Negative Syndrome Scale (PANSS): rationale and standardisation. Br J Psychiatry Suppl 1989(7):59–67. [PubMed] [Google Scholar]
- 53. Woods SW. Chlorpromazine equivalent doses for the newer atypical antipsychotics. J Clin Psychiatry. 2003;64:663–667. [DOI] [PubMed] [Google Scholar]
- 54. Chan RC, Di X, McAlonan GM, Gong QY. Brain anatomical abnormalities in high-risk individuals, first-episode, and chronic schizophrenia: an activation likelihood estimation meta-analysis of illness progression. Schizophr Bull. 2011;37:177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fraguas D, Díaz-Caneja CM, Pina-Camacho L, Janssen J, Arango C. Progressive brain changes in children and adolescents with early-onset psychosis: a meta-analysis of longitudinal MRI studies. Schizophr Res. 2016;173:132–139. [DOI] [PubMed] [Google Scholar]
- 56. Fan Y, Gur RE, Gur RC et al. . Unaffected family members and schizophrenia patients share brain structure patterns: a high-dimensional pattern classification study. Biol Psychiatry. 2008;63:118–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ren W, Lui S, Deng W et al. . Anatomical and functional brain abnormalities in drug-naive first-episode schizophrenia. Am J Psychiatry. 2013;170:1308–1316. [DOI] [PubMed] [Google Scholar]
- 58. Wolf DH. Anhedonia in schizophrenia. Curr Psychiatry Rep. 2006;8:322– 328. [DOI] [PubMed] [Google Scholar]
- 59. Bleich-Cohen M, Jamshy S, Sharon H, Weizman R, Intrator N, Poyurovsky M, Hendler T. Machine learning fMRI classifier delineates subgroups of schizophrenia patients. Schizophr Res. 2014;160:196–200. [DOI] [PubMed] [Google Scholar]
- 60. Ivleva EI, Bidesi AS, Keshavan MS et al. . Gray matter volume as an intermediate phenotype for psychosis: Bipolar-Schizophrenia Network on Intermediate Phenotypes (B-SNIP). Am J Psychiatry. 2013;170:1285–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Narayanan B, Ethridge LE, O’Neil K et al. . Genetic sources of subcomponents of event-related potential in the dimension of psychosis analyzed from the B-SNIP study. Am J Psychiatry. 2015;172:466–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Koutsouleris N, Gaser C, Patschurek-Kliche K et al. . Multivariate patterns of brain-cognition associations relating to vulnerability and clinical outcome in the at-risk mental states for psychosis. Hum Brain Mapp. 2012;33:2104–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Karageorgiou E, Schulz SC, Gollub RL et al. . Neuropsychological testing and structural magnetic resonance imaging as diagnostic biomarkers early in the course of schizophrenia and related psychoses. Neuroinformatics. 2011;9:321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Koutsouleris N, Davatzikos C, Bottlender R et al. . Early recognition and disease prediction in the at-risk mental states for psychosis using neurocognitive pattern classification. Schizophr Bull. 2012;38:1200–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pina-Camacho L, Garcia-Prieto J, Parellada M et al. . Predictors of schizophrenia spectrum disorders in early-onset first episodes of psychosis: a support vector machine model. Eur Child Adolesc Psychiatry. 2015;24:427–440. [DOI] [PubMed] [Google Scholar]
- 66. Satterthwaite TD, Wolf DH, Calkins ME et al. . Structural brain abnormalities in youth with psychosis spectrum symptoms. JAMA Psychiatry. 2016;73:515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dong A, Toledo JB., Nicolas H. et al. . Heterogeneity of neuroanatomical patterns in prodromal Alzheimer’s disease: links to cognition, progression, and biomarkers. Brain. 2016;140(3):735–747. doi:10.1093/brain/aww319 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.