Abstract
Exposure to UV light can result in severe DNA damage. The alternative general transcription factor (GTF) TFB3 has been proposed to play a key role in the UV stress response in the thermoacidophilic crenarchaeon Sulfolobus acidocaldarius. Reporter gene assays confirmed that tfb3 is upregulated 90–180 min after UV treatment. In vivo tagging and immunodetection of TFB3 confirmed the induced expression at 90 min. Analysis of a tfb3 insertion mutant showed that genes encoding proteins of the Ups pili and the Ced DNA importer are no longer induced in a tfb3 insertion mutant after UV treatment, which was confirmed by aggregation assays. Thus, TFB3 plays a crucial role in the activation of these genes. Genome wide transcriptome analysis allowed a differentiation between a TFB3-dependent and a TFB3-independent early UV response. The TFB3-dependent UV response is characterized by the early induction of TFB3, followed by TFB3-dependent expression of genes involved in e.g. Ups pili formation and the Ced DNA importer. Many genes were downregulated in the tfb3 insertion mutant confirming the hypothesis that TFB3 acts as an activator of transcription. The TFB3-independent UV response includes the repression of nucleotide metabolism, replication and cell cycle progression in order to allow DNA repair.
INTRODUCTION
Although many processes, which occur in the archaeal cell are exclusively present in this unique third domain of life, other processes are often a mosaic of bacterial and eukaryotic properties. This is especially the case for information processing, like the regulation of transcription. While transcriptional regulators resemble their bacterial counterparts, the basal transcription apparatus is considered a simplified version of the more complex system found in eukaryotes (1,2). In contrast to eukaryotes, archaea exhibit a much smaller set of general transcription factors (GTFs). The basal archaeal transcription machinery comprises a multi-subunit RNA-polymerase (RNAP) and the GTFs representing homologs of the eukaryotic TATA box binding protein (TBP) and the transcription factor TFIIB (TFB). Most of the archaeal promoters show structural similarity to the eukaryotic RNA polymerase II system (3), as they contain the typical sequences for TATA box, TFB responsive element (BRE) and initiator element (Inr).
The archaeal transcription machinery, including the initiation of transcription, has been extensively studied (4–6). Briefly, TBP recognizes and binds to the TATA box. Subsequently, TFB associates with the TBP–DNA complex and forms sequence specific interactions with the BRE site followed by a conformational change, i.e. bending, of the DNA, which determines the direction of transcription. Finally, the RNAP is recruited by the N-terminal part of TFB (7–10). In vitro analyses showed that only TBP, TFB and the RNAP are required and sufficient to initiate transcription (11,12). Furthermore, archaea possess a homolog of the eukaryotic GTF TFIIE (TFE). TFE is involved in the stimulation of transcription processivity, since it is able to increase the transcription of certain promoters, especially those, which show suboptimal interaction with TBP (13,14). Additionally, it has been shown that TFE plays an important role in promoter escape as it competes with the elongation factor Spt4/5 for RNAP binding (15,16).
Most of the sequenced archaeal genomes encode multiple homologs of the GTFs (i.e. TBP and TFB) and the numbers vary depending on the species. Recent structural comparisons revealed similarity between eukaryotic TFIIB, archaeal TFB and bacterial σ factors (17). The multiplicity of GTFs is extensively studied in euryarchaea and a role in the adaptation to changing environmental conditions has been proposed for several organisms. The function of multiple GTFs has best been characterized in the halophilic euryarchaeon Halobacterium salinarum NRC. H. salinarum NRC encodes seven TFBs and six TBPs, which are used in different combinations to regulate the transcription of genes essential during the heat shock response (18), oxidative stress (19) and adaptation to low temperatures (20). The euryarchaeon Pyrococcus furiosus contains two TFBs and one TBP. Transcript levels of tfb2 increased dramatically upon heat shock, while tfb1 levels remained stable, suggesting a role of TFB2 in the response to higher temperatures (21). In contrast to the detailed studies, which are available for euryarchaea, information about the role of multiple GTFs in crenarchaea is still limited. The thermoacidophilic crenarchaeal model organisms Sulfolobus solfataricus and Sulfolobus acidocaldarius both possess three tfb genes and one tbp gene (22,23). In general, TFBs have a conserved structure with two major domains (Supplementary Figure S1). The N-terminal domain harbors a zinc ribbon motif and a conserved B-finger and is required for the recruitment of RNAP (11,24). The C-terminal domain, which represents two third of the protein, is essential for the interaction with the TBP–DNA complex and contains a helix-turn-helix (HTH) motif, which is used to form sequence specific contacts with the BRE site (25). In Sulfolobus species, TFB3 is significantly smaller compared to TFB1 and TFB2. It comprises only a short C-terminal domain and lacks the B-finger and the HTH motif (Supplementary Figure S1). While TFB1, which supports transcription initiation in vitro (26), functions as a classical TFIIB homolog fulfilling widespread roles in the regulation of housekeeping genes (11,27), the alternative transcription factors TFB2 and TFB3 show much lower transcript levels and are therefore proposed to have specialized functions (28). Genome wide microarray experiments in which transcription levels were studied during the cell cycle of S. acidocaldarius, showed that tfb2 is induced during the transition from the G1 to the S phase, indicating that expression is cell cycle dependent and that TFB2 plays a role in cell-cycle regulation (29).
Two independent transcriptome studies using microarrays of S. acidocaldarius and S. solfataricus revealed that tfb3 is one of the most upregulated genes following UV irradiation (30,31). To further explore the role of TFB3 in UV stress response, Paytubi and White performed in vitro transcription assays with purified proteins from S. solfataricus (32). In this study, DNA fragments containing promotors that were previously shown to be either unresponsive to UV stress (T6, ssb) or to be activated (dps, thsB) or repressed (sta1) after UV stress were used. The experiments revealed that TFB3 had a stimulating effect on the transcription of all analyzed promoters and that transcription required TFB1 bound at the promoter. Therefore, TFB3 was proposed to interact with the TBP–TFB1–DNA complex and to stimulate RNAP recruitment and thus to function as a transcriptional activator (32).
In this study, the role of TFB3 in early UV stress response in S. acidocaldarius is addressed by in vivo tagging of the protein for immunodetection and co-immunoprecipitation as well as promoter activity studies, creation of a tfb3 insertion mutant and a comprehensive transcriptome analysis.
MATERIALS AND METHODS
Growth conditions
Sulfolobus acidocaldarius strains MW001, MW001pyrEF+, MW1104, tfb3::pyrEF, tfb3CSF and MW001_tfb3p were grown aerobically at 75°C in basal Brock medium (33) pH 3.5, supplemented with 0.1% (w/v) NZ- amine, 0.2% (w/v) dextrin and for strains MW001, MW004 and tfb3CSF with 0.2% (w/v) uracil. The growth of the cells was monitored by measuring the optical density at 600 nm (OD600).
Creation of the genomic HA-tagged and Strep-/FLAG-tagged versions of tfb3
The tfb3-C terminal HA strain and the tfb3-C terminal Strep-/FLAG strain were obtained using the ‘pop in-pop out’ method described previously (34). To generate the HA-tagged strain, specific overlapping primer pairs (7518/7519, Supplementary Table S1) were used to amplify the upstream and downstream regions of the stop codon of tfb3. These regions were connected by overlap polymerase chain reaction (PCR) using the external primers 7516/7517 (Supplementary Table S1). The PCR product was purified and cloned into plasmid pSVA407 by ApaI and BamHI resulting in the plasmid pSVA3640. For the construction of the Strep-/FLAG-tagged strain, upstream and downstream regions of the tfb3 gene were amplified using S. acidocaldarius DSM639 genomic DNA as a template and Strep-/FLAG tag sequences were introduced via PCR using the primer pairs SF01/SF03 and SF02/SF04 (Supplementary Table S1). Overlap extension PCR was performed to fuse the DNA fragments using primer pair SF01/SF04. The PCR product was inserted into pSVA406 via BamHI and NcoI restrictions sites resulting in the plasmid pSVA406-tfb3CSF. After verification by DNA sequencing, Escherichia coli ER1821 was used for the methylation of the plasmids. S. acidocaldarius MW001 cells were transformed with the respective methylated insertion-construct. Mutant screening was performed as described previously (34). The resulting strains MW1104 and tfb3CSF comprise the HA tag and the Strep-/FLAG tag, respectively, at the C-terminus of TFB3.
Generation of the tfb3 insertion mutant
The tfb3 mutant strain tfb3::pyrEF was created via double homologous recombination and insertion of the Sso-pyrEF cassette in the tfb3 gene. A total of 86 bp of the tfb3 gene and 106 bp of the tfb3 gene were fused upstream and downstream to the pyrEF cassette, which was amplified via PCR using pSVA406 as a template. S. acidocaldarius MW001 cells were transformed with 300 ng of the PCR product via electroporation. The resulting cells were selected on plates and liquid media lacking uracil but containing 0.1% (w/v) NZ-amine. Clones were tested for the presence of the pyrEF cassette via colony PCR and positive clones were confirmed by sequencing. Attempts to create a marker-less deletion mutant via the ‘pop in-pop out’ method (34) failed. Whereas the recombination step in which the Sso-pyrEF cassette containing plasmid was integrated in the chromosome was successful, all colonies (over 200 colonies were screened) that lost the plasmid in the second recombination step still contained the complete tfb3 gene despite approximately equally sized regions for the two possible recombination steps. This phenomenon has been observed previously, e.g. (35) and suggests that either the resulting marker-less mutant is harmful to the cell, or that secondary or tertiary DNA structures strongly favor the recombination resulting in the original genomic arrangement.
Generation of the tfb3 reporter gene construct
In order to determine the activity of the tfb3 promoter after UV stress, the maltose promoter in the shuttle vector pSVA1450 (34) was replaced with the tfb3 promoter including 200 bp of its upstream promoter region. The DNA fragment was obtained via PCR using S. acidocaldarius DSM639 genomic DNA as a template and the primer pair RGA01/RGA02 (Supplementary Table S1). The resulting PCR product was inserted into pSVA1450 via the NcoI and SacII restriction sites, yielding the reporter gene construct pSVA1450-tfb3p.
UV treatment and aggregation assays
Exposure of S. acidocaldarius cells to UV light was performed as described previously (36). Briefly, 10 ml of a culture (OD600 = 0.2–0.3) was induced with 100 J/m² UV light (254 nm, Spectroline UV-crosslinker) in a Petri dish. After that, cultures were incubated at 75°C for 3 h. Subsequently, 5 μl of each culture (diluted to OD600 = 0.2) was spotted on a microscope slide coated with 1% (w/v) agar. Single and aggregated cells (n ≥ 3) were analyzed by the ImageJ cell counter plug-in (NIH, Bethesda, MD, USA) from at least three independent experiments. The percentage of cells found in aggregates was subsequently calculated.
Quantitative real-time-polymerase chain reaction (qRT-PCR)
To compare the expression of genes of the ups operon and genes regulated by UV before and 3 h after UV induction, total RNA was isolated from a 10 ml culture grown to an OD600 of 0.4 using Trizol (Sigma) followed by DNAse I treatment. cDNAs were synthesized with the First Strand cDNA synthesis Kit (Thermo Scientific) following the instructions of the manufacturer. Quantitative real-time-PCR (qRT-PCR) was performed using the Maxima SYBR green master mix in a Rotor-Gene Q qPCR cycler (Qiagen). Gene-specific primer sets were designed for the following genes: UpsX (Saci_1493), upsE (Saci_1494), upsA (Saci_1496), cedB (Saci_0748) (for details, see Supplementary Table S1). The housekeeping gene secY served as a control (primer set 1480/1481). The threshold cycle (CT) values obtained were determined to compare the non-UV-induced expression with the UV-induced expression of the tested genes. Furthermore, expression levels of tested genes of MW001pyrEF+ and tfb3::pyrEF were compared. Differences in expression are displayed as log2 changes.
ONPG assay
The o-nitrophenyl-β--D-galactopyranoside (ONPG) assay was performed as described previously with few modifications (37). Briefly, 50 and 350 ml Brock medium containing 0.1% (w/v) NZ-amine and 0.2% (w/v) dextrin were used for the pre-cultures and the main cultures, respectively, and the main cultures were grown until an OD600 of 0.2–0.4 was reached. UV treatment was performed as described above. Samples before treatment (control) as well as after treatment with UV irradiation (+UV) and without UV irradiation (−UV) were collected by centrifugation at 5000 × g for 20 min (4°C). Cell disruption was performed using a Precellys cell homogenizer (Peqlab) and the lysate was centrifuged (16 000 × g, 45 min, 4°C) to remove cell debris. The protein concentration in the soluble fraction was determined using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). The ONPG assay was performed in a total volume of 500 μl containing 5 μg protein of the lysate in the Z-buffer. In all steps, 2-mercaptoethanol and Triton X-100 were omitted from the Z-buffer. The measurements were performed at 75°C using a spectrophotometer with a 1 cm cuvet (Specord). Specific activities were determined using the following equation:
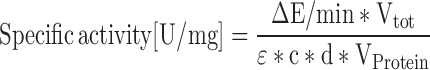 |
with ΔE/min, extinction change per minute; Vtot, total volume of the assay [ml]; VProtein, volume of the protein solution applied in the assay [ml]; ϵ, extinction coefficient; c, concentration of the protein solution applied in the assay [mg/ml]; d, thickness of the cuvette [cm].
Generation and purification of polyclonal antibodies
Antibodies were generated against purified recombinant 6x-His-TFB1. 1 mg of protein was used for the immunization of one rabbit (28 days Speedy protocol, Eurogentec, Belgium). The antibodies were purified from the final bleed antiserum using protein A agarose (Roche) following the manufacturer’s instructions.
Western blotting and immunodetection
Cell pellets from UV treated and control cultures were resuspended in modified N-buffer (25 mM Tris–HCl pH 7.5, 10 mM MgCl2, 100 μM ZnSO4, 1 mM Tris(2-carboxyethyl)phosphin supplemented with 500 mM NaCl and protease inhibitor cocktail and subsequently disrupted using a Precellys cell homogenizer (Peqlab). To remove unbroken cells, the lysate was centrifuged at 16 000 × g and 4°C for 45 min. The protein concentration was determined according to the method by Bradford (38) using the Bio Rad protein assay following the manufacturer's instructions. Following separation via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (39), the proteins were analyzed via western blotting and immunodetection using an anti-FLAG-tag antibody (NEB, dilution 1:1000) and a polyclonal anti-TFB1 antibody (Eurogentec, dilution 1:10 000) as primary antibodies. As a secondary antibody, a horseradish peroxidase (HRP)-conjugated anti-rabbit-IgG raised in mouse (NEB) was used with a dilution of 1:10 000. Signals were detected using the SignalFire™ Elite ECL Reagent (NEB) and visualized by autoradiography.
Co-immunoprecipitation
The strains MW001 and MW1104 were grown in Brock medium to OD600 = 0.4. Cells were exposed to 100 J/m² UV light (254 nm, Spectroline UV-crosslinker) in 10 ml aliquots and then incubated at 75°C for 3 h. Formaldehyde crosslinking (1% (v/v) final concentration) was carried out in a 35 ml culture for 10 min. The reaction was quenched by addition of 125 mM glycine. After that, cells were harvested by centrifugation at 5000 × g for 20 min (4°C) and stored at −20°C. The cell pellets were resuspended in Pierce IP Lysis/Wash Buffer (0.025 M Tris–HCL pH 7.4, 0.15 M NaCl, 0.001 M ethylenediaminetetraacetic acid, 1% (v/v) NP40, 5% (v/v) glycerol) and then lysed by sonication (40% amplitude, 15-s pulse, 15-s pause) using a KE76 tip (Bandelin). Soluble proteins were obtained by ultracentrifugation at 100 000 × g for 45 min at 4°C. The pulldown assay was performed using the Pierce Anti-HA Magnetic Beads kit (Thermo Scientific) following the manufacturer's instructions. Protein identification was performed by the Mass spectrometry facility of the Center for Biological Systems Analysis (ZBSA) Freiburg.
RNA isolation for RNA sequencing
RNA was isolated for all samples and replicates with Trizol (ThermoFisher, Waltham, USA) as described previously (40). The obtained RNA samples were treated with RNase-free DNase (Qiagen) according to the protocol and afterward purified by ethanol precipitation. Ribosomal RNA was depleted using a RiboZero magnetic kit for bacteria (Illumina, San Diego, USA) with a modified protocol. Only 90 μl instead of 225 μl magnetic beads were used and for the rRNA removal reaction and only 1 μg RNA was mixed with 4 μl removal solution in a total volume of 20 μl instead of 40 μl.
RNA sequencing and data analysis
Sequencing libraries for all samples and replicates were prepared with the TruSeq® Stranded mRNA HT kit (Illumina) starting with the RNA fragmentation step after elution of precipitated RNA in 19 μl of the Fragment-Prime-Mix. Sequencing libraries were quantified with a High-Sensitivity Chip on a Bioanalyzer (Agilent, Böblingen, Germany) and a measurement with a Quant-iT PicoGreen® dsDNA Assay Kit (Invitrogen, Carlsbad, USA) on a Microplate Reader Tecan Infinite 200 (Tecan, Männedorf, Switzerland). Sequencing was performed on a HiSeq1500 instrument (Illumina) in rapid mode with a read length of 2 × 25 nt. Sequencing reads were mapped with Bowtie2 (41) against the reference genome. Since S. acidocaldarius MW001 is a deletion mutant of S. acidocaldarius DSM639, the latter one was chosen as a reference (S. acidocaldarius DSM639, genome size: 2 225 959 nt, RefSeq ID: NC_007181.1). The data were visualized in the software ReadXplorer and mapped reads were counted for each gene (42). Subsequently RPKM values (43) were manually calculated for each gene. In contrast to the original value, only reads mapping to coding sequences were included in order to get the total number of reads. For determination of regulated genes, a statistical analysis was performed using DESeq2 (44). Sequences relating to tfb3 found in the tfb3::pyrEF strain are related to the small 5′ and 3′ regions of tfb3 that remained intact after insertion of the pyrEF cassette, but do not represent expression of the complete gene.
Identification of a non-palindromic hexanucleotide motif
All possible sequence patterns for the desired motif were searched with Bowtie2 (Version: 2.3.0) [(41); 38 in draft] in S. acidocaldarius DSM639. No mismatches were allowed for the alignment. The results were filtered by a custom Python script to only contain features where the complete motif was between 30 and 80 bp upstream of a gene start. All steps were automated via Snakemake (Version: 3.13.3) (45). The logo plot was created with WebLogo (Version: 2.8.2) (46).
RESULTS
TFB3 protein levels are increased upon UV irradiation
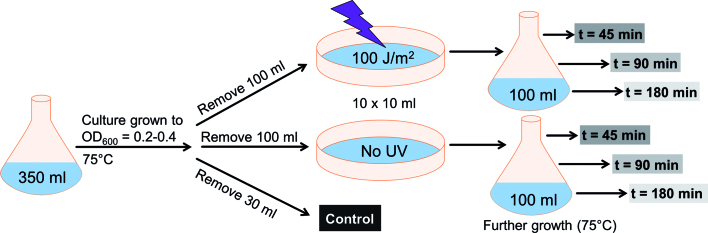
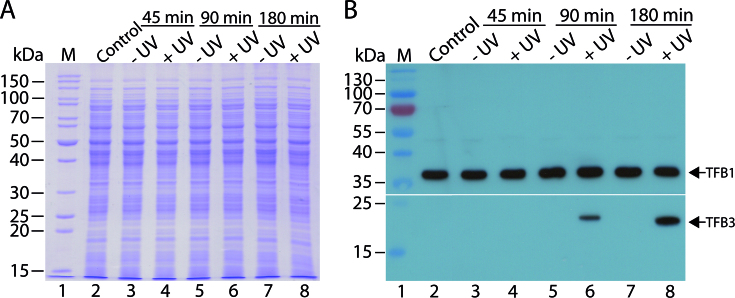
In order to follow the expression of TFB3 in vivo, the MW001 derived strain tfb3CSF in which a C-terminally Strep-/FLAG-tag tagged TFB3 is expressed from the native promoter was constructed. To study the expression levels of TFB3 after UV treatment in vivo, tfb3CSF was cultured in shaking flasks (350 ml) and grown until an OD600 of 0.2–0.4 was reached. The experimental setup is shown in Figure 1. First, 30 ml of the culture were removed as untreated control sample. The remaining culture was split (10 ml portions each) and transferred to petri dishes. Ten petri dishes were either exposed to UV light (100 J/m2) or were handled in the same manner but without UV treatment. Subsequently, the cells were transferred to fresh Erlenmeyer flasks with complex medium and growth was continued at 75°C. In contrast to previous studies focusing on DNA repair mechanisms (30,31) cells were not maintained in the dark after UV treatment to prevent photolyase activity. Samples for total protein extraction were taken before (control) as well as 45, 90 and 180 min after the treatment with UV irradiation (+UV) or without UV irradiation (−UV). The total protein extracts were analyzed via SDS-PAGE (Figure 2A) and immunodetection using an anti-FLAG-tag antibody (Figure 2B). As shown in Figure 2B, no signal for TFB3 was detected before UV treatment nor in any of the samples that were not treated with UV. TFB3 was also not detected at 45 min after UV irradiation (Figure 2B, lane 4), but could be detected 90 min after UV irradiation (Figure 2B, compare lanes 5 and 6). TFB3 levels increased further until 180 min (Figure 2B, compare lanes 7 and 8). Our data confirm two previous studies (30,31) and show that TFB expression is induced by UV irradiation.
Figure 1.
Experimental setup for the growth, UV treatment and sampling of tfb3CSF. Cells were grown until an OD600 of 0.2–0.4 was reached. After collecting the cells before exposure (control), cells were split into portions of 10 ml and 10 petri dishes were either UV-irradiated with 100 J/m2 or were not irradiated, but treated the same way. Afterward the cells were further cultivated and samples for immunodetection were taken 45, 90 and 180 min after the treatment. The experiment was carried out with three biological replicates.
Figure 2.
Expression levels of the endogenously tagged TFB3 in UV-treated and untreated S. acidocaldarius (tfb3cSF) cells. (A) SDS-PAGE (12.5% (w/v) polyacrylamide; Coomassie staining) of crude extracts of tfb3CSF before the treatment (control) and 45, 90 and 180 min after the treatment with 100 J/m2 UV irradiation (+UV) or without UV irradiation (−UV). A total of 30 μg of protein extract was loaded per lane. M, PageRuler unstained protein ladder (Thermo Fisher Scientific). (B) Autoradiograph showing the expression of TFB3 (23 kDa) and TFB1 (35 kDa) before the treatment (control) and 45, 90 and 180 min after the treatment with (+UV) or without UV irradiation (−UV). To monitor both proteins, which show significant differences in size and expression level, the membrane was cut after the transfer and the parts were handled separately. For TFB3, an anti-FLAG-tag antibody was used as primary antibody with a dilution of 1:1000. As an internal loading control, TFB1 was detected using a polyclonal antibody raised in rabbit (dilution: 1:10 000). Both TFB1 and TFB3 were visualized using a HRP-coupled anti-rabbit-IgG as secondary antibody. The exposition time was 2 min for TFB3 and 5 s for TFB1. M, PageRuler prestained protein ladder (Thermo Fisher Scientific).
The activity of the tfb3 promoter is induced by UV irradiation
To test the activity of the tfb3 promoter under the influence of UV stress, a 200 bp fragment of the tfb3 promoter was fused before the lacS gene in the shuttle vector pSV1450. MW001 transformants were grown in shaking flasks under standard growth conditions until an OD600 of 0.2–0.4 was reached and the UV treatment was performed as described above. Samples were taken at 45, 90 and 180 min after the treatment. The β-galactosidase activity in the total protein extracts was determined using the ONPG assay.
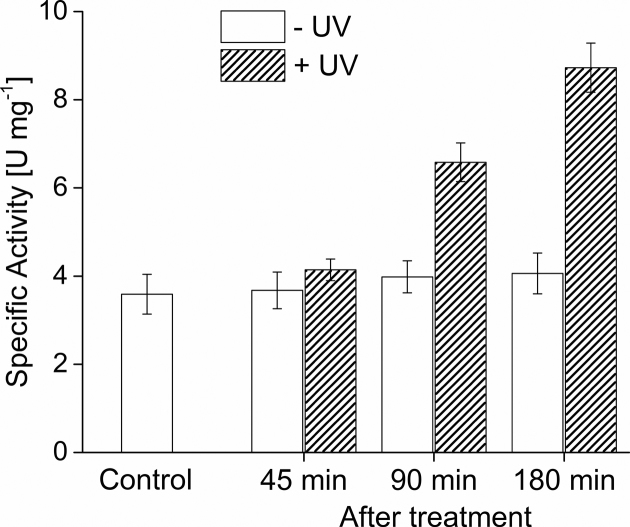
As shown in Figure 3, a basal activity of 3.7 U/mg was determined in the sample before the treatment (control). While no significant increase of the activity was observed in all samples without UV irradiation, higher β-galactosidase activities were observed in the UV irradiated samples increasing from 4.1 U/mg to 6.7 U/mg and 8.7 U/mg after 45, 90 and 180 min, respectively. These results were in agreement with the observed expression of the TFB3 protein after 90 min in UV treated cells (Figure 2B). The β-galactosidase activity in the sample before treatment indicates a basal expression from the tfb3 promoter. Indeed analysis of increased amounts of crude extracts (100 μg protein compared to 30 μg protein extract loaded on SDS-PAGE previously (Figure 2)) allowed for detection of TFB3 in non UV treated cells (Supplementary Figure S2).
Figure 3.
Activity of the tfb3 promoter after UV irradiation using the lacS reporter gene assay. MW001-tfb3p was grown in a shaking flask in Brock media supplemented with 0.1% (w/v) NZ- amine, 0.2% (w/v) dextrin at 75°C until an OD600 of 0.2–0.4 was reached. The control sample was taken before treatment. Afterwards, the remaining cells were split in 10 ml portions and the UV treatment was carried out in petri dishes. Subsequently, growth was continued in shaking flasks at 75°C. Specific β-galactosidase activities of samples before (control) and 45, 90 and 180 min after the treatment with (+UV, 100 J/m2) or without (−UV) UV irradiation were determined using ONPG as substrate and are given as specific activities (U mg−1 protein). Medians were calculated from three biological replicates (n = 3) and error bars represent the respective standard deviation.
TFB3 in vivo protein–protein interaction
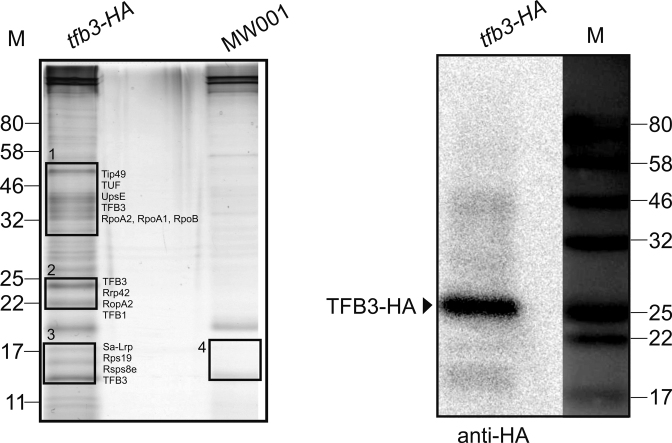
In order to identify proteins that interact with TFB3 in vivo, S. acidocaldarius strain MW1104, which carries the endogenously HA-tagged tfb3 gene was constructed. Soluble proteins were extracted from MW001 and MW1104, were immune-precipitated using an anti-HA antibody and these fractions were analyzed on a SDS-PAGE followed by silver staining. Mass spectrometry was further used to identify proteins (indicated by black rectangles, Figure 4) that were observed in MW1104 but not in MW001. As shown in Figure 4, candidates specifically identified in the TFB3-HA immunoprecipitation fractions could be identified (Supplementary Table S2). Notably, TFB1 and the RNAP subunits RpoA2, RpoA1, RpoB and the TBP-binding protein (Tip49) were detected. TFB3 was previously reported to interact with the RNAP and probably TFB1 in the pre-initiation complex in vitro (32). Remarkably, also Sa-Lrp (Saci_1588) (47) was specifically detected in MW1104. Sa-Lrp was previously shown to bind to the promoters of the ups genes and the deletion mutant of Sa-Lrp showed abnormal sizes of the UV-induced aggregates (47).
Figure 4.
Co-immunoprecipitation using an endogenously tfb3-HA-tagged Sulfolobus acidocaldarius strain. The SDS polyacrylamide gel (silver-staining) and the immunodetection with Anti-HA antibody (Sigma) of the protein fractions after the pull-down experiment using Anti-HA magnetic beads are shown. The elution fractions of MW1104 (tfb3-HA) and MW001 (control) cell extracts were analyzed by SDS gel. Black rectangles with numbers indicate the bands excised for mass spectrometry. Identified peptides are listed in Supplementary Table S1: Tip49, TBP interacting protein 49kDa; TUF, Elongation factor 1-α; UpsE, ATPase of the ups pilus; RopA(1,2,B), RNAP subunits; Rrp42, Exosome complex component; Rps19, 30S ribosomal protein S19; Rsps8e, 30S ribosomal protein S8e; M, marker in kDa.
Cellular aggregation after UV treatment
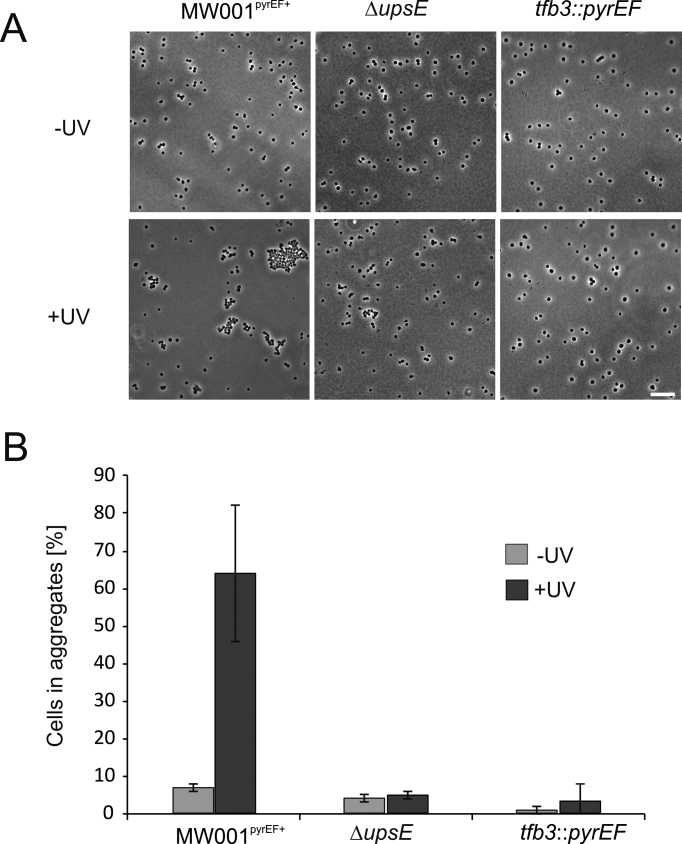
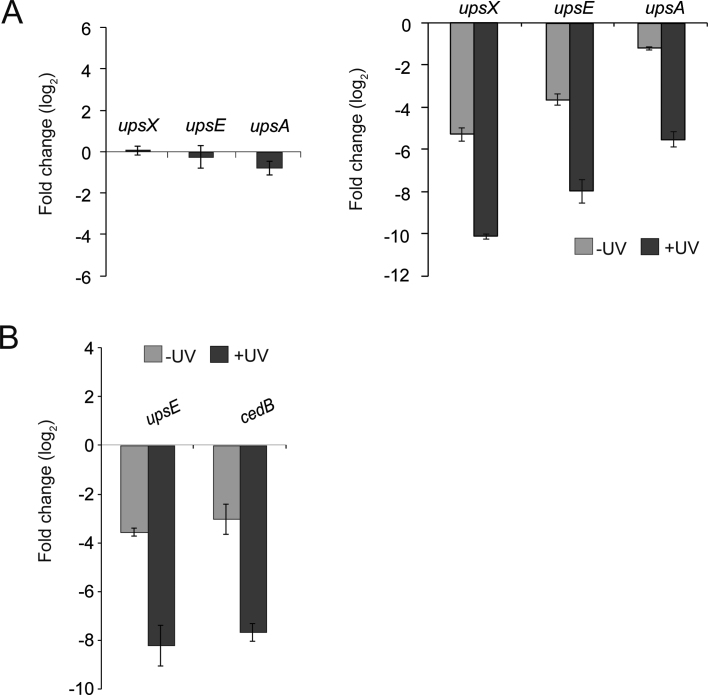
Upon UV stress, S. acidocaldarius cells express ups pili (UV-inducible pili operon of Sulfolobus), which result in the formation of aggregates in which subsequently DNA exchange can occur (36,48–50). This DNA exchange is an important mechanism to repair DNA damage via homologous recombination. Deletion of e.g. upsE that encodes the secretion adenosinetriphosphatase (ATPase) involved in ups pili formation results in a strain (MW109), which does no longer aggregate after UV treatment (49). In order to further investigate the involvement of TFB3 in this UV stress response, the tfb3 insertion mutant strain tfb3::pyrEF was generated. In this strain, a cassette containing the S. solfataricus pyrEF genes is inserted into the tfb3 gene resulting in disruption of the tfb3 gene. As a control the S. acidocaldarius strain MW001pyrEF+ (MW001 complemented with the pyrEF cassette (35)), was used. Aggregation assays performed with MW001pyrEF+ showed that 180 min after UV treatment, 45–85% of the cells were present in aggregates, whereas in the ΔupsE strain MW109 <10% of the cells were present in aggregates (Figure 5). Interestingly, the tfb3 disruption mutant tfb3::pyrEF also showed almost no aggregates after UV treatment (5%) (Figure 5B) suggesting that TFB3 might be involved in the regulation of ups pili formation. To study this possibility, qRT-PCR was performed on selected genes involved in cellular aggregation and DNA exchange (36,49,51). This showed that the upsX, upsE and upsA genes, contrary to the reference strain MW001pyrEF+, were not induced in the tfb3 mutant 180 min after UV stress. Moreover, when expression of these three genes was compared to that in MW001pyrEF+ (Figure 6A, on the right side), their expression was also significantly lower in non-UV-treated samples. Beside the ups genes, also the transcription of the cedB gene, which encodes the membrane protein involved in DNA transport in aggregates after UV treatment (51) was dramatically decreased in the tfb3 mutant compared to that of MW001pyrEF+ (Figure 6B) (30,31). Therefore, it is concluded that TFB3 is involved in the regulation of the ups and ced genes.
Figure 5.
Aggregation assay with MW001 tfb3::pyrEF. (A) Cells without UV (−UV) and 180 min after UV treatment (100 J/m2) (+UV) were visualized using phase-contrast microscopy. MW001pyrEF+ was used as positive control and ΔupsE (MW109) as a negative control (scale bar: 10 μm). (B) Quantification of cellular aggregation of non-UV-treated cells (−UV) and cells 180 min after UV treatment (+UV). Single and aggregated cells (n > 3) were counted. Average amounts of cells in aggregates were calculated from three individual experiments.
Figure 6.
qRT-PCR of selected genes involved in cellular aggregation and DNA exchange. (A) On the left: changes of transcription levels of upsX, upsE and upsA in the tfb3 insertion mutant tfb3::pyrEF upon UV treatment compared to non-induced cells displayed as log2 fold changes. On the right: transcription levels of upsX, upsE and upsA before (light gray bars) and 180 min after (dark gray bars) UV stress in tfb3::pyrEF in comparison to that of the reference strain MW001pyrEF+. (B) Transcription levels of upsE and cedB in not induced (light gray bars) and UV induced (dark gray bars) conditions in tfb3::pyrEF in comparison to that of MW001pyrEF+. Changes in transcription levels are displayed as log2.
Early transcriptional response to UV irradiation
To investigate the role of TFB3 in the early response to UV treatment, gene expression in the MW001pyrEF+ and the tfb3::pyrEF strain were studied by a genome wide transcriptome analysis. To exclude an effect of the pyrEF cassette on the differential transcription of genes, strain MW001pyrEF+ was used as reference. Cells (see Supplementary Figure S3 for detailed transcript levels) were cultivated until an OD600 of 0.2–0.4 was reached and were subsequently UV-treated with 100 J/m2 as shown in Figure 1. RNA was extracted from samples taken before as well as 45 and 90 min after UV treatment. A detailed overview of the transcription of the S. acidocaldarius genes in the MW001pyrEF+ strain and the tfb3::pyrEF strain at the different timepoints is given in Supplementary Table S3. Supplementary Tables S4–6 show a detailed presentation of all significant differences in gene expression (P-value ≤ 0.01). In MW001pyrEF+, 45 min after UV irradiation, 12 genes were, when compared to the not UV treated cells, regulated more than 4-fold and 100 genes more than 2-fold. Ninety minutes after UV irradiation, 45 genes were regulated more than 4-fold and 191 genes more than 2-fold (for details see Supplementary Figures S4, 5 and 8). In the tfb3::pyrEF strain, after 45 min, 8 genes were regulated more than 4-fold and 122 genes more than 2-fold and after 90 min 24 genes were regulated more than 4-fold and 190 genes more than 2-fold, when compared to non UV treated cells (for details see Supplementary Figures S4, 5 and 8).
When the tfb3::pyrEF strain was compared with strain MW001pyrEF+, 7, 11 and 15 genes were regulated more than 4-fold (46, 30 and 45 genes more than 2-fold) at the timepoints before UV treatment and at 45 and 90 min after UV treatment, respectively (for details see Supplementary Figures S6 and 7). The regulated genes are distributed evenly over the whole genome (Supplementary Figure S11). To further analyze the identified genes, they were classified according their arCOG (Archaeal Clusters of Orthologous Genes) classes. This identified major changes in the classes describing genes involved in transcription (K), replication (L), cell-cycle control and division (D), as well as nucleotide metabolism and transport (F), indicating that the organism adapts to UV stress by regulating a variety of biological processes (Supplementary Figures S10 and 11).
The comparison of the UV response in both strains allows distinguishing between UV-responsive genes whose expression depends on the presence of TFB3 and thus is lower in the tfb3::pyrEF strain compared to the MW001+pyrEF strain and genes whose transcription is independent of TFB3 and therefore shows no significant difference in the tfb3::pyrEF and MW001+pyrEF strains. The data also enables to differentiate between genes that show an early response (45 min) and a delayed response (90 min). Tables 1 and 2 show a selection of genes, which are affected by the UV treatment but are either dependent or independent on the presence of TFB3.
Table 1.
TFB3-dependent regulation of selected genes following UV irradiation in S. acidocaldarius MW001pyrEF+ (WT) and tfb3::pyrEF (Mut)
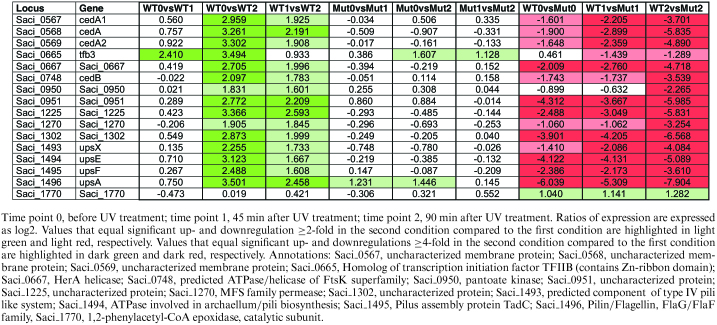
|
Table 2.
TFB3-independent regulation of selected genes following UV irradiation in S. acidocaldarius MW001pyrEF+ (WT) and tfb3::pyrEF (Mut)
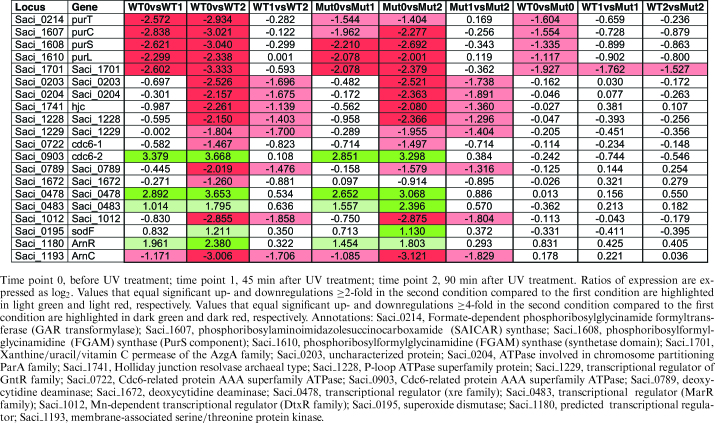
|
TFB3-dependent UV stress response
As expected, tfb3 is one of the five genes that are more than 4-fold upregulated 45 min after UV treatment (WT0 versus WT1), while upregulation of all other genes, which are expressed differentially in a TFB3-dependent manner, follows after 90 min (WT0 versus WT2) (Table 1). We found that tfb3 is slightly upregulated as a later response to UV stress in the tfb3 mutant strain (Mut0 versus Mut2). Furthermore, a slight down regulation could be observed in the irradiated tfb3 mutant strain compared to the reference strain (WT1 versus Mut1, WT2 versus Mut2). This is due to the fact that in the insertion mutant transcription of the gene occurs and short reads of the disrupted gene might be mapped during sequencing despite the pyrEF insertion. Five and 22 genes are differentially regulated more than 4-fold 45 and 90 min after UV treatment in the reference strain MW001+pyrEF, while no response was observed in the tfb3 insertion mutant tfb3::pyrEF (see Supplementary Figure S4C and D for details). Accordingly, the expression of a subset of these genes was significantly different in the untreated and/or irradiated reference compared to the tfb3::pyrEF mutant strain (WT0 versus Mut0, WT1 versus Mut1, WT2 versus Mut2, see Supplementary Figure S6 for details).
Comparing the expression in the reference and the mutant strain almost all genes show a strong downregulation in the mutant strain and only one gene is slightly upregulated (2-fold) under all conditions (Supplementary Figure S7), Saci_1770 annotated as 1,2-phenylacetyl-CoA epoxidase, catalytic subunit (arCOG Q). The enzyme shows significant structural similarity to the recently characterized E. coli phenylacetic acid degradation protein (probability 99.96; E-value 2e-29; (HHPRED, (52)); 3pvt_A, (52)) that is involved in the aerobic and anaerobic hybrid degradation pathway for aromatic compounds. More distant structural similarity is found to ribonucleotide reductases (probability 99,79; E-value 3.1e-20, 4M1H_A (HHPRED), (53)), which catalyze the formation of deoxyribonucleotides from ribonucleotides suggesting a possible role of Saci_1770 in the regulation of DNA synthesis in S. acidocaldarius. All other genes showed an upregulation in the MW001pyrEF+ strain when compared to the tfb3::pyrEF strain. Most regulation was observed 90 min after UV irradiation, suggesting that this is the result of the expression of TFB3. Indeed upregulation of tfb3 was observed after 45 min and the TFB3 protein was detected after 90 min.
Among the TFB3-dependent UV responsive genes identified, the ups gene cluster (upsX, Saci_1493; upsE, Saci_1494; upsF, Saci_1495; upsA/B, Saci_1496) was most highly upregulated (more than 4-fold). In addition, also the cedA1, cedA2 and cedB genes encoding the membrane and the ATPase components of the Ced (crenarchaeal system for exchange of DNA) DNA importer were more than 4-fold upregulated. Remarkably, also the membrane-bound VirB4/HerA/CedB homolog Saci_0667, which is encoded in close vicinity of the Ced system is highly upregulated. It has been proposed that Saci_0667 either encodes an additional ATPase involved in the Ced system or might function as a part of a putative export machinery on the donor site of DNA transfer events (51).
Furthermore, Saci_0950 shows more than 2-fold TFB3 dependent upregulation at 90 min after UV treatment. Saci_0950 shows 29% sequence identity (e-value = 8e−18) to the characterized pantoate kinase from Thermococcus kodakarensis (TK2141) involved in coenzyme A biosynthesis (54). Other genes, which show a TFB3-dependent UV stress response are the putative transporter protein encoding Saci_1270 (annotated as a MFS permease) and two genes of unknown function, Saci_0951 and Saci_1225, which were also previously observed to be upregulated upon UV irradiation (31).
TFB3-independent UV stress response
To find genes that showed a TFB3 independent response to UV treatment, genes that were similarly regulated in the MW001pyrEF+ strain and the tfb3::pyrEF strain were identified. Seven genes were identified that were at least 4-fold up- or downregulated at 45 min after UV treatment and 23 genes were identified after 90 min (Supplementary Figure S4C and D).
Among the genes that show an upregulation is one of the three Cdc6 (cell division cycle 6) proteins, which are key players in the initiation step of DNA replication. Archaeal homologs of proteins involved in the eukaryotic origin recognition complex have been studied in Methanothermobacter thermautotrophicus and it was proposed that Cdc6 proteins directly interact with the replicative minichromosome maintenance helicase MCM and bind to replication origins (55). Cdc6-2 (Saci_0903) was one of the most highly induced genes upon UV stress, whereas cdc6-3 (Saci_0001) remained unaffected and cdc6-1 (Saci_0722) was slightly downregulated. This is in accordance with previous studies (30,31). For S. solfataricus a putative role of Cdc6-2 as a repressor of DNA replication was suggested based on the presence of Cdc6-2 only in the post-replicative G2 phase of the cell cycle and the binding of Cdc6-2 to DNA sequences that overlap with Cdc6-1 or Cdc6-3 binding site (56). In contrast, Cdc6-1 and Cdc6-3 have been discussed to act on the origins in a positive manner to facilitate replication initiation.
Also the Saci_0478 and Saci_0483 genes, which are annotated as conserved conjugative plasmid proteins possessing HTH DNA-binding motifs similar to that of the transcriptional regulator XRE and MarR families, respectively, show upregulation after UV irradiation. The XRE (xenobiotic response element) family of transcriptional regulators is present in all three domains of life (57) and for MarR like regulators a role in antibiotic resistance, oxidative stress response, virulence or catabolism of aromatic compounds is reported (58). A new member of this family, BldR2, involved in aromatic stress response was characterized in S. solfataricus (59). One can speculate that the regulators have a possible role in conjugative processes induced in the course of oxidative stress response upon formation of pyrimidine and purine photoproducts. This is in accordance with the observed 2-fold upregulation of the predicted superoxide dismutase (Saci_0195) in the delayed UV response of both strains.
At last, also the ArnR encoding gene Saci_1180 is highly induced upon UV irradiation. ArnR is together with ArnR1 (Saci_1181) one of the two transcriptional regulators (i.e. activators) of the archaellum regulatory network (Arn), which induce the expression of the filament forming archaeallins encoded by flaB. The archaellum operon of S. acidocaldarius is divided into two loci, flaB and flaX-J, which encode essential genes for the biosynthesis of the archaellum, a rotating type IV pilus-like structure, which acts as the motility structure of this organism (60,61). No upregulation of FlaB expression was observed in our experiments. Indeed a delay between ArnR induction and the expression of FlaB was observed under starvation condition where the assembly of the archaellum is strongly induced after 4 h of starvation (62).
Among the genes, which were downregulated, predicted transcriptional regulators are found. Saci_1012, which is strongly downregulated upon UV stress, encodes a putative Mn-dependent transcriptional regulator of the DtxR family and might be involved in metal homeostasis. Manganese-responsive DtxR family regulators were studied in many bacteria, including the MntR protein of E. coli (63) and Bacillus subtilis (64). An MntR mediated repression of a manganese transporter has been reported for Mycobacterium tuberculosis (65). Manganese plays an important role in free radical-detoxifying enzymes, including superoxide dismutase and catalase (66). Therefore, the downregulation of a possible repressor of manganese transport (Saci_1012) might also aid oxidative stress response mediated by the upregulated predicted superoxide dismutase (Saci_0195).
Remarkably, the gene encoding ArnC (Saci_1193), one of the Hanks-type protein kinases (eSTKs) (besides ArnD, Saci_1694) that have been shown to be involved in the regulation of the archaellum (61,67,68), is downregulated 2-fold after 45 min and more than 4-fold after 90 min after UV treatment in both investigated strains. Deletion mutants of ArnC were only slightly affected in their motility but no direct effect on FlaB expression was observed, the filament protein of the archaellum, which indicated that they might be involved in other regulatory pathways possibly interconnected with the archaellum regulatory network (68).
Several genes involved in purine biosynthesis (e.g. Saci_0214, formate-dependent phosphoribosylglycinamide formyl-transferase (purT)); Saci_1607, purC, phosphoribosylaminoimidazolesuccinocarboxamide synthase and Saci_1608 and Saci_1610, which encode the PurS and the PurL domains of the phosphoribosylformylglycinamidine synthase complex) were also strongly downregulated. Also Saci_1701, which encodes a conserved protein annotated as a xanthine/uracil/vitamin C permease of the AzgA family is downregulated after UV stress in a TFB3 independent manner. Its amino acid sequence shows 30% identity (e-value: 2e−43) with the AzgA protein of Aspergillus nidulans, which has been characterized as a purine transporter with high affinity to hypoxanthine, guanine and adenine (69). Saci_0789 and Saci_1672, which both encode predicted deoxycytidine deaminases were also found to be repressed 90 min after UV irradiation. However, whereas Saci_0789 was downregulated 4- and 2-fold in the WT and tfb3::pyrEF strain, respectively, for Saci_1672 only a 2-fold change in the WT strain was observed. Saci_0789 possesses similarity to the characterized bifunctional dCTP deaminase-dUTPase (DCD-DUT) from Methanocaldococcus jannaschii MJ0430 (28% protein sequence identity, e-value = 6e−10) (70,71) and is therefore assumed to play an essential role in pyrimidine metabolism.
Further genes that show a downregulation at 90 min after UV irradiation are Saci_0203 and Saci_0204, which are annotated as an uncharacterized protein and as a ParA ATPase involved in chromosome partitioning, respectively. The two genes show high similarity to SSO0035/SegB (sequence identity = 53%, e-value = 4e−38) and SSO0034/SegA (sequence identity = 71%, e-value = 1e−111), which were shown to play an essential role in chromosome partitioning (72). While SegA is an ortholog of bacterial Walker-type ParA proteins, SegB is considered an archaea specific factor (72). In addition, also the annotated Holliday junction (HJ) resolvase Hjc, encoded by Saci_1741 was strongly downregulated. The corresponding gene product shows high similarity to several other archaeal HJ resolvases, e.g. Hjc (SSO0575, sequence identity = 42%, residues 8 to 79, e-value = 2e−14) and Hje (SSO1176, sequence identity = 73%, e-value = 4e−66) from S. solfataricus. For the genes Saci_1228 annotated as a predicted P-loop ATPase superfamily protein and the adjacent gene Saci_1229, which is annotated as a transcriptional regulator of the GntR family, a 4- and 2-fold downregulation, was observed. Saci_1228 shows distant homology to ATP-dependent HrpA-like helicases, which are absent in archaeal genomes. Hence, the Saci_1228 gene product might represent a HrpA counterpart involved in cell cycle progression in archaea. (29,73). Thus our data suggest that the TFB3-independent UV response includes the repression of nucleotide biosynthesis and transport, replication and cell cycle progression in order to allow DNA repair.
Identification of a non-palindromic hexanucleotide motif
Previous motif searches identified a non-palindromic hexanucleotide motif in S. acidocaldarius (5′-A(N)TTTC-3′) that was supposed to be important in UV-inducible promoters (74). The motif is located 30 to 80 bp upstream of the transcription start, and a so far unknown regulator was supposed to bind to the motif and to interact with TFB3, which lacks a DNA binding domain. It is speculated that TFB3 then interacts with the RNAP and thus activates transcription via bridging the RNAP and TFB1–TBP–DNA complex (32,74). In our study, we performed an automated motif search and identified in total 299 genes in S. acidocaldarius. Among these genes were 31 and 24 (log2 fold change ≥ 2: 11 and 2) significantly upregulated genes and 9 and 15 (log2 fold change ≤ −2: 3 and 3) down regulated genes after UV treatment in the reference strain and tfb3 mutant strain, respectively (for details see Supplementary Table S7 and Figure S12). Two of the 11 highly upregulated genes (more than 4-fold) in the reference strain, with the searched motif in its upstream region, were also highly upregulated in the tfb3 insertion mutant indicating that the presence of TFB3 is not required for the observed regulation. One of these genes was cdc6-2 (Saci_0903) (Table 2). From the highly downregulated genes (more than 4-fold) all three were found in the reference strain and the tfb3 insertion mutant. We, therefore, speculate that the motif is important for binding of a so far unknown factor or factors that regulate TFB3-independent transcription probably also under other growth or stress conditions.
DISCUSSION
In Sulfolobus, UV-induced DNA damage leads to the formation of photoproducts like cyclobutane pyrimidine dimers (CPD) and 6,4-pyrimidine-pyrimidones (6-4 PP) as well as single and double strand breaks (30). Interestingly, contrary to what has been previously observed in eukaryotes and bacteria, no significant transcriptional induction of DNA repair genes was observed in Sulfolobales (30,31). Instead, upregulated genes were mainly involved in cell cycle control (cdc6-2), oxidative stress (dps) and transcription regulation (tfb3). In contrast, DNA replication proteins are repressed during UV stress (31).
Two independent DNA microarray studies in Sulfolobus demonstrated that the gene encoding the alternative TFIIB homolog TFB3 is one of the highest upregulated genes upon UV irradiation (30,31). Our studies with chromosomally tagged TFB3 and promoter activity assays (Figures 2 and 3) showed that the previously observed changes in transcript levels (31), as expected, result in a significant increase in the cellular TFB3 concentration after UV irradiation, while the concentrations of TFB1 remained stable. Remarkably, changes in transcript level are observed already at 45 min, whereas an increase in the TFB3 protein concentration is only observed at 90 min after UV treatment indicating that changes in tfb3 transcript numbers are manifested only after about 45 min in changes in the TFB3 protein concentration (Figure 2).
This study also provided first evidence of the in vivo function of TFB3. The tfb3::pyrEF strain was impaired in functional Ups pili formation and UV induced cell aggregation (Figures 5 and 6), a process, which normally leads to DNA exchange. This demonstrated that TFB3 is required for Ups pili expression after UV treatment.
Furthermore, several interaction partners of TFB3 were identified. Using the tfb3-HA-tagged strain MW1104, we were able to show the interaction of TFB3 with TFB1 and the RNA polymerase (represented by the large subunit RpoA2) in vivo by co-immunoprecipitation (Figure 4). This was consistent with previous in vitro studies where co-immunoprecipitation of TFB3 with the ternary complex of DNA–TBP–TFB1 and an interaction of TFB3 with the completely assembled pre-initiation complex was observed (32). Co-immunoprecipitation with TFB3-HA also identified Sa-Lrp (Saci_1588). Sa-Lrp is a member of the leucine-responsive regulatory protein (Lrp)-like family of transcriptional regulators in S. acidocaldarius (47). Sa-Lrp binds to promoter regions of genes with a variety of functions including ammonia assimilation, transcriptional control and UV-induced pili synthesis. In vivo studies with the Sa-lrp deletion mutant showed that the absence of Sa-Lrp lead to reduced UV-induced cell aggregation. A differential effect of Sa-Lrp on the expression of the upsA gene before and after UV exposure was observed and it was suggested that Sa-Lrp acts in conjunction with other transcriptional regulators in driving the transcription of the ups operon (47). Our pulldown experiments provided first evidence that TFB3 might be the interacting factor of Sa-Lrp.
The genome wide transcriptome analysis using the MW001+pyrEF and the tfb3::pyrEF strains revealed two separate UV stress response pathways. While the regulation of several genes depends on the presence of TFB3, other genes are differentially transcribed in a TFB3-independent manner. The TFB3-dependent response showed a strict chronological order. Only after expression of TFB3 (45 min), an effect on target gene expression (90 min) is observed, demonstrating that the presence of high levels of TFB3 is a prerequisite for the enhanced transcription of the TFB3-dependent target genes. The gene encoding the classical housekeeping GTF tfb1 was found not to be regulated upon UV stress, confirming the stable expression that was observed in all samples during the immunodetection experiment with the chromosomally tagged strain tfb3CSF (Figure 2B).
Among the processes that are under control of TFB3 (Table 1) is the exchange of DNA via the Ups and Ced systems after UV treatment (49,51). DNA exchange between the cells occurs in order to repair UV-induced DNA damages via homologous recombination. The observed TFB3-dependent regulation is in accordance with the results of two previous studies on the response of S. solfataricus and S. acidocaldarius to UV damage. In both microarray studies, the ups genes as well as the ced genes were upregulated upon UV stress (30,31). The fact that all genes, which exhibit a differential expression depending on the presence of TFB3, are downregulated in the tfb3::pyrEF strain compared to MW001+pyrEF strain and no upregulation was observed in this comparison, confirms that TFB3 acts as an activator of transcription (32).
Analysis of the TFB3-independent response, which is observed in both the MW001+pyrEF strain and the tfb3::pyrEF strain(Table 2) showed that especially the transcription of genes, which drive replication, genome segregation and cell division, is significantly reduced upon UV stress. Consistently, a large number of these genes shows a cell cycle-dependent transcriptional induction during the respective cell cycle stage under standard growth conditions as demonstrated by studies with cell cycle-synchronized S. acidocaldarius cultures (29,73). For example, the predicted chromosome partitioning ATPase (parA, SegA, Saci_0204) and its preceding gene Saci_0203 (SegB) (72), which in our transcriptome study both are downregulated more than 4-fold upon UV treatment, were among the very first genes to be induced in the synchronized cultures under standard growth conditions. This confirms their function in genome segregation, the first cell-cycle process to be initiated after release from G2 arrest (29). In addition, the annotated HJ resolvase (Saci_1741) shows a significant downregulation. The biochemical and structural properties and function of these endonucleases, which catalyze the cleavage/resolution of HJs and are of fundamental importance in chromosome segregation, have been described in detail (75,76). One might speculate that the accumulation of HJs, because of the lower abundance of HJ resolvases, may intentionally impair cell-cycle progression to allow DNA repair. The predicted HrpA counterpart Saci_1228 and the neighboring gene Saci_1229, which are strongly repressed upon UV stress in both strains have also been reported to be highly induced at the mitosis/cytokinesis stage in cell cycle-synchronized cultures and a function in cell-cycle progression was proposed (29,73).
Furthermore cdc6-2, which encodes one of the three identified Cdc6s in S. acidocaldarius, was among the early, highly upregulated genes, whereas cdc6-1 was slightly downregulated at 90 min. For cdc6-2, a function as repressor of DNA replication initiation was proposed, whereas Cdc6-1 and Cdc6-3 seem to promote initiation of DNA replication (56). However, our studies indicate that Cdc6-2 might also operate as an inducible transcription factor, like TFB3. Thus, our data show that upon UV stress, mechanisms that inhibit DNA replication and cell division are initiated to allow DNA repair. Notably all these mechanisms are part of the TFB3-independent UV response in S. acidocaldarius and except cdc6-2 all genes are downregulated as part of the delayed UV response. The inhibition of DNA replication following UV-induced DNA damage has been reported previously both for bacteria and eukaryotes (77,78).
Exposure to UV light is accompanied by DNA damage, e.g. misincorporation of bases during the replication process, deamination of bases, depurination and depyrimidination as well as oxidative stress caused by UV radiation-induced free radicals or reactive oxygen species (79,80). Although the most observed consequences of UV radiation are pyrimidine photoproducts, e.g. CPDs, characterized by covalent linkage between adjacent pyrimidines and (6-4) pyrimidone photoproducts (6–4 PPs), several purine photoproducts have also been reported. These include the adenine dimer, the Porschke photoproduct and the T-A photoadduct (79,81,82). Accordingly, we found that processes, which are involved in synthesis and uptake of nucleotides, are strongly affected upon UV treatment in the MW001+pyrEF strain and the tfb3::pyrEF strain. The genes involved in purine biosynthesis and uptake are highly downregulated in the early UV response and two predicted deoxycytidine deaminases for pyrimidine biosynthesis in the delayed UV response. In previous studies the amount of transcripts of the pyrimidine synthesis gene cluster (Saci_1607-Saci_1613) and the uptake system (Saci_0214) have been shown to be coordinately increased in the chromosome replication stage of the cell cycle (S-phase), where high amounts of DNA building blocks are required (29).
Our data show that multiple predicted proteins, which share structural relationships to well-known families of transcriptional regulators (Saci_0478 (XRE family) and Saci_0483 (MarR family) (both annotated conserved conjugative plasmid proteins with HTH motif); Saci_1012 (DtxR family), Saci_1180 (ArnR)), exhibit a strong UV stress response in both investigated MW001 strains. In addition, the expression of the gene encoding the Hanks-type protein kinase ArnC (Saci_1193) is drastically reduced upon UV treatment in both investigated strains suggesting a general regulatory role in cellular UV response.
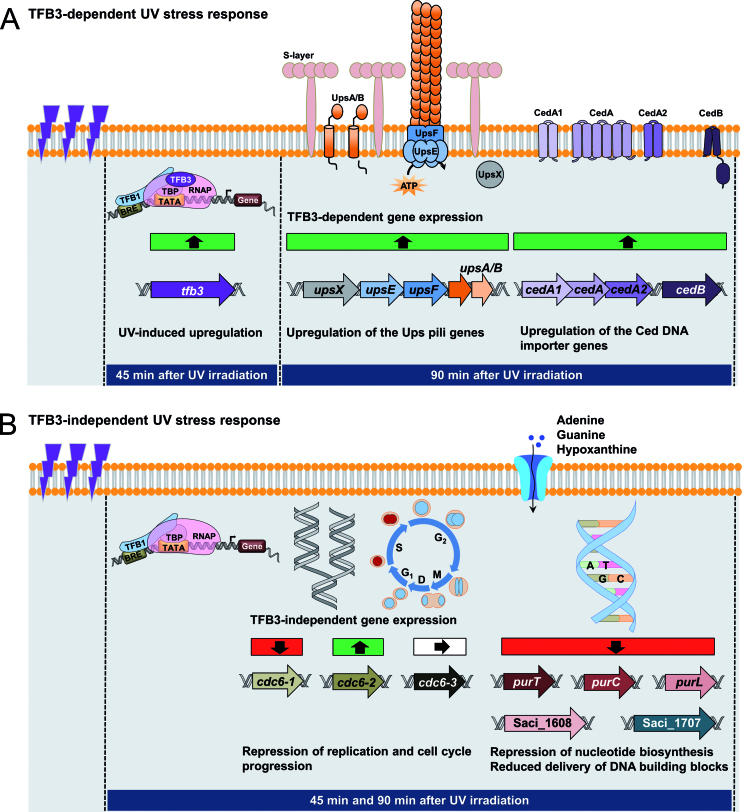
In summary, our results confirm the UV-dependent expression of TFB3 on both protein and gene level. Using the tfb3::pyrEF strain, we identified genes, whose differential gene expression upon UV irradiation depends on the presence of TFB3 and demonstrated the importance of TFB3 in UV-mediated cellular aggregation and DNA exchange (i.e. ups pili operon and the Ced DNA importer) in order to allow for DNA repair via homologous recombination. Our findings confirm TFB3 as a key player and activator of transcription in early UV stress response, since all genes are downregulated in the tfb3::pyrEF strain compared to the MW001+pyrEF strain. However, we observed an additional clear TFB3-independent response that targets major cellular processes resulting in repression of DNA replication, cell-cycle control as well as nucleotide biosynthesis in order to allow for DNA repair (Figure 7).
Figure 7.
Model of early UV stress response in Sulfolobus acidocaldarius. The increased and decreased transcription of genes is depicted by green and red squares, respectively, while white squares represent unchanged transcription. (A) TFB3-dependent UV stress response. 45 min after UV irradiation (100 J/m2), tfb3 is highly up-regulated and acts as an activator of transcription. The delayed (90 min after UV irradiation) response includes the enhanced transcription of the tfb3-dependent target genes (ups genes of the UV inducible pili operon, ced genes of the Crenarchaeal Exchange of DNA importer), leading to increased cellular aggregation and DNA exchange between the cells in order to allow DNA repair via homologous recombination. (B) TFB3-independent response. Apart from the TFB3-dependent response, S. acidocaldarius shows a UV stress response (45 and 90 min after UV irradiation), which does not depend on the presence of TFB3. This is characterized by the repression of DNA replication and cell cycle progression as well as the inhibition of nucleotide biosynthesis, leading to reduced delivery of DNA building blocks. Downregulation of these processes allow the DNA repair, which is mediated by the TFB3-dependent features described above to take place. For detailed discussion see text.
Supplementary Material
ACKNOWLEDGEMENTS
F.S. and B.R. were supported by the German Research Foundation, DFG Graduate Training Program 1431 ‘Transcription, chromatin structure and DNA repair in development and differentiation’. BS, A.A., J.K. A.G. and P.B. acknowledge funding by the Ferderal Ministery of Education and Research (BMBF) within the e:Bio initiative HotSysAPP. A.G. and P.B. acknowledge technical assistance by the Bioinformatics Core Facility/Professorship of Systems Biology at JLU Giessen and access to resources financially supported by the BMBF grant FKZ 031A533 to the BiGi center within the de.NBI network. T.N.L. acknowledges funding by the Minstry of Education and Training, Vietman. S.V.A. acknowledges funding by the European Research councli (ERC starting grant, ARCHAELLUM). We acknowledge support by the Open Access Publication Fund of the University of Duisburg-Essen.
Author Contributions’: B.S. conceived the study. F.S., B.S., S.V.A., A.G. and J.K. designed the experiments. F.S., B.R., T.L., P.B. and A.A. performed the experiments. Computational motif Search P.B., A.G.. F.S. and B.S. wrote the manuscript with input from A.A., T.L., C.D., S.V.A. All authors approved the final manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Research Foundation [DFG Graduate Training Program 1431 to F.S., B.R.]; Federal Ministry of Education and Research (BMBF) [e:Bio initiative, HotSysAPP, 03120078A to B.S., 031L0078C to A.A., J.K., 031L0078D to A.G., P.B., de.NBI network FKZ 031A533 to A.G., P.B.]; Ministry of Education and Training, Vietnam [911 grant to T.N.L.]; European Research council [ERC starting grant, ARCHAELLUM, 311523 to S.V.A.]. Funding for open access charge: University Duisburg-Essen (Bettina Siebers).
Conflict of interest statement. None declared.
REFERENCES
- 1. Langer D., Hain J., Thuriaux P., Zillig W.. Transcription in Archaea: similarity to that in eucarya. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:5768–5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bell S.D., Jackson S.P.. Transcription and translation in Archaea: a mosaic of eukaryal and bacterial features. Trends Microbiol. 1998; 6:222–228. [DOI] [PubMed] [Google Scholar]
- 3. Reiter W.D., Palm P., Zillig W.. Analysis of transcription in the archaebacterium Sulfolobus indicates that archaebacterial promoters are homologous to eukaryotic pol II promoters. Nucleic Acids Res. 1988; 16:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gehring A.M., Walker J.E., Santangelo T.J.. Transcription regulation in Archaea. J. Bacteriol. 2016; 198:1906–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Werner F., Grohmann D.. Evolution of multisubunit RNA polymerases in the three domains of life. Nat. Rev. Microbiol. 2011; 9:85–98. [DOI] [PubMed] [Google Scholar]
- 6. Peeters E., Driessen R.P., Werner F., Dame R.T.. The interplay between nucleoid organization and transcription in archaeal genomes. Nat. Rev. Microbiol. 2015; 13:333–341. [DOI] [PubMed] [Google Scholar]
- 7. Thomm M. Archaeal transcription factors and their role in transcription initiation. FEMS Microbiol. Rev. 1996; 18:159–171. [DOI] [PubMed] [Google Scholar]
- 8. Soppa J. Basal and regulated transcription in archaea. Adv. Appl. Microbiol. 2001; 50:171–217. [DOI] [PubMed] [Google Scholar]
- 9. Bell S.D., Jackson S.P.. Mechanism and regulation of transcription in archaea. Curr. Opin. Microbiol. 2001; 4:208–213. [DOI] [PubMed] [Google Scholar]
- 10. Geiduschek E.P., Ouhammouch M.. Archaeal transcription and its regulators. Mol. Microbiol. 2005; 56:1397–1407. [DOI] [PubMed] [Google Scholar]
- 11. Bell S.D., Jackson S.P.. The role of transcription factor B in transcription initiation and promoter clearance in the archaeon Sulfolobus acidocaldarius. J. Biol. Chem. 2000; 275:12934–12940. [DOI] [PubMed] [Google Scholar]
- 12. Bell S.D., Jackson S.P.. Transcription in archaea. Cold Spring Harb. Symp. Quant. Biol. 1998; 63:41–51. [DOI] [PubMed] [Google Scholar]
- 13. Bell S.D., Brinkman A.B., van der Oost J., Jackson S.P.. The archaeal TFIIEalpha homologue facilitates transcription initiation by enhancing TATA-box recognition. EMBO Rep. 2001; 2:133–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hickey A.J., Conway de Macario E., Macario A.J.. Transcription in the archaea: basal factors, regulation, and stress gene expression. Crit. Rev. Biochem. Mol. Biol. 2002; 37:199–258. [DOI] [PubMed] [Google Scholar]
- 15. Grohmann D., Nagy J., Chakraborty A., Klose D., Fielden D., Ebright R.H., Michaelis J., Werner F.. The initiation factor TFE and the elongation factor Spt4/5 compete for the RNAP clamp during transcription initiation and elongation. Mol. Cell. 2011; 43:263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Blombach F., Ausiannikava D., Figueiredo A.M., Soloviev Z., Prentice T., Zhang M., Zhou N., Thalassinos K., Allers T., Werner F.. Structural and functional adaptation of Haloferax volcanii TFEalpha/beta. Nucleic Acids Res. 2018; 46:2308–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Burton S.P., Burton Z.F.. The sigma enigma: bacterial sigma factors, archaeal TFB and eukaryotic TFIIB are homologs. Transcription. 2014; 5:e967599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Coker J.A., DasSarma S.. Genetic and transcriptomic analysis of transcription factor genes in the model halophilic Archaeon: coordinate action of TbpD and TfbA. BMC Genet. 2007; 8:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kaur A., Van P.T., Busch C.R., Robinson C.K., Pan M., Pang W.L., Reiss D.J., DiRuggiero J., Baliga N.S.. Coordination of frontline defense mechanisms under severe oxidative stress. Mol. Syst. Biol. 2010; 6:393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Turkarslan S., Reiss D.J., Gibbins G., Su W.L., Pan M., Bare J.C., Plaisier C.L., Baliga N.S.. Niche adaptation by expansion and reprogramming of general transcription factors. Mol. Syst. Biol. 2011; 7:554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shockley K.R., Ward D.E., Chhabra S.R., Conners S.B., Montero C.I., Kelly R.M.. Heat shock response by the hyperthermophilic archaeon Pyrococcus furiosus. Appl. Environ. Microbiol. 2003; 69:2365–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen L., Brugger K., Skovgaard M., Redder P., She Q., Torarinsson E., Greve B., Awayez M., Zibat A., Klenk H.P. et al. . The genome of Sulfolobus acidocaldarius, a model organism of the Crenarchaeota. J. Bacteriol. 2005; 187:4992–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. She Q., Singh R.K., Confalonieri F., Zivanovic Y., Allard G., Awayez M.J., Chan-Weiher C.C., Clausen I.G., Curtis B.A., De Moors A. et al. . The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:7835–7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhu W., Zeng Q., Colangelo C.M., Lewis M., Summers M.F., Scott R.A.. The N-terminal domain of TFIIB from Pyrococcus furiosus forms a zinc ribbon. Nat. Struct. Biol. 1996; 3:122–124. [DOI] [PubMed] [Google Scholar]
- 25. Littlefield O., Korkhin Y., Sigler P.B.. The structural basis for the oriented assembly of a TBP/TFB/promoter complex. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:13668–13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Qureshi S.A., Bell S.D., Jackson S.P.. Factor requirements for transcription in the Archaeon Sulfolobus shibatae. EMBO J. 1997; 16:2927–2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bell S.D., Cairns S.S., Robson R.L., Jackson S.P.. Transcriptional regulation of an archaeal operon in vivo and in vitro. Mol. Cell. 1999; 4:971–982. [DOI] [PubMed] [Google Scholar]
- 28. Bini E., Dikshit V., Dirksen K., Drozda M., Blum P.. Stability of mRNA in the hyperthermophilic archaeon Sulfolobus solfataricus. RNA. 2002; 8:1129–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lundgren M., Bernander R.. Genome-wide transcription map of an archaeal cell cycle. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:2939–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Frols S., Gordon P.M., Panlilio M.A., Duggin I.G., Bell S.D., Sensen C.W., Schleper C.. Response of the hyperthermophilic archaeon Sulfolobus solfataricus to UV damage. J. Bacteriol. 2007; 189:8708–8718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gotz D., Paytubi S., Munro S., Lundgren M., Bernander R., White M.F.. Responses of hyperthermophilic crenarchaea to UV irradiation. Genome Biol. 2007; 8:R220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Paytubi S., White M.F.. The crenarchaeal DNA damage-inducible transcription factor B paralogue TFB3 is a general activator of transcription. Mol. Microbiol. 2009; 72:1487–1499. [DOI] [PubMed] [Google Scholar]
- 33. Brock T.D., Brock K.M., Belly R.T., Weiss R.L.. Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Microbiol. 1972; 84:54–68. [DOI] [PubMed] [Google Scholar]
- 34. Wagner M., van Wolferen M., Wagner A., Lassak K., Meyer B.H., Reimann J., Albers S.V.. Versatile genetic tool box for the crenarchaeote Sulfolobus acidocaldarius. Front. Microbiol. 2012; 3:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Orell A., Peeters E., Vassen V., Jachlewski S., Schalles S., Siebers B., Albers S.V.. Lrs14 transcriptional regulators influence biofilm formation and cell motility of Crenarchaea. ISME J. 2013; 7:1886–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ajon M., Frols S., van Wolferen M., Stoecker K., Teichmann D., Driessen A.J., Grogan D.W., Albers S.V., Schleper C.. UV-inducible DNA exchange in hyperthermophilic archaea mediated by type IV pili. Mol. Microbiol. 2011; 82:807–817. [DOI] [PubMed] [Google Scholar]
- 37. Wagner M., Wagner A., Ma X., Kort J.C., Ghosh A., Rauch B., Siebers B., Albers S.V.. Investigation of the malE promoter and MalR, a positive regulator of the maltose regulon, for an improved expression system in Sulfolobus acidocaldarius. Appl. Environ. Microbiol. 2014; 80:1072–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976; 72:248–254. [DOI] [PubMed] [Google Scholar]
- 39. Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970; 227:680–685. [DOI] [PubMed] [Google Scholar]
- 40. Hottes A.K., Meewan M., Yang D., Arana N., Romero P., McAdams H.H., Stephens C.. Transcriptional profiling of Caulobacter crescentus during growth on complex and minimal media. J. Bacteriol. 2004; 186:1448–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hilker R., Stadermann K.B., Doppmeier D., Kalinowski J., Stoye J., Straube J., Winnebald J., Goesmann A.. ReadXplorer–visualization and analysis of mapped sequences. Bioinformatics. 2014; 30:2247–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B.. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008; 5:621–628. [DOI] [PubMed] [Google Scholar]
- 44. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Koster J., Rahmann S.. Snakemake–a scalable bioinformatics workflow engine. Bioinformatics. 2012; 28:2520–2522. [DOI] [PubMed] [Google Scholar]
- 46. Crooks G.E., Hon G., Chandonia J.M., Brenner S.E.. WebLogo: a sequence logo generator. Genome Res. 2004; 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vassart A., Van Wolferen M., Orell A., Hong Y., Peeters E., Albers S.V., Charlier D.. Sa-Lrp from Sulfolobus acidocaldarius is a versatile, glutamine-responsive, and architectural transcriptional regulator. Microbiologyopen. 2013; 2:75–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Frols S., Ajon M., Wagner M., Teichmann D., Zolghadr B., Folea M., Boekema E.J., Driessen A.J., Schleper C., Albers S.V.. UV-inducible cellular aggregation of the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by pili formation. Mol. Microbiol. 2008; 70:938–952. [DOI] [PubMed] [Google Scholar]
- 49. van Wolferen M., Ajon M., Driessen A.J., Albers S.V.. Molecular analysis of the UV-inducible pili operon from Sulfolobus acidocaldarius. Microbiologyopen. 2013; 2:928–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. van Wolferen M., Ma X., Albers S.V.. DNA processing proteins involved in the UV-induced stress response of Sulfolobales. J. Bacteriol. 2015; 197:2941–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. van Wolferen M., Wagner A., van der Does C., Albers S.V.. The archaeal Ced system imports DNA. PNAS. 2016; 113:2496–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Grishin A.M., Ajamian E., Tao L., Zhang L., Menard R., Cygler M.. Structural and functional studies of the Escherichia coli phenylacetyl-CoA monooxygenase complex. J. Biol. Chem. 2011; 286:10735–10743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dassama L.M., Krebs C., Bollinger J.M. Jr., Rosenzweig A.C., Boal A.K.. Structural basis for assembly of the Mn(IV)/Fe(III) cofactor in the class Ic ribonucleotide reductase from Chlamydia trachomatis. Biochemistry. 2013; 52:6424–6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tomita H., Yokooji Y., Ishibashi T., Imanaka T., Atomi H.. Biochemical characterization of pantoate kinase, a novel enzyme necessary for coenzyme A biosynthesis in the Archaea. J. Bacteriol. 2012; 194:5434–5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kasiviswanathan R., Shin J.H., Kelman Z.. Interactions between the archaeal Cdc6 and MCM proteins modulate their biochemical properties. Nucleic. Acids. Res. 2005; 33:4940–4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Robinson N.P., Dionne I., Lundgren M., Marsh V.L., Bernander R., Bell S.D.. Identification of two origins of replication in the single chromosome of the archaeon Sulfolobus solfataricus. Cell. 2004; 116:25–38. [DOI] [PubMed] [Google Scholar]
- 57. Barragan M.J., Blazquez B., Zamarro M.T., Mancheno J.M., Garcia J.L., Diaz E., Carmona M.. BzdR, a repressor that controls the anaerobic catabolism of benzoate in Azoarcus sp. CIB, is the first member of a new subfamily of transcriptional regulators. J. Biol. Chem. 2005; 280:10683–10694. [DOI] [PubMed] [Google Scholar]
- 58. Perera I.C., Grove A.. Molecular mechanisms of ligand-mediated attenuation of DNA binding by MarR family transcriptional regulators. J. Mol. Cell Biol. 2010; 2:243–254. [DOI] [PubMed] [Google Scholar]
- 59. Fiorentino G., Del Giudice I., Bartolucci S., Durante L., Martino L., Del Vecchio P.. Identification and physicochemical characterization of BldR2 from Sulfolobus solfataricus, a novel archaeal member of the MarR transcription factor family. Biochemistry. 2011; 50:6607–6621. [DOI] [PubMed] [Google Scholar]
- 60. Albers S.V., Jarrell K.F.. The archaellum: how Archaea swim. Front. Microbiol. 2015; 6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Esser D., Hoffmann L., Pham T.K., Brasen C., Qiu W., Wright P.C., Albers S.V., Siebers B.. Protein phosphorylation and its role in archaeal signal transduction. FEMS Microbiol. Rev. 2016; 40:625–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Haurat M.F., Figueiredo A.S., Hoffmann L., Li L., Herr K., A J.W., Beeby M., Schaber J., Albers S.V.. ArnS, a kinase involved in starvation-induced archaellum expression. Mol. Microbiol. 2017; 103:181–194. [DOI] [PubMed] [Google Scholar]
- 63. Yamamoto K., Ishihama A., Busby S.J., Grainger D.C.. The Escherichia coli K-12 MntR miniregulon includes dps, which encodes the major stationary-phase DNA-binding protein. J. Bacteriol. 2011; 193:1477–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Que Q., Helmann J.D.. Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol. Microbiol. 2000; 35:1454–1468. [DOI] [PubMed] [Google Scholar]
- 65. Pandey R., Russo R., Ghanny S., Huang X., Helmann J., Rodriguez G.M.. MntR(Rv2788): a transcriptional regulator that controls manganese homeostasis in Mycobacterium tuberculosis. Mol. Microbiol. 2015; 98:1168–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Leyn S.A., Rodionov D.A.. Comparative genomics of DtxR family regulons for metal homeostasis in Archaea. J. Bacteriol. 2015; 197:451–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Reimann J., Lassak K., Khadouma S., Ettema T.J., Yang N., Driessen A.J., Klingl A., Albers S.V.. Regulation of archaella expression by the FHA and von Willebrand domain-containing proteins ArnA and ArnB in Sulfolobus acidocaldarius. Mol. Microbiol. 2012; 86:24–36. [DOI] [PubMed] [Google Scholar]
- 68. Hoffmann L., Schummer A., Reimann J., Haurat M.F., Wilson A.J., Beeby M., Warscheid B., Albers S.V.. Expanding the archaellum regulatory network—the eukaryotic protein kinases ArnC and ArnD influence motility of Sulfolobus acidocaldarius. Microbiologyopen. 2017; 6:e00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cecchetto G., Amillis S., Diallinas G., Scazzocchio C., Drevet C.. The AzgA purine transporter of Aspergillus nidulans. Characterization of a protein belonging to a new phylogenetic cluster. J. Biol. Chem. 2004; 279:3132–3141. [DOI] [PubMed] [Google Scholar]
- 70. Bjornberg O., Neuhard J., Nyman P.O.. A bifunctional dCTP deaminase-dUTP nucleotidohydrolase from the hyperthermophilic archaeon Methanocaldococcus jannaschii. J. Biol. Chem. 2003; 278:20667–20672. [DOI] [PubMed] [Google Scholar]
- 71. Johansson E., Bjornberg O., Nyman P.O., Larsen S.. Structure of the bifunctional dCTP deaminase-dUTPase from Methanocaldococcus jannaschii and its relation to other homotrimeric dUTPases. J. Biol. Chem. 2003; 278:27916–27922. [DOI] [PubMed] [Google Scholar]
- 72. Kalliomaa-Sanford A.K., Rodriguez-Castaneda F.A., McLeod B.N., Latorre-Rosello V., Smith J.H., Reimann J., Albers S.V., Barilla D.. Chromosome segregation in Archaea mediated by a hybrid DNA partition machine. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:3754–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bernander R., Lundgren M., Ettema T.J.. Comparative and functional analysis of the archaeal cell cycle. Cell Cycle. 2010; 9:794–806. [PubMed] [Google Scholar]
- 74. Le T.N., Wagner A., Albers S.V.. A conserved hexanucleotide motif is important in UV-inducible promoters in Sulfolobus acidocaldarius. Microbiology. 2017; 163:778–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wyatt H.D., West S.C.. Holliday junction resolvases. Cold Spring Harb. Perspect. Biol. 2014; 6:a023192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Agmon N., Yovel M., Harari Y., Liefshitz B., Kupiec M.. The role of Holliday junction resolvases in the repair of spontaneous and induced DNA damage. Nucleic Acids Res. 2011; 39:7009–7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Verma M., Moffat K.G., Egan J.B.. UV irradiation inhibits initiation of DNA replication from oriC in Escherichia coli. Mol. Gen. Genet. 1989; 216:446–454. [DOI] [PubMed] [Google Scholar]
- 78. Edenberg H.J. Inhibition of DNA replication by ultraviolet light. Biophys. J. 1976; 16:849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Rastogi R.P., Richa K.A., Tyagi M.B., Sinha R.P.. Molecular mechanisms of ultraviolet radiation-induced DNA damage and repair. J. Nucleic Acids. 2010; 2010:592980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. de Jager T.L., Cockrell A.E., Du Plessis S.S.. Ultraviolet light induced generation of reactive oxygen species. Adv. Exp. Med. Biol. 2017; 996:15–23. [DOI] [PubMed] [Google Scholar]
- 81. Duker N.J., Gallagher P.E.. Purine photoproducts. Photochem. Photobiol. 1988; 48:35–39. [PubMed] [Google Scholar]
- 82. Cadet J., Sage E., Douki T.. Ultraviolet radiation-mediated damage to cellular DNA. Mutat. Res. 2005; 571:3–17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.