Abstract
Seventeen years after the sequencing of the human genome, the human proteome is still under revision. One in eight of the 22 210 coding genes listed by the Ensembl/GENCODE, RefSeq and UniProtKB reference databases are annotated differently across the three sets. We have carried out an in-depth investigation on the 2764 genes classified as coding by one or more sets of manual curators and not coding by others. Data from large-scale genetic variation analyses suggests that most are not under protein-like purifying selection and so are unlikely to code for functional proteins. A further 1470 genes annotated as coding in all three reference sets have characteristics that are typical of non-coding genes or pseudogenes. These potential non-coding genes also appear to be undergoing neutral evolution and have considerably less supporting transcript and protein evidence than other coding genes. We believe that the three reference databases currently overestimate the number of human coding genes by at least 2000, complicating and adding noise to large-scale biomedical experiments. Determining which potential non-coding genes do not code for proteins is a difficult but vitally important task since the human reference proteome is a fundamental pillar of most basic research and supports almost all large-scale biomedical projects.
INTRODUCTION
Before the human genome was sequenced, most researchers estimated that human protein coding gene numbers would be between 25 000 and 40 000 (1), with some estimates closer to 100 000 genes (2,3). However, the accumulation of experimental data has progressively brought this estimate down. The ‘finished’ version of the human genome revised the estimates to between 20 000 and 25 000 coding genes (4).
The gradual downward trend of the human protein gene count has been mirrored in the reference human gene set. The annotation of human coding genes began with the Ensembl project (5) and the initial release included more than 24 000 coding genes. This number soon decreased to 22 000 as the genome assembly improved and automatic predictions were refined (6). Until recently there were still gene loci in the reference set defined as coding based on the initial automatic predictions, and a number of these had little support as coding genes beyond their initial prediction. After the merge with the GENCODE manual annotations (7) in 2009, 1004 poorly supported automatic annotation models were removed from the Ensembl annotation set.
These refinements and intensive manual annotation have brought the number of annotated protein coding genes down to slightly over 20 000 genes in the Ensembl/GENCODE (7,8) reference, and indeed the three maintained manual reference databases, Ensembl/GENCODE, RefSeq (9) and UniProtKB (10), have converged on similar numbers of protein coding genes [f1000research: doi: 10.12688/f1000research.11119.1], a number that is in line with the prediction by Clamp et al. using evolutionary comparisons (11).
However, the human gene sets are in certain state of flux with coding genes being added and reclassified with each new release, and it is important to note that these 20 000 plus coding genes are not the same in each database. Indeed, as we show in this paper, the number of annotated coding genes in the union of the three reference sets exceeds 22 000.
The task of manually inspecting >20 000 annotated coding genes is enormous and the process has taken many years (7). Manual annotators have to accomplish two difficult tasks, detecting the remaining hard-to-find coding genes, and separating bona fide coding genes from misannotated pseudogenes and non-coding genes. Curators determine the status of the gene models based on transcript (ESTs and mRNAs) and protein data (from the main protein databases) available for each gene (12). Protein-coding potential depends first on whether an open reading frame (ORF) can be defined. However, the definition of ORFs is complicated by the fact that many noncoding transcripts may contain long ORFs by chance, particularly in GC-rich regions (11). In order to get round this problem, annotators also require some sort of protein evidence, such as whether the locus has sequence similarity to orthologues from other species, whether the resulting gene product contains Pfam functional domains (13), or whether experimental data is available from published papers, large-scale interaction studies (14) or mass spectrometry experiments (15).
Genes and transcripts may change their status between releases as annotators adjust the annotation to the available evidence. A gene's status is updated based on the available evidence and this evidence can change over time. For example GENCODE manual annotators recently decided to reclassify as non-coding approximately 200 ‘orphan’ protein coding genes [GENCODE blog, https://gencodegenes.wordpress.com, April 2018]. Most of these genes were early in silico predictions.
A number of studies have put an estimation on the number of human coding genes, including several that have estimated the number to be close to or below 20 000 (11,16–18). Two of the more comprehensive studies into the coding complement of the human genome, Clamp et al. (11) and Church et al. (17), were carried out before GENCODE and other groups began the systematic manual reannotation of the genes in the human gene set. Both analyses assumed that most novel genes, defined as genes that arose from scratch in the primate lineage, are not protein coding. According to the Clamp analysis, the vast majority of novel ORFs did not have evolutionary conservation and had features that resembled non-coding RNA rather than coding genes. After discarding these orphan DNA sequences, as well as genes that appeared to be transposons, pseudogenes, and other miscellaneous artefacts, the authors ended up with a gene count of 20 500, roughly 4000 fewer than were annotated at that time. Church et al. carried out a comparison between the human and the mouse genomes and found that there were very few truly novel human genes, and that almost all protein-coding genes gained in the mammalian lineage were generated from whole gene duplications. They estimated that the number of protein coding genes was <20 000.
Many of the genes tagged as non-coding in these two analyses have since been removed from the reference set after manual annotation, though a number of genes identified in both studies as orphans or pseudogenes are nevertheless still annotated as protein coding, including the predicted pseudogenes DHFRL1, which has experimental evidence for a protein, and HIGD2B, which does not.
In 2014, we predicted that the human genome was likely to have just 19 000 protein coding genes based on the identification of 2001 ‘potential non-coding’ genes (18). GENCODE manual annotators have since withdrawn or reclassified almost half of these genes from the human reference set. Most recently Southan [f1000research: doi: 10.12688/f1000research.11119.1] contrasted gene numbers in the three manually annotated reference sets with those of the HUGO Gene Nomenclature Committee [HGNC, (19)], noting the differences in coding gene counts and showing that UniProtKB proteins missing in RefSeq and Ensembl were enriched for elements classified by HGNC as endogenous retrovirus, long non-coding RNA or pseudogene.
Here, we expand our previous analysis to incorporate an analysis of the RefSeq and UniProtKB proteomes. We find that these two references databases and Ensembl/GENCODE annotate 22 210 genes as coding but only agree on 86% of the genes they annotate. In order to determine whether all 22 210 genes will code for proteins we contrasted the experimental evidence for genes annotated as coding in all three reference sets with those that are classified differently.
MATERIALS AND METHODS
Comparison of Ensembl/GENCODE, RefSeq and UniProtKB gene sets
We merged the coding genes in the three main versions of the reference human proteome, the Ensembl/GENCODE reference set (GENCODE v24, which is the equivalent of Ensembl 83), the RefSeq gene set (RefSeq 107) and the UniProtKB proteome (UniProtKB June 2016).
The UniProtKB reference proteome contained more than 70 612 SwissProt (reviewed) and TrEMBL (non-reviewed) entries. In order to compare UniProtKB with RefSeq and Ensembl/GENCODE, we merged these entries where possible by gene name. In UniProtKB genes can have more than one entry and UniProtKB entries may have more than one gene. After the initial merge the many orphan transcripts were merged first by their associated Ensembl identifier and then by hand where possible. This set of UniProtKB genes were then merged with the RefSeq and Ensembl genes using Ensembl's BioMart, UniProtKB’s mapping tools and the HGNC gene names provided by the three reference sets. We carried out a painstaking manual reannotation of the more than 2700 genes where HGNC gene names, BioMart and UniProtKB correspondences did not agree.
Finally, for the 2764 genes not classified as coding in all the three reference databases we manually cross-referenced their status in the reference sets in which they were not annotated as coding.
Possible non-coding features
We have shown that a number of protein features, such as gene family age and cross-species conservation, are correlated with the detection of peptides in mass spectrometry experiments (18). These features can also be used to predict whether peptides will be detected in proteomics experiments and to flag protein-coding genes as potentially non-coding. The features are listed below.
UniProtKB uncertain, predicted, homology and missing evidence codes
Protein evidence codes are taken from the UniProtKB database. UniProtKB carries out manual annotation of proteins and human proteins in particular are well annotated and a large majority are annotated with the highest evidence score ‘protein evidence’. The other four evidence codes in decreasing order are: ‘Transcript evidence’, ‘Homology’, ‘Predicted’ and ‘Uncertain’.
Where there was more than one UniProtKB entry associated to an Ensembl/GENCODE gene we chose the UniProtKB entry with the highest ranked evidence to represent the gene. Genes annotated with ‘Homology’, ‘Predicted’ or ‘Uncertain’ evidence, and those genes for which we could not detect any evidence code at all, had very little evidence of protein expression; the four features between them covered 1599 genes and we found peptide evidence for 52.
UniProtKB cautions
UniProtKB appends cautions to many of their protein entries. Several of these cast doubt on whether they are expressed as proteins. We did not select all UniProtKB cautions, just those that suggested that the gene might be non-coding, non-functional or a pseudogene. The two most common cautions were: ‘Product of a dubious gene prediction’, ‘Could be the product of a pseudogene’. There were 86 genes tagged with these cautions. We found peptide evidence for just three of these genes.
GENCODE
We took the translated GENCODE sequences as the coding gene set. The 20,266 genes in this set included not just protein coding genes, but also immunoglobulin receptors, nonsense mediated decay (NMD) transcripts and polymorphic pseudogenes. 13 148 of the coding genes are also annotated with non-coding transcripts, but these were not analysed.
Polymorphic pseudogenes
Polymorphic pseudogenes are loci that are pseudogenes in the reference genome that are intact in other individuals, and may represent coding genes that are undergoing a process of pseudogenization. There are 58 polymorphic pseudogenes in the reference gene set, of which 43 are olfactory receptors. It is particularly difficult to determine whether olfactory receptors are pseudogenes or code for functional proteins (20). We find peptide evidence for two of these polymorphic pseudogenes, GBA3 and PNLIPRP2. Unlike most genes annotated with the polymorphic pseudogene tag, these two genes were annotated with both coding and polymorphic pseudogene transcripts.
Nonsense-mediated decay genes
A number of genes in the reference gene set only have NMD and non-coding transcripts. There were 204 genes annotated just with NMD and/or non-coding transcripts in the GENCODE v24 reference set. As might be expected, we did not find peptides for any of these genes.
Read-through transcripts
Read-through genes are genes in which all coding or NMD transcripts are tagged as read-through transcripts. There are also genes that have a mix of read-through and coding transcripts, though these are gradually being cleaned up. Read-through transcripts usually occur when a transcript skips the 3′ exon and reads through to exons from the neighbouring gene (which is usually coding but may be non-coding or pseudogene too). If translated, read-through transcripts would produce fusion proteins.
Read-through variants are annotated as part of the human coding gene set for technical reasons. While it is possible that the splicing together of two neighbouring genes is one way for proteins to gain new domains (21), it appears that very few of these read-through transcripts produce proteins at detectable levels. While we found peptide evidence for one of these genes (IQCJ-SCHIP1), there is enough evidence to suggest that it may actually be a single gene rather than two separate genes with read-through transcripts
Because read-through transcripts and proteins overlap with transcripts and proteins from known coding genes, these transcripts introduce a number of technical problems to genome-scale analysis. For example we had to map the spectra from the MS analyses to the GENCODE v24 database twice, once including the read-through proteins and once excluding them.
The numbers of read-through genes in the coding gene set is ever increasing. There were 470 read-through genes annotated either by GENCODE or in the Ensembl description.
Ensembl
Pseudogenes, non-functional genes, non-coding genes, antisense/opposite strand genes, miscellaneous RNA
We manually curated genes with tags from the Ensembl gene descriptions. Genes that were annotated as ‘pseudogene’, ‘read-through’, ‘non-coding’, ‘non-functional’, ‘antisense’, ‘opposite strand’ and ‘long non-coding RNA’ were tagged as potentially non-coding. There were 131 genes described as pseudogenes by Ensembl, 70 were olfactory receptors. We found peptide evidence for 4 of these genes. Another 93 genes were described as ‘non-functional’, ‘antisense’ or ‘opposite strand’. We found peptide evidence for 6 of these genes. Finally 6 genes were described as ‘non-coding’ or ‘long non-coding RNA’. We found peptides for three of these genes.
Primate gene family
These were genes from families that evolved in the primate lineage according to our analysis of data from Ensembl Compara (22). The primate lineage was here defined as all strata more recent than the boroeutheria class. Gene birth dating was carried out used the phylogenetic reconstructions of Ensembl Compara v84. We estimated a gene family age and an individual gene age for all coding genes annotated in GENCODE v24. The analysis was identical to that carried out in the previous paper (18), which itself was based on earlier study of gene ages (23) and is detailed below. Ensembl Compara v84 is constructed from genes from 70 different species; here we focused on phylostrata that represented the last common ancestors of Homo sapiens and that had at least 5× coverage. Inconsistencies between gene trees and species phylogeny have been described for the Euarchontoglires phylostratum (24,25), so this was collapsed into the Eutherian level. Human coding genes were classified in the following age classes: Fungi/Metazoa, Bilateria, Chordata, Vertebrata, Euteleostomi, Sarcopterygii, Tetrapoda, Amniota, Mammalia, Theria, Eutheria, Boreoeutheria, Primates, Simiiformes, Catarrhini, Hominoidea, Hominidae, HomoPanGorilla and H. sapiens. In the analysis, all classes from Boreoeutheria to H. sapiens formed the ‘Primate’ class. The Sarcopterygii class was later clustered with Euteleostomi class because it contained few genes.
Compara classifies speciation and duplication nodes in family trees by the phylogenetic level in which the event took place (26) and our pipeline uses this information to define the gene family age and the gene age of each coding gene. Gene family age is the phylostratum at the root of the family tree (the earliest common ancestor that has a member of the gene family) while gene age is the phylostratum in which the genomic event leading to an extant gene takes place. For singleton genes the family gene age is always the same as the gene age, for duplicated genes the gene age represents the species in which the last duplication took place. Only duplication events with a consistency score (27) >0.3 were considered in the gene age analysis. Nodes with zero scores were trimmed out of the analysis. Duplication nodes with consistencies between 0 and 0.3 were labelled as ‘unclear’ and gene age was not assigned.
To our surprise we found more primate family genes in this study (700) than in our previous study (563). We found protein evidence for just 27 primate family genes.
Curiously there are sixteen coding genes that Compara tags as novel (non-duplicated) human genes in GENCODE v24. All are single exon genes predicted by Ensembl automatic prediction programs (e.g. see Supplementary Figure S1). None of these novel human genes have their coding status supported in any other reference set or any by peptide or antibody evidence.
PhyloCSF Score
We used exon-based PhyloCSF scores (27) to represent a measure of conservation for each gene. PhyloCSF was run using the 58 mammals parameters and the ‘mle’ and ‘bls’ option on the coding portion of each exon, trimmed to codon boundaries and excluding the final stop codon. Alignments were extracted from the 100-vertebrate MULTIZ hg38 alignment, with species restricted to the 58 placental mammals.
The conservation score was the PhyloCSF score of the highest scoring exon, counting only exons at least 42 bases in length and for which the relative branch length of the local alignment reported by PhyloCSF’s ‘bls’ option was at least 0.1, since PhyloCSF scores are unreliable if there is insufficient branch length. Genes having no exons satisfying these conditions were flagged as having exons that were too short or with too few relatives to return a PhyloCSF score.
Genes with a maximum PhyloCSF exon score of less than −16 or genes that had a relative branch length of less than 0.1 were flagged as having a poor PhyloCSF score. We found peptide evidence in PeptideAtlas for 28 of the 453 genes with poor branch length and 2 of the 132 genes with a maximum PhyloCSF exon score of less than −16.
APPRIS
All Ensembl genes are annotated with protein data in the APPRIS database (28). APPRIS annotates the following protein-based features: homology to proteins with known structure is mapped onto variants using HHsearch (29); functionally important residues and protein functional domain mapping comes from firestar (30) and pfamscan (31); trans-membrane helices are mapped using three separate trans-membrane predictors (32–34) and signal peptides are predicted by SignalP (35). A module of APPRIS calculates a measure of conservation by mapping vertebrate orthologues present in the protein databases. While APPRIS calculates features for all annotated coding variants, we took the mapping from the principal isoforms for each gene.
Protein features were calculated for all genes. Genes that did not have functional information, structural information or conservation in formation were tagged as potential non-coding when they had a PhyloCSF score below 2. There were just 17 genes with no protein information but with peptide evidence in our analysis.
Transcript expression from Human Protein Atlas
We downloaded data from the RNAseq experiments carried out for the Human Protein Atlas (36). The Human Protein Atlas RNAseq experiments were carried out on 36 tissues using Ensembl v83 (equivalent to GENCODE v24). For each gene, we counted the number of tissues in which the expression level was measured to be at least 1 transcript per million (TPM). Genes were binned by the number of tissues in which they were detected with at least 1 TPM.
Peptide data from PeptideAtlas
We downloaded all peptides identified in the January 2016 build of the human PeptideAtlas (15), in total 1 166 164 peptides. 880 101 peptides (75.5%) were semi-tryptic with respect to the GENCODE v24 human reference set, even though trypsin is used to cleave the proteins in the vast majority of proteomics experiments. We have previously found that semi-tryptic peptides are considerably less reliable than tryptic peptides (18), though most of these peptides were by-products of wholly tryptic peptides.
Including semi-tryptic peptides would have identified 711 more genes, 13.5% of which would have been potential non-coding genes. Less than 1% of the genes identified with tryptic peptides were potential non-coding genes. There is no reason why semi-tryptic peptides should identify 10 times as many potential non-coding genes than tryptic peptides, so semi-tryptic peptides were excluded on the grounds of accuracy.
We also eliminated peptides shorter than nine residues and peptides that mapped to more than one gene. Finally, we eliminated nested peptides; where two peptides had the same sequence but one was shorter than the other, we eliminated the shorter peptide. We mapped the remaining 153 913 peptides to the genes in GENCODE v24. At least two peptides had to map to each gene in order to identify it.
Obtaining and filtering of CNV maps
Whole genome copy number variation (CNV) maps were downloaded from five different publications (37–41). In order to homogenize the different maps, we selected autosomal and not private CNVs. Additionally, we removed CNVs marked as low quality from Handsaker et al. (40) and all the variants from two of the individuals (NA07346 and NA11918) because we were not confident about their genotype. From the maps in Zarrei et al., (39) we selected the stringent map that considered CNVs that appeared in at least two individuals and in two studies. Homozygous whole gene losses were calculated for all maps except for Abyzov et al., (41) which did not specify the copy number of the deletions.
Genetic variation
We compared rates of genetic variation for genes with potential non-coding features against the genetic variation rates for likely coding genes using data from 2504 individuals in phase 3 of the 1000 Genomes Project (42). We remapped these variants from GRCh37 to GRCh38 using dbSNP v149 (43). Most of the variants could be mapped from GRCh37 to 38 by using dbSNP identifiers (rsIDs). The exceptions were 186 854 variants with no rsID in dbSNP v149, and 256,769 variants for which the reference base has changed between GRCh37 and GRCh38. The rest of the variants (99.47%; 84 358 257/84 801 880) were successfully mapped. When available, ancestral allele information from the 1000 Genomes Project was used to translate allele frequencies into derived allele frequencies.
We ran VEP (variant_effect_predictor.pl, (44)) v84 using either the Ensembl v84 cache (for Ensembl/GENCODE) or a cache built locally using gene annotations from RefSeq v107 to predict the effects of variants. We calculated the percentage of high-impact variants and the ratio of non-synonymous to synonymous variants for rare and common allele frequencies. High impact variants were splice acceptor, splice donor, stop gain and stop loss variants. Common alleles were those with an allele frequency higher than 0.005 (equivalent to >25 allele counts in autosomes), while rare alleles were those with an allele frequency <0.005.
Only variant effects corresponding to the APPRIS principal isoform (28) of each coding gene were considered. Variants were considered only for strictly defined protein coding genes, not for the immunoglobulin and t-cell receptor fragment genes to exclude the possibility of positive selection.
RESULTS
Coding genes in the three main reference sets
We compared the coding genes in the three main versions of the human proteome, the merged Ensembl/GENCODE reference set, the RefSeq gene set and the UniProtKB proteome. The comparison was based on GENCODE v24 (Ensembl 83), UniProtKB June 2016, and RefSeq 107. RefSeq 107 annotates 20 450 coding genes, and the Ensembl/GENCODE merge contains 20,266 coding genes. The UniProtKB proteome is based around proteins rather than genes. UniProtKB June 2016 proteins mapped to 21 212 coding genes.
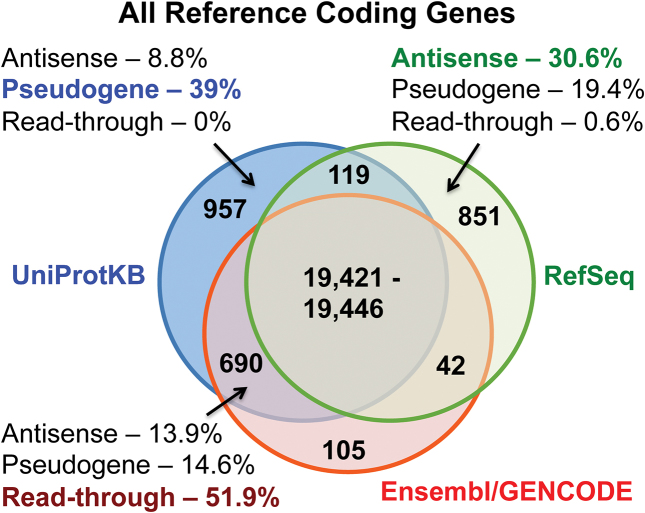
In total the three reference sets annotate 22 210 protein-coding genes. There are a maximum of 19 446 genes annotated as coding in the intersection of the three sets (Figure 1). This is a maximum because boundaries are disputed for a small number of genes. There are eight cases where Ensembl/GENCODE has two genes but RefSeq annotates one gene and sixteen cases where single Ensembl/GENCODE genes are annotated as multiple genes in RefSeq (PTPRQ is three RefSeq coding genes and only one in Ensembl/GENCODE). If all 24 genes were single genes rather than being split, there would be 19 421 coding genes common to the three reference sets. Beyond the intersection of the three reference databases, 851 genes are supported by two of the three reference sets and 1903 genes are annotated in just one of the three reference sets.
Figure 1.
The overlap between Ensembl/GENCODE, RefSeq and UniProtKB genes. The number of genes classified as coding in each of the three reference databases and the intersection between them. The number of genes in the intersection of A is variable because RefSeq and Ensembl/GENCODE disagree on gene boundaries for a number of genes. For three subsets of genes, we show the percentage of coding genes annotated as antisense, pseudogene or read-through in another database.
Ensembl/GENCODE has the fewest unique coding genes (105). This is for technical reasons. Most genes annotated by Ensembl/GENCODE are automatically included in UniProtKB. Given the near automatic transmission of coding genes between Ensembl/GENCODE and the UniProtKB proteome, the 690 genes annotated as coding by Ensembl/GENCODE and UniProtKB might also be regarded as singleton coding genes.
Almost a quarter of coding genes not present in all three reference sets are annotated as pseudogenes by manual annotators from other databases (Supplementary Table S1) and this rises to 39% of coding genes annotated in UniProtKB only (Figure 1). Potential ‘antisense’ genes, non-coding genes on the opposite strand to protein-coding loci, form the second largest group of differently annotated genes; 17% of coding genes not annotated in all three sets and 31% of genes classified as coding in RefSeq only are antisense. More than 50% of genes that are coding in Ensembl/GENCODE and UniProtKB but not in RefSeq are read-through genes. Read-through genes (genes made up entirely of transcripts that skip the last exon of one coding gene to read through to exons from the neighbouring gene or pseudogene) are currently annotated as coding by both the RefSeq and Ensembl/GENCODE annotations even though there is little indication that they code for proteins.
Each reference set has its own biases and idiosyncrasies. UniProtKB annotates 26 retroviral genes and a large number of T-cell receptor and immunoglobulin genes as part of the human reference proteome and include 84 genes that are part of alternative loci in the haploid assembly. RefSeq annotates 44 genes as sense overlapping (i.e. the locus of the gene overlaps with a known protein coding gene in the same sense), while Ensembl/GENCODE has 41 genes that are exact duplicates of annotated coding genes due to technical problems with the merge between GENCODE and Ensembl (Supplementary Table S1).
Are there 22 210 coding genes in the human genome?
There is a remarkable discrepancy between the number of genes classified as coding by all three reference sets and the number of genes classified as coding by at least one of the individual reference sets; 14.4% more genes are classified as coding in the union of the three reference sets than in the intersection. How many of these 2764 extra genes annotated by just one or two of the reference databases are protein coding?
UniProtKB annotation
Genes classified as coding solely by UniProtKB are unique in that they do not come with reference coordinates. Indeed many UniProtKB proteins are annotated as unplaced because the annotators do not know where in the genome the gene is found. However, the UniProtKB database provides an evidence scale for their manual annotations, ranging from the most reliable (‘supported by protein evidence’) to the most dubious (‘uncertain’). We used these classifications to compare genes classified as coding by UniProtKB. For each gene, we took the protein with the most reliable evidence as the representative.
The evidence codes of genes classified as coding in the coding gene subsets (UniProtKB and RefSeq, UniProtKB and Ensembl/GENCODE and solely UniProtKB) are clearly distinct from those classified as coding in all three reference databases (Supplementary Figure S2). More than 80% of the genes classified as coding across all three reference databases are annotated with the highest UniProtKB evidence score, ‘supported by protein evidence’. Outside of this intersection the proportion of genes supported by protein evidence is much smaller; those genes annotated by UniProtKB only have the next highest level of confirmation with just 19% of proteins supported by protein evidence, and three quarters of these are immunoglobulin genes, T-cell receptors, viral proteins and proteins from alternative loci that are not in the reference genome-based databases. By contrast >50% of the coding genes unique to UniProtKB are supported by the ‘uncertain’ evidence code, while over half the genes classified as coding by UniProtKB and RefSeq are supported by transcript evidence alone, and more than two thirds of genes that are classified as coding by UniProtKB and Ensembl/GENCODE are annotated as being supported by ‘predicted’ evidence. Genes annotated as coding in just one or two reference databases clearly have much weaker evidence in UniProtKB.
Potential non-coding features
In a previous work we flagged 2001 coding genes from the GENCODE v12 gene set as potentially non-coding (18) based on a set of features that were more typical of non-coding genes than coding genes (potential non-coding features). These features were all associated with extremely poor detection rates in mass spectrometry analyses. Manual annotators have since reclassified 908 of these genes as pseudogenes or non-coding RNA. Since genes annotated as coding in just one or two reference sets have less evidence in UniProtKB, it seems logical that many of these genes will also be enriched potential non-coding features.
Using the Ensembl/GENCODE coding genes we defined a set of potential non-coding features. The features included the weakest three UniProtKB evidence codes and manually added caution notes from the UniProtKB manual annotators, read-through, nonsense mediated decay and polymorphic pseudogenes tags from the GENCODE manual annotation, labels indicating pseudogene or non-coding gene from the Ensembl database and four measures of conservation, poor PhyloCSF (21) maximum score and relative branch length (which indicates that evolutionary coding potential within placental mammals is low), absence of conserved protein structure, function or conservation according to the APPRIS (28) database and those genes that have evolved within the primate clade according to Ensembl Compara (22).
The 16 potential non-coding features, the numbers of genes that were tagged with each feature and the number of these genes that had peptide evidence from large-scale proteomics analyses are listed in Table 1 and the features themselves are detailed in the Materials and Methods section.
Table 1.
The 16 potential non-coding features used to select the 2278 potential non-coding genes
| Features | Genes G24 | No. Detected in MS |
|---|---|---|
| No protein features [A] | 586 | 17 |
| Primate gene [C] | 700 | 27 |
| Pseudogene [E] | 131 | 4 |
| Non-functional [E] | 74 | 6 |
| Antisense/Opposite Strand [E] | 19 | 3 |
| Non-coding [E] | 6 | 3 |
| Read-through gene [G] | 467 | 1 |
| Nonsense mediated decay [G] | 204 | 0 |
| Polymorphic pseudogene [G] | 56 | 2 |
| PhyloCSF branch length [M] | 453 | 28 |
| PhyloCSF maximum [M] | 132 | 2 |
| Predicted evidence [U] | 853 | 12 |
| Homology evidence [U] | 613 | 39 |
| No evidence code [U] | 101 | 0 |
| Caution note [U] | 86 | 3 |
| Uncertain evidence [U] | 32 | 1 |
The abbreviations show the source of each annotation: A – APPRIS, C- Ensembl Compara, E – Ensembl annotations, G – GENCODE annotations, M - MIT, U – UniProtKB annotations.
A total of 2278 Ensembl/GENCODE coding genes were tagged with at least one of the 16 potential non-coding features. These genes were labelled as ‘potential non-coding genes’. The remaining 17 988 coding genes are referred to as the ‘likely coding gene’ set in this analysis. The correspondence between the potential coding genes tagged in Ensembl/GENCODE and in GENCODE v12 is shown in Supplementary Figure S3.
Potential non-coding genes are not distributed evenly between the intersection of three reference sets and the Ensembl/GENCODE gene subsets (Ensembl/GENCODE and UniProtKB, Ensembl/GENCODE and RefSeq, and Ensembl/GENCODE alone). While there were 1471 potential non-coding genes in the intersection of the three sets, this was just 7.6% of the genes. By contrast potential non-coding genes made up 96.5% of the genes (808 of 837) in the Ensembl/GENCODE gene subsets (Supplementary Figure S4).
The fact that almost all the genes outside the intersection of the three reference sets have potential non-coding features suggests that many of them may not code for proteins under normal cellular conditions. As a first step to testing this hypothesis we analyzed the experimental expression of potential non-coding genes using available experimental transcriptomics, proteomic and antibody binding data and compared this to likely coding genes.
Transcript evidence
We downloaded RNA expression data from the Human Protein Atlas. The Human Protein Atlas details RNAseq experiments carried out on 36 tissues using Ensembl83 (equivalent to GENCODE v24). For each gene we looked at the maximum expression in any one tissue and counted the number of tissues in which expression was at least 1 transcript per million (TPM). We binned genes by maximum expression and by number of tissues and compared the tissue distributions of likely coding genes and potential non-coding genes in both the intersection and subsets of coding genes annotated in Ensembl/GENCODE, but not in both other reference sets.
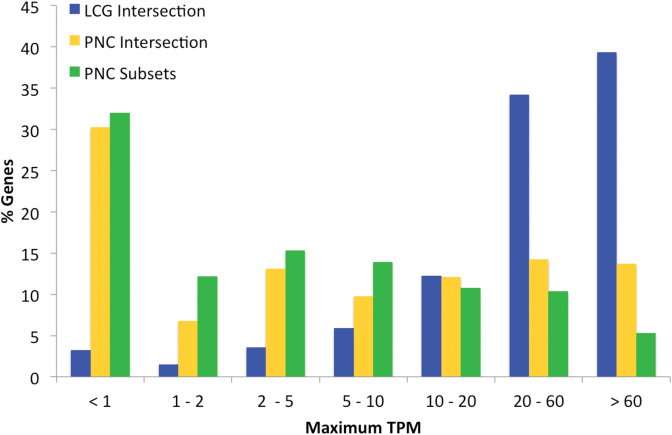
There was considerably more evidence for the expression of likely coding genes: 73.5% of likely coding genes had a maximum TPM of 20 or more against just 24.3% of potential non-coding genes (Figure 2). In fact 52.9% of potential non-coding genes had a maximum TPM of fewer than 5. The median expression level for potential non-coding genes was just 4.1 TPM, compared to 43.4 TPM in the likely coding set. Potential non-coding genes from the Ensembl/GENCODE coding subsets have a very similar distribution to potential non-coding genes from the intersection of the three sets. There were too few likely coding genes in the Ensembl/GENCODE coding subsets (29) to show in the graphic.
Figure 2.
Maximum transcript expression of potential non-coding genes and likely coding genes. The percentage of genes in seven different maximum TPM bins. Maximum TPM comes from the 36 tissues of the Human Protein Atlas RNAseq experiments. Tissue distribution shown for the likely coding genes (LCG Intersection) as well as potential non-coding genes annotated by all three reference sets (PNC Intersection) and by just one or two sets of annotators (PNC Subsets).
Likely coding genes also have entirely different tissue-specific characteristics from potential non-coding genes. While likely coding genes tend to be expressed in detectable quantities over most tissues (62.7% of these genes are detected in at least 30 tissues), the majority of potential non-coding genes are found in few tissues (Supplementary Figure S5). More than two thirds of potential non-coding genes (66.5%) have detectable expression in five or fewer tissues.
The skewed tissue-distribution of both sets of possible non-coding genes (Supplementary Figure S5) might suggest that these genes are more tissue-specific, and it is true that a higher proportion of potential non-coding genes are olfactory receptors and would be expected to be expressed in limited tissues. However, potential non-coding genes still have much lower expression levels even when olfactory receptors are removed (Supplementary Figure S6). Most potential non-coding genes had a maximum expression of fewer than 5 TPM, so differences in tissue expression might also be a reflection of generally low expression levels in which the 1 TPM threshold is crossed only in few tissues.
Protein expression
We carried out two analyses to identify gene products, an analysis of the collected peptides from the PeptideAtlas proteomics database and an investigation of the antibody information housed in the Human Protein Atlas.
We culled peptides from the PeptideAtlas database (January 2016), which contains 238 402 discriminating tryptic peptides. We required protein detection to be supported by two or more distinct uniquely-mapping, non-nested peptide sequences of at least 9 amino acids as suggested by Human Proteome Project consortium (45).
We detected at least two non-nested peptides for 13 360 of the 17 988 likely coding genes (74.3%). By way of contrast genes with potential non-coding features had extremely low levels of peptide detection [Table 1]. In total we detected peptides for just 142 of the 2278 potential non-coding genes (6.2%). Less than 1% of the genes identified by PeptideAtlas were potential non-coding genes.
Human Protein Atlas antibodies
The Human Protein Atlas has been developed to validate tissue-specific protein expression. We downloaded antibody-specific protein expression information from normal tissues from the Human Protein Atlas (Version 16, January 2016). We excluded expression data for antibodies that identified more than one gene and identifications tagged as ‘uncertain’.
The remaining antibodies detected a higher proportion of protein expression for the genes in the likely coding set (9896 of 17 988 genes, 55%) than for the genes in the potential non-coding set (just 79 of the 2278 genes, 3.5%). Potential non-coding genes that were validated by Human Protein Atlas antibodies included primate genes STATH (statherin), HTN3 (histatin-3) and SCT (secretin), all of which code for secreted proteins.
Genes detected by PeptideAtlas peptides and Human Protein Atlas antibodies are shown in Supplementary Figure S7. 8794 genes were detected in both analyses, only 46 of which were potential non-coding genes (0.52%). At the same time 2101 of the 5681 genes not detected in either analysis (37%) were potential non-coding genes.
There is quite clearly less evidence for the expression of potential non-coding genes both at the transcript and protein level. Chi-squared tests show that expression patterns of potential non-coding genes are significantly different from those of likely coding genes in all three sets of experimental observations.
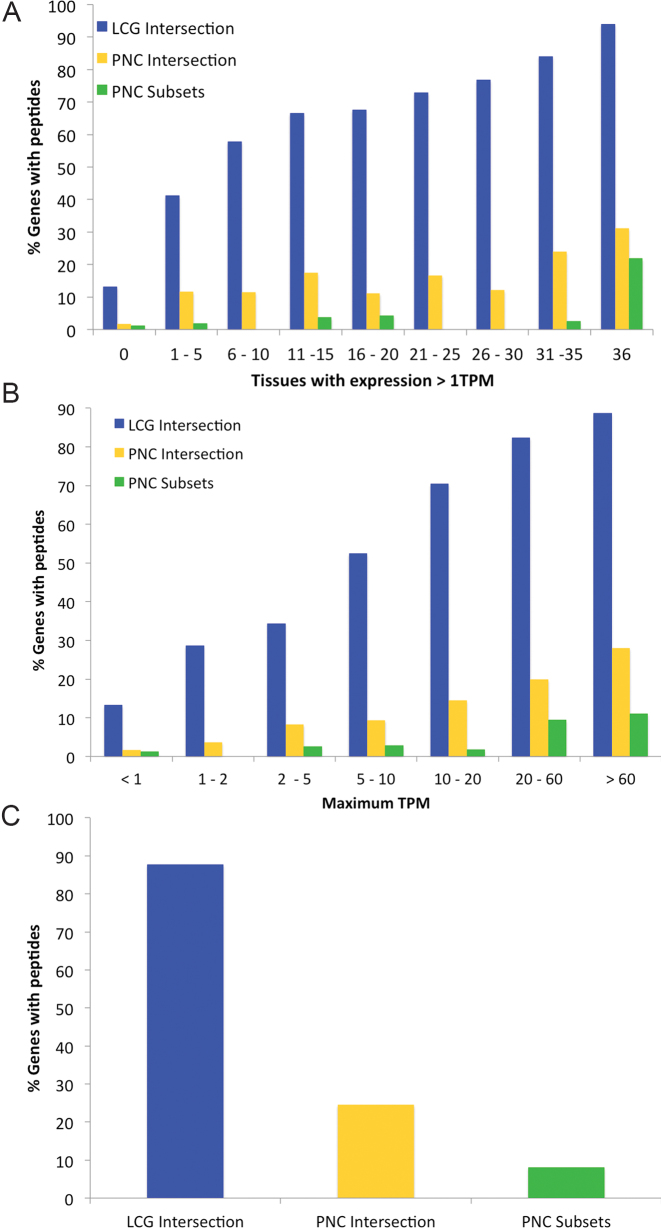
Potential non-coding genes even have even less protein evidence than one would expect from the RNAseq levels. Peptides can be found in PeptideAtlas for 92% of likely coding genes that have RNAseq expression of at least 1TPM in all 36 Human Protein Atlas tissues (Figure 3), but the peptide support falls to just 25% for potential non-coding genes. A similar pattern can be seen when genes are binned by maximum TPM across all 36 Human Protein Atlas tissues. Proportionally we found 5–10 times more likely coding genes than potential non-coding genes (Figure 3) in each bin. Even in the most widely expressed genes, which we defined those genes that are expressed in at least 10 tissues with a minimum of 10 TPM, there is still much more peptide evidence for likely coding genes than potential non-coding genes. We detected peptides for 85.6% of likely coding genes, 19.4% of potential non-coding genes annotated by all three reference sets and just 6.1% of potential non-coding genes annotated in two or fewer sets (Figure 3).
Figure 3.
The relation between peptides in proteomics experiments and transcript expression. (A) The percentage of genes for which peptides are detected in PeptideAtlas across nine different bins. The bins are based on the number of tissues in which the transcripts are detected with a TPM of >1 in the Human Protein Atlas RNAseq experiments. (B) The percentage of genes for which peptides are detected in PeptideAtlas divided across seven bins of maximum TPM for each gene taken from the 36 tissues of the Human Protein Atlas RNAseq experiments. (C) The percentage of genes for which peptides are detected for those genes that have RNAseq expression in at least 10 tissues with a TPM of 10 or more. In each case the percentage of genes for the likely coding genes (LCG Intersection, blue bars) as well as potential non-coding genes annotated by all three reference sets (PNC Intersection, yellow) and by just one or two sets of annotators (PNC Subsets, green).
Genetic variation
Human genetic variation can be used to shed light on whether or not potential non-coding genes code for proteins. The rate of copy number variation and the proportion of damaging high impact variants can provide clues to the functional relevance of coding (or non-coding) genes. Because of the effects of purifying selection coding genes should have substantially lower non-synonymous to synonymous variant ratios than non-coding genes that are misannotated as coding.
Copy number variation
We downloaded genome copy number variations (CNV) maps from five different publications (37–41). The CNVs were mapped to the GRCh37 build of the human genome, so we compared rates of gene gain and loss in the subset of Ensembl/GENCODE genes that were also annotated GENCODE v12. We also looked at CNVs in those GENCODE v12 genes that have since been removed from the coding reference set and reclassified as non-coding, pseudogene or artefact.
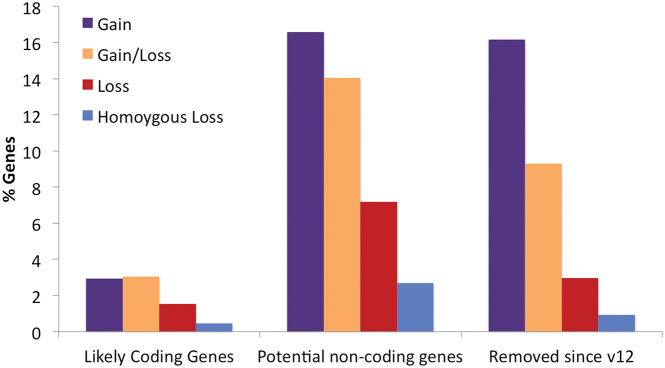
The rate of gene loss and homozygous gene loss through CNVs for each set are shown in Figure 4. Potential non-coding genes from Ensembl/GENCODE have more than five times as much gene gain as likely coding genes and almost five times as much genes loss. The distribution of CNVs in potential non-coding genes is similar to that of GENCODE v12 genes that are no longer classified as coding, though potential non-coding genes have even more evidence of gene loss.
Figure 4.
Whole gene gains and losses for likely coding and potential non-coding gene in GENCODE v12. The percentage of genes that have undergone gene gain/loss (purple), whole gene gain (orange), whole gene loss (red) or homozygous gene loss (blue) in at least one of the five different analyses. Potential non-coding genes present in both GENCODE v12 and v24 undergo a similar proportion of gene gain and loss to GENCODE v12 genes that have since been reclassified as not coding.
Genetic variation within the human population
The patterns from the CNV study suggest that potential non-coding genes are under weaker selection than likely coding genes. To further characterize the strength of selection we analysed the patterns of genetic variation in the human population using data from 2504 individuals in phase 3 of the 1000 Genomes Project (42). For the calculation we separated variants by allele frequency: common alleles were those with an allele count of more than 25, equivalent to an allele frequency of 0.005, while rare alleles were those with an allele count of fewer than 25. Variant effects were determined using the main protein isoform to represent each GENCODE v24 coding gene (28).
The percentage of high-impact variants and the ratio of non-synonymous to synonymous variants for rare and common allele frequencies were calculated using the results from VEP (44). For the large-scale comparison high impact variants included splice acceptor, splice donor, stop gain, stop loss, but not indel variants. This is because indels are generally validated only with higher allele counts and are therefore almost always overrepresented in common alleles.
If purifying selection is preventing high impact or missense substitutions, these variants should be depleted from higher allele frequencies. Hence, differences in the patterns of high-impact and missense substitutions between rare and common alleles can be used to determine whether there is purifying selection or neutral evolution in large sets of protein coding genes. We have used this method previously to show that the majority of alternative exons are not undergoing purifying selection (46,47).
The percentage of high impact variants at rare allele frequencies is 1.88% for likely coding genes and this drops to 0.61% for common alleles. Within the likely coding gene set there are genes undergoing positive selection and there may even be genes that are not functionally important within this set. When we filter out immune system genes from the likely coding gene set and calculate high impact variants just for those genes that we detect peptides for, the difference is even starker: 1.6% for rare allele frequencies and just 0.36% for common allele frequencies (Figure 5). Likely coding genes with peptide support also have a much lower non-synonymous to synonymous ratio in common alleles, as would be expected for protein-coding genes evolving under negative selection.
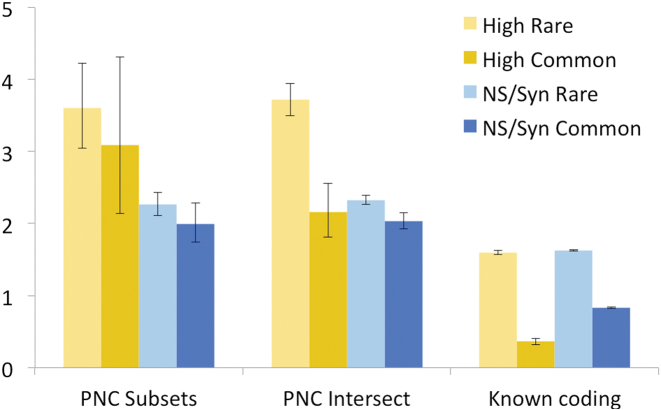
Figure 5.
Genomic variation in likely coding genes and possible non-coding genes. Percentage high impact variants (yellow) and non-synonymous/synonymous ratios (blue) for known coding genes (likely coding genes with peptide evidence, see text) and for possible non-coding genes (PNC) from the intersection of the three sets (Intersect) or annotated by two or fewer reference sets (Subsets). Read-through genes were removed when calculating variants because they always overlap known coding genes. The darker colours show the values for common variants and the lighter shades show the values for rare variants. 95% confidence intervals are shown.
By way of contrast potential non-coding genes annotated in all three sets have proportionally more high impact variants (3.72% at rare allele frequencies and 2.16% at common allele frequencies) and non-synonymous to synonymous ratios (2.33 for rare allele frequencies and 2.03 for common allele frequencies), and the results for potential non-coding genes annotated as coding in just one or two sets are similar (Figure 5). The fact that potential non-coding genes have a much higher proportion of high impact variants and greater non-synonymous to synonymous ratios than likely coding genes, suggests that many potential non-coding genes are unlikely to code for functionally important proteins.
With the genes annotated in Ensembl/GENCODE and RefSeq it is possible to generate human population data for all subsets of genes in Figure 1 with the exception of those genes annotated as coding only by UniProtKB. The percentage of high-impact variants and the ratio of non-synonymous to synonymous variants for these subsets are shown in Supplementary Figure S8. The contrast between genes classified as coding in all three reference databases and those in two or fewer sets is clear. Genes classified as coding in just one or two sets have much higher rates of high impact variants than genes classified as coding across all three databases. There are also no significant differences in non-synonymous to synonymous ratios between rare and common allele frequencies in any set of genes that are classified differently across the three reference sets.
The genetic variation for individual potential non-coding features sheds some light on which of the potential non-coding genes are more likely to code for functional proteins. With the exception of read-through genes (most read-through genes are two known coding genes joined together) all features have genetic variant distributions that are very different from likely coding genes (Supplementary Figure S9). Primate genes, genes with ‘predicted’ UniProtKB evidence and genes with poor PhyloCSF scores have much higher non-synonymous to synonymous ratios and percentages of high impact variants than likely coding genes. However, the non-synonymous to synonymous ratios are lower for common allele frequencies and the differences between rare and common allele frequencies are significant. This suggests that a certain number of genes in these three categories may be functionally important protein-coding genes.
By contrast there are no significant differences in non-synonymous to synonymous ratios between rare and common allele frequencies for genes tagged with the potential non-coding features ‘pseudogene’, ‘uncertain’ UniProtKB evidence and ‘UniProtKB caution’, which suggests that a large majority of these genes are undergoing neutral evolution and are not functionally important.
Another subset of genes with high rates of damaging mutations and little differences between rare and common allele frequency non-synonymous to synonymous ratios are those genes populated entirely by automatically predicted transcript models. There were more than 800 genes predicted automatically in RefSeq and more than 200 in Ensembl/GENCODE. In sets of automatically predicted genes non-synonymous to synonymous ratios are practically identical for rare and common allele frequencies (Supplementary Figure S10), suggesting that most of these genes are also subject to neutral evolution.
Genes with high rates of missense variants
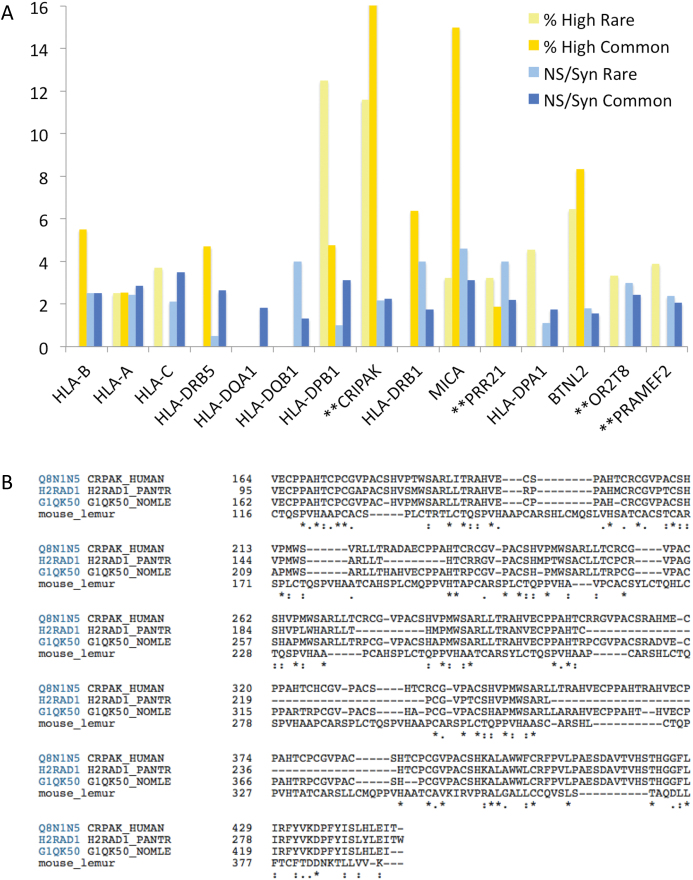
Genetic variation data is useful for pinpointing probable neutral evolution in large cohorts of genes, but the sparseness of the variants means that it is difficult to make conclusions about most individual genes. A number of coding genes do have remarkably high rates of missense and damaging variants though. We looked at the 15 genes with the highest proportion of non-synonymous variants (minimum 30 common allele variants). Nine were HLA histocompatibility antigens (Figure 6), which is not surprising since these genes are known to have many missense variants. Two of the other six genes might also be expected to have higher levels of missense variants because of their likely function. MICA (MHC class I polypeptide-related sequence A) a self-recognising antigen from the major histocompatibility complex class I locus and has more than 50 known alleles, several of which are truncating. Similarly, BTNL2 (Butyrophilin-like protein 2) is a known polymorphic locus bordering the major histocompatibility complex class II and class III regions.
Figure 6.
Genes with the highest proportion of high impact and non-synonymous variants. In (A), the percentage of high impact variants (yellow) and non-synonymous/synonymous ratios (blue) for the 15 genes with the highest rate of common non-synonymous variants. Minimum 30 common variants per gene. The darker colors show the values for common variants and the lighter shades show the values for rare variants. In (B), the alignment between human CRIPAK gene product and primate homologues annotated as CRIPAK in UniProtKB. There is very little evidence of conservation.
The remaining four genes (Figure 6) are CRIPAK, PRAMEF2, PRR21 and OR2T8.
PRAMEF2 and OR2T8 are likely to be pseudogenes; olfactory receptors are highly duplicated and many of these duplications may be pseudogenes, while PRAMEF2 has 22 almost identical paralogues, none of which is supported by protein evidence. PRR21 (Putative proline-rich protein 21) is a single exon gene, which was annotated as ‘uncertain’ by UniProtKB but has since been removed from the UniProtKB proteome. It has an orthologue in chimpanzee, but little other supporting evidence and no evidence of transcript expression. CRIPAK (Cysteine-rich PAK1 inhibitor) was described in a 2006 paper (48) in which CRIPAK constructs appeared to bind and block PAK1 activity. It was described as having 13 zinc finger domains, but the zinc finger domains are not real domains, merely degenerate cysteine-rich repeats (Supplementary Figure S11). Meanwhile CRIPAK is primate-specific and has practically no cross-species conservation at all, as can be seen from the partial alignment of the few orthologous sequences that can be found in UniProtKB (Figure 6). Although transcript expression is ubiquitous, there is no evidence for its expression as protein. Curiously it has the same expression pattern as the upstream coding gene, UVSSA (Supplementary Figure S11). CRIPAK is highly unlikely to be a coding gene and has been reclassified by GENCODE annotators.
Annotation of coding genes based on conflicting evidence
Manual annotators determine the status of genes based on the balance of the available evidence. For most genes, the available evidence is in agreement and the designation of coding or non-coding status is fairly straightforward. However for those genes that might be considered edge cases at the boundary between coding and non-coding, the evidence can often be contradictory.
There are a number of genes in the potential non-coding gene set that are supported by published studies, but that have little other evidence to support their translation to protein in normal tissues. One example is CRIPAK (see above), annotated as coding based on a single published study. At the other end of the spectrum is ARMS2, a gene that evolved in the primate clade from an L2 transposon. Since ARMS2 has been linked to macular degeneration, it has >200 publications, many of which are association studies. The exact role of ARMS2 in macular degeneration is not clear. Experiments carried out with a plasmid-induced protein show that if ARMS2 were expressed in retinal cells, it would be secreted via an unconventional route (49).
Ensembl/GENCODE and UniProtKB annotate DLEU1 (deleted in leukaemia 1) as encoding a short protein as well as 33 non-coding transcripts (Supplementary Figure S12). RefSeq annotate the DLEU1 as non-coding. DLEU1 was added to UniProtKB in 1997, and in 2007 it was annotated as having ‘protein evidence’ because it appeared to interact with other proteins in large-scale protein-protein interaction experiments (50). There is little other evidence that DLEU1 codes for a protein. There is no proper proteomics evidence, very poor cross species conservation, practically no conservation of the reading frame (12) and coding exons of DLEU1 overlap with a SINE (MIRb) element. While UniProtKB annotators use evidence from large-scale protein-protein interaction experiments to label proteins with the evidence code, ‘protein evidence’, evidence from large-scale protein-protein interaction experiments is not always sufficient to confirm protein-coding status. Large-scale interaction experiments construct proteins artificially and use these artificially generated proteins to see if they stick to other proteins. Proteins are generally sticky, even artificial ones, so binding between artificial constructs and real proteins is possible. While a great many of the detected in vitro interactions may also take place in vivo, a number will not. DLEU1 is almost certainly a non-coding gene rather than a coding gene and will be reclassified as non-coding by Ensembl/GENCODE manual annotators.
There are many potential non-coding genes annotated with ‘protein evidence’ because of protein-protein interaction studies. These include DRICH1 (which has a dN/dS above 1 between human and primates and a higher non-synonymous/synonymous ratio for common allele frequencies), FAM218A (which has very little evidence of homologues, even in primates), PRR20C (which has a dN/dS above 1 and homologues in primates only) and RP11-511P7.5 (which appears to be a pseudogene at the 3′ end of ZNF755).
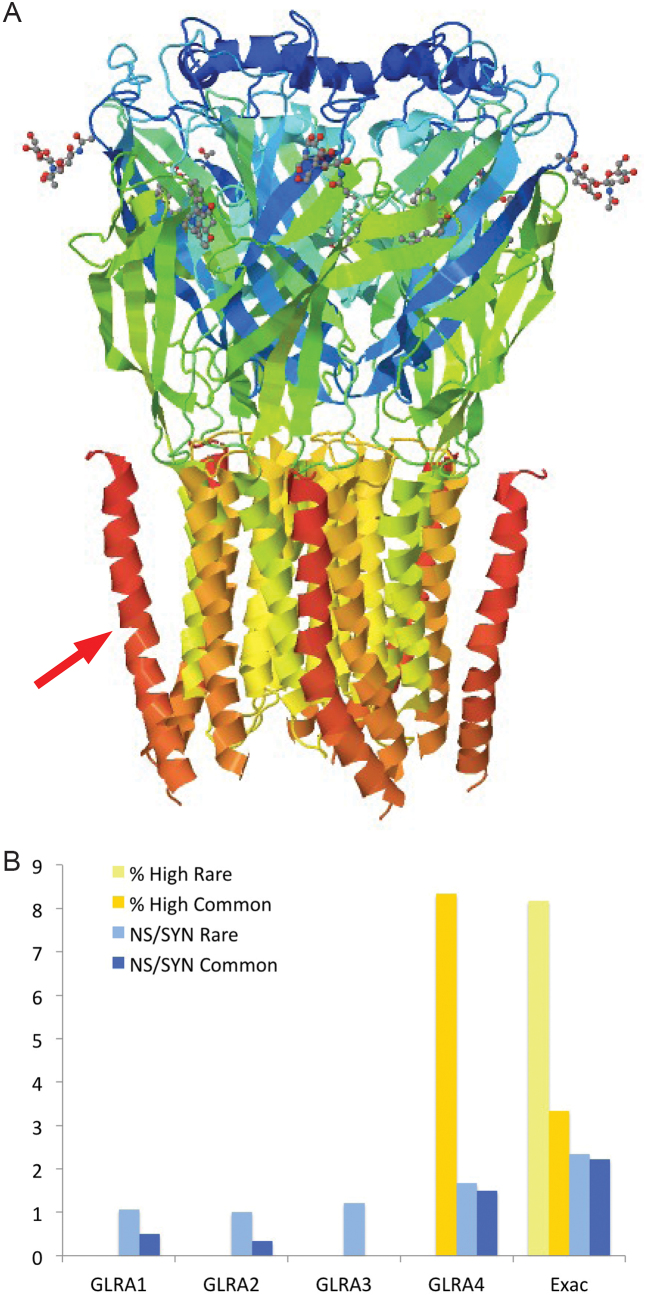
Some genes within the likely coding gene set also have conflicting evidence for their coding capability. Polo-like kinase 5 (PLK5) is detailed in Supplementary Figure S13. Glycine receptor subunit alpha-4 (GLRA4) is interesting because it is one of a number of coding genes that have human-specific stop codons. Glycine receptors are ligand-gated chloride channels and are highly conserved (chicken and mouse GLRA4 are 94% identical over all but the first 40 residues). A mutation in the human version of GLRA4 generates a protein that is truncated 39 amino acids from the C-terminus of the protein, removing the C-terminal trans-membrane helix (Figure 7). This would almost certainly destabilize any pore, and would probably have considerable effect on the function.
Figure 7.
GLRA4 loss of trans-membrane helix and genetic variation. (A) The cryo-EM structure of the GLRA1 kinase domain from Danio rerio (PDB code: 3JAD), which is 80% sequence identical to human GLRA4. In GLRA4, the premature stop codon would lead to the loss of the dark orange trans-membrane helices in the figure (one of which is marked with a red arrow). From the point of view of the pore, this would mean the loss of five of the twenty helices, albeit the helices which are furthest away from the inside of the pore. This would almost certainly destabilize the pore, and would probably have considerable effect on the function. It would also leave the C-terminals of the protein on the cytoplasmic side instead of the extra-cellular side. (B) The percentage of high impact variants (yellow) and non-synonymous/synonymous ratios (blue) for the GLRA gene family. The percentage of high impact variants and non-synonymous/synonymous ratios for GLRA4 from Exac are marked as "Exac". The darker colors show the values for common variants and the lighter shades show the values for rare variants. GLRA4 does not have the same variation pattern as the other four genes.
The genetic variation for the four human glycine receptor genes is shown in Figure 7 along with data for GLRA4 from the Exac experiment (51). The family members with intact structure ( GLRA1, GLRA2 and GLRA3) have no high impact mutations and the non-synonymous to synonymous ratio is higher for rare alleles than for common allelesIn contrast, 3% of common alleles variants in GLRA4 are high impact and the non-synonymous to synonymous ratios are high for both rare and common alleles. The variation data suggest that GLRA4 is not under selective pressure and is likely to be a unitary pseudogene.
The propagation of erroneous annotations
GVQW1 and GVQW2 are short primate-specific genes with poor conservation (Supplementary Figure S14) that are classified as coding in all three reference databases, but that are tagged as potential non-coding in our study. GVQW1 originated from an Alu SINE element, while GVQW2 was annotated as coding recently. Pfam domains (13) are often used to help distinguish coding genes from non-coding genes and both genes seem to have been annotated as coding based on the presence of the domain GVQW.
Pfam annotators have recently removed the transposon-derived GVQW domain from the database as part of a revision of Pfam families because they no longer believe it is a true protein family. Unfortunately, when domains are removed from Pfam, there are no mechanisms to revise genes that were validated as coding based on these Pfam domains.
The now defunct domain seems to have been instrumental in the prediction of 1178 novel human coding genes by the CHESS database [BioRxiv: https://doi.org/10.1101/332825]. These novel predictions were based on RNAseq evidence and similarity to known proteins. More than half of these novel genes were similar to one of just nine UniProtKB proteins (eight human and one chimp). The alignment of the nine proteins (Supplementary Figure S15) shows that they are all closely related.
Two of these proteins came from GVQW1 and GVQW2. Although GVQW1 and GVQW2 are in the process of being reclassified by GENCODE, they are still present in the UniProtKB and RefSeq reference sets. GVQW1 and GVQW2 are transposon-based, so it is reasonable to assume that all nine sequences are derived from Alu sequences (indeed isoforms from both C16orf89 and C9orf85 are among the nine proteins) and are therefore erroneously annotated as coding. This in turn suggests that the novel sequences in CHESS predicted as coding because of their similarity to the nine proteins (more than half of the novel coding genes in CHESS) will also be transposon-related.
Clearly, misannotating genes as protein coding can have important downstream effects on a wide range of databases that depend on reliable predictions of coding genes. The CHESS database's prediction of hundreds of new coding genes based on a defunct, transposon-linked Pfam domain underscores how easily misclassifications can proliferate.
A number of other dead Pfam domains may have been used to help validate the potential non-coding genes, for example C19orf48 and C1orf145. We also ran the Pfam-based tool Antifam (52) to check whether any genes had similarity to known non-coding domains and we found evidence for two more genes, AC079355.1 and AC118758.1, which mapped to the same ‘spurious ORF’ domain. Both coding genes are automatic predictions.
DISCUSSION
There are >22 000 genes annotated as coding across the Ensembl/GENCODE, RefSeq and UniProtKB human proteomes. While manual annotators agree on >19 000 genes, one in eight of these genes are classified differently in at least one of the reference sets. Evidence from various sources suggests that many of the genes classified differently across the three reference sets are unlikely to code for essential proteins; these genes have poor UniProtKB evidence scores, a higher proportion of the most damaging germline variants and non-synonymous to synonymous substitution ratios that suggest many are under neutral selection.
To study differences between these genes and genes annotated as coding in all three reference sets we defined a set of 16 potential non-coding features from the Ensembl/GENCODE reference set. More than 11% of Ensembl/GENCODE coding genes had at least one potential non-coding feature and there were profound differences between these genes and the remaining 89% of genes. Only a handful of potential non-coding genes had reliable proteomics or antibody evidence, most had significantly lower transcript expression and their transcripts were detected in very few tissues. Non-coding genes are known to have much lower levels of expression than coding genes (53), so the fact that so many potential non-coding genes had low or negligible RNAseq expression levels supports the possibility that many will not code for proteins.
Data from genetic variation studies showed that potential non-coding genes had many more copy number variants, a much higher rate of potentially damaging variants, and larger non-synonymous to synonymous substitution ratios. The pattern of variants suggested that many of these genes are under neutral selection. Since neutral selection is not typical of coding genes, this reinforces the likelihood that many potential non-coding genes will not code for functional proteins.
There are 4234 coding genes that could be considered potentially non-coding across the three reference sets. These genes are either annotated differently across the three reference sets or were flagged as potential non-coding (Supplementary Figure S4). If the majority do not code for proteins, as the genetic variation patterns suggest, the number of coding genes will be much closer to the 19 446 genes common to the three reference sets than to the 22 210 genes in the union of those sets. However, it is still early to speculate on the precise number of coding genes because it is impossible to know how many potential non-coding genes will be reclassified by manual annotators, and because there is a steady trickle of new coding genes being annotated (54).
Human population variation data shows that two types of genes in particular appeared not under selection pressure and were therefore unlikely to code for functional proteins. The first are automatic gene predictions, genes in which all gene models are predicted, which make up approximately 1% of Ensembl/GENCODE coding genes and more than 4% of RefSeq coding genes. Our results suggest that these genes are adding little to the human reference annotation. The second group of genes are likely pseudogenes. Pseudogenes form the largest group of non-coding annotations and are especially difficult to distinguish from coding genes but have the clearest evidence for neutral selection of all the potential non-coding features. Likely pseudogenes are particularly prevalent in the UniProtKB unique subset.
Pseudogenes highlight the difficulties that manual annotators face when interpreting the available data (55). Most pseudogenes derive from protein coding genes, either by duplication or retrotransposition, and as a result often have large intact ORFs and protein-like features. In addition recent duplications usually have few obviously deleterious mutations, making the distinction between coding and pseudogene even more difficult. The Ubiquitin carboxyl-terminal hydrolase 17 family has 26 close to identical members, but while non-synonymous to synonymous ratios suggest that most or all are pseudogenes, they are all annotated as coding because there is no clear way of discriminating between them.
Experimental evidence is often ambiguous for many pseudogenes. Negative evidence (evidence to show that a gene does not code for proteins) does not exist, antibodies are rarely sufficiently specific to distinguish similar proteins and proteomics experiments can easily confuse similar peptides because of single-amino acid variations or post-translational modifications. Indeed, a number of the potential non-coding genes detected in the proteomics experiments may be false positive identifications. For example PeptideAtlas validates two peptides for potential non-coding gene FO538757.2. The two peptides identified for FO538757.2 are just one amino acid different from the equivalent peptides from WASH1, a likely coding gene. Indeed UniProtKB annotates both these single amino acid differences as known WASH1 sequence conflicts. It is more than probable that the peptides we detected for FO538757.2 really came from WASH1. Here we should point out that although some identifications will be false positives, many potential non-coding genes, such as SEMG1 and SEMG2, sperm-specific potential non-coding genes with a primate origin, were identified with strong peptide evidence.
The increase in genetic variation data (42,51) should provide valuable support for manual annotators in this sense, though genetic diversity is not infinite and it will not be suitable for all genes. Most bona fide coding genes should have very few high impact variants in common alleles and should have non-synonymous to synonymous ratios that are lower for common allele than they are for rare alleles. We have used genetic variation data to flag a number of possible pseudogenes that were not caught by our potential non-coding features (for example PLK5, GLRA4).
Over the years since the human genome sequence was released (4) rigorous manual annotation has brought us considerably closer to a final catalogue of human coding genes and annotators agree for more than 85% of coding genes. The final 12% of genes, those with the most conflicting evidence, will be more difficult to classify. One useful source of information to discriminate coding from non-coding genes makes use of the recent increase in the number of annotated mammalian genomes (27). With time and with more extensive data, large-scale genetic variation studies could also be a powerful tool to aid in the annotation of coding genes.
In order to flag potential non-coding genes we have built a pipeline that updates with the Ensembl/GENCODE reference set. This approach is a highly practical means of informing the curation of the human genome. The set of human coding genes needs to be as complete as possible for biomedical experiments, but inevitably some genes will be misannotated as coding. Once a gene has entered a reference set it may be propagated in large-scale databases and its coding potential may end up being validated via circular annotation. Detecting errors, retracing steps and rescinding the coding status of a gene once it is annotated as coding is a difficult process, so a system to catch and label genes that have conflicting or insufficient coding support is useful. The pipeline will be used to help pinpoint potential non-coding genes in the Ensembl/GENCODE human reference set. However, the approach could be made available for use by other annotation initiatives and could be extended to the annotation of other species. In fact, a pipeline has already been developed for the mouse reference set. Future releases of these analyses will be made publicly available.
Manual curators from the three main reference databases will investigate and debate the coding potential of these potential non-coding genes. It is important to note that while many potential non-coding genes will be reclassified, those that have evidence of coding capability will be maintained in the reference set. In addition a number of genes with conflicting evidence or insufficient evidence to determine coding status one way or another are also likely to be remain in the reference set. It may be possible to flag this second set of genes as potentially non-coding or pseudogene, while maintaining them as coding in the reference set.
Even if just half of these the potential non-coding genes we have highlighted turn out to be non-coding, this would clearly have a substantial impact on a range of fields. In particular, overestimating the number of coding genes inevitably complicates large-scale biomedical experiments, especially those that involve the mapping of disease-related variations to human genes. The more potential non-coding genes that are classified as coding as part of any analytical process, the noisier the results.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Iakes Ezkurdia and Jon Mudge for their input on this paper.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [2 U41 HG007234 to I.J., L.M., J.M.R. and M.L.T., R01 HG004037 to I.J.]. Funding for open access charge: NIH [2 U41 HG007234].
Conflict of interest statement. None declared.
REFERENCES
- 1. Harrison P.M., Kumar A., Lang N., Snyder M., Gerstein M.. A question of size: the eukaryotic proteome and the problems in defining it. Nucleic Acids Res. 2002; 30:1083–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liang F., Holt I., Pertea G., Karamycheva S., Salzberg S.L., Quackenbush J.. Gene index analysis of the human genome estimates approximately 120,000 genes. Nat. Genet. 2000; 24:239–240. [DOI] [PubMed] [Google Scholar]
- 3. Wright F.A., Lemon W.J., Zhao W.D., Sears R., Zhuo D., Wang J.P., Yang H.Y., Baer T., Stredney D., Spitzner J. et al. A draft annotation and overview of the human genome. Genome Biol. 2001; 2:RESEARCH0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004; 431:931–945. [DOI] [PubMed] [Google Scholar]
- 5. Hubbard T., Barker D., Birney E., Cameron G., Chen Y., Clark L., Cox T., Cuff J., Curwen V., Down T. et al. The Ensembl genome database project. Nucleic Acids Res. 2002; 30:38–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Southan C. Has the yo-yo stopped? An assessment of human protein-coding gene number. Proteomics. 2004; 4:1712–1726. [DOI] [PubMed] [Google Scholar]
- 7. Aken B.L., Achuthan P., Akanni W., Amode M.R., Bernsdorff F., Bhai J., Billis K., Carvalho-Silva D., Cummins C., Clapham P. et al. Ensembl 2017. Nucleic Acids Res. 2017; 45:D635–D642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S. et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012; 22:1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D. et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016; 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clamp M., Fry B., Kamal M., Xie X., Cuff J., Lin M.F., Kellis M., Lindblad-Toh K., Lander E.S.. Distinguishing protein-coding and noncoding genes in the human genome. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:19428–19433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harrow J., Denoeud F., Frankish A., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D. et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006; 7(Suppl 1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A. et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016; 44:D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rolland T., Taşan M., Charloteaux B., Pevzner S.J., Zhong Q., Sahni N., Yi S., Lemmens I., Fontanillo C., Mosca R. et al. A proteome-scale map of the human interactome network. Cell. 2014; 159:1212–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Desiere F., Deutsch E.W., King N.L., Nesvizhskii A.I., Mallick P., Eng J., Chen S., Eddes J., Loevenich S.N., Aebersold R.. The PeptideAtlas project. Nucleic Acids Res. 2006; 34:D655–D658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goodstadt L., Ponting C.P.. Phylogenetic reconstruction of orthology, paralogy, and conserved synteny for dog and human. PLoS Comput. Biol. 2006; 2:e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Church D.M., Goodstadt L., Hillier L.W., Zody M.C., Goldstein S., She X., Bult C.J., Agarwala R., Cherry J.L., DiCuccio M. et al. Mouse Genome Sequencing Consortium. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol. 2009; 7:e1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ezkurdia I., Juan D., Rodriguez J.M., Frankish A., Diekhans M., Harrow J., Vazquez J., Valencia A., Tress M.L.. Multiple evidence strands suggest that there may be as few as 19,000 human protein-coding genes. Hum. Mol. Genet. 2014; 23:5866–5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yates B., Braschi B., Gray K., Seal R., Tweedie S., Bruford E.. Genenames.org: the HGNC and VGNC resources in 2017. Nucleic Acids Res. 2017; 45:D619–D625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Menashe I., Aloni R., Lancet D.. A probabilistic classifier for olfactory receptor pseudogenes. BMC Bioinformatics. 2006; 7:393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Buljan M., Frankish A., Bateman A.. Quantifying the mechanisms of domain gain in animal proteins. Genome Biol. 2010; 11:R74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Herrero J., Muffato M., Beal K., Fitzgerald S., Gordon L., Pignatelli M., Vilella A.J., Searle S.M., Amode R., Brent S. et al. Ensembl comparative genomics resources. Database. 2016; 2016:baw053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roux J, Robinson-Rechavi M. Age-dependent gain of alternative splice forms and biased duplication explain the relation between splicing and duplication. Genome Res. 2011; 21:357–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huerta-Cepas J., Dopazo H., Dopazo J., Gabaldón T.. The human phylome. Genome Biol. 2007; 8:R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cannarozzi G., Schneider A., Gonnet G.. A phylogenomic study of human, dog, and mouse. PLoS Comput. Biol. 2007; 3:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vilella A.J., Severin J., Ureta-Vidal A., Heng L., Durbin R., Birney E.. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009; 19:327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lin M.F., Jungreis I., Kellis M.. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011; 27:i275–i282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rodriguez J.M., Rodriguez-Rivas J., Di Domenico T., Vázquez J., Valencia A., Tress M.L.. APPRIS 2017: principal isoforms for multiple gene sets. Nucleic Acids Res. 2018; 46:D213–D217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005; 21:951–960. [DOI] [PubMed] [Google Scholar]
- 30. Lopez G., Maietta P., Rodriguez J.M., Valencia A., Tress M.L.. firestar–advances in the prediction of functionally important residues. Nucleic Acids Res. 2011; 39:W235–W241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li W., Cowley A., Uludag M., Gur T., McWilliam H., Squizzato S., Park Y.M., Buso N., Lopez R.. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 2015; 43:W580–W584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jones D.T. Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics. 2007; 23:538–544. [DOI] [PubMed] [Google Scholar]
- 33. Käll L., Krogh A., Sonnhammer E.L.. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004; 338:1027–1036. [DOI] [PubMed] [Google Scholar]
- 34. Viklund H., Elofsson A.. Best alpha-helical transmembrane protein topology predictions are achieved using hidden Markov models and evolutionary information. Protein Sci. 2004; 13:1908–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Emanuelsson O., Brunak S., von Heijne G., Nielsen H.. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007; 2:953–971. [DOI] [PubMed] [Google Scholar]
- 36. Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A. et al. Tissue-based map of the human proteome. Science. 2015; 347:1260419. [DOI] [PubMed] [Google Scholar]
- 37. Sudmant P.H., Rausch T., Gardner E.J., Handsaker R.E., Abyzov A., Huddleston J., Zhang Y., Ye K., Jun G., Fritz M.H. et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015; 526:75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sudmant P.H., Mallick S., Nelson B.J., Hormozdiari F., Krumm N., Huddleston J., Coe B.P., Baker C., Nordenfelt S., Bamshad M. et al. Global diversity, population stratification, and selection of human copy-number variation. Science. 2015; 349:aab3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zarrei M., MacDonald J.R., Merico D., Scherer S.W.. A copy number variation map of the human genome. Nat. Rev. Genet. 2015; 16:172–183. [DOI] [PubMed] [Google Scholar]
- 40. Handsaker R.E., Van Doren V., Berman J.R., Genovese G., Kashin S., Boettger L.M., McCarroll S.A.. Large multiallelic copy number variations in humans. Nat. Genet. 2015; 47:296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Abyzov A., Li S., Kim D.R., Mohiyuddin M., Stütz A.M., Parrish N.F., Mu X.J., Clark W., Chen K., Hurles M. et al. Analysis of deletion breakpoints from 1,092 humans reveals details of mutation mechanisms. Nat. Commun. 2015; 6:7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012; 491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. NCBI Resource Coordinators Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016; 44:D7–D19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McLaren W., Gil L., Hunt S.E., Riat H.S, Ritchie G.R., Thormann A., Flicek P., Cunningham F.. The Ensembl Variant Effect Predictor. Genome Biol. 2016; 17:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Deutsch E.W., Overall C.M., Van Eyk J.E., Baker M.S., Paik Y.K., Weintraub S.T., Lane L., Martens L., Vandenbrouck Y., Kusebauch U. et al. Human proteome project mass spectrometry data interpretation guidelines 2.1. J. Proteome Res. 2016; 15:3961–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tress M.L., Abascal F., Valencia A.. Alternative splicing may not be the key to proteome complexity. Trends Biochem. Sci. 2017; 42:98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tress M.L., Abascal F., Valencia A.. Most alternative isoforms are not functionally important. Trends Biochem. Sci. 2017; 42:408–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Talukder A.H., Meng Q., Kumar R.. CRIPak, a novel endogenous Pak1 inhibitor. Oncogene. 2006; 25:1311–1319. [DOI] [PubMed] [Google Scholar]
- 49. Kortvely E., Hauck S.M., Behler J., Ho N., Ueffing M.. The unconventional secretion of ARMS2. Hum. Mol. Genet. 2016; 25:3143–3151. [DOI] [PubMed] [Google Scholar]
- 50. Stelzl U., Worm U., Lalowski M., Haenig C., Brembeck F.H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S. et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005; 122:957–968. [DOI] [PubMed] [Google Scholar]
- 51. Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B. et al. Exome Aggregation Consortium. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016; 536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Eberhardt R.Y., Haft D.H., Punta M., Martin M., O’Donovan C., Bateman A.. AntiFam: a tool to help identify spurious ORFs in protein annotation. Database. 2012; 2012:bas003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012; 22:1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wright J., Mudge J.M., Weisser H., Barzine M.P., Gonzalez J.M., Brazma A., Choudhary J.S., Harrow J.. Improving GENCODE reference gene annotation using a high-stringencyproteogenomics workflow. Nat. Commun. 2016; 7:11778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bruford E.A., Lane L., Harrow J.. Devising a consensus framework for validation of novel human coding loci. J. Proteome Res. 2015; 14:4945–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.