Abstract
Small-molecule compounds that target mismatched base pairs in DNA offer a novel prospective for cancer diagnosis and therapy. The potent anticancer antibiotic echinomycin functions by intercalating into DNA at CpG sites. Surprisingly, we found that the drug strongly prefers to bind to consecutive CpG steps separated by a single T:T mismatch. The preference appears to result from enhanced cooperativity associated with the binding of the second echinomycin molecule. Crystallographic studies reveal that this preference originates from the staggered quinoxaline rings of the two neighboring antibiotic molecules that surround the T:T mismatch forming continuous stacking interactions within the duplex. These and other associated changes in DNA conformation allow the formation of a minor groove pocket for tight binding of the second echinomycin molecule. We also show that echinomycin displays enhanced cytotoxicity against mismatch repair-deficient cell lines, raising the possibility of repurposing the drug for detection and treatment of mismatch repair-deficient cancers.
INTRODUCTION
DNA bases can be damaged by various mechanisms, such as errors in DNA replication, heteroduplex formation during homologous recombination, spontaneous deamination of cytosine, and base damage by mutagens as well as ionising radiation (1). Such damage may alter the interaction between DNA bases, resulting in incorrect base pairing and mutations. There are eight non-Watson–Crick alternatives, or ‘mismatches’, which include the purine-pyrimidine G:T and A:C pairings, the purine-purine G:G, A:A and G:A pairings, and the pyrimidine-pyrimidine C:C, T:T and C:T mismatches (2–4). These mismatched base pairs occasionally occur in the DNA duplex during the replication process, but are quickly corrected by a mismatch repair system because they could be potentially mutagenic as well as causing local structural deformations (5,6). Deficiencies in the mismatch repair system increase the mutation rate and consequently the risk of cancer (7,8). Given the pleiotropic nature of carcinogenesis and the steady march towards targeted therapy, an ability to pinpoint whether a particular cancer arises from a deficiency of the mismatch repair system or from other causes could be valuable in cancer diagnosis and therapy. Small ligands capable of recognising mismatches within the DNA may thus become important tools for research and even for clinical purposes (9–16).
Echinomycin (Figure 1A) is a natural depsipeptide produced by Streptomyces echinatus with notable antibiotic activity. It consists of two quinoxaline rings linked to a cross-bridged cyclic octapeptide dilactone (17). Echinomycin has been shown to inhibit topoisomerase II, DNA helicase, and partially inhibit DNA methyltransferase 1 in vitro (18–20). It has also been shown to disrupt the function of hypoxia-inducible factor-1α (HIF-1α) and may act as a potent HIF-1α inhibitor (21). The biological activity of echinomycin is related to its ability to interfere with DNA replication and transcription by bis-intercalation of the two quinoxaline rings into the DNA duplex, with a preference for CpG sites (22,23). Structural studies have shown that its CpG binding preference can be attributed to hydrogen bonding between the alanine residues of echinomycin and the guanine base in CpG. Sequences flanking the CpG site are also known to influence the binding affinity of the antibiotic for the DNA duplex (22,24). In the present study, we found that echinomycin preferred to bind to consecutive CpG sequences harbouring a single T:T mismatch at the centre compared to other mismatched and Watson–Crick base pairs. We also solved the crystal structures of echinomycin-d(ACGTCG(5-BrU))2 and echinomycin-d(ACGACGT)/d(ACGTCGT) in order to investigate the structural basis behind this preference. Binding assays in vitro together with cell-based proliferation tests suggest that the binding affinity may correlate with inhibition of growth in cancer cell lines. Our results provide valuable information on the mechanism of echinomycin binding to T:T mismatches that could lead to potential applications in cancer diagnosis as well as the development of a novel therapeutic protocol.
Figure 1.
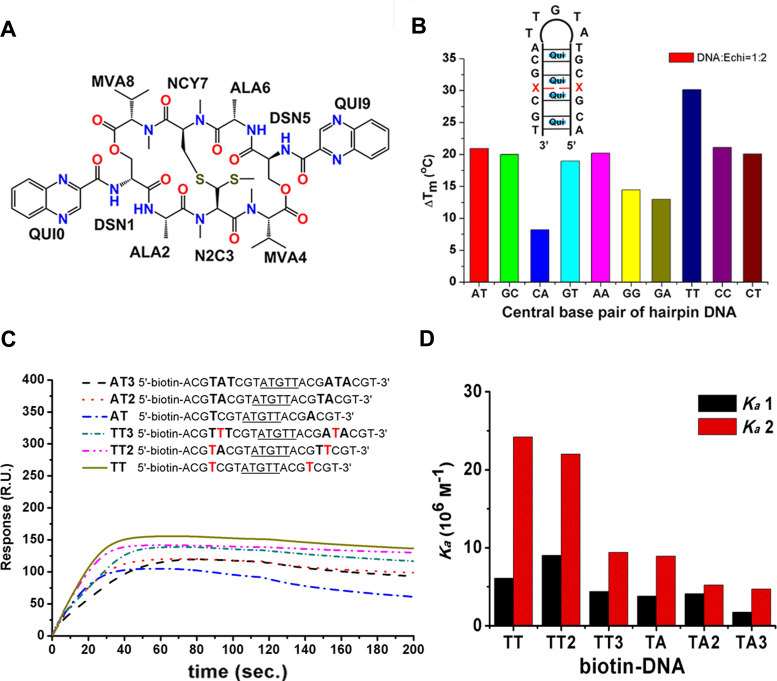
The stabilising effects and binding affinity of echinomycin to DNA duplexes. (A) Chemical structure of echinomycin. The abbreviations as shown in the figure represent D-serine (DSN), l-alanine (ALA), N-dimethyl-l-cysteine (N2C), N-methyl-l-cysteine (NCY), N-methyl-l-valine (MVA) and quinoxaline (QUI), respectively. The numbers indicate the order of the cyclic pentapeptide and quinoxaline rings. (B) The effects of echinomycin on the Tm values of various hairpin DNA fragments shown in the schematic diagram, where ‘X’ indicates the bases A, T, C or G that compose various central base pairs, including Watson–Crick or mismatch base pairing, measured in PBE buffer with DNA and echinomycin at 1:2 molar ratio. (C) Binding affinity analysis of echinomycin–DNA complexes by SPR. Sensorgrams of the binding of antibiotic to the immobilized 5′ biotin-labelled hairpin DNAs illustrated in the schematic. Underlining indicates the hairpin loop, with capital letters showing the bases between the two CpG steps. The reactions were carried out in 50 mM sodium cacodylate buffer at pH 7. The resonance unit (RU) is defined as 1 RU = 1 pg/mm2. The dissociation phase starts at 120 s. (D) Bar chart representing the association equilibrium constants (Ka1 and Ka2) for echinomycin binding to the immobilized hairpin DNAs.
MATERIALS AND METHODS
Drug and oligonucleotides
All chemicals used were of reagent grade and were obtained from Sigma Chemical Co. (MO, USA). The PAGE-purified synthetic DNA oligomers were purchased from Genomics (New Taipei City, Taiwan). Absorbance measurements to determine echinomycin and oligonucleotide concentrations were performed at 325 and 260 nm, respectively, using a V-630 UV-Vis spectrophotometer (JASCO, MD, USA) with a 1 cm path length quartz cuvette. The concentration of echinomycin was calculated from the optical density (ϵ325 nm = 11 500 M−1 cm−1) (25), and oligomer extinction coefficients were calculated on the basis of the tabulated values for monomer and dimer extinction coefficients (26) at the IDT Biophysics website (http://biophysics.idtdna.com/).
Melting temperature (Tm) experiment
The Tm values of the DNA duplexes were analysed as previously described using a JASCO UV/VIS spectrophotometer by monitoring the sample absorption at 260 nm (27). The hairpin DNAs (illustrated in Figure 1B) were dissolved in BPE buffer (6 mM Na2HPO4, 2 mM NaH2PO4 and 1 mM Na2EDTA, pH 7.0), then heated at 95°C for 5 min and reannealed by cooling on ice for 30 min. To form the echinomycin–DNA complex, 3 μM hairpin DNA was incubated with echinomycin at 1:0 and 1:2 molar ratios at 4°C overnight. UV melting curves of the hairpin DNA was obtained by recording absorbance at 260 nm every 30 s while the temperature was increased from 4 to 95°C at a rate of 1°C min−1. The Tm values were obtained from the observed curves using polynomial curve fitting by Varian Cary WinUV Thermal application software (version 3.00).
Surface plasmon resonance (SPR) analysis
The affinity, association and dissociation between the ligand and the DNA duplexes were measured using a BIAcore™ T100 SPR instrument (BIAcore, Uppsala, Sweden) with a S-series sensor chip SA from GE Life Sciences that monitored changes in the refractive index at the sensor chip's surface. The sequences of the hairpin DNAs used in the SPR experiment are illustrated in Figure 1C. The 5′-biotinylated hairpin DNAs were immobilised on the sensor chip (GE Healthcare, Uppsala, Sweden) until 400 resonance units (RU) was reached. Echinomycin were prepared in 50 mM sodium cacodylate buffer, pH 7.0 and were passed over the surface of the chip for 120 s at a flow rate of 50 μl min−1. The buffer solution described above was then passed over the chip for an additional 600 s. Finally, the chip was regenerated with a solution of 1000 mM NaCl and 50 mM NaOH. The kinetic data for the association and dissociation phases were analysed to allow a reasonable range of binding constants using a bivalent ligand model created in the BIAevaluation software (version 3.0) as described previously (28). This model, shown below, usually considers cooperative effects and achieves an acceptable fit when the chi-square value is <5.
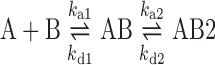 |
Crystallization
To obtain the echinomycin–TT complex crystals, the d(ACGTCG(5-BrU))2 DNA oligomer was co-crystallized with echinomycin in 1:2 molar ratios. Initially, 0.25 mM single-strand DNA was heated at 95°C for 5 min, reannealed by cooling on ice for 5 min to form duplex DNA, and then incubated with 0.275 mM echinomycin at 4°C for 3 days. Tiny rod-shaped crystals were obtained within 2–7 days in a 5 μl drop containing 20 mM MES (pH 6.0), 10 mM MgCl2, 2 mM spermine 4 HCl, 2% 2-methyl-2,4-pentanediol (MPD), 10 mM MnSO4 and 10 mM KBr by equilibrating at 20°C against 500 μl of 30% MPD reservoir solution using the sitting-drop vapor diffusion technique. To obtain the echinomycin–AT DNA complex crystals, 0.125 mM of d(ACGACGT/ACGTCGT) DNA oligomer was co-crystallized with echinomycin at a 1:2 molar ratio. Crystals of the echinomycin–AT complex were obtained within 2 days (diamond shaped) in the same condition described above.
X-ray data collection, phasing, and structure refinement
X-ray diffraction data of the echinomycin–TT and echinomycin–AT complexes were collected at beamlines TPS 05A and BL15A1 of the National Synchrotron Radiation Research Center (Hsinchu, Taiwan), respectively. Diffraction data integration and data reduction were processed with the HKL-2000 program package (29). The phase of the echinomycin–TT complex structure was determined by molecular replacement with Phaser MR in CCP4i (version 7.0.012) (30) using the partial structure of the Echi-d(ACGTACGT)2 complex (PDB ID: 3go3) as a template. The initial model was built with the molecular graphics program MIFit (version 2010.10) and WinCoot (version 0.8.4). The refined echinomycin–TT complex structure was used as a template to solve the phase of the echinomycin–AT complex structure. Structure refinements were performed by phenix.refine in PHENIX (version 1.10.1) (31) and Refmac5 in CCP4i (32), and the crystallographic and refinement statistics of the echinomycin–DNA complexes are listed in Supplementary Table S1. The final 2Fo – Fc electron density maps were created with the fast Fourier transform (FFT) in CCP4i (33) and PyMOL (version 1.8) was used to draw the graphical representations of the refined structures. DNA helical parameters were analysed with Web 3DNA (34) and the CURVES+ program (35). The hydrophobic interaction between DNA and echinomycin was analysed by LigPlot+ (36).
Cell viability assay
Human MMR-deficient (Hct116) and MMR-proficient (Hct116+ch3) colorectal cancer cell lines were kindly provided by Dr. Kunkel (NIEHS/NIH). The Hct116 cell line with a hemizygous mutation of hMLH1 results in dysfunction of DNA mismatch repair (MMR), and the MMR function of the Hct116+ch3 cell line is complete because it is complemented by chromosome 3, which contains the wild-type gene of hMLH1. The SW620 colon carcinoma cell line, which has normal MMR activity, was used for comparison. Briefly, cells were maintained in DMEM/F12 and DMEM medium containing 10% foetal bovine serum (FBS) for Hct116 and SW620, respectively, and DMEM/F12 with the addition of 400 μg/ml G418 (Geneticin) for Hct116+ch3, at 37°C in a humidified atmosphere containing 5% CO2. 1 × 105 cells were seeded into 96-well culture plates and incubated for 24 hrs. Echinomycin was dissolved in the fresh DMEM/F12 medium and added to the wells at final concentrations in the range 0–100 nM after the original culture medium was removed. After an additional incubation for 24 or 48 h, the anticancer efficiency of echinomycin in cancer cell lines with varying MMR status was evaluated by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay as described previously (37). The LD50 value, the concentration of echinomycin that caused the 50% death of cancer cells, was evaluated with more than three independent experiments.
RESULTS
Preferential binding to consecutive CpG sites separated by a T:T mismatch
The stabilising effects of echinomycin on non-canonical duplex structures containing two consecutive CpG sites separated by one mismatched base pair were determined by measuring the difference in the melting temperature (ΔTm) of a hairpin DNA (Figure 1B) at fixed echinomycin:DNA stoichiometry of 2:1. The stoichiometry was chosen to provide the best signal-to-noise ratio. Remarkably, the binding of echinomycin to hairpin DNA containing a single T:T mismatch dramatically increased the Tm by 30°C compared to increases of ∼12–21°C for a Watson–Crick base pair as well as most other mismatch sequences. The weakest stabilization effect was observed for the A:C mismatch with a ΔTm of 8°C. These results indicate that in the context of consecutive CpG binding sites separated by a single base pair, the T:T mismatch provides an environment far more conducive to echinomycin binding compared to other base pairings at the same position.
To characterise further the affinity and cooperativity of echinomycin binding to CpG sites with a T:T mismatch, we compared the surface plasmon resonance (SPR) sensorgrams for binding of echinomycin to hairpins containing two CpG sites separated by the mismatch (TT), a single A:T Watson–Crick base pair (AT), two A:T base pairs (AT2), three A:T base pairs (AT3), one T:T and one A:T (TT2), and one T:T sandwiched between two A:T base pairs (TT3) (Figure 1C). The sensorgrams were fitted to a 2-to-1 binding model characterised by two equilibrium association constants (Ka1 and Ka2, Figure 1D). The Ka1 values for the binding of echinomycin to the various sequences were segregated into two groups: the ones which contained the T:T mismatch (TT, TT2 and TT3) generally had higher affinities than those without (AT, AT2 and AT3). This suggests that echinomycin does possess an intrinsic preference towards T:T mismatches even during the initial binding event. The same trend was observed for Ka2. The Ka2/Ka1 ratio, an indicator of cooperativity, ranged from ∼4 for TT to ∼1.2 for AT2. With the exception of the TT3/AT3 pair, sequences containing a T:T mismatch generally had higher binding cooperativity than those containing a standard Watson–Crick base pair. The comparable cooperativity of TT3 and AT3 may be due to the position of the T:T mismatch in TT3, which is not immediately adjacent to any CpG site. Increasing the distance between the two CpG sites generally led to diminished binding affinity and cooperativity (compare AT with AT2 and AT3; TT with TT2 and TT3), suggesting that both the number and sequence of the base pairs separating the two CpG sites affected the binding behavior of echinomycin towards these sequences.
Binding of echinomycin forces deformation of the T:T mismatch DNA duplex
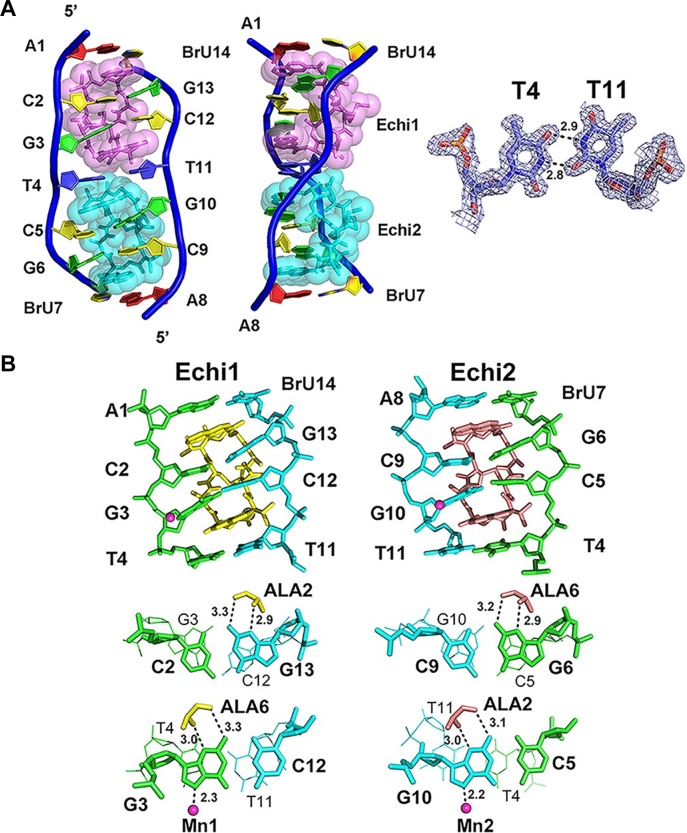
To understand the structural basis behind the preference of echinomycin for T:T mismatches, we solved the crystal structure of echinomycin complexed to the palindromic duplex d(ACGTCG(5-BrU))2 DNA, denoted TT, at 1.55 Å resolution (Supplementary Table S1). Each asymmetric unit contains only one DNA duplex bound by two echinomycin molecules, which are labeled Echi1 (pink) and Echi2 (cyan) in Figure 2A. We also observed several metal ions which stabilised the crystal packing of the complex (Supplementary Figure S1A). Two Mn2+ ions, each with ∼50% occupancy, were associated with N7 of G3, G10 of a symmetry-related molecule as well as four waters with perfect octahedral geometry. One K+ cation was found to interact with two symmetry-related echinomycin molecules at the oxygen atoms of MVA4 and N2C3, again including four water molecules with octa-coordination. The sheared T:T mismatch separating the two CpG sites adopts a wobble configuration, creating a cavity at the GpT step which allows the N-methyl group of Echi1 MVA4 to fit snugly in (Figure 2A). The rest of the base pairs retained the standard Watson–Crick pairings. The two planar quinoxaline rings of each echinomycin molecule intercalate on both sides flanking the CpG steps in the minor groove. The alanine residues within the cyclic depsipeptide ring of each echinomycin form hydrogen bonds to the guanine bases, resulting in expansion of the minor groove width and forcing the DNA backbone to unwind at the CpG steps, whilst the DNA duplex still retains a right-handed helical twist. The neighboring quinoxaline rings of Echi1 and Echi2 on both sides of the T:T mismatch are staggered relative to each other to avoid steric hindrance. These quinoxaline rings are stacked with the thymine bases of the sheared T:T base pair and guanine bases of the flanking G:C base pairs to form stable and continuous stacking interactions, distorting the DNA backbone and forcing it to assume an S-shape.
Figure 2.
Crystal structure of the echinomycin–TT duplex. (A) The asymmetric unit of the echinomycin–TT complex structure in front (left) and side (middle) views. Two echinomycin molecules bind to a duplex DNA containing one T:T mismatch. The 2Fo-Fc electron density of the T:T mismatch in the refined structure is shown at right, contoured at 1.0 σ. Hydrogen bonds are represented by dotted lines, with numbers indicating the distance between the two contributing atoms in Ångströms. (B) Skeletal models showing the antibiotic binding sites in the echinomycin–TT complex. The diagrams illustrate the stacking interactions and hydrogen bonding at the CpG steps. Mn2+ ions are shown as magenta spheres. Dotted lines represent the direct interactions between atoms, and numbers show the distances.
Structural details of the echinomycin binding sites
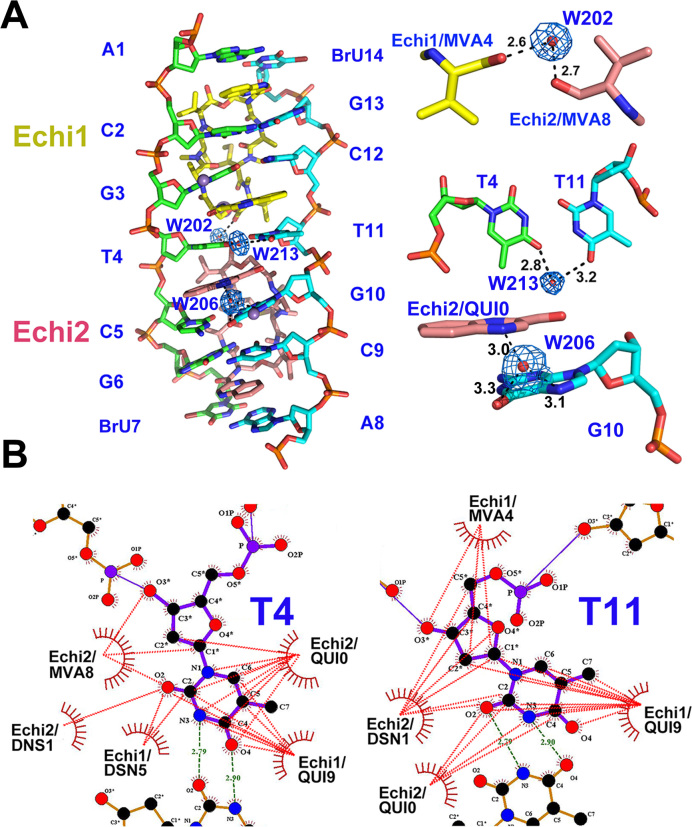
Close-up views of the echinomycin binding sites are shown in Figure 2B. The two palindromic halves of the echinomycin–TT complex that contain the d(ACGT)2 tetranucleotide sequence are related by a local 2-fold symmetry with a root mean square deviation (r.m.s.d.) of 0.136 Å between the heavy atoms. The strong intermolecular hydrogen bonds between N2/N3 of G3 (G13) and CO/NH of Echi1 ALA6 (ALA2), N2/N3 of G6 (G10) and CO/NH of Echi2 ALA6 (ALA2) explain the strong sequence preference for two adjacent 3′-G nucleotides from opposite strands in the DNA duplex, which are prominent features of CpG steps in the duplex. Three water molecules are involved in stabilizing interactions within the echinomycin–TT complex (Figure 3A). One, W202, acts as a bridge between MVA4 of Echi1 and MVA8 of Echi2. The second, W213, stabilizes the interactions between the two thymine bases within the T:T mismatch, and the last water molecule, W206, mediates the interaction between the N4 of Echi2 QUI0 and O6/N7 of G10. LigPlot+ analysis showed that the QUI, DNS and MVA residues of both Echi1 and Echi2 are in close van der Waals contact with the two thymine bases of the T:T wobble base pair, which may contribute towards the specific recognition of T:T mismatches by echinomycin (Figure 3B). The two neighbouring echinomycin molecules in the minor groove also form van der Waals contacts with each other through the carbonyl groups of their respective MVA moieties which would further stabilize the complex.
Figure 3.
Water-mediated and the hydrophobic interactions in the echinomycin–TT complex. (A) Overall structure (left) and close-up views (right) showing the water molecules that directly mediate the hydrogen-bonding interactions in the echinomycin–TT complex. The Mn2+ ions and water molecules are shown as purple and red spheres, respectively. The 2Fo – Fc electron density map is contoured at 1.0 σ to show the coordinated waters in the refined structure. Dashed lines show the direct hydrogen bonds between atoms, with the distance in Ångströms. (B) LigPlot+ 2D plots to illustrate the hydrophobic interaction (red dashed lines) between echinomycin molecules and the T4 (T11) base in the complex.
Comparison with the echinomycin–DNA complex containing only Watson–Crick base pairs
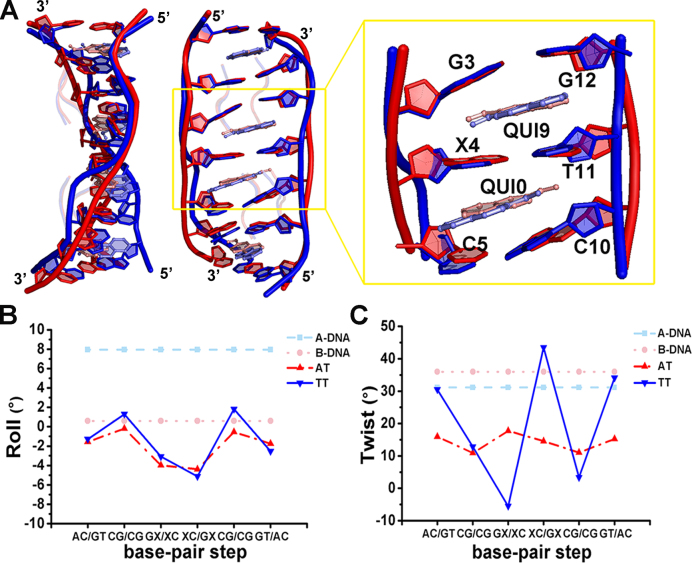
The crystal structure of echinomycin in complex with the palindromic DNA duplex harbouring one A:T base pair between two CpG sequences, d(ACGACGT)/d(ACGTCGT), denoted by AT, at 1.58 Å resolution was solved for comparison (Supplementary Figure S2A). Just as in echinomycin–TT, each asymmetric unit of echinomycin–AT contains only one DNA duplex bound by two antibiotic molecules, in which the central A:T base pair adopts a standard Watson–Crick configuration. The octahedral coordination of Mn2+ is completed by the N7 of a symmetry-related guanine with four water molecules in crystal packing (Supplementary Figure S1B). Superimposing the structures of the echinomycin–TT and echinomycin–AT complexes (Figure 4A) indicated that the two structures are similar overall (r.m.s.d. 0.558 Å for all heavy atoms) but with significant differences surrounding the central X4-T11 base pair. The central GpXpC segment in both echinomycin–DNA complexes has a similar negative roll angle of around −3° to −5°, which causes the DNA to bend slightly towards the minor groove (Figure 4B and Supplementary Table S2). Moreover, the sugar pucker of most bases in both echinomycin–DNA complexes reveals that they prefer the C2′-endo and C3′-endo conformations, whereas most of the cytosines adopt an unexpected C4′-exo sugar pucker - probably induced by the intercalation of the quinoxaline rings into the DNA duplex (Supplementary Table S3).
Figure 4.
Structural comparisons between echinomycin–TT and echinomycin–AT complexes. (A) Superimposition of the echinomycin–AT and echinomycin–TT complexes viewed from the side (left) and front (middle). The refined structures of the echinomycin–AT and echinomycin–TT complexes are coloured in red and blue, respectively. The close-up view (right) shows the environment of the central base pair with two intercalated quinoxaline rings, and an ‘X’ indicates the A4 or T4 base paired with T11. Prominent variations in the roll (B) and twist (C) DNA parameters were observed in the echinomycin–AT and echinomycin–TT structures by w3DNA analysis. The canonical values for A-DNA (cyan) and B-DNA (pink) are included for comparison.
A major point of difference between the two complexes, however, is manifested in the DNA backbone structure. In echinomycin–TT, the DNA helix is unwound by 20–30° at both CpG steps (twist angles of 12.8 and 3.3°) (Figure 4C and Supplementary Table S2). In addition, the intercalation of Echi1 QUI9 causes unwinding at the GpT step to generate a left-handed helix with a twist angle of −5.5° and opening up the T:T mismatch site by 14.8°. Interestingly, this left-handed rotation is a characteristic of Z-DNA. A larger DNA helical twist (43.4°) at the TpC step caused by the angled intercalation (39.2°) of Echi2 QUI0 indicates that the TpC step has become overwound. By contrast, the average twist angles of echinomycin–AT at GpTpC steps are ∼16° due to the parallel intercalation of the quinoxaline rings, resulting in a global unwinding of the DNA conformation. These differences correlate with the distinctly shorter inter-strand distance of 15.8 Å at the T:T mismatch site compared to 17.6 Å for the echinomycin–AT complex (Supplementary Figure S3A). We propose that the overwinding of the DNA backbone when Echi2 is bound to TT may compensate the released torsional energy caused by the left-handed rotation of the GpT step upon Echi1 binding. The detailed stacking interaction between echinomycin and the DNA duplex is also different between echinomycin–TT and echinomycin–AT (Supplementary Figure S3B). In echinomycin–TT, the quinoxaline rings are skewed relative to each other, whereas in echinomycin–AT the rings are aligned with respect to the centre of the major axis of the central A:T base pair. The overall result of these structural differences is reflected in the number of interactions between echinomycin and the DNA duplex: the number of hydrogen bonds between the alanine residues and guanine bases in echinomycin–AT is less than in the echinomycin–TT complex (Supplementary Figure S2B). The total number of van der Waals contacts and hydrogen bonds between echinomycin and the DNA duplex is also less in the echinomycin–AT structure compared to echinomycin–TT (Supplementary Figure S4 and Supplementary Table S4). These findings are consistent with our Tm and SPR results showing that echinomycin binding to TT is substantially more favourable than to AT (Figure 1).
Mismatch repair-deficient cells are more sensitive to echinomycin
The cytotoxic effects of echinomycin on MMR-deficient (Hct116) and MMR-proficient (Hct116+ch3 and SW620) colorectal cancer cell lines were determined by standard MTT assay. The Hct116 line has been extensively used to assess the anti-proliferative effects of a variety of DNA intercalators (10). The full dose-response of echinomycin treatment of these cells after 24 and 48 h were plotted in Supplementary Figure S5, and the LD50 values calculated from three independent cytotoxicity experiments are shown in Table 1. The growth of all cell lines was inhibited after 48 h. Echinomycin was revealed to be substantially more toxic to the MMR-deficient Hct116 line with an LD50 value as low as 31 nM, suggesting that the antibiotic might be selectively cytotoxic towards MMR-deficient cell lines in general compared to MMR-proficient cells.
Table 1.
Toxicity of echinomycin towards colorectal cancer cell lines
| Cell line | Hct116 | Hct116+ch3 | SW620 |
|---|---|---|---|
| LD50/24 h | >100 nM | >100 nM | >100 nM |
| LD50/48 h | 31.6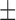 0.1 nM 0.1 nM |
86.4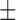 0.1 nM 0.1 nM |
93.8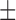 0.2 nM 0.2 nM |
| MMR status | Deficient | Proficient | Proficient |
DISCUSSION
Several small-molecule ligands have been found to bind to mismatched DNA which may have potential applications in the diagnosis and therapy of cancer as well as neurological diseases (2). Barton's group showed that rhodium metalloinsertors can recognise DNA mismatches with high affinity and specificity in vitro, and may target MMR-deficient cells over MMR-proficient cells (11,38). Their findings led them to propose the use of rhodium metalloinsertors as a basis for the development of new types of chemotherapeutic agents active against MMR-deficient cancers. We have also reported that many venerable antibiotic small-molecules may recognise mismatch sites within the non-canonical structures of repetitive DNA sequences (28,39,40). Since such sequences are hallmarks of many neurodegenerative diseases, we proposed that these antibiotic molecules could serve as a basis for the development of novel compounds that may either block DNA expansion or provide a read-out for disease detection (23). Our results show that echinomycin is another member of this select group by virtue of its ability to recognise consecutive CpG sites separated by a T:T mismatched base pair.
The structural bases behind the bis-intercalation of echinomycin and its biosynthetic precursor triostin A into the CpG steps of perfect DNA duplexes have been extensively studied by both NMR and X-ray crystallography (42–46). Although the overall mode of binding among the different studies appears to be similar, later NMR analyses have shown that the bases flanking the bis-intercalation sites may be conformationally unstable and capable of forming Hoogsteen or Watson–Crick base pairs (47,48). Consistent with this idea, our analysis of the echinomycin–AT and echinomycin–TT structures has revealed a higher B-factor for the A:T base pair compared to the T:T base pair (Supplementary Figure S6). Since binding of echinomycin to DNA is driven by hydrophobic interactions (43), the conformational flexibility of A:T within the complex may be detrimental to the binding affinity since interactions between echinomycin and the A:T base pair may need to be constantly broken and re-formed. This might conceivably explain the lower stability implied by the Tm of the echinomycin–AT complex compared to the echinomycin–TT complex (Figure 1B).
Also in agreement with previous studies (24), we have shown that the binding of echinomycin to d(ACGTCG(5-BrU))2 occurs via a pairwise cooperative binding model. Whilst the binding of Echi1 is little affected by the nature and length of the base pair(s) separating the two CpG sites, the association constant of Echi2 (Ka2) was significantly modulated by both the type and length of these base pairs. The structure of the echinomycin–TT complex may provide some clues as to the mechanistic basis behind this cooperativity. The helices of the DNA in the complex structure exhibit a smooth bend caused by the stacking interactions starting at the thymines of the sheared T:T base pair through the quinoxaline ring of echinomycin and ending at the guanines of the flanking G:C base pairs. The bending generates a negative roll in the base pairs along their long axis and compresses the major groove of the double helix. This is accompanied by a concomitant widening of the minor groove, which would assist Echi2 to fit in. Indeed, more generally the width of the minor groove could be a major determinant of DNA shape recognition by ligands. DNA intercalators such as octahedral ruthenium complexes and actinomycin D induce DNA unwinding and local deformation of the DNA helix, thereby changing its overall organisation including the width of the grooves and the length of the DNA strand (39,49). In the case of actinomycin D bound to CGG repeats, it also induced the formation of a left-handed helix (28). The average twist angle varies between echinomycin–AT (∼14.2°) and echinomycin–TT (∼17.6°), causing the DNA to unwind by ∼20° and ∼17° respectively, and may produce the different crystal packing superstructures observed for the two complexes that were never previously observed for DNA or DNA–ligand structures (Supplementary Figure S7). Echinomycin-TT formed a pseudo-continuous double helix organised as a right-handed plectonemic supercoil with a diameter of ∼20 Å and a rise of 115 Å in length per turn (∼24 bp) (Supplementary Figure S7A). However, the echinomycin–AT complex formed a barrel consisting of four pseudo-continuous double helices with a diameter of ∼40 Å and a rise of 150 Å in length per turn, needing eleven additional base pairs (∼35 bp) to compensate for one turn of supercoiled structure (Supplementary Figure S7B).
Echinomycin is known to be highly cytotoxic and consequently has failed most pre-clinical trials. Nevertheless, it may still serve as a valuable lead for the development of compounds with clinical potential. The differential inhibition of cellular proliferation by echinomycin in MMR-deficient cells may be correlated to its mismatch-binding affinity; similar to what was observed for rhodium DNA intercalators (11,50). The preference for T:T mismatches of echinomycin, a feature that has not previously been observed, suggests that it could be repurposed for the detection of MMR-deficient cancers, possibly by leveraging DNA binding-induced fluorescence quenching of echinomycin (41). Since echinomycin appears to bind more tightly to T:T mismatches, it may exhibit stronger quenching (and thus lower fluorescence emission) than normal Watson–Crick pairing under appropriate conditions. The structural basis for T:T mismatch recognition described in this work may also facilitate the development of lower-toxicity derivatives to treat MMR-deficient cancers.
To summarize, we have shown through X-ray and SPR analyses that adjacent CpG sequences flanking a T:T mismatch provide excellent sites for echinomycin to bind with high cooperativity. We also show that this preference could have potential applications against MMR-deficient cancers.
DATA AVAILABILITY
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data bank under accession number 5YTY for echinomycin–AT complex, 5YTZ for echinomycin–TT complex.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the NSRRC staff for data collection, and Dr. Thomas A. Kunkel (NIH, USA) for providing the colon cancer cell lines.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology, Taiwan [106-2628-M-005-001-MY3 to M.-H.H.]; National Chung Hsing University, Taiwan; Chung Shan Medical University, Taiwan [NCHU-CSMU-10506 and NCHU-CSMU-10601]. Funding for open access charge: Ministry of Science and Technology, Taiwan.
Conflict of interest statement. None declared.
REFERENCES
- 1. Lord C.J., Ashworth A.. The DNA damage response and cancer therapy. Nature. 2012; 481:287–294. [DOI] [PubMed] [Google Scholar]
- 2. Granzhan A., Kotera N., Teulade-Fichou M.P.. Finding needles in a basestack: recognition of mismatched base pairs in DNA by small molecules. Chem. Soc. Rev. 2014; 43:3630–3665. [DOI] [PubMed] [Google Scholar]
- 3. Tikhomirova A., Beletskaya I.V., Chalikian T.V.. Stability of DNA duplexes containing GG, CC, AA, and TT mismatches. Biochemistry. 2006; 45:10563–10571. [DOI] [PubMed] [Google Scholar]
- 4. Huang T.Y., Chang C.K., Kao Y.F., Chin C.H., Ni C.W., Hsu H.Y., Hu N.J., Hsieh L.C., Chou S.H., Lee I.R. et al. . Parity-dependent hairpin configurations of repetitive DNA sequence promote slippage associated with DNA expansion. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:9535–9540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kunkel T.A., Erie D.A.. Eukaryotic mismatch repair in relation to DNA replication. Annu. Rev. Genet. 2015; 49:291–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sharma M., Predeus A.V., Mukherjee S., Feig M.. DNA bending propensity in the presence of base mismatches: implications for DNA repair. J. Phys. Chem. B. 2013; 117:6194–6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xiao X., Melton D.W., Gourley C.. Mismatch repair deficiency in ovarian cancer – molecular characteristics and clinical implications. Gynecol. Oncol. 2014; 132:506–512. [DOI] [PubMed] [Google Scholar]
- 8. Sameer A.S., Nissar S., Fatima K.. Mismatch repair pathway: molecules, functions, and role in colorectal carcinogenesis. Eur. J. Cancer Prev. 2014; 23:246–257. [DOI] [PubMed] [Google Scholar]
- 9. Keane P.M., Poynton F.E., Hall J.P., Sazanovich I.V., Towrie M., Gunnlaugsson T., Quinn S.J., Cardin C.J., Kelly J.M.. Reversal of a single Base-Pair step controls guanine Photo-Oxidation by an intercalating ruthenium(II) dipyridophenazine complex. Angew. Chem. Int. Ed. Engl. 2015; 54:8364–8368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fung S.K., Zou T., Cao B., Chen T., To W.P., Yang C., Lok C.N., Che C.M.. Luminescent platinum(II) complexes with functionalized N-heterocyclic carbene or diphosphine selectively probe mismatched and abasic DNA. Nat. Commun. 2016; 7:10655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boyle K.M., Barton J.K.. Targeting DNA mismatches with rhodium metalloinsertors. Inorg. Chim. Acta. 2016; 452:3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mukherjee S., Dohno C., Asano K., Nakatani K.. Cyclic mismatch binding ligand CMBL4 binds to the 5′-T-3′/5′-GG-3′ site by inducing the flipping out of thymine base. Nucleic Acids Res. 2016; 44:7090–7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pierre V.C., Kaiser J.T., Barton J.K.. Insights into finding a mismatch through the structure of a mispaired DNA bound by a rhodium intercalator. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:429–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peng T., Dohno C., Nakatani K.. Mismatch-binding ligands function as a molecular glue for DNA. Angew. Chem. Int. Ed. Engl. 2006; 45:5623–5626. [DOI] [PubMed] [Google Scholar]
- 15. Bai Y., Nguyen L., Song Z., Peng S., Lee J., Zheng N., Kapoor I., Hagler L.D., Cai K., Cheng J. et al. . Integrating display and delivery functionality with a cell penetrating peptide mimic as a scaffold for intracellular multivalent multitargeting. J. Am. Chem. Soc. 2016; 138:9498–9507. [DOI] [PubMed] [Google Scholar]
- 16. Yamada T., Miki S., Ul’Husna A., Michikawa A., Nakatani K.. Synthesis of naphthyridine carbamate dimer (NCD) derivatives modified with alkanethiol and binding properties of G-G mismatch DNA. Org. Lett. 2017; 19:4163–4166. [DOI] [PubMed] [Google Scholar]
- 17. Sato M., Nakazawa T., Tsunematsu Y., Hotta K., Watanabe K.. Echinomycin biosynthesis. Curr. Opin. Chem. Biol. 2013; 17:537–545. [DOI] [PubMed] [Google Scholar]
- 18. Yoshinari T., Okada H., Yamada A., Uemura D., Oka H., Suda H., Okura A.. Inhibition of topoisomerase II by a novel antitumor cyclic depsipeptide, BE-22179. Jpn. J. Cancer Res. 1994; 85:550–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bachur N.R., Johnson R., Yu F., Hickey R., Applegren N., Malkas L.. Antihelicase action of DNA-binding anticancer agents: relationship to guanosine-cytidine intercalator binding. Mol. Pharmacol. 1993; 44:1064–1069. [PubMed] [Google Scholar]
- 20. Adams R.L., Rinaldi A.. Effect of echinomycin on DNA methylation. FEBS Lett. 1987; 215:266–268. [DOI] [PubMed] [Google Scholar]
- 21. Kong D., Park E.J., Stephen A.G., Calvani M., Cardellina J.H., Monks A., Fisher R.J., Shoemaker R.H., Melillo G.. Echinomycin, a small-molecule inhibitor of hypoxia-inducible factor-1 DNA-binding activity. Cancer Res. 2005; 65:9047–9055. [DOI] [PubMed] [Google Scholar]
- 22. Van Dyke M.M., Dervan P.B.. Echinomycin binding sites on DNA. Science. 1984; 225:1122–1127. [DOI] [PubMed] [Google Scholar]
- 23. Dawson S., Malkinson J.P., Paumier D., Searcey M.. Bisintercalator natural products with potential therapeutic applications: isolation, structure determination, synthetic and biological studies. Nat. Prod. Rep. 2007; 24:109–126. [DOI] [PubMed] [Google Scholar]
- 24. Bailly C., Hamy F., Waring M.J.. Cooperativity in the binding of echinomycin to DNA fragments containing closely spaced CpG sites. Biochemistry. 1996; 35:1150–1161. [DOI] [PubMed] [Google Scholar]
- 25. Takusagawa H.L., Takusagawa F.. Crystallization and preliminary X-ray diffraction studies of d(ACGTAGCTACGT)2:[actinomycin D, (echinomycin)2] and d(ACGTAGCTACGT)2:[actinomycin D, (triostin A)2] complexes. Acta Crystallogr. D Biol. Crystallogr. 2000; 56:344–347. [DOI] [PubMed] [Google Scholar]
- 26. Murphy J.H., Trapane T.L.. Concentration and extinction coefficient determination for oligonucleotides and analogs using a general phosphate analysis. Anal. Biochem. 1996; 240:273–282. [DOI] [PubMed] [Google Scholar]
- 27. Lin Y.H., Chuang S.M., Wu P.C., Chen C.L., Jeyachandran S., Lo S.C., Huang H.S., Hou M.H.. Selective recognition and stabilization of new ligands targeting the potassium form of the human telomeric G-quadruplex DNA. Sci. Rep. 2016; 6:31019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lo Y.S., Tseng W.H., Chuang C.Y., Hou M.H.. The structural basis of actinomycin D-binding induces nucleotide flipping out, a sharp bend and a left-handed twist in CGG triplet repeats. Nucleic Acids Res. 2013; 41:4284–4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 30. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. . PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Murshudov G.N., Vagin A.A., Dodson E.J.. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997; 53:240–255. [DOI] [PubMed] [Google Scholar]
- 33. Teneyck L.F. Crystallographic fast Fourier-transforms. Acta Crystallogr. A. 1973; A 29:183–191. [Google Scholar]
- 34. Zheng G.H., Lu X.J., Olson W.K.. Web 3DNA-a web server for the analysis, reconstruction, and visualization of three-dimensional nucleic-acid structures. Nucleic Acids Res. 2009; 37:W240–W246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Blanchet C., Pasi M., Zakrzewska K., Lavery R.. CURVES+ web server for analyzing and visualizing the helical, backbone and groove parameters of nucleic acid structures. Nucleic Acids Res. 2011; 39:W68–W73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Laskowski R.A., Swindells M.B.. LigPlot+: Multiple Ligand-Protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011; 51:2778–2786. [DOI] [PubMed] [Google Scholar]
- 37. Hou M.H., Lu W.J., Huang C.Y., Fan R.J., Yuann J.M.P.. Effects of polyamines on the DNA-Reactive properties of dimeric mithramycin complexed with cobalt(II): Implications for anticancer therapy (vol 48, pg 4691, 2009). Biochemistry. 2009; 48:10192–10192. [DOI] [PubMed] [Google Scholar]
- 38. Komor A.C., Schneider C.J., Weidmann A.G., Barton J.K.. Cell-selective biological activity of rhodium metalloinsertors correlates with subcellular localization. J. Am. Chem. Soc. 2012; 134:19223–19233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hou M.H., Robinson H., Gao Y.G., Wang A.H.. Crystal structure of actinomycin D bound to the CTG triplet repeat sequences linked to neurological diseases. Nucleic Acids Res. 2002; 30:4910–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tseng W.H., Chang C.K., Wu P.C., Hu N.J., Lee G.H., Tzeng C.C., Neidle S., Hou M.H.. Induced-Fit recognition of CCG trinucleotide repeats by a Nickel-Chromomycin complex resulting in Large-Scale DNA deformation. Angew. Chem. Int. Ed. Engl. 2017; 56:8761–8765. [DOI] [PubMed] [Google Scholar]
- 41. Jarikote D.V., Li W., Jiang T., Eriksson L.A., Murphy P.V.. Towards echinomycin mimetics by grafting quinoxaline residues on glycophane scaffolds. Bioorg. Med. Chem. 2011; 19:826–835. [DOI] [PubMed] [Google Scholar]
- 42. Wang A.H., Ughetto G., Quigley G.J., Hakoshima T., van der Marel G.A., van Boom J.H., Rich A.. The molecular structure of a DNA-triostin A complex. Science. 1984; 225:1115–1121. [DOI] [PubMed] [Google Scholar]
- 43. Ughetto G., Wang A.H., Quigley G.J., van der Marel G.A., van Boom J.H., Rich A.. A comparison of the structure of echinomycin and triostin A complexed to a DNA fragment. Nucleic Acids Res. 1985; 13:2305–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gilbert D.E., Feigon J.. Proton NMR study of the [d(ACGTATACGT)]2-2echinomycin complex: conformational changes between echinomycin binding sites. Nucleic Acids Res. 1992; 20:2411–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pfoh R., Cuesta-Seijo J.A., Sheldrick G.M.. Interaction of an echinomycin–DNA complex with manganese ions. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2009; 65:660–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cuesta-Seijo J.A., Weiss M.S., Sheldrick G.M.. Serendipitous SAD phasing of an echinomycin-(ACGTACGT)2 bisintercalation complex. Acta Crystallogr. D Biol. Crystallogr. 2006; 62:417–424. [DOI] [PubMed] [Google Scholar]
- 47. Gilbert D.E., van der Marel G.A., van Boom J.H., Feigon J.. Unstable Hoogsteen base pairs adjacent to echinomycin binding sites within a DNA duplex. Proc. Natl. Acad. Sci. U.S.A. 1989; 86:3006–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Leng F., Chaires J.B., Waring M.J.. Energetics of echinomycin binding to DNA. Nucleic Acids Res. 2003; 31:6191–6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gill M.R., Thomas J.A.. Ruthenium(II) polypyridyl complexes and DNA–from structural probes to cellular imaging and therapeutics. Chem. Soc. Rev. 2012; 41:3179–3192. [DOI] [PubMed] [Google Scholar]
- 50. Ernst R.J., Komor A.C., Barton J.K.. Selective cytotoxicity of rhodium metalloinsertors in mismatch repair-deficient cells. Biochemistry. 2011; 50:10919–10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data bank under accession number 5YTY for echinomycin–AT complex, 5YTZ for echinomycin–TT complex.