Abstract
The N-terminal domain (NTD) of nuclear human uracil DNA glycosylase (hUNG2) assists in targeting hUNG2 to replication forks through specific interactions with replication protein A (RPA). Here, we explored hUNG2 activity in the presence and absence of RPA using substrates with ssDNA–dsDNA junctions that mimic structural features of the replication fork and transcriptional R-loops. We find that when RPA is tightly bound to the ssDNA overhang of junction DNA substrates, base excision by hUNG2 is strongly biased toward uracils located 21 bp or less from the ssDNA–dsDNA junction. In the absence of RPA, hUNG2 still showed an 8-fold excision bias for uracil located <10 bp from the junction, but only when the overhang had a 5′ end. Biased targeting required the NTD and was not observed with the hUNG2 catalytic domain alone. Consistent with this requirement, the isolated NTD was found to bind weakly to ssDNA. These findings indicate that the NTD of hUNG2 targets the enzyme to ssDNA–dsDNA junctions using RPA-dependent and RPA-independent mechanisms. This structure-based specificity may promote efficient removal of uracils that arise from dUTP incorporation during DNA replication, or additionally, uracils that arise from DNA cytidine deamination at transcriptional R-loops during immunoglobulin class-switch recombination.
INTRODUCTION
Human nuclear uracil DNA glycosylase (hUNG2) is the primary enzyme for excising uracil bases from genomic DNA. This critical function initiates base excision repair pathways that help maintain genomic sequence integrity during and after DNA replication (1). Conversely, the same uracil excision activity, in conjunction with activation-induced DNA cytidine deaminase, promotes programmed DNA mutagenesis during class-switch recombination and somatic hypermutation in B cells (2–4). Localization of hUNG2 to replication forks involves interactions with two proteins, proliferating cell nuclear antigen (PCNA) and replication protein A (RPA), both of which bind to distinct segments of hUNG2’s largely disordered N-terminal domain (NTD) that precedes its globular catalytic domain (1,5–7). Although the action of hUNG2 in class-switch recombination has not yet been associated with specific protein–protein interactions, the process requires active transcription and the formation of three-stranded DNA:RNA structures known as R-loops that recruit RPA (8). The importance of protein–protein interactions involving the NTD of hUNG2 is highlighted by the fact that these interactions are highly regulated by post-translational modifications (9,10), including phosphorylation events that promote binding to RPA (10).
This study was specifically designed to investigate whether the catalytic activity of hUNG2 was altered through interactions of its NTD with RPA or with ssDNA–dsDNA junctions that resemble a structural feature of replication forks or R-loops. These studies were facilitated by the sub-nanomolar affinity of RPA for ssDNA (11), which allows preparation of stoichiometric complexes of RPA with the ssDNA overhangs of junction substrates that contain uracil in the duplex region. These specific RPA-bound substrates allowed us to directly measure the effect of RPA on hUNG2’s excision of uracil bases spaced at specific distances from the junction. In this context, we hypothesized that RPA might capture and tether hUNG2 in the vicinity of the junction through the known interaction between the NTD of hUNG2 and the ‘winged-helix’ domain of RPA32 (5,9), which is separated from the DNA binding core of RPA by a disordered linker (Figure 1A) (12,13). Although several studies have reported inhibitory effects of RPA on the catalytic activity of hUNG2 using uracilated single- or double-stranded DNA substrates (5,6,9), these previous findings are counterintuitive given the targeting function of RPA in cellular base excision repair (12,14). Thus, we speculated that the use of junction substrates might reveal novel properties of hUNG2 and its NTD that promote spatial targeting of uracil excision in the presence RPA. Quite surprisingly, in the course of this study we also discovered that the NTD itself serves as a targeting domain that confers a strong kinetic bias towards excision of uracils within ∼10 bp of the junction. These findings suggest models for the action of hUNG2 at replication forks and R-loops.
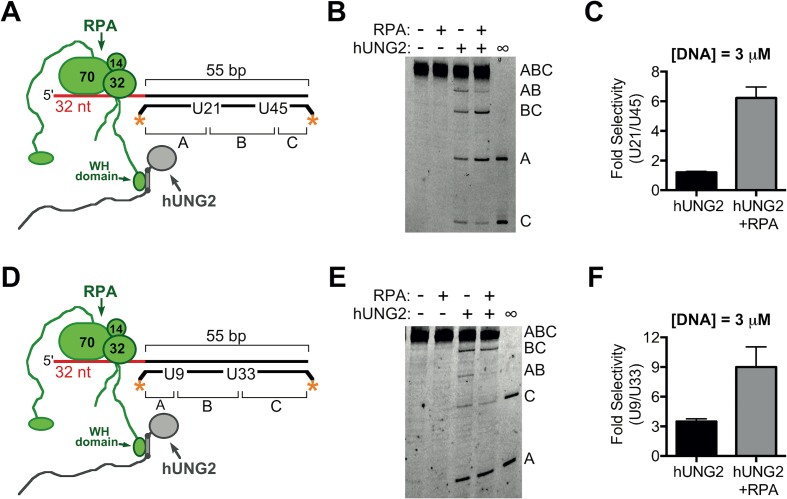
Figure 1.
Biased uracil excision by hUNG2 with junction DNA substrates in the presence and absence of RPA. (A) Junction DNA substrates have a 5′ ssDNA overhang, two uracil bases in the duplex region, and fluorescein end-labels indicated by the asterisks. In this case, the uracil bases are 21 and 45 bp from the ssDNA–dsDNA junction and excision of one or both sites can produce labeled DNA fragments A, AB, BC or C. hUNG2 assays were performed with and without RPA. (B) Image of reaction products, separated using denaturing PAGE, after reacting 3 μM of DNA substrate from panel A with 900 pM hUNG2. In the presence of RPA, fragments BC and A are enriched over fragments AB and C, which indicates preferred excision at U21. On all panels, lanes marked with ∞ have an arbitrary amount of DNA substrate that was entirely processed by hUNG2 and serves as a band marker. (C) Relative selectivity for U21 compared to U45. (D) Junction substrate with uracil bases at 9 and 33 bp from the ssDNA–dsDNA junction. (E) Image of hUNG2 reaction products in the presence and absence of RPA using the substrate in panel D after separation using denaturing PAGE. Enzyme and substrate concentrations were 900 pM and 3 μM, respectively. (F) Relative selectivity for U9 and U33 with and without RPA.
MATERIALS AND METHODS
DNA oligonucleotides
All oligonucleotides were purchased from Integrated DNA Technologies (IDT) and were purified by PAGE before use (Supplementary Information). Oligonucleotides were annealed by mixing complementary strands, heating to 95°C, then slowly cooling to room temperature. The annealing buffer contained 10 mM Tris–HCl (pH 8.0), 100 mM NaCl, and 0.1 mM EDTA, and the formation of duplex DNA was confirmed using native PAGE.
The sequences of the oligonucleotides were generated by a random sequence generator then manually modified to contain ∼50% GC content with minimal likelihood of secondary structure, as determined by the IDT OligoAnalyzer tool (15). Additionally, oligonucleotides were designed such that the uracil bases were flanked by an identical palindromic sequence GAATCGAUAGCTAAG. An adenine base was opposite to the uracil on the complementary strand, as would be found after dUTP incorporation during replication. The 5′ and 3′ fluorescein end-labels were conjugated to the phosphate of the terminal nucleotide as phosphodiester linkages with a six-carbon spacer between the phosphate and the fluorescein.
Recombinant proteins
The procedures for the expression and purification of full-length hUNG2 (10) and the hUNG2 catalytic domain (amino acids 92–313) have been described previously (16,17). Heterotrimeric RPA was expressed and purified using the p11d-tRPA plasmid as described previously (10,18,19). The p11d-tRPA plasmid was a generous gift from Dr. Marc Wold.
To generate the isolated hUNG2 NTD (amino acids 1–91), a stop codon was inserted after residue 91 of hUNG2 in the pET21a vector described previously for the expression and purification of full-length hUNG2 (10). The encoded construct expressed in Escherichia coli contained an 8xHis-tag and the SUMO protein fused in-frame to the NTD of hUNG2. Protein expression in BL21(DE3)pLysS cells, Ni2+ column purification steps, and proteolytic removal of the 8xHis-SUMO tag was performed identically as for full-length hUNG2 (10). An additional column purification step was performed using a Superdex 75 10/300 GL column (GE Healthcare) with a mobile phase of 25 mM HEPES–NaOH (pH 7.4), 200 mM NaCl, 0.01% Triton X-100 and 1 mM TCEP. Peak fractions containing the hUNG2 NTD were pooled, glycerol was added to a final concentration of 10%, and protein was concentrated to ∼0.4 mg/ml with an Amicon 3-kDa molecular-mass cutoff centrifugal filter. The protein was snap frozen in liquid N2 before storage at −80°C. To confirm protein identity, the mass of the intact NTD was measured using electrospray ionization mass spectrometry (predicted mass: 9404 Da; observed mass: 9407 ± 1 Da) using a published protocol (10).
Uracil excision assays
All in vitro assays were performed at 22°C in 10 mM Tris–HCl (pH 8.0), 100 mM NaCl, 1 mM DTT and 0.1 mM EDTA, and were designed to capture the initial rate of the enzymatic reaction. The DNA substrates had a 55 bp duplex region containing one or two uracil bases and, in some cases, a ssDNA overhang region that was 4, 16 or 32 nt long. For hUNG2 assays in which two uracil bases were present in the substrate, reactions were run for 5 minutes, and the enzyme concentration was 180 pM or 900 pM using DNA substrate concentrations of 0.1 or 3 μM, respectively. Identical assays were run with the hUNG2 catalytic domain, using enzyme concentrations of 540 or 900 pM for the 0.1 or 3 μM DNA concentrations. For hUNG2 and catalytic domain assays using substrates with a single uracil base, the enzyme concentration (180 pM–1.6 nM), reaction time (2–4 minutes), and DNA concentration (30 nM–12 μM) were varied as appropriate to generate full Michaelis-Menten saturation curves. Assay time points were quenched by diluting the reaction at least 10-fold with 200 mM NaOH then heating for 15 min at 70°C; this NaOH treatment also cleaves the abasic sites generated by enzymatic removal of uracil bases, while leaving the unprocessed DNA substrate intact (20). The quenched samples were diluted 5-fold with formamide containing 5 mM EDTA, and the DNA fragments were resolved on 15% polyacrylamide–8 M urea TBE gels. The fluorescein-labeled fragments were imaged in the gel with a Typhoon 9500 imager, and the gel bands were quantified using Fiji/ImageJ (21). For the two-site substrates, the extent of uracil excision at each site was determined in the initial rate regime and the site preference was reported as a fold-selectivity (kfast site/kslow site).
To confirm that the results were not affected by the fluorescein end-labels, a control experiment was performed using a DNA substrate with 5′ and 3′ 32P labels. For this, ATP-[γ-32P] and 3′-deoxyadenosine 5′-triphosphate (cordycepin 5′-triphosphate)-[α-32P] were purchased from Perkin Elmer. The 5′ end was labeled using ATP-[γ-32P] and T4 polynucleotide kinase (New England Biolabs), and the 3′ end was labeled using cordycepin 5′-triphosphate-[α-32P] and terminal deoxynucleotidyl transferase (Thermo Fisher). After labeling, reactions were desalted and unreacted nucleoside phosphates were removed by sequentially filtering the reactions through Bio-Spin P-30 and Bio-Spin P-6 columns (Bio-Rad). The labeled uracil-containing oligonucleotide was then annealed to its complementary strand. Kinetic assays were performed as described above, and after electrophoresis, 32P-labeled DNA fragments were analyzed by exposing the gel to a phosphor screen then scanning with the Typhoon 9500.
Uracil excision in the presence of RPA
Uracil excision assays in the presence of RPA were performed using junction substrates containing a 32 nt ssDNA overhang with two uracil bases located in the duplex region and fluorescein end-labels on the uracil-containing strand. The reaction buffer was 10 mM Tris–HCl (pH 8.0), 100 mM NaCl, 1 mM DTT and 0.1 mM EDTA, and assays were performed at 22°C. Stochiometric complexes between RPA and the single-stranded overhang of the junction substrate were formed by mixing molar equivalent amounts of RPA with the DNA (final concentrations of 3 μM each). Reactions were initiated by the addition of 900 pM UNG2 and were terminated after 5 min. The 3 μM concentration of RPA in these reactions is sufficient to bind 50% of the total hUNG2 based on previous equilibrium binding measurements (10). Reaction quenching, abasic site cleavage, and analysis via electrophoresis and imaging were performed identically as described above for the assays without RPA.
Electrophoretic mobility shift assays (EMSA) established that RPA bound the fluorescein-labeled junction DNA with a 1:1 stoichiometry. The absorbance (A280) of separate protein samples denatured in 6 M guanidinium–HCl was used to measure protein concentration. For EMSA, we equilibrated 3 μM of the DNA with varying concentrations of RPA for 5 min in 10 mM Tris–HCl (8.0), 100 mM NaCl, 1 mM DTT, 0.1 mM EDTA and 10% glycerol. Four microliters of this mixture were loaded onto a 0.9% agarose–0.1× TAE native electrophoresis gel. After electrophoresis, the free and bound DNA bands were imaged with the Typhoon 9500 imager and quantified using Fiji/ImageJ.
Fluorescence anisotropy assays
All binding assays were performed at 23°C, and the concentration of fluorescein-labeled nonspecific DNA was 50 nM with variable protein concentrations as indicated on the Figure axis. For experiments with hUNG2 and its catalytic domain, the assay buffer contained 25 mM HEPES–NaOH (pH 7.4), 10% glycerol, 100 mM NaCl, 1 mM MgCl2, 1 mM DTT and 0.01% Triton X-100. Because the hUNG2 N-terminal domain bound very weakly in this buffer, we also performed experiments at a lower NaCl concentration of 20 mM. Individual solutions of protein and DNA (150 μl) were equilibrated for at least 5 min before measuring the fluorescence anisotropy (16,17). The anisotropy values were plotted against total protein concentration and fit to a quadratic binding isotherm as described previously (10,17,22). No corrections for changes in emission intensities upon binding were required.
Data statistics
The kinetic and binding data are reported as means with standard errors derived from three to five independent experiments for each condition. In many cases for DNA binding, the error bars were smaller than the plotted point. Steady-state kinetic data were plotted using GraphPad Prism 6, which was also used for determination of Km and kcat, and the corresponding errors associated with these parameters represent standard errors from the curves fit to the data.
RESULTS
RPA targets hUNG2 to ssDNA–dsDNA junctions
To examine the effects of RPA on uracil excision by hUNG2, we generated DNA substrates that contained a duplex DNA region with an adjacent single-stranded section (‘junction substrates’). This simple structure can be found at moving or stalled replication forks, including after fork uncoupling (23), and also at R-loops that enable class-switch recombination (24,25). For initial experiments, we prepared a 55 bp duplex DNA that contained a 32 nt ssDNA overhang with two uracils positioned 21 and 45 bp from the ssDNA–dsDNA junction. The DNA contained fluorescein labels on both ends of the uracilated strand to allow simultaneous quantification of the excision efficiencies at both uracil sites (U21 and U45) (Figure 1A). Excision at U21 produces two unique fragments designated A and BC, while excision at U45 produces fragments C and AB.
In the absence of RPA, we reacted 3 μM of this substrate with hUNG2 under low conversion conditions and then chemically cleaved the resulting abasic sites to yield the distinct DNA fragments, which were resolved by electrophoresis and quantified (Figure 1B). In the presence of hUNG2 alone, all of the DNA fragments were approximately equal in intensity, indicating that U21 and U45 were kinetically equivalent under these conditions. The lack of discrimination between uracil sites was anticipated based on previous studies that measured hUNG2 activity on DNA duplexes containing two uracil bases at various spacings (16,17,20,26).
We then performed the same experiment except that one equivalent of RPA was added to the DNA substrate to form a 1:1 complex with the ssDNA overhang prior to the addition of hUNG2 (11,27) (Supplementary Figure S1). The concentration of RPA used in these experiments approximated its nuclear concentration (0.4–3.7 μM) as well as its Kd for binding to hUNG2 (10,28–30), which ensured that a substantial fraction of hUNG2 was bound to RPA in the assays (see Supplementary Information). In contrast to the results in the absence of RPA, uracil excision now showed a striking 6-fold selectivity for U21 rather than U45 (i.e. fragments A and BC were the dominant products) (Figure 1B and C). Compared to experiments without RPA, the rate of uracil excision increased for position 21 and decreased for position 45. Therefore, the overall effect of RPA on hUNG2 activity can be characterized as having stimulatory and inhibitory components. We further explored RPA-induced selectivity by performing an excision assay using a substrate that contained uracils positioned closer to the ssDNA–dsDNA junction while maintaining a 24 bp spacing between the two uracils (U9 and U33) (Figure 1D and Supplementary Figure S1). With this substrate in the presence of RPA, an even greater 9-fold selectivity was observed for the uracil located closest to the ssDNA–dsDNA junction (Figure 1E and F). Possible mechanisms accounting for these effects of RPA are presented in the Discussion.
Although hUNG2 alone showed no selectivity for excision of U21 or U45 (Figure 1B), a surprising observation was that the enzyme showed a 3.5-fold selectivity for excising U9 as compared to U33 when the DNA substrate was present at 3 μM (Figure 1E and F). To exclude that this selectivity might arise from preferential interaction of hUNG2 with the 3′ fluorescein label, we repeated the site selectivity experiment with hUNG2 alone using an identical substrate that was end-labeled with 32P and again observed biased excision at U9 compared to U33 (Supplementary Figure S2). This result was unexpected because uracil excision selectivity has only been observed previously for uracils in different sequence contexts or where steric/structural features affected its accessibility (31–34). In contrast, our substrates were designed to have an identical sequence flanking the U/A bp to avoid potential sequence bias. We therefore investigated further the basis for the selectivity of hUNG2 for uracil sites near the ssDNA–dsDNA junction in the experiments that follow.
NTD targets hUNG2 to ssDNA–dsDNA junctions in the absence of RPA
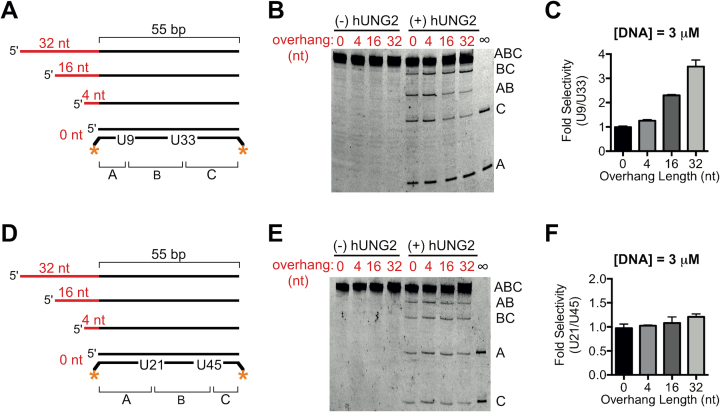
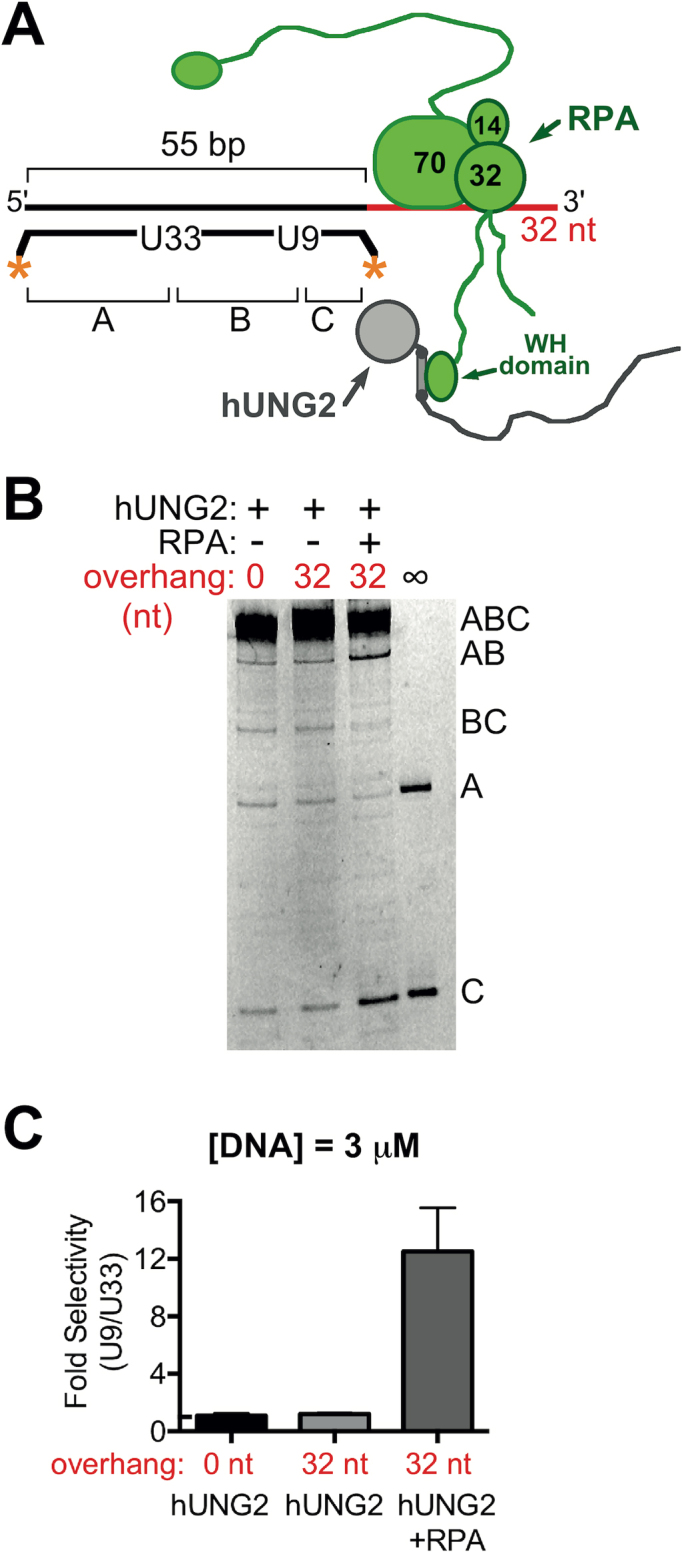
To explore the substrate features that led to preferential targeting of hUNG2 to U9 located near the ssDNA–dsDNA junction, we prepared 55 bp duplexes with uracils at positions 9 and 33, but with shorter ssDNA overhangs or no overhang at all (Figure 2A). Using 3 μM DNA substrate, we found that hUNG2 preferentially removed U9 only when a ssDNA overhang was present, and that the selectivity for U9 increased with longer ssDNA overhangs (Figure 2B and C). Compared to the 55 bp duplex without a ssDNA section, where the rates of U9 and U33 excision were equal, the 3.5-fold selectivity observed with the 32 nt overhang was driven by a ∼2-fold enhanced rate for excising U9 and a ∼2-fold decreased rate for excising U33 (Supplementary Figure S3). Because hUNG2 is equally efficient at removing each uracil in the absence of an overhang, the divergence of enzymatic rates suggests that the overhang partitions the enzyme upon its initial encounter with DNA to increase its effective concentration near U9 and deplete its proximity to U33. The correlation between ssDNA length and uracil selectivity also suggested that the ssDNA overhang was being recognized by hUNG2 and not the ssDNA–dsDNA junction itself. We further established that the excision selectivity of hUNG2 alone did not extend to U21 or U45 because substrates with 0, 4, 16 and 32 nt ssDNA overhangs showed no excision bias (Figure 2D–F and Supplementary Figure S3). Thus, compared to hUNG2 alone, one apparent function of RPA is to extend the excision selectivity of hUNG2 to uracils that are located farther from the ssDNA–dsDNA junction.
Figure 2.
Biased uracil excision by hUNG2 with junction DNA substrates containing different 5′ ssDNA overhang lengths in the absence of RPA. (A) Junction substrates with uracils at positions 9 and 33 bp from the ssDNA–dsDNA junction. The 5′ overhang length was varied in the range of zero to 32 nt. (B) Image of hUNG2 reaction products using junction substrates from panel A after separation using denaturing PAGE. For these assays, hUNG2 concentration was 900 pM, and substrate concentration was 3 μM. (C) Relative selectivity for U9 compared to U33 for different overhang lengths. (D) Junction substrates with different 5′ overhang lengths and uracil bases at 21 and 45 bp from the ssDNA–dsDNA junction. (E) Image of hUNG2 reaction products using the substrates in panel D. (F) hUNG2 shows no selectivity for U21 or U45. The selectivity was calculated from the band intensities in panel E.
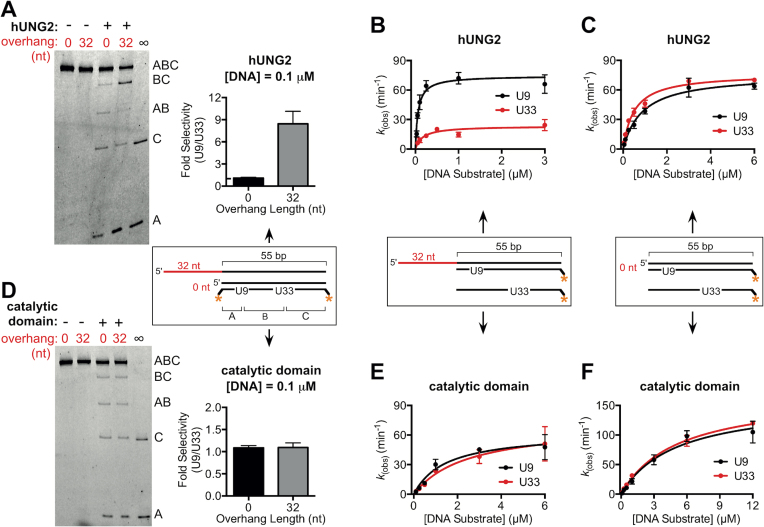
We examined the concentration dependence of the site selectivity using the junction substrate with a 32 nt 5′ overhang and uracils at positions 9 and 33 bp in the duplex (Figure 3A). Decreasing the substrate concentration to 0.1 μM amplified the selectivity for U9 to ∼8-fold. This result prompted us to generate full Michaelis–Menten saturation curves for hUNG2 using essentially identical duplex substrates with and without a 32 nt overhang, but containing only a single U/A bp at position 9 or 33 (Figure 3B and C). With the overhang present, we measured a 3-fold larger kcat and an 8-fold larger kcat/Km for excision of U9 as compared to U33, which was entirely consistent with the selectivity measurements with two uracils at low and high substrate concentrations (Figure 3B and Table 1). Further kinetic assays again established that hUNG2 had no site bias when the overhang was not present (Figure 3C and Table 1). Thus, the selectivity of hUNG2 with overhang substrates is maintained when each uracil site is present by itself, establishing that the operative mechanism does not involve inhibition of U33 excision by the product arising from excision of U9.
Figure 3.
Uracil excision by hUNG2 or its catalytic domain with junction DNA substrates containing one or two uracils. (A) Image of hUNG2 reaction products using the indicated U9–U33 junction substrates with or without a 32 nt 5′ overhang. The substrate concentration (0.1 μM) is 30-fold lower than in Figures 1 and 2, and the hUNG2 concentration was 180 pM. (B) Steady-state kinetic measurements for hUNG2 acting on the indicated junction substrates with a single U/A bp (U9 or U33). (C) Steady-state kinetic measurements for hUNG2 acting on duplex substrates without overhangs and containing a single U/A bp (U9 or U33). (D) Image of reaction products derived from the reaction of the catalytic domain with the indicated U9–U33 junction substrates with or without a 32 nt 5′ overhang. The substrate concentration was 0.1 μM, and the concentration of the catalytic domain was 540 pM. (E) and (F), Steady-state kinetic measurements for the catalytic domain acting on the indicated junction substrates with a single U/A bp (U9 or U33).
Table 1.
Kinetic parameters of hUNG2 and its catalytic domain with 55 bp duplex substrates containing single uracil bases
| hUNG2 | Catalytic domain | ||||||
|---|---|---|---|---|---|---|---|
| ssDNA length (nt) | Uracil site | k cat (min−1) | K m (μM) | k cat/Km (μM−1min−1) | k cat (min−1) | K m (μM) | k cat/Km (μM−1min−1) |
| 32 | U9 | 74 ± 3 | 0.06 ± 0.01 | 1233 | 63 ± 6 | 1.46 ± 0.41 | 43 |
| 32 | U33 | 23 ± 2 | 0.16 ± 0.05 | 144 | 72 ± 12 | 2.48 ± 0.92 | 29 |
| 0 | U9 | 76 ± 5 | 0.88 ± 0.18 | 86 | 158 ± 15 | 5.02 ± 1.03 | 31 |
| 0 | U33 | 77 ± 2 | 0.50 ± 0.06 | 154 | 169 ± 10 | 5.13 ± 0.66 | 33 |
To assess whether the uracil excision selectivity was conferred by the NTD, we repeated the above experiments using the catalytic domain of hUNG2 that lacks the 91 residue NTD. In contrast to the full-length enzyme, the catalytic domain alone had no selectivity for uracils located at position 9 or 33 regardless of the 5′ overhang length or substrate concentration (Figure 3D and Supplementary Figure S4). The absence of uracil excision selectivity by the catalytic domain was further confirmed by performing steady-state kinetic measurements with the substrates containing a single uracil in the presence and absence of the 32 nt overhang (Figure 3E, F, and Table 1). Comparing the steady-state kinetic parameters of the catalytic domain with the full-length enzyme revealed that the presence of the NTD reduced the Km for overhang-containing substrates by 16- to 24-fold, suggesting that the NTD interacts with DNA to enhance affinity. Comparisons of kcat/Km values showed that the presence of the NTD enhanced excision efficiency of U9 by ∼30-fold, but excision of U33 was enhanced by only 5-fold (Table 1). Together, these data indicate that the presence of the NTD confers a strong preference for uracils positioned close to ssDNA–dsDNA junctions, and that this specificity was enabled by a 5′ single-stranded overhang.
Effects of the hUNG2 NTD on DNA binding
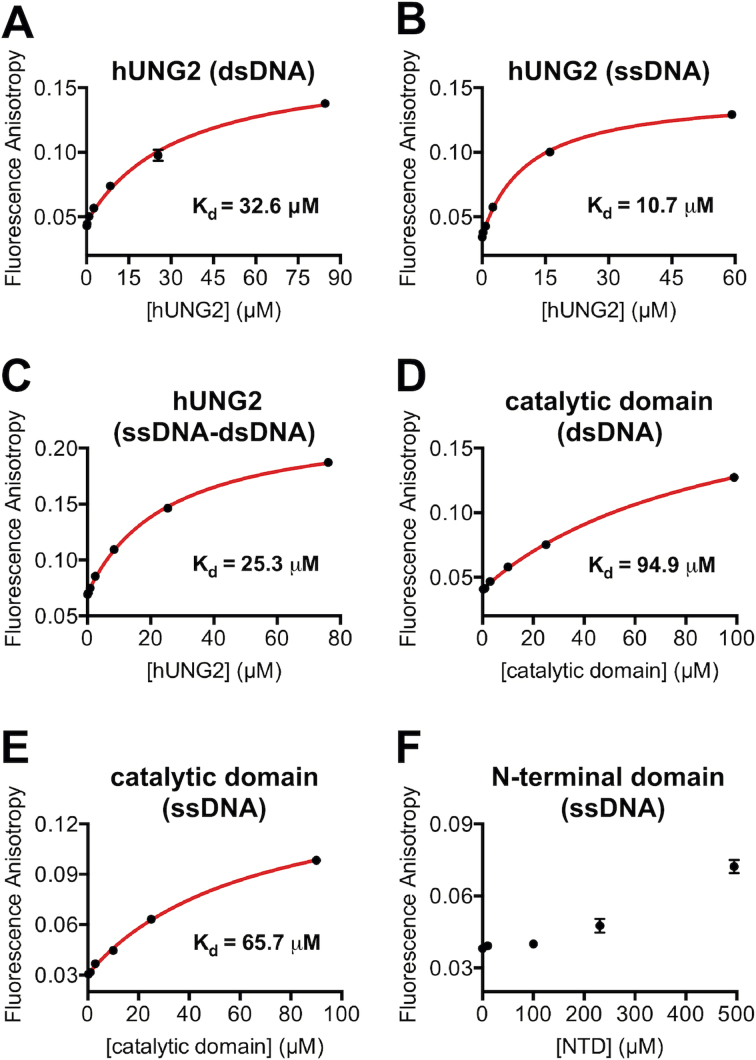
The steady-state kinetic measurements indicated that the presence of the NTD significantly lowered the Km values for substrates that contained a ssDNA overhang, which led us to speculate that the NTD might contribute to preferential binding to ssDNA. To examine this under equilibrium binding conditions, we performed fluorescence anisotropy measurements to assess the binding affinity of hUNG2 for ssDNA, dsDNA, and a hybrid ssDNA–dsDNA construct. Although these experiments used non-specific DNA and do not exactly match the kinetic experiments with uracil-containing substrates, it was reasonable to expect that similar trends might be observed. This expectation was realized because hUNG2 bound with 3-fold greater affinity for ssDNA compared to dsDNA, and the overhang-containing duplex bound with an intermediate affinity (Figure 4A–C). In comparison, the catalytic domain protein without the NTD had 6-fold and 3-fold reduced affinities for ssDNA and dsDNA relative to the full-length enzyme, further indicating a secondary interaction between the NTD and DNA that affects ssDNA binding to a larger degree (Figure 4D and E). Finally, we tested the ssDNA binding activity of the isolated NTD (amino acids 1–91), which we recombinantly expressed and purified (Supplementary Figure S5). However, we were unable to detect binding to ssDNA using a buffer similar to that used in our activity assays with NTD concentrations of ∼500 μM (data not shown). However, reducing the ionic strength of the buffer from 100 to 20 mM NaCl allowed us to measure a concentration-dependent increase in the fluorescence anisotropy, suggesting a weak ssDNA binding affinity for the isolated NTD that is at least partially driven by electrostatic interactions (Figure 4F). Although DNA binding of the isolated NTD is weak, this may be explained by a large entropic penalty for binding of a disordered peptide to ssDNA, which in the context of the full-length protein would be offset by the intrinsic binding affinity of the structured catalytic domain for DNA.
Figure 4.
Fluorescence anisotropy binding assays. Except for panel F, the buffer consisted of 25 mM HEPES–NaOH (pH 7.4), 10% glycerol, 100 mM NaCl, 1 mM MgCl2, 1 mM DTT, and 0.01% Triton X-100. In all assays, the fluorescein-labeled DNA was 50 nM. (A) Binding of hUNG2 to a 29 bp duplex. (B) Binding of hUNG2 to a 29 nt ssDNA. (C) Binding of hUNG2 to a hybrid ssDNA–dsDNA duplex containing a 29 nt 5′ ssDNA overhang and a 29 bp duplex region. (D) Binding of the catalytic domain to a 29 bp duplex. (E) Binding of the catalytic domain to a 29 nt ssDNA. (F) Binding of the isolated N-terminal domain (residues 1–91) to a 29 nt ssDNA in a buffer with 20 mM NaCl.
Relevance of DNA strand polarity on hUNG2 targeting
The junction DNA substrates used in the activity and binding measurements above had single-stranded DNA overhangs 5′ to the duplex section. To test the opposite orientation, we prepared uracilated DNA substrates with a 32 nt 3′ overhang that maintained uracil spacing 9 and 33 bp from the ssDNA–dsDNA junction (Figure 5A). Remarkably, in this 3′ overhang context, hUNG2 alone had no preference for U9 or U33 (Figure 5B and C, and Supplementary Figure S6). The rates of uracil excision for both sites were approximately equal to those measured above for the duplex substrate without an overhang when U9 and U33 were oriented with the opposite polarity with respect to the overhang.
Figure 5.
Effect of ssDNA overhang polarity on RPA-dependent and RPA-independent uracil excision selectivity. (A) Junction substrate with a 32 nt 3′ ssDNA overhang. The polarity of this substrate is reversed compared to those used in Figures 1–3, but the uracils were maintained 9 and 33 bp from the ssDNA–dsDNA junction. (B) Reaction of 900 pM hUNG2 with the 3′ ssDNA overhang junction substrate in panel A in the presence and absence of RPA. A reaction was also performed in the absence of RPA with an identical duplex substrate that had no overhang. For all assays, DNA concentration was 3 μM. (C) Selectivity of hUNG2 for U9 and U33 in the presence and absence of RPA, and also in the absence of RPA without an overhang on the duplex. The selectivity was calculated from the band intensities in panel B.
Finally, we tested whether RPA could still target hUNG2 to U9 using the substrate with a 32 nt 3′ ssDNA overhang. As before, gel-shift assays clearly indicated that RPA bound tightly and stoichiometrically to the 3′ overhang (Supplementary Figure S7). In contrast to the results with hUNG2 alone, the complex with RPA preserved its capacity to preferentially target hUNG2 to U9 with essentially identical selectivity and kinetics that were observed with the 5′ overhang (Figure 5B and C).
DISCUSSION
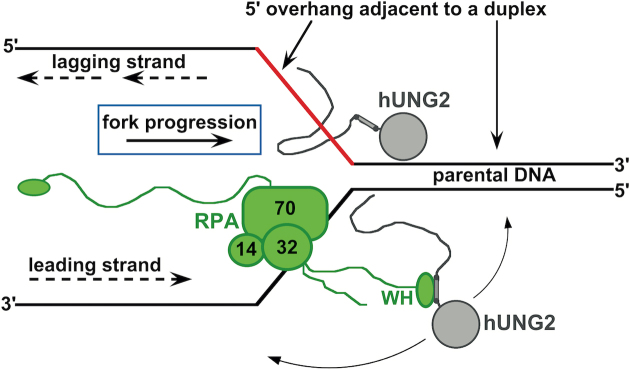
Inside cells, the vast majority of DNA is undamaged, which presents a large reservoir of non-specific DNA binding sites for hUNG2 that could hinder the rapid removal of uracil from transient hot spots that appear in specific locations of the genome (20). Plausible mechanisms that could ensure hUNG2 is targeted to DNA regions with elevated potential for containing uracil is for the enzyme to interact with specific proteins or DNA structures that are hallmarks of these sites. Two such examples are RPA and the ssDNA–dsDNA junctions that are associated with replication forks and R-loops. Association of hUNG2 with the replication fork is likely critical under conditions where cellular dUTP levels are elevated and the replicative polymerases insert dUMP opposite to adenine in the template strand (35). Such conditions prevail during chemotherapy treatment with antimetabolite drugs (36), but also in the general population when folic acid is deficient in the diet (37). During replication, stretches of ssDNA are transiently exposed during DNA unwinding at moving replication forks and can accumulate during fork stalling (38). In addition, during the early stages of class-switch recombination in B cells, ssDNA becomes displaced from the double-stranded helix at R-loops and becomes accessible to activation-induced DNA cytidine deaminase (24,39). The presence of ssDNA–dsDNA junctions during these processes provides a plausible mechanism for localizing hUNG2 in the presence and absence of RPA (Figure 6).
Figure 6.
Hypothetical targeting of hUNG2 at a simplified replication fork using its NTD. We propose that hUNG2 by itself could be targeted by transient protein-free ssDNA located on the 5′ end of a duplex region. This targeting could function to efficiently excise uracils prior to new strand synthesis. In the presence of RPA, which can bind to 5′ or 3′ ssDNA overhangs, we speculate that hUNG2 could be targeted to uracil bases before or after polymerase synthesis. When tethered to the winged-helix (WH) domain of RPA, hUNG2 can act on DNA strands in any direction. This arrangement could allow hUNG2 to preemptively remove uracils ahead of the fork, but could also allow hUNG2 to serve as a proofreading enzyme by acting on the newly synthesized strand. We note that other replication fork proteins (e.g. PCNA) were excluded for simplicity.
Regardless of whether the RPA-dependent or RPA-independent pathway is utilized, our data indicate that the NTD is a key determinant for efficient targeting to such sites. The discovery that hUNG2 uses the NTD to localize near ssDNA–dsDNA junctions by two different mechanisms suggests a strong selection pressure to evolve such a capacity. Although closely related family 1 bacterial uracil DNA glycosylases do not contain NTD extensions, these appendages linked to the catalytic domain are found in homologs from other eukaryotes, additionally serving as mitochondrial and nuclear targeting domains (40).
RPA-dependent targeting
The different orientational and spatial aspects of RPA-dependent and RPA-independent uracil excision near DNA junctions suggest that the architecture of the RPA-hUNG2 complex is an important determinant of function. Compared to targeting by ssDNA alone, hUNG2’s interaction with anchored RPA gives rise to greater capacity for the removal of uracil bases near the ssDNA–dsDNA junction and also extends biased uracil excision by an additional ∼10 bp into the duplex region. RPA also promotes biased targeting when positioned on either a 5′ or 3′ overhang. These characteristics hint that the maximal distance hUNG2 can extend away from the ssDNA binding core of RPA is limited by the linker length between the DNA binding domain and the winged-helix domain of RPA32. This disordered linker of 32 residues has a maximal extended contour length of 115 Å (10,41), although unstructured polypeptides typically extend only ∼60% of their contour length in solution (42). Thus, the complex between the winged-helix domain and hUNG2 might extend as far as ∼69 Å away from the DNA binding core of RPA, which corresponds to ∼20 bp of duplex DNA at 3.4 Å/bp. This maximal distance is consistent with the observed biased excision at U21 in the junction substrate as opposed to U33 and U45 (Figure 1). Targeting of hUNG2 when RPA is bound to either 5′ or 3′ overhangs suggests that the flexibility of the RPA32 winged-helix linker could serve to tether hUNG2 to any junction present at replication forks. We also suspect that RPA could efficiently target hUNG2 to uracils in nearby ssDNA regions provided that the ssDNA substrate is exposed and not already occupied by additional RPA molecules.
RPA-dependent targeting of hUNG2 has distinct structural and orientational aspects compared to RPA targeting of the ERCC1-XPF endonuclease complex using similar ssDNA–dsDNA substrates (43–45). In contrast to hUNG2, ERCC1-XPF is thought to interact with the RPA70 subunit rather than the winged-helix domain of RPA32, and RPA stimulation of ERCC1-XPF occurred only when RPA was bound to a 5′ overhang (43). The DNA binding domains of RPA orient with a defined polarity on ssDNA (Supplementary Figure S8), with the RPA70 subunit positioned upstream on the 5′ end and RPA32 on the 3′ end (46). The available structural information suggests that the flexible linker of RPA32 that tethers hUNG2 to its winged-helix domain is positioned favorably to extend in both directions from the DNA binding core of RPA (Supplementary Figure S8) (46).
RPA-independent targeting
This study and previous work strongly suggest that the NTD of hUNG2 interacts with duplex and ssDNA. We previously proposed that the NTD interacted with dsDNA based on the observation that full-length hUNG2 translocated along duplex DNA more efficiently than the isolated catalytic domain (17). Results from the present study are consistent with this proposal because hUNG2 has a slightly higher affinity for duplex DNA than the catalytic domain alone (Figure 4). Surprisingly, our results with two uracil junction substrates indicate that excision is strongly favored at the site near the junction (U9), and that the full-length enzyme is thwarted in its ability to excise the other site (U33) in the presence of a 5′ overhang. To our knowledge, such behavior has never been observed previously with duplex substrates lacking overhangs and indicates that the NTD interacts with ssDNA segments of sufficient length.
How does the presence of a 5′ overhang target a robust enzyme like hUNG2 to one uracil and prevent excision of the other uracil site during the same encounter? Any plausible answer requires two components: preferential encounter with the uracil proximal to the junction and a biased departure pathway from the DNA that reduces the probability of encountering the distal uracil. An attractive mechanism is one where the flexible ssDNA overhang is captured by the NTD, facilitating excision of the proximal uracil. Furthermore, based on microscopic reversibility, the exit of the enzyme from the single-excised product should follow the same biased pathway, followed by capture of a new DNA substrate by the same mechanism. In this mechanism, the second site would only react after multiple encounters with an enzyme molecule. The present data provide support for this biased partitioning of the enzyme during each encounter and departure from the DNA. First, preferential targeting to the proximal site is indicated by the ∼10-fold larger kcat/Km for U9 as compared to U33 (Table 1). Second, biased departure is supported by the 3.5-fold larger kcat for the proximal uracil. This follows because steady-state turnover with saturating substrate is limited by a step following uracil excision (47). Finally, we also observed that targeting of hUNG2 was enhanced when longer overhangs were present. The greater length and flexibility of longer ssDNA polymers would provide a larger target area for the NTD. Thus, we propose that transient NTD binding to the ssDNA overhang partitions the enzyme during each encounter. In accord with our binding measurements, such a kinetic partitioning mechanism would not necessarily be strongly manifested in equilibrium binding affinity. Since hUNG2 reacts with uracils in both ssDNA and dsDNA (48,49), the enzyme might also be able to scan ssDNA regions before encountering the duplex area.
The strict spatial and orientational requirements for biased uracil excision near junctions in the absence of RPA suggests that the NTD directs the catalytic domain towards the junction and the uracil site. This targeting may derive from two components. First, the orientation of the NTD with respect to the polarity of the ssDNA region could serve to orient the catalytic domain in the direction of the uracil site (5′ overhang), or away from the site (3′ overhang). The second component is the orientation of the catalytic domain with respect the polarity of the uracil site in the substrate DNA strand. Our data indicates that the NTD alone, when bound to a 5′ overhang, can orient the enzyme preferentially to a uracil base that is in a 3′-5′ orientation relative to the overhang. In contrast, the tail bound to a 3′ overhang cannot productively orient the catalytic domain to a uracil base oriented in the 5′-3′ direction. These data indicate that the NTD tether serves to constrain the activity of the catalytic domain in terms of distance and orientation of the uracils with respect to the overhang. Apparently, when RPA tethers the enzyme to the 3′ overhang, the favorable orientation of the enzyme is restored. The structural basis for this is not discernible, but may simply reflect that a longer flexible tether allows the catalytic domain to achieve the required productive orientation.
In summary, we have presented RPA-dependent and RPA-independent mechanisms that target hUNG2 to uracil sites near ssDNA–dsDNA junctions. This study unveils several new properties of the NTD of hUNG2 and reinforces the notion that the disordered segments on many DNA binding proteins are critical for tuning the kinetic and thermodynamic properties of enzymes (50,51).
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01-GM056834 to J.T.S., R01-CA74305 to P.A.C., F32-GM119230 to B.P.W., T32-CA9110, T32-GM8403]. Funding for open access charge: Rowan University School of Osteopathic Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1. Otterlei M., Warbrick E., Nagelhus T.A., Haug T., Slupphaug G., Akbari M., Aas P.A., Steinsbekk K., Bakke O., Krokan H.E.. Post-replicative base excision repair in replication foci. EMBO J. 1999; 18:3834–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rada C., Williams G.T., Nilsen H., Barnes D.E., Lindahl T., Neuberger M.S.. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 2002; 12:1748–1755. [DOI] [PubMed] [Google Scholar]
- 3. Kavli B., Andersen S., Otterlei M., Liabakk N.B., Imai K., Fischer A., Durandy A., Krokan H.E., Slupphaug G.. B cells from hyper-IgM patients carrying UNG mutations lack ability to remove uracil from ssDNA and have elevated genomic uracil. J. Exp. Med. 2005; 201:2011–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zan H., White C.A., Thomas L.M., Mai T., Li G., Xu Z., Zhang J., Casali P.. Rev1 recruits Ung to switch regions and enhances dU glycosylation for immunoglobulin class switch DNA recombination. Cell Rep. 2012; 2:1220–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nagelhus T.A., Haug T., Singh K.K., Keshav K.F., Skorpen F., Otterlei M., Bharati S., Lindmo T., Benichou S., Benarous R. et al. . A sequence in the N-terminal region of human uracil-DNA glycosylase with homology to XPA interacts with the C-terminal part of the 34-kDa subunit of replication protein A. J. Biol. Chem. 1997; 272:6561–6566. [DOI] [PubMed] [Google Scholar]
- 6. Torseth K., Doseth B., Hagen L., Olaisen C., Liabakk N.-B., Græsmann H., Durandy A., Otterlei M., Krokan H.E., Kavli B. et al. . The UNG2 Arg88Cys variant abrogates RPA-mediated recruitment of UNG2 to single-stranded DNA. DNA Repair (Amst.). 2012; 11:559–569. [DOI] [PubMed] [Google Scholar]
- 7. Buchinger E., Wiik S.Å., Kusnierczyk A., Rabe R., Aas P.A., Kavli B., Slupphaug G., Aachmann F.L.. Backbone 1H, 13C and 15N chemical shift assignment of full-length human uracil DNA glycosylase UNG2. Biomol. NMR Assign. 2017; 12:15–22. [DOI] [PubMed] [Google Scholar]
- 8. Yamane A., Resch W., Kuo N., Kuchen S., Li Z., Sun H., Robbiani D.F., McBride K., Nussenzweig M.C., Casellas R.. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nat. Immunol. 2011; 12:62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hagen L., Kavli B., Sousa M.M.L., Torseth K., Liabakk N.B., Sundheim O., Pena-Diaz J., Otterlei M., Hørning O., Jensen O.N. et al. . Cell cycle-specific UNG2 phosphorylations regulate protein turnover, activity and association with RPA. EMBO J. 2008; 27:51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weiser B.P., Stivers J.T., Cole P.A.. Investigation of N-terminal phospho-regulation of uracil DNA glycosylase using protein semisynthesis. Biophys. J. 2017; 113:393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim C., Paulus B.F., Wold M.S.. Interactions of human replication protein A with oligonucleotides. Biochemistry. 1994; 33:14197–14206. [DOI] [PubMed] [Google Scholar]
- 12. Fanning E., Klimovich V., Nager A.R.. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic. Acids. Res. 2006; 34:4126–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brosey C.A., Soss S.E., Brooks S., Yan C., Ivanov I., Dorai K., Chazin W.J.. Functional dynamics in replication protein A DNA binding and protein recruitment domains. Structure. 2015; 23:1028–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dianov G.L., Jensen B.R., Kenny M.K., Bohr V.A.. Replication protein A stimulates proliferating cell nuclear antigen-dependent repair of abasic sites in DNA by human cell extracts. Biochemistry. 1999; 38:11021–11025. [DOI] [PubMed] [Google Scholar]
- 15. Owczarzy R., Tataurov A.V., Wu Y., Manthey J.A., McQuisten K.A., Almabrazi H.G., Pedersen K.F., Lin Y., Garretson J., McEntaggart N.O. et al. . IDT SciTools: a suite for analysis and design of nucleic acid oligomers. Nucleic Acids Res. 2008; 36:W163–W169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cravens S.L., Hobson M., Stivers J.T.. Electrostatic properties of complexes along a DNA glycosylase damage search pathway. Biochemistry. 2014; 53:7680–7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rodriguez G., Esadze A., Weiser B.P., Schonhoft J.D., Cole P.A., Stivers J.T.. Disordered N-terminal domain of human uracil DNA glycosylase (hUNG2) enhances DNA translocation. ACS Chem. Biol. 2017; 12:2260–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Henricksen L.A., Umbricht C.B., Wold M.S.. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 1994; 269:11121–11132. [PubMed] [Google Scholar]
- 19. Binz S.K., Dickson A.M., Haring S.J., Wold M.S.. Functional assays for replication protein A (RPA). Methods Enzymol. 2006; 409:11–38. [DOI] [PubMed] [Google Scholar]
- 20. Esadze A., Rodriguez G., Weiser B.P., Cole P.A., Stivers J.T.. Measurement of nanoscale DNA translocation by uracil DNA glycosylase in human cells. Nucleic Acids Res. 2017; 45:12413–12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cardona A., Schmid B., Rueden C., White D.J., Frise E., Arganda-Carreras I., Tinevez J.-Y., Schindelin J., Eliceiri K., Longair M. et al. . Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Seamon K.J., Sun Z., Shlyakhtenko L.S., Lyubchenko Y.L., Stivers J.T.. SAMHD1 is a single-stranded nucleic acid binding protein with no active site-associated nuclease activity. Nucleic Acids Res. 2015; 43:6486–6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hedglin M., Benkovic S.J.. Replication protein A prohibits diffusion of the PCNA sliding clamp along single-stranded DNA. Biochemistry. 2017; 56:1824–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Daniels G.A., Lieber M.R.. RNA:DNA complex formation upon transcription of immunoglobulin switch regions: implications for the mechanism and regulation of class switch recombination. Nucleic Acids Res. 1995; 23:5006–5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Roy D., Zhang Z., Lu Z., Hsieh C.-L., Lieber M.R.. Competition between the RNA transcript and the nontemplate DNA strand during R-loop formation in vitro: a nick can serve as a strong R-Loop initiation site. Mol. Cell. Biol. 2010; 30:146–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cravens S.L., Stivers J.T.. Comparative effects of ions, molecular crowding, and bulk DNA on the damage search mechanisms of hOGG1 and hUNG. Biochemistry. 2016; 55:5230–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pestryakov P.E., Khlimankov D.Y., Bochkareva E., Bochkarev A., Lavrik O.I.. Human replication protein A (RPA) binds a primer-template junction in the absence of its major ssDNA-binding domains. Nucleic Acids Res. 2004; 32:1894–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wiśniewski J.R., Hein M.Y., Cox J., Mann M.. A ‘proteomic ruler’ for protein copy number and concentration estimation without spike-in standards. Mol. Cell. Proteomics. 2014; 13:3497–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gibb B., Ye L.F., Gergoudis S.C., Kwon Y., Niu H., Sung P., Greene E.C.. Concentration-dependent exchange of replication protein A on single-stranded DNA revealed by single-molecule imaging. PLoS One. 2014; 9:e87922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ghaemmaghami S., Huh W.-K., Bower K., Howson R.W., Belle A., Dephoure N., O’Shea E.K., Weissman J.S.. Global analysis of protein expression in yeast. Nature. 2003; 425:737–741. [DOI] [PubMed] [Google Scholar]
- 31. Eftedal I., Guddal P.H., Slupphaug G., Volden G., Krokan H.E.. Consensus sequences for good and poor removal of uracil from double stranded DNA by uracil-DNA glycosylase. Nucleic Acids Res. 1993; 21:2095–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nilsen H., Yazdankhah S.P., Eftedal I., Krokan H.E.. Sequence specificity for removal of uracil from U.A pairs and U.G mismatches by uracil-DNA glycosylase from Escherichia coli, and correlation with mutational hotspots. FEBS Lett. 1995; 362:205–209. [DOI] [PubMed] [Google Scholar]
- 33. Bellamy S.R., Baldwin G.S.. A kinetic analysis of substrate recognition by uracil-DNA glycosylase from herpes simplex virus type 1. Nucleic Acids Res. 2001; 29:3857–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ye Y., Stahley M.R., Xu J., Friedman J.I., Sun Y., McKnight J.N., Gray J.J., Bowman G.D., Stivers J.T.. Enzymatic excision of uracil residues in nucleosomes depends on the local DNA structure and dynamics. Biochemistry. 2012; 51:6028–6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Grogan B.C., Parker J.B., Guminski A.F., Stivers J.T.. Effect of the thymidylate synthase inhibitors on dUTP and TTP pool levels and the activities of DNA repair glycosylases on uracil and 5-fluorouracil in DNA. Biochemistry. 2011; 50:618–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Longley D.B., Harkin D.P., Johnston P.G.. 5-fluorouracil: mechanisms of action and clinical strategies. Nat. Rev. Cancer. 2003; 3:330–338. [DOI] [PubMed] [Google Scholar]
- 37. Blount B.C., Mack M.M., Wehr C.M., MacGregor J.T., Hiatt R.A., Wang G., Wickramasinghe S.N., Everson R.B., Ames B.N.. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: Implications for cancer and neuronal damage. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:3290–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Toledo L.I., Altmeyer M., Rask M.-B., Lukas C., Larsen D.H., Povlsen L.K., Bekker-Jensen S., Mailand N., Bartek J., Lukas J.. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013; 155:1088–1103. [DOI] [PubMed] [Google Scholar]
- 39. Yu K., Huang F.-T., Lieber M.R.. DNA substrate length and surrounding sequence affect the activation-induced deaminase activity at cytidine. J. Biol. Chem. 2004; 279:6496–6500. [DOI] [PubMed] [Google Scholar]
- 40. Otterlei M., Haug T., Nagelhus T.A., Slupphaug G., Lindmo T., Krokan H.E.. Nuclear and mitochondrial splice forms of human uracil-DNA glycosylase contain a complex nuclear localisation signal and a strong classical mitochondrial localisation signal, respectively. Nucleic Acids Res. 1998; 26:4611–4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ainavarapu S.R.K., Brujić J., Huang H.H., Wiita A.P., Lu H., Li L., Walther K.A., Carrion-Vazquez M., Li H., Fernandez J.M.. Contour length and refolding rate of a small protein controlled by engineered disulfide bonds. Biophys. J. 2007; 92:225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chin A.F., Toptygin D., Elam W.A., Schrank T.P., Hilser V.J.. Phosphorylation increases persistence length and end-to-end distance of a segment of tau protein. Biophys. J. 2016; 110:362–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. de Laat W.L., Appeldoorn E., Sugasawa K., Weterings E., Jaspers N.G., Hoeijmakers J.H.. DNA-binding polarity of human replication protein A positions nucleases in nucleotide excision repair. Genes Dev. 1998; 12:2598–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fisher L.A., Bessho M., Wakasugi M., Matsunaga T., Bessho T.. Role of interaction of XPF with RPA in nucleotide excision repair. J. Mol. Biol. 2011; 413:337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Krasikova Y.S., Rechkunova N.I., Maltseva E.A., Petruseva I.O., Lavrik O.I.. Localization of xeroderma pigmentosum group A protein and replication protein A on damaged DNA in nucleotide excision repair. Nucleic Acids Res. 2010; 38:8083–8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fan J., Pavletich N.P.. Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev. 2012; 26:2337–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schonhoft J.D., Kosowicz J.G., Stivers J.T.. DNA translocation by human uracil DNA glycosylase: role of DNA phosphate charge. Biochemistry. 2013; 52:2526–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schonhoft J.D., Stivers J.T.. DNA translocation by human uracil DNA glycosylase: the case of ssDNA and clustered uracils. Biochemistry. 2013; 52:2536–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kavli B., Sundheim O., Akbari M., Otterlei M., Nilsen H., Skorpen F., Aas P.A., Hagen L., Krokan H.E., Slupphaug G.. hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J. Biol. Chem. 2002; 277:39926–39936. [DOI] [PubMed] [Google Scholar]
- 50. Vuzman D., Levy Y.. Intrinsically disordered regions as affinity tuners in protein-DNA interactions. Mol. Biosyst. 2012; 8:47–57. [DOI] [PubMed] [Google Scholar]
- 51. Coey C.T., Malik S.S., Pidugu L.S., Varney K.M., Pozharski E., Drohat A.C.. Structural basis of damage recognition by thymine DNA glycosylase: key roles for N-terminal residues. Nucleic Acids Res. 2016; 44:10248–10258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.