Abstract
Protein synthesis is a fundamental requirement of all cells for survival and replication. To date, vast numbers of genetic and biochemical studies have been performed to address the mechanisms of translation and its regulation in Escherichia coli, but only a limited number of studies have investigated these processes in other bacteria, particularly in slow growing bacteria like Mycobacterium tuberculosis, the causative agent of human tuberculosis. In this Review, we highlight important differences in the translational machinery of M. tuberculosis compared with E. coli, specifically the presence of two additional proteins and subunit stabilizing elements such as the B9 bridge. We also consider the role of leaderless translation in the ability of M. tuberculosis to establish latent infection and look at the experimental evidence that translational regulatory mechanisms operate in mycobacteria during stress adaptation, particularly focussing on differences in toxin-antitoxin systems between E. coli and M. tuberculosis and on the role of tuneable translational fidelity in conferring phenotypic antibiotic resistance. Finally, we consider the implications of these differences in the context of the biological adaptation of M. tuberculosis and discuss how these regulatory mechanisms could aid in the development of novel therapeutics for tuberculosis.
INTRODUCTION
Life processes have historically been considered to be well conserved amongst all domains of life. However, there is increasing evidence that even fundamental biological processes can differ between organisms. One example is translation, the process by which the sequence of nucleotides in a messenger RNA (mRNA) molecule directs the synthesis of polypeptides. With growing evidence that translational regulation associated with heterogeneity in ribosomal composition makes an important contribution to biological adaptation (1,2), it is important to better understand the extent to which selective translation can shape the bacterial proteome, particularly during pathogenesis.
Translation takes place on the ribosome, a ribonucleoprotein composed of a small and a large subunit. The small, 30S, subunit reads the mRNA whilst the large, 50S, subunit catalyses peptide bond formation between incoming amino acids and the growing nascent chain. The ribosome has three binding sites for transfer RNA (tRNA): the A- (aminoacyl) site, the P- (peptidyl) site and the E- (exit) site, formed in the inter-subunit interface. Translation is an intricate process that involves many cellular components, with about half of the energy expenditure of growing cells being used towards protein synthesis (3).
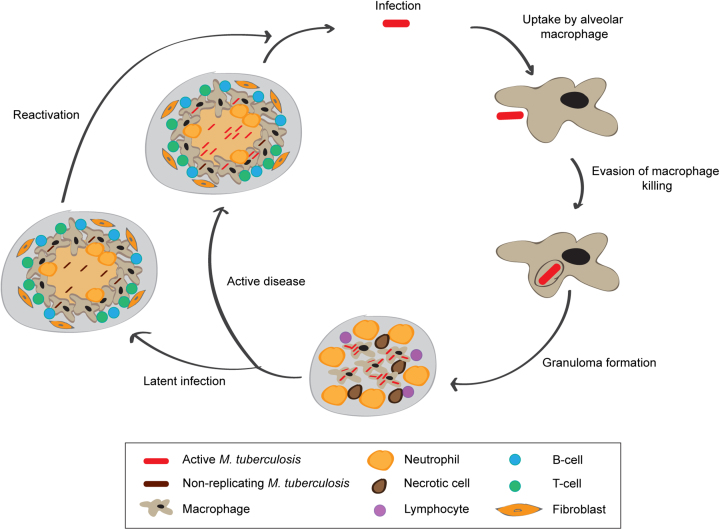
Mycobacterium tuberculosis is the causative agent of human tuberculosis (TB), one of humankind's deadliest diseases. Tuberculosis remains a threat to the health of people worldwide. The World Health Organization (WHO) estimated 10.4 million new cases of TB and 1.4 million deaths caused by this disease in 2016 (4). Furthermore, it is estimated that one-quarter of the world's population harbours the bacterium in the form of an asymptomatic infection referred to as latent TB (5). Person-to-person transmission occurs by inhalation of aerosolized droplets generated by an individual with active disease. Among infected individuals, only a minor percentage will develop clinical manifestations of active TB whilst most will remain latently infected, with bacteria contained within clusters of immune cells called granulomas in the lung (Figure 1). The immunology of TB is complex and multifaceted and even today it is not well known which immunological parameters or biomarkers predict who will control the infection and who will develop clinical disease (6).
Figure 1.
Schematic representation of the life cycle of M. tuberculosis. Person-to-person transmission occurs by inhalation of aerosolized droplets generated by a person with active disease. Bacteria travel to the lungs, where they are taken up by alveolar macrophages. Inside the alveolar macrophages bacteria are exposed to reactive oxygen (ROS) and nitrogen (NOS) species generated by macrophages. M. tuberculosis is able to evade macrophage killing by inhibiting phagosome-lysosome fusion. This leads to recruitment of immune cells, which contributes to the formation of granulomas which can contain M. tuberculosis. In 90% of the cases, infected individuals contain the infection within the granuloma, where the bacteria are able to survive in a non-replicating state, probably triggered by hypoxic and nutrient starved conditions. In around 10% of cases, the disease will progress and develop to active disease, which can lead to release of M. tuberculosis from the granulomas. In a small percentage of latently infected individuals, the disease can reactivate later in life leading to the development of active disease.
Latent infection not only presents an enormous reservoir of potential reactivation of TB but also reflects the complex life cycle of the bacteria that can involve prolonged periods of non-replicating persistence (where the host is able to control the infection but not completely eradicate the bacteria) (Figure 1) (7). During non-replicating persistence, protein synthesis is globally down-regulated and bacteria are correspondingly less susceptible to antibiotics (8). Still today, the mechanisms underlying persistence in the human host are poorly understood. Adaptive responses have been extensively analysed by transcriptional profiling in well-defined experimental models (9–13) and in combination with ChIP-seq analysis, this information is being used to construct networks of genes that have the potential to predict transcriptional responses that occur in environments encountered during in vivo infection (14–17). However, the extent to which the transcriptome can be used to predict changes in the proteome and metabolome—and ultimately drug susceptibility—remains undetermined. For example, immediate survival of M. tuberculosis under nitric oxide stress is likely to be driven by selective degradation of specific proteins and rapid metabolic adjustments rather than by transcriptional regulation itself (18). The recent identification of an extensive leaderless transcriptome in M. tuberculosis (19), characterized by genes lacking a Shine-Dalgarno (SD) sequence, together with the identification of novel features in the mycobacterial ribosome (20–23) highlight a potentially important layer of translational regulation, which could reflect the plasticity of this pathogen to adapt to diverse environmental conditions. The basic biology of M. tuberculosis is clearly different to the model organism Escherichia coli and fundamental biological processes in the single bacterial cell should be re-assessed in light of these differences.
In this review, we summarize what is known about the translational machinery in mycobacteria, discussing the latest evidence that supports the existence of non-canonical translation mechanisms that could contribute to the phenotypic adaptation of M. tuberculosis. We begin with an overview of translation, before discussing differences between the translational machinery in E. coli and M. tuberculosis. We then look at the effects of cellular stress, including the production of toxins and exposure to antibiotics on translation. Finally, we offer perspectives on how a better understanding of translational regulation could be directly relevant to the understanding of M. tuberculosis pathogenesis and potentially applied to the rational design of drug combinations for TB treatment
TRANSLATION INITIATION IN BACTERIA: FROM THE CANONICAL MECHANISM TO LEADERLESS TRANSLATION
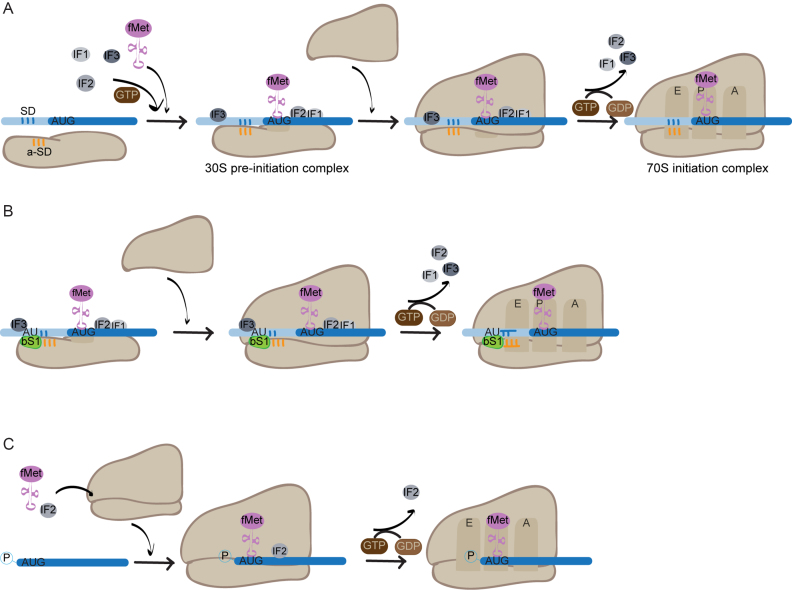
Translation occurs in four steps: initiation, elongation, termination and ribosome recycling. Each of these steps is highly regulated and subject to control mechanisms that ensure the fidelity of the translated product. However, probably the most stringent regulatory mechanisms operate at the level of initiation. In bacteria, canonical translation initiation involves the binding of mRNA, the three initiation factors (IFs) and the initiator tRNA (N-formylmethionine-charged tRNA; fMet-tRNAfMet) to the small ribosomal subunit to form the 30S pre-initiation complex. In prokaryotes, this involves direct interaction between the transcript mRNA and the ribosomal RNA (rRNA). The binding generally occurs between the SD sequence, a purine-rich region upstream of the start codon of mRNA and the complementary anti-Shine-Dalgarno (a-SD) sequence on 16S rRNA. The 30S subunit may bind to any single-stranded region of the mRNA near the translation start site, unwinding any secondary structure. A realignment then occurs to position the start codon in the P-site of the 30S subunit. Next, the large ribosomal subunit docks onto the preinitiation complex, promoted by IF2, which forces the fMet-tRNAfMet into the correct orientation at the P-site (24). The efficiency of docking is dependent on the nature of the mRNA ribosome binding site (RBS) including the sequence of the start codon, any mRNA secondary structure present, the presence of an SD sequence and AU-rich elements in the mRNA. The strength of the RBS is a reasonably good predictor of translation efficiency (25). Lastly, the three IFs dissociate, catalysed by GTP hydrolysis by IF2, and the resulting 70S complex is committed for translation (Figure 2A).
Figure 2.
Mechanisms of translation initiation in bacteria. (A) Mechanism of canonical SD initiation involving recruitment of the 30S subunit through the SD - a-SD interaction. Formation of the 30S pre-initiation complex occurs by binding of IFs1–3 and the initiator tRNA, fMet-tRNAfMet, to the 30S subunit of the ribosome. The SD sequence (represented in blue) is then selected and binds the a-SD sequence (represented in orange) in the 16S rRNA. Recognition of the AUG start codon triggers the stabilisation of fMet-tRNAfMet binding. The 50S subunit docks on the pre-initiation complex and GTP hydrolysis by IF2 results in dissociation of the IFs and tightening of the 70S complex ready for translation. (B) Mechanism of initiation involving the interaction between ribosomal protein bS1 and AU rich elements in the mRNA. This mechanism operates when either a weak SD sequence is present, or it is totally absent. Formation of the 30S pre-initiation complex is mediated by direct interaction of ribosomal protein bS1 with AU-rich elements situated upstream from the SD sequence. (C) Mechanism of initiation of leaderless translation. IF2 and the initiator tRNA bind independently to the 70S ribosome, leaderless mRNA is recruited through recognition of its 5′ phosphate group and AUG start codon. Following GTP hydrolysis by IF2, the IF dissociates, and the complex is committed for translation.
The interaction between the SD and the a-SD motifs is thought to be the canonical mechanism for translation initiation in bacteria, as alterations of the SD sequence strongly inhibit protein synthesis (26–28). However, representation of genes with SD motifs in their leader sequence is highly variable across bacterial genomes (29) and genes lacking a leader sequence (known as leaderless) are widespread among bacteria (30).
At least two alternative mechanisms of translation initiation have been described to operate in bacteria. The first is mediated by the ribosomal protein bS1, a component of the 30S subunit. In E. coli, bS1 can directly interact with AU-rich sequences upstream of the SD sequence in the mRNA resulting in efficient initiation of translation (31,32) (Figure 2B). There is no similar protein to the ribosomal protein bS1 in Eukaryotes, but it is well conserved among Gram-negative bacteria, with six distinct domains present that play defined roles during the translation process in E. coli. More distantly related forms have been described in Gram-positive bacteria that vary in the presence and conservation of the protein domains. Interestingly, members of the phylum Actinobacteria, which includes M. tuberculosis, have a bS1 that has an uncharacterized C-terminal domain that could influence the specificity of bS1-mediated initiation (33).
The second mechanism operates when mRNAs lack the 5′ elements usually required by the ribosome for binding. Such mRNAs are described as leaderless and without the presence of a 5′ untranslated region (UTR) or ribosome recognition signals, they are thought to represent an early evolutionary relic of translation (34). Despite their suspected ancestral origin, leaderless mRNAs are found in all domains of life (30).
In bacteria initiation of translation of leaderless mRNAs occurs by a mechanism involving pre-formed 70S ribosomes (35) (Figure 2C). It has been shown that in E. coli leaderless translation requires a 70S ribosome and an mRNA transcript equipped with a 5′ terminal phosphate (36,37). In fact, in the presence of an initiator tRNA, 70S ribosomes have a ∼10-fold higher affinity for 5′ AUGs than 30S subunits have (35 and references therein). In E. coli, initiation by 70S ribosomes also requires an AUG start codon situated in close proximity to the 5′ terminus (38); alteration of the AUG start codon to other naturally occurring initiation codons is insufficient for the translation of leaderless mRNA (39). Initiation then proceeds via binding of the initiator fMet-tRNAfMet to the 70S ribosome followed by the leaderless mRNA binding. The ratio of IF2 and IF3 plays a decisive role in the initiation of leaderless mRNAs, with higher relative concentrations of IF2 promoting leaderless translation (40).
The distribution of leaderless transcripts has been predicted to be diverse amongst Eubacteria, with a notably high representation amongst the Actinobacteria (41,42). Few leaderless mRNAs are expressed in E. coli during optimal growth, and 70S-initiation yields less efficient translation than canonical SD-mediated initiation. Thanks to advances in next-generation sequencing technologies, mapping of transcriptional start sites (TSSs) at a genome-wide level is now possible, revealing the primary transcriptome of bacterial pathogens (43). This technique has allowed the quantification of leaderless transcripts at a transcriptome-wide level, confirming their high abundance amongst members of the Actinobacteria (44,45). In particular, genome-wide mapping of TSSs in M. tuberculosis has revealed that approximately a quarter of the transcriptome is expressed as leaderless (19,46), highlighting a fundamental difference with the transcriptome of other bacterial pathogens where only one to two percent of the transcriptome is leaderless (43,47,48). Interestingly, the contribution of leaderless translation to the shaping of the E. coli proteome varies when environmental conditions change, and bacteria face different stresses, highlighting the potential implication of this alternative translation initiation mechanism in re-shaping the bacterial proteome during stress adaptation.
PREFERENTIAL TRANSLATION OF LEADERLESS mRNAS IN E. coli
In E. coli, efficient translation of leaderless transcripts is performed by specialized ribosomes. The concept of specialized ribosomes emerged after the identification of heterogeneity in the composition of ribosomes amongst a broad range of organisms and cell types (reviewed in (1)). This heterogeneity can result from differential expression or post-translational modifications of ribosomal proteins, differences in the rRNA sequence or variation in the activity of ribosome associated factors.
There are two mechanisms described in E. coli that generate specialized ribosomes that either lack specific ribosomal proteins or have structural changes in the 16S rRNA. Both mechanisms are triggered by stress conditions such as the presence of antibiotics or bacterial toxins (49,50). The first mechanism can be induced following exposure to kasugamycin, an aminoglycoside antibiotic that inhibits formation of the initiation complex by binding to the 30S subunit and inducing dissociation of the P-site-bound fMet-tRNAfMet through perturbation of the mRNA. This results in inhibition of the translation of SD but not leaderless transcripts (51,52). In addition to changes induced by direct binding to the ribosome, kasugamycin also induces the formation of 61S ribosomes in E. coli, both in vitro and in vivo. These specialized ribosomes are devoid of several small subunit proteins, including bS1 and bS21, have structural changes in the 16S rRNA (in particular within helix 45 at the 3′-terminus) and are proficient in selectively translating leaderless mRNAs (49).
The second mechanism is mediated by the MazF toxin. MazEF is a stress-induced toxin-antitoxin (TA) module implicated in stress survival and persistence (53). Under optimal growth conditions, the toxin, MazF, is neutralized by binding to the antitoxin, MazE. However, under stress conditions, the unstable antitoxin is degraded and, MazF inhibits translation by cleaving both mRNA and rRNA. In E. coli, Vesper and colleagues have shown that MazF cleaves at ACA sequences within the 5′UTR of mRNAs, removing the SD sequences and hence generating leaderless transcripts (50). The system also targets 16S rRNA, cleaving off the 3′-terminal 43 nucleotides of the 16S rRNA that include the a-SD sequence and resulting in the formation of 70SΔ43 stress-ribosomes that selectively translate the newly cleaved leaderless mRNAs. Further studies combining RNA sequencing with polysome profiling have identified a MazF regulon comprising over 300 genes in the E. coli transcriptome that are selectively targeted (54). Interestingly, the set of MazF-processed transcripts identified encode proteins with a broad variety of functions, leading the authors to conclude that translational reprogramming in E. coli serves as a fast and effective mechanism for shaping the proteome during stress to guarantee bacterial survival. Furthermore, it has been recently shown that the 70SΔ43 stress-ribosomes generated as a result of the post-transcriptional response generated by MazF can be repaired when optimal growth conditions resume (55). During stress conditions, both the 70SΔ43 ribosome and the cleaved RNA43 are stable, but upon recovery from stress the stable RNA43 can be re-ligated to the 16SΔ43 rRNA via the RNA ligase RtcB, generating repaired 70S ribosomes that are able to translate canonical mRNAs. This reversibility of ribosome heterogeneity opens a new layer on the control of translational regulation in bacteria, equipping the bacterial cell with a dynamic mechanism to modulate the proteome in response to varying conditions.
As previously mentioned, M. tuberculosis has an extensive leaderless transcriptome (19,46). In M. tuberculosis, leaderless genes are not evenly distributed throughout the transcriptome, but genes encoding proteins with secondary adaptive functions, such as TA systems, are over-represented amongst leaderless mRNAs (19). Using translational reporters, Shell and colleagues were able to demonstrate that leaderless translation in Mycobacterium smegmatis, a non-pathogenic and fast-growing mycobacteria frequently used as a surrogate model for M. tuberculosis, can be efficient when either an AUG or GUG codon is present at the 5′end of the mRNA (46), revealing an important difference from the E. coli model, in which an AUG codon is strongly preferred (56). Furthermore, the observation that under conditions of nutrient starvation, the abundance of leaderless transcripts increases (19) highlights a potential target for translational regulation in M. tuberculosis. It is not yet known whether a similar specialized subpopulation of ribosomes as that described in E. coli, is responsible for leaderless translation in mycobacteria, or whether mycobacterial ribosomes are more versatile and the same population can translate either canonical leadered or leaderless mRNAs. Taken together with the observations that kasugamycin treatment and the presence of endogenous toxins also influence not only the translatome but also the translational machinery of the cell, this suggests that leaderless translation may be an important aspect of the adaptive response of bacteria (19,46). During the next sections, we will review evidence that could support this hypothesis.
RIBOSOMAL HETEROGENEITY IN MYCOBACTERIA: INSIGHTS FROM STRUCTURAL STUDIES
It is anticipated that structural studies of ribosomes from M. tuberculosis may help to shed light on the mechanism of leaderless translation in mycobacteria, specifically on whether all ribosomes are equally capable of translating all types of transcript, or whether a specialized subpopulation of ribosomes preferentially translates leaderless mRNAs.
Prokaryotic ribosomes, exemplified by the model organism E. coli, are around 20 nm in diameter with a molecular weight of ∼2.5 MDa and comprise two subunits. In E. coli, the 30S subunit consists of 16S rRNA (1542 nucleotides) and 21 proteins (S1–S21). The large subunit consists of 5S rRNA (120 nucleotides), 23S rRNA (2904 nucleotides) and 33 proteins (L1-L36, where the previously annotated bL12 has been shown to be a complex of bL7/bL12, L8 to be absent and L26 to be bS20). With the exception of bS21, which is implicated in SD-mediated initiation (57), all E. coli ribosomal proteins have homologues in M. tuberculosis, although there are some differences in size and charge.
In general, ribosomal proteins from M. tuberculosis are larger than those from E. coli, with some (uS2, uS3, uS5, uS9, bS16, uS17, bS18, uL4, uL10, bL17, uL22, bL25 and uL29) thirteen or more residues longer than their E. coli homologues. The larger size of M. tuberculosis proteins compared with E. coli proteins is in good agreement with data from M. smegmatis, in which the same proteins are also larger than their E. coli homologues (20). Furthermore, ribosomal proteins from M. tuberculosis also tend to be more positively charged (have a higher isoelectric point) than those from E. coli, especially bS6, bL9 and uL22. The absence of bS21 in M. tuberculosis is interesting because it represents a point of similarity between mycobacterial ribosomes and 61S ribosomes from E. coli that are both able to translate leaderless transcripts with high efficiency (49). In addition, the ribosomal protein bS1 differs between ribosomes from E. coli and M. tuberculosis. In E. coli, bS1 comprises six domains; domains 1 and 2 bind the protein to the ribosome, whilst domains 3–6 are involved in interactions with the mRNA, including mediating mRNA unfolding and docking (58) with the sixth domain having been shown to be dispensable for translation initiation. In M. tuberculosis, the sixth domain of bS1 is replaced by a domain of unknown function of approximately 100 residues, which seems to be specific to Actinobacteria and renders bS1 functionally inequivalent to bS1 from E. coli (33). In addition to the set of ribosomal proteins required for assembly of the bacterial ribosome, M. tuberculosis has at least four duplicated or alternative ribosomal proteins, that differ from their primary partner in lacking a Cys-rich zinc binding motif. Interestingly, the ratio of primary to alternative ribosomal protein bS18 in M. tuberculosis varies during conditions of zinc deprivation, with an increased production of the alternative protein that gets assembled into alternative ribosomes. This mechanism might provide the bacteria with a mechanism to ensure protein production in the zinc depleted extracellular environment, following release from macrophages (59,60).
The near-atomic resolution structures of ribosomes from M. smegmatis and M. tuberculosis were published recently, yielding important insights into the translational machinery of this pathogen (21,23). Compared with other bacterial ribosomes, the overall architecture of M. tuberculosis ribosomes is conserved, with all the usual structural landmarks (the central protuberance, the L1 stalk etc.) in place. However, some regions of additional electron density were noted, which correspond to structures that appear to be unique to mycobacterial ribosomes.
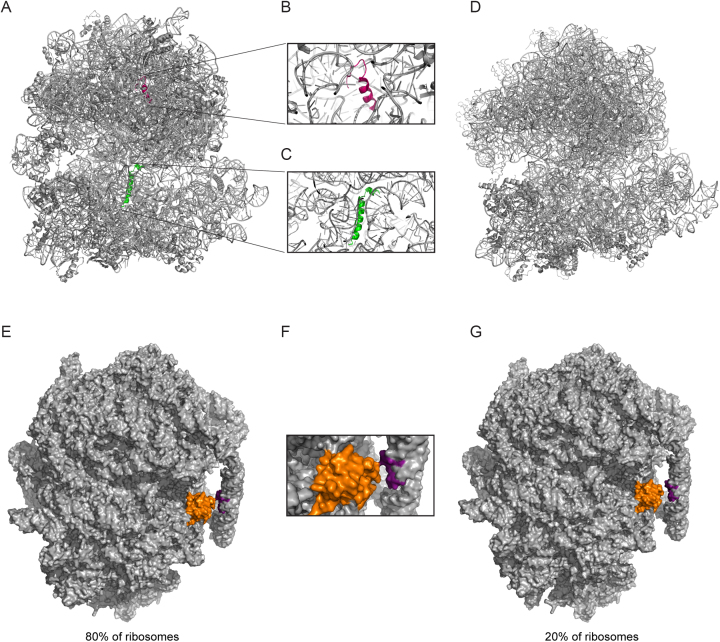
Most notably, two new protein densities were discovered near the decoding centre and the peptidyl transferase centre (PTC) (Figure 3A). The first of these proteins, designated bL37, sits close to the PTC and appears to be unique to mycobacteria. It consists of ∼20 residues and adopts an alpha-helix-coiled loop structure. The protein resides in a pocket formed by helices H39 and H89 of the 23S rRNA (Figure 3B). The presence of this protein is corroborated by evidence from the structure of the ribosome from M. smegmatis, in which a similar pattern of electron density was observed (20,21). The second protein, named bS22, closely resembles the eukaryotic protein eL41, in terms of both fold and location. The protein lies in a highly conserved pocket that is found empty in E. coli (61) (Figure 3C) but occupied by an alpha-helical protein in both cytosolic and mitochondrial ribosomes from Saccharomyces cerevisiae and Homo sapiens (62–65). This protein is also present in the M. smegmatis ribosome where it is hypothesized to play a major role in the stabilization of the assembled 70S complex (21).
Figure 3.
Structural differences in mycobacterial ribosomes. (A) Structure of M. tuberculosis ribosome showing the two novel proteins bL37 (pink) and bS22 (green). Images generated from alignment of PDB files 5V93 (M. tuberculosis 70S ribosome) and 5O61 (M. smegmatis 70S ribosome; to show bL37). Zoomed in views of bL37 (B) and bS22 (C). (D) Structure of E. coli ribosome lacking bL37 and bS22 (generated from PDB file 4V4A). (E) The intersubunit bridge, B9, formed between protein bS6 (orange) and nucleotides 1576–1578 and 1628–1630 of the handle (purple) is present in about 80% of M. tuberculosis ribosomes. A zoomed in view of this region is shown in (F). (G) 20% of ribosomes lack the B9 bridge, with implications for subunit assembly.
Other interesting features of M. tuberculosis ribosomes were also uncovered, including the presence of a novel inter-subunit bridge, B9, present in about 80% of the ribosomes, formed from a 100-nucleotide expansion in the 23S rRNA, that undergoes a dramatic conformational change upon association of the 30S and 50S subunits (Figure 3E and F). The authors propose that this structure may play a role in coordinating translation initiation in M. tuberculosis, perhaps by preventing the premature formation of the 70S ribosome until the initiation complex is properly formed. Intriguingly, the study also found that M. tuberculosis ribosomes exhibit significant structural heterogeneity, raising the possibility that there may be subpopulations of ribosomes that differ from canonical ribosomes—perhaps these specialized ribosomes are responsible for the translation of leaderless mRNAs in M. tuberculosis. For example, the absence of the B9 bridge in 20% of the ribosomes could render these ribosomes more amenable to 70S association, which is a prerequisite for the translation of leaderless mRNAs (Figure 3G).
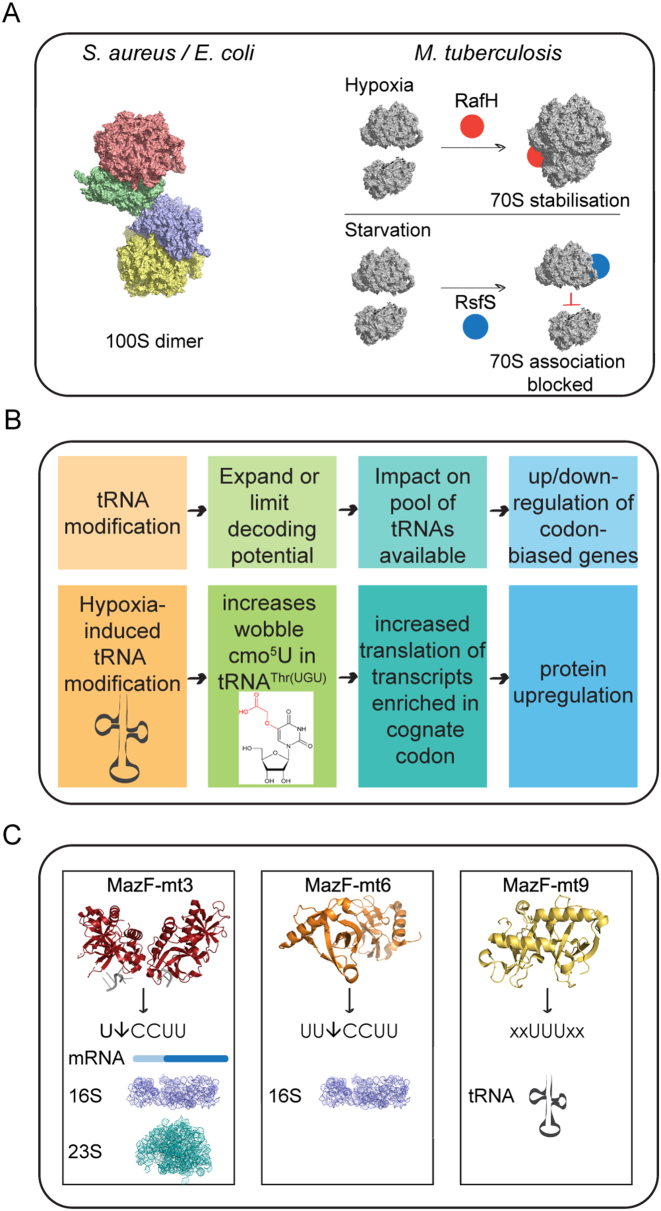
Ribosomal heterogeneity can also be achieved at the level of the stabilized ribosome. Ribosome stabilization can be defined as the association of ribosomal subunits or ribosomes rendering the ribosome translationally inactive. In E. coli, when cells stop growing a proportion of the ribosomal population undergoes dimerization, generating 100S ribosomal dimers that are translationally inactive and considered to be in a hibernation state (reviewed in (66)) (Figure 4A). This provides another example for reversible bacterial stress management, so when cellular conditions become favourable again the hibernating ribosomes can be disassembled and recycled for new rounds of translation. In contrast to E. coli, it has been shown that under conditions of hypoxic stress, M. tuberculosis ribosomes do not form 100S dimers but become stabilized in the associated 70S form and do not readily dissociate into their 30S and 50S subunits (67). This stabilization is mediated by the protein RafH, a member of the DosR regulon (67). The DosR regulon comprises over 50 genes and is involved in inducing dormancy in M. tuberculosis (68). In M. smegmatis, the RafH protein becomes associated with the ribosome under hypoxic conditions (67) (Figure 4A). This is one of the two long hibernation promoting factors (HPF) described in M. tuberculosis that are related to the RMF/short-HPF/YfiA proteins of E. coli (69) and bind to ribosomes in a way that modifies subunit interactions and translational efficiency. In addition, a recent study has identified structural similarities for the gene Rv1738 to HPFs from E. coli using racemic protein crystallography (70). This gene is highly up-regulated in M. tuberculosis during conditions thought to trigger the onset of dormancy, like hypoxia and exposure to nitric oxide (68,71), providing a new ribosome-associated protein that could selectively modify the proteome by interacting with ribosomes. Ribosome silencing factors also bind to the ribosome under unfavourable growth conditions to shut down translation. Combining structural studies and biochemical analysis, Li et al. have shown that during periods of nutrient limitation the ribosomal silencing factor RsfS from M. tuberculosis binds to the 50S inhibiting the association of the small 30S subunit to block protein synthesis (72) (Figure 4A). RsfS interaction does not affect pre-formed 70S monosomes, potentially unaltering translation of leaderless transcripts during nutrient starving conditions.
Figure 4.
Mechanisms by which translational regulation can act in mycobacteria. (A) Stabilization of ribosomes during non-optimal growth. During non-optical growth conditions, both E. coli and Staphylococcus aureus 70S monosomes form dimers to generate 100S hibernating ribosomes through the interaction of their 30S subunits. This hibernation is reversible when conditions turn favourable again. Here the 100S hibernating ribosome from S. aureus is shown (PDB 6FXC) that slightly differs from the E. coli counterpart (98). In mycobacteria, under hypoxic stress, RafH promotes the stabilization of 70S ribosomes, whereas under conditions of starvation, 50S ribosomal subunits are bound by the ribosome silencing factor, RsfS - crystal structure shown here (PBD 4WCW) - blocking association of monomer via interaction with the ribosomal protein uL14. RsfS interaction does not affect pre-formed 70S monosomes. (B) Impact of tRNA modifications on translation. Top boxes show a schematic of the impact that tRNA modifications have on translation. Bottom boxes illustrate the example of uridine 5-oxyacetic acid modification in tRNAThr(UGU) induced by hypoxia in M. bovis. (C) Role of TA systems in M. tuberculosis translation. MazF-mt3 (PDB 5HK0) cleaves around 20% of M. tuberculosis mRNAs and also cleaves within the anti-Shine-Dalgarno (aSD) sequence at the 3′ end of 16S rRNA and U*CCUU in 23S rRNA (in the A-site). MazF-mt3 may possess dual functionality, with the potential to either completely inactivate the ribosome via 23S rRNA cleavage or alter the specificity of the ribosome by removing the aSD sequence. MazF-mt6 (PDB 5UCT) cleaves UU*CCU and inactivates the ribosome by cleaving helix/loop 70 in 23S rRNA in the same position as mt3. MazF-mt9 (PDB 5WYG) is the only MazF member to specifically degrade tRNA in M. tuberculosis. The consensus UUU triplet needs to be situated in the middle of the 7-nt anticodon loop (flanked by 2 nucleotides at either side).
Altogether, this detailed structural information is proving a valuable resource for understanding the different levels at which translational regulation can occur and also the modes of action of numerous antibiotics that target the ribosome. In particular, it may help shed light on the mechanisms by which M. tuberculosis becomes resistant to antibiotics and aid in the design of novel compounds to inhibit translation. For example, the newly identified protein bL37 has the potential to influence the orientation of the 23S rRNA close to the linezolid binding site, an oxazolidinone antibiotic used to treat M. tuberculosis that is resistant to first-line antibiotics, altering the conformation of nucleotides involved in conferring resistance to this antibiotic (21). A further species-specific alteration in the rRNA in the region of the PTC causes remodeling of a tRNA-binding loop and leads to greater flexibility of the 23S rRNA near the PTC, with implications for tolerance of mutations that are lethal in E. coli but confer linezolid resistance in mycobacteria (21).
EXPERIMENTAL EVIDENCE FOR TRANSLATIONAL REGULATION IN MYCOBACTERIA DURING STRESS ADAPTATION
Having looked at differences in initiation, in the types of mRNA transcripts that can be decoded and in the structures of ribosomes from M. tuberculosis and E. coli, in this section we now consider features of elongation and stress responses.
Codon preference and mistranslation
M. tuberculosis has a GC-rich genome (65%) and this is reflected in a strong bias towards G/C at the third base position of preferred codons for every amino acid (73). In contrast, the preference for G-starting codons appears to be a general feature of bacteria, regardless of their overall GC content (74). Preferred codons are those that are recognized by the most abundant tRNAs and tend to occur in highly expressed genes, whereas in genes expressed at lower levels, codon usage is more uniform. Codon usage also has implications for mRNA stability, with mRNAs rich in optimal codons being more stable than those rich in rare codons (75).
Like the ribosome, tRNA is not a passive player in translation, but rather its role as an adapter molecule between mRNA and proteins is central to the process. tRNAs have highly conserved secondary and tertiary structures, crucial for their interactions with proteins and other RNA molecules but must also possess a certain degree of flexibility. However, misreading can occur. A study in M. smegmatis quantified all possible misreading errors at a defined location in a firefly luciferase reporter protein and found that during stationary phase there was a significant increase in mistranslation by tRNALys(CCU) from 0.05% mis-incorporation in exponential growth to 0.2% in stationary phase. Furthermore, treatment with streptomycin increased misreading errors at several codons whereas oxidative stress had no effect on translational fidelity (76). Despite the intrinsic need for high translational fidelity, quality control appears to vary depending on changes in growth conditions. It is possible that mistranslation can even be beneficial to cells, increasing proteomic diversity and facilitating adaptive protein evolution (77). In fact, high rates of substitution of glutamate for glutamine and aspartate for asparagine during specific growth conditions, have been shown to lead to phenotypic resistance to rifampicin in M. smegmatis (78) and in M. tuberculosis the GatCAB complex mediates the translational fidelity of glutamine and asparagine codons, also with implications for rifampicin-specific phenotypic resistance (79).
Amongst all RNA species, tRNAs have the highest density of post-transcriptional modifications, reviewed in Lorenz et al. (80). These modifications are important for the fine-tuning of the 3D structure of tRNA and range from simple methylations of the base or the ribose to large-scale hypermodifications that result in significant structural changes or stabilization. tRNA modifications form a dynamic system that can respond to cellular stress by expanding or limiting the decoding capabilities of tRNAs, leading to the selective up- or downregulation of codon-biased genes (81) (Figure 4B). This has recently been shown in a model of hypoxic stress that mimics the environment M. tuberculosis encounters in granulomas. Chionh et al. grew cultures of the M. tuberculosis surrogate, M. bovis Bacille Calmette Guérin (BCG), and subjected them to the slow withdrawal of oxygen before extracting total RNA and analyzing the tRNAs for modifications. They identified a number of hypoxia-induced tRNA modifications, some of which were accompanied by significant changes in the number of copies of particular tRNAs, leading to up- or downregulation of translation of mRNAs with biased cognate codons for these tRNAs (Figure 4B). Furthermore, they found that the BCG genome exhibits biased use of synonymous codons that are read by hypoxia-induced reprogrammed tRNAs, leading to selective translation of proteins from these genes during hypoxic stress and that the concentration of the DosR protein, a transcription factor that mediates the hypoxic response, was closely paralleled by changes in the level of tRNAThr(cmo5UGU) but not in the levels of dosR mRNA (82).
Taken together, a complex picture of codon matching and preference begins to emerge in which the stability of the mRNA, the efficiency of its translation and the modification of tRNAs all play a role in determining the amount of protein produced.
Toxin-antitoxin systems
Toxin-antitoxin (TA) systems are small genetic modules encoding both a toxin and its corresponding antitoxin and are best characterized in E. coli. The antitoxin tends to be less stable than the toxin and is readily degraded under conditions of environmental stress, freeing the toxin, which then targets cellular processes such as DNA replication, cell wall biosynthesis or translation. If further antitoxins are produced, they will bind to the toxins preventing them from causing further disruption. Thus activation of TA systems could facilitate bacterial survival until the environmental stress is removed and conditions become favourable for growth again (83). There are five types of TA system categorized by the mode of action of the toxin. Here, we focus on type II systems (in which both toxin and antitoxin are proteins) affecting translation. There are differences not only in the numbers of TA systems in E. coli and M. tuberculosis, but also in their modes of action and effects on translation.
E. coli has eleven type II TA loci, all of which encode mRNA endoribonucleases that inhibit global cellular protein synthesis, inducing persistence. M. tuberculosis has undergone a large expansion in the number of TA systems (79 putative or confirmed) (84), which belong to six families: VapBC, MazEF, YefM/YoeB, RelBE, HigBA and ParDE. It has been proposed that active toxins may play a role in persistence as at least ten TA systems have been shown to be up-regulated under conditions of antibiotic-induced persistence (85) and conditions that result in arrest of translation (86). With the exception of the ParDE family, which blocks DNA replication by inhibiting DNA gyrase, and the DarTG system that modifies thymidines on single-stranded DNA by ADP-ribosylation (87), the other toxin families all act on RNA and influence protein synthesis. In the VapBC system, the toxin, VapC, is a ribonuclease that cleaves CG-rich RNAs. There are 50 VapBC systems in M. tuberculosis and 11 of these have been implicated in persistence due to their elevated concentrations in a nutrient starvation model (88). Recently, the cellular targets of these toxins have been identified in M. tuberculosis (89). These toxins inhibit protein translation by cleaving RNAs that are essential for decoding at the ribosomal A-site. In particular, at least six VapCs cleave specific tRNAs and the VapC20 toxin cleaves the conserved Sarcin-Ricin loop of the 23S rRNA (89).
As described above, the MazF toxin from E. coli plays an important role in the translational reprogramming of the cell following stress (50,54). M. tuberculosis produces nine MazF family members TA modules. The toxin MazF-mt6 has been shown to disrupt protein synthesis by cleaving 23S rRNA in free 50S subunits at a conserved site in the ribosomal active centre, the helix/loop 70, disrupting tRNA binding at the A-site (90,91) (Figure 4C). This cleavage is sufficient to inhibit protein synthesis in the absence of mRNA cleavage. Furthermore, MazF-mt6 destabilizes the association of the 30S and 50S ribosomal subunits (90). In addition to this role and in similarity with the mechanism described in E. coli, the toxin MazF-mt3 of M. tuberculosis also has the ability to cleave the a-SD sequence of 16S rRNA, both from the precursor and mature forms of 16S rRNA and also within the context of the 70S ribosome (92) (Figure 4C). Based on the recognition sequence for cleavage, it is predicted that only around 20% of M. tuberculosis genes are susceptible to cleavage by MazF-mt3. Due to the high proportion of leaderless transcripts already present in the M. tuberculosis genome, it is tempting to speculate that specific cleavage of mRNAs to create de novo leaderless transcripts is not as important in this pathogen as it is in E. coli but that cleavage of the a-SD sequence from the 16S rRNA is a high priority to ensure the availability of a pool of ribosomes capable of translating leaderless transcripts. Finally, a recent study has shown that in addition to cleavage by MazF within rRNA and mRNAs, the MazF-mt9 system from M. tuberculosis preferentially targets a subset of tRNAs, mainly tRNAPro14 and tRNALys43 but also tRNALys19, tRNAVal22, tRNALeu13 and tRNAAsn36 (93) (Figure 4C). Following a computational analysis to identify proteins enriched in proline, lysine and asparagine codons they predict that the MazF-mt9 system can inhibit or reduce the rate of protein synthesis for transcripts enriched in these three amino acids, mainly ribosomal proteins and members of the PE/PPE families, a group of proteins only found in M. tuberculosis that have been suggested to be a source of antigenic and genetic variation (94).
Both YoeB and RelE toxins from E. coli are ribosome-dependent ribonucleases that cleave mRNA at the A-site, albeit by different mechanisms. This feature of RelE has recently been exploited to improve the resolution of ribosome profiling studies in E. coli (95). M. tuberculosis encodes two RelBE systems and one YefM/YoeB system and their overexpression leads to an increase in the proportion of bacilli surviving rifampicin treatment (96). The HigB toxin is also a ribosome-dependent ribonuclease that binds the 50S subunit and cleaves AAA sequences on the mRNAs being translated. M. tuberculosis possesses three HigBA toxin systems, with HigBA1 and HigBA2 amongst the ten most up-regulated toxin-antitoxin systems in drug-tolerant persisters (85). The HigBA1 system has been shown to cleave and degrade transfer-messenger RNA (tmRNA), the stable RNA product of the ssrA gene, in M. tuberculosis (97).
TA systems thus play a vital role in inducing down- or selective-regulation of cellular processes, including translation, in response to environmental cues. As we have seen, emerging evidence implicates TA systems in inducing persistence, surviving antibiotic treatment and promoting bacterial survival until environmental conditions improve. As such, TA systems are potential targets for therapeutic interventions against bacterial infection.
Perspectives
M. tuberculosis responds to environmental change by altering the expression of critical genes that favour its growth and survival. As demonstrated in this Review, these changes operate not only at the level of transcription, but also at the level of translation, with certain types of transcript changing in abundance and presumably also in translational efficiency as the ribosome also undergoes changes that favour particular mechanisms of translation, such as stabilization of 70S ribosomes.
Both the improvement of new drug treatment regimens and the development of new therapeutics against TB are current imperatives, but progress is hampered by our incomplete understanding of the fundamental biology of the pathogen and an over-dependence on the knowledge of E. coli. In particular, the molecular mechanisms underlying the regulation of translation, which ultimately lead to changes in the metabolic profile of the bacterium, require clarification. The complex lifestyle of this bacterium renders M. tuberculosis populations phenotypically heterogeneous, suggesting a connection between differences in the phenotypic state of bacteria and selective mechanisms of translation. Thus, a deeper understanding of the translational responses of M. tuberculosis should provide clues to better understand the adaptive response and identify therapeutic targets that are susceptible even in persisting bacteria.
CONCLUSIONS
As we have seen in this Review, there is considerable scope for diversity of fine mechanistic detail and regulation in life processes. Translation is a highly conserved process that is fundamental to life, yet seemingly, small variations in this process even between bacteria can lead to important differences in the way bacteria are able to adapt and survive, particularly in the face of environmental stress. In this Review, we have considered differences in protein synthesis in the model organism E. coli and the pathogen M. tuberculosis, with an emphasis on the role and mechanism of leaderless translation. We have examined differences in the structure and composition of the ribosomes from these organisms and discussed how differences in codon preference and the toxin-antitoxin systems present influence the translational landscape of the cell under both optimal and stress conditions.
With the ribosome the target of many antibiotics and antibiotic resistance on the rise, this is an important time to consider how differences in the translational machinery in E. coli and M. tuberculosis may have significant implications for treatment of TB, particularly latent TB infection.
ACKNOWLEDGEMENTS
We thank Brendan Wren from the London School of Hygiene and Tropical Medicine and Joy Fleming from the Institute of Biophysics at the Chinese Academy of Sciences for their constructive critical reading of the manuscript.
FUNDING
European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme [637730]. Funding for open access charge: H2020 European Research Council [637730].
Conflict of interest statement. None declared.
REFERENCES
- 1. Xue S., Barna M.. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol. 2012; 13:355–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Byrgazov K., Vesper O., Moll I.. Ribosome heterogeneity: Another level of complexity in bacterial translation regulation. Curr. Opin. Microbiol. 2013; 16:133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Russell J.B., Cook G.M.. Energetics of bacterial growth: balance of anabolic and catabolic reactions. Microbiol. Rev. 1995; 59:48–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. World Health Organization Global tuberculosis report 2017. 2017; Geneva: World Health Organization; 2017. Licence: CC BY-NC- SA 3.0 IGO. [Google Scholar]
- 5. Houben R.M.G.J., Dodd P.J.. The global burden of latent tuberculosis infection: A Re-estimation using mathematical modelling. PLOS Med. 2016; 13:e1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nunes-Alves C., Booty M.G., Carpenter S.M.. In search of a new paradigm for protective immunity to TB. Nat. Rev. Microbiol. 2014; 12:289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barry C.E., Boshoff H.I., Dartois V., Dick T., Ehrt S., Flynn J., Schnappinger D., Wilkinson R.J., Young D.. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat. Rev. Microbiol. 2009; 7:845–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gomez J.E., McKinney J.D.. M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis. 2004; 84:Churchill Livingstone; 29–44. [DOI] [PubMed] [Google Scholar]
- 9. Rustad T.R., Sherrid A.M., Minch K.J., Sherman D.R.. Hypoxia: a window into Mycobacterium tuberculosis latency. Cell. Microbiol. 2009; 11:1151–1159. [DOI] [PubMed] [Google Scholar]
- 10. Rohde K.H., Veiga D.F.T., Caldwell S., Balázsi G., Russell D.G.. Linking the transcriptional profiles and the physiological states of Mycobacterium tuberculosis during an extended intracellular infection. PLoS Pathog. 2012; 8:e1002769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Betts J.C., Lukey P.T., Robb L.C., McAdam R.A., Duncan K.. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 2002; 43:717–731. [DOI] [PubMed] [Google Scholar]
- 12. Stewart G.R., Wernisch L., Stabler R., Mangan J.A., Hinds J., Laing K.G., Young D.B., Butcher P.D.. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology. 2002; 148:3129–3138. [DOI] [PubMed] [Google Scholar]
- 13. Deb C., Lee C.M., Dubey V.S., Daniel J., Abomoelak B., Sirakova T.D., Pawar S., Rogers L., Kolattukudy P.E.. A novel in vitro multiple-stress dormancy model for Mycobacterium tuberculosis generates a lipid-loaded, drug-tolerant, dormant pathogen. PLoS One. 2009; 4:e6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Uplekar S., Rougemont J., Cole S.T., Sala C.. High-resolution transcriptome and genome-wide dynamics of RNA polymerase and NusA in Mycobacterium tuberculosis. Nucleic Acids Res. 2013; 41:961–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Galagan J.E., Minch K., Peterson M., Lyubetskaya A., Azizi E., Sweet L., Gomes A., Rustad T., Dolganov G., Glotova I. et al. The Mycobacterium tuberculosis regulatory network and hypoxia. Nature. 2013; 499:178–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Peterson E.J.R., Reiss D.J., Turkarslan S., Minch K.J., Rustad T., Plaisier C.L., Longabaugh W.J.R., Sherman D.R., Baliga N.S.. A high-resolution network model for global gene regulation in Mycobacterium tuberculosis. Nucleic Acids Res. 2014; 42:11291–11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Minch K.J., Rustad T.R., Peterson E.J.R., Winkler J., Reiss D.J., Ma S., Hickey M., Brabant W., Morrison B., Turkarslan S. et al. The DNA-binding network of Mycobacterium tuberculosis. Nat. Commun. 2015; 6:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cortes T., Schubert O.T., Banaei-Esfahani A., Collins B.C., Aebersold R., Young D.B.. Delayed effects of transcriptional responses in Mycobacterium tuberculosis exposed to nitric oxide suggest other mechanisms involved in survival. Sci. Rep. 2017; 7:8208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cortes T., Schubert O.T., Rose G., Arnvig K.B., Comas I., Aebersold R., Young D.B.. Genome-wide mapping of transcriptional start sites defines an extensive leaderless transcriptome in Mycobacterium tuberculosis. Cell Rep. 2013; 5:1121–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shasmal M., Sengupta J., Rees B., Moras D., Yusupov M.. Structural diversity in bacterial ribosomes: mycobacterial 70S ribosome structure reveals novel features. PLoS One. 2012; 7:e31742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hentschel J., Burnside C., Mignot I., Leibundgut M., Boehringer D., Ban N.. The complete structure of the Mycobacterium smegmatis 70S ribosome. Cell Rep. 2017; 20:149–160. [DOI] [PubMed] [Google Scholar]
- 22. Li Z., Ge X., Zhang Y., Zheng L., Sanyal S., Gao N.. Cryo-EM structure of Mycobacterium smegmatis ribosome reveals two unidentified ribosomal proteins close to the functional centers. Protein Cell. 2018; 9:384–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang K., Chang J.-Y., Cui Z., Li X., Meng R., Duan L., Thongchol J., Jakana J., Huwe C.M., Sacchettini J.C. et al. Structural insights into species-specific features of the ribosome from the human pathogen Mycobacterium tuberculosis. Nucleic Acids Res. 2017; 45:10884–10894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Allen G.S., Zavialov A., Gursky R., Ehrenberg M., Frank J.. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell. 2005; 121:703–712. [DOI] [PubMed] [Google Scholar]
- 25. Vellanoweth R.L., Rabinowitz J.C.. The influence of ribosome‐binding‐site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Mol. Microbiol. 1992; 6:1105–1114. [DOI] [PubMed] [Google Scholar]
- 26. Jacob W.F., Santer M., Dahlberg A.E.. A single base change in the Shine-Dalgarno region of 16S rRNA of Escherichia coli affects translation of many proteins. Proc. Natl. Acad. Sci. U.S.A. 1987; 84:4757–4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Band L., Henner D.J.. Bacillus subtilis requires a ‘stringent’ Shine-Dalgarno region for gene expression. DNA. 1984; 3:17–21. [DOI] [PubMed] [Google Scholar]
- 28. Jianhua Z., Petracca R.. Influence of single base change in Shine-Dalgarno sequence on the stability of B. subtilis plasmid PSM604. J. Tongji Med. Univ. 2000; 20:183–185. [DOI] [PubMed] [Google Scholar]
- 29. Nakagawa S., Niimura Y., Miura K., Gojobori T.. Dynamic evolution of translation initiation mechanisms in prokaryotes. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:6382–6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Janssen G.R. Baltz RH, Hegeman G, Skatrud PL. Eubacterial, archaebacterial and eucaryotic genes that encode leaderless mRNA. Industrial Microorganisms: Basic and Applied Molecular Genetics. 1993; Washington, D.C.: American Society for Microbiology; 59–67. [Google Scholar]
- 31. Boni I. V, Isaeva D.M., Musychenko M.L., Tzareva N. V. Ribosome-messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res. 1991; 19:155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Komarova A. V., Tchufistova L.S., Dreyfus M., Boni I. V.. AU-Rich sequences within 5′ untranslated leaders enhance translation and stabilize mRNA in Escherichia coli. J. Bacteriol. 2005; 187:1344–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Salah P., Bisaglia M., Aliprandi P., Uzan M., Sizun C., Bontems F.. Probing the relationship between gram-negative and gram-positive S1 proteins by sequence analysis. Nucleic Acids Res. 2009; 37:5578–5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Londei P. Evolution of translational initiation: new insights from the archaea. FEMS Microbiol. Rev. 2005; 29:185–200. [DOI] [PubMed] [Google Scholar]
- 35. Moll I., Hirokawa G., Kiel M.C., Kaji A., Bläsi U.. Translation initiation with 70S ribosomes: an alternative pathway for leaderless mRNAs. Nucleic Acids Res. 2004; 32:3354–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Giliberti J., O’Donnell S., Van Etten W.J., Janssen G.R.. A 5′-terminal phosphate is required for stable ternary complex formation and translation of leaderless mRNA in Escherichia coli. RNA. 2012; 18:508–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moll I., Grill S., Gualerzi C.O., Bläsi U.. Leaderless mRNAs in bacteria: Surprises in ribosomal recruitment and translational control. Mol. Microbiol. 2002; 43:239–246. [DOI] [PubMed] [Google Scholar]
- 38. Krishnan K.M., Van Etten W.J., Janssen G.R.. Proximity of the start codon to a leaderless mRNA’s 5′ terminus is a strong positive determinant of ribosome binding and expression in Escherichia coli. J. Bacteriol. 2010; 192:6482–6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Van Etten W.J., Janssen G.R.. An AUG initiation codon, not codon-anticodon complementarity, is required for the translation of unleadered mRNA in Escherichia coli. Mol. Microbiol. 1998; 27:987–1001. [DOI] [PubMed] [Google Scholar]
- 40. Grill S., Gualerzi C.O., Londei P., Bläsi U.. Selective stimulation of translation of leaderless mRNA by initiation factor 2: Evolutionary implications for translation. EMBO J. 2000; 19:4101–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zheng X., Hu G.-Q.Q., She Z.-S.S., Zhu H.. Leaderless genes in bacteria: clue to the evolution of translation initiation mechanisms in prokaryotes. BMC Genomics. 2011; 12:361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nakagawa S., Niimura Y., Miura K., Gojobori T.. Dynamic evolution of translation initiation mechanisms in prokaryotes. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:6382–6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sharma C.M., Hoffmann S., Darfeuille F., Reignier J., Findeiß S., Sittka A., Chabas S., Reiche K., Hackermüller J., Reinhardt R. et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010; 464:250–255. [DOI] [PubMed] [Google Scholar]
- 44. Jeong Y., Kim J.-N., Kim M.W., Bucca G., Cho S., Yoon Y.J., Kim B.-G., Roe J.-H.H., Kim S.C., Smith C.P. et al. The dynamic transcriptional and translational landscape of the model antibiotic producer Streptomyces coelicolor A3(2). Nat. Commun. 2016; 7:11605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pfeifer-Sancar K., Mentz A., Rückert C., Kalinowski J.. Comprehensive analysis of the Corynebacterium glutamicum transcriptome using an improved RNAseq technique. BMC Genomics. 2013; 14:888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shell S.S., Wang J., Lapierre P., Mir M., Chase M.R., Pyle M.M., Gawande R., Ahmad R., Sarracino D.A., Ioerger T.R. et al. Leaderless transcripts and small proteins are common features of the mycobacterial translational landscape. PLoS Genet. 2015; 11:e1005641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kroger C., Dillon S.C., Cameron A.D.S., Papenfort K., Sivasankaran S.K., Hokamp K., Chao Y., Sittka A., Hebrard M., Handler K. et al. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E1277–E1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Seo J.-H., Hong J., Kim D., Cho B.-K., Huang T.-W., Tsai S.-F., Palsson B.O., Charusanti P.. Multiple-omic data analysis of Klebsiella pneumoniae MGH 78578 reveals its transcriptional architecture and regulatory features. BMC Genomics. 2012; 13:679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kaberdina A.C., Szaflarski W., Nierhaus K.H., Moll I.. An unexpected type of ribosomes induced by kasugamycin: a look into ancestral times of protein synthesis. Mol. Cell. 2009; 33:227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vesper O., Amitai S., Belitsky M., Byrgazov K., Kaberdina A.C., Engelberg-Kulka H., Moll I.. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell. 2011; 147:147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chin K., Shean C.S., Gottesman M.E.. Resistance of λ cI translation to antibiotics that inhibit translation initiation. J. Bacteriol. 1993; 175:7471–7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Moll I., Bläsi U.. Differential inhibition of 30S and 70S translation initiation complexes on leaderless mRNA by kasugamycin. Biochem. Biophys. Res. Commun. 2002; 297:1021–1026. [DOI] [PubMed] [Google Scholar]
- 53. Gerdes K., Maisonneuve E.. Bacterial persistence and toxin-antitoxin loci. Annu. Rev. Microbiol. 2012; 66:103–123. [DOI] [PubMed] [Google Scholar]
- 54. Sauert M., Wolfinger M.T., Vesper O., Müller C., Byrgazov K., Moll I.. The MazF-regulon: a toolbox for the post-transcriptional stress response in Escherichia coli. Nucleic Acids Res. 2016; 44:6660–6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Temmel H., Müller C., Sauert M., Vesper O., Reiss A., Popow J., Martinez J., Moll I.. The RNA ligase RtcB reverses MazF-induced ribosome heterogeneity in Escherichia coli. Nucleic Acids Res. 2017; 45:4708–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. O’Donnell S.M., Janssen G.R.. The initiation codon affects ribosome binding and translational efficiency in Escherichia coli of cI mRNA with or without the 5′ untranslated leader. J. Bacteriol. 2001; 183:1277–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Czernilofsky A.P., Kurland C.G., Stöffler G.. 30S Ribosomal proteins associated with the 3′-terminus of 16S RNA. FEBS Lett. 1975; 58:281–284. [DOI] [PubMed] [Google Scholar]
- 58. Duval M., Korepanov A., Fuchsbauer O., Fechter P., Haller A., Fabbretti A., Choulier L., Micura R., Klaholz B.P., Romby P. et al. Escherichia coli ribosomal protein S1 unfolds structured mRNAs onto the ribosome for active translation initiation. PLoS Biol. 2013; 11:e1001731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Prisic S., Hwang H., Dow A., Barnaby O., Pan T.S., Lonzanida J.A., Chazin W.J., Steen H., Husson R.N.. Zinc regulates a switch between primary and alternative S18 ribosomal proteins in Mycobacterium tuberculosis. Mol. Microbiol. 2015; 97:263–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dow A., Prisic S.. Alternative ribosomal proteins are required for growth and morphogenesis of Mycobacterium smegmatis under zinc limiting conditions. PLoS One. 2018; 13:e0196300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fischer N., Neumann P., Konevega A.L., Bock L. V., Ficner R., Rodnina M. V., Holger S.. Structure of the E. coli ribosome-EF-Tu complex at. Nature. 2015; 520:567–570. [DOI] [PubMed] [Google Scholar]
- 62. Ben-Shem A., Garreau de Loubresse N., Melnikov S., Jenner L., Yusupova G., Yusupov M.. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011; 334:1524–1529. [DOI] [PubMed] [Google Scholar]
- 63. Desai N., Brown A., Amunts A., Ramakrishnan V.. The structure of the yeast mitochondrial ribosome. Science. 2017; 355:528–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Khatter H., Myasnikov A.G., Natchiar S.K., Klaholz B.P.. Structure of the human 80S ribosome. Nature. 2015; 520:640–645. [DOI] [PubMed] [Google Scholar]
- 65. Amunts A., Brown A., Toots J., Scheres S.H.W., Ramakrishnan V.. The structure of the human mitochondrial ribosome. Science. 2015; 348:95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gohara D.W., Yap M.-N.F.. Survival of the drowsiest: the hibernating 100S ribosome in bacterial stress management. Curr. Genet. 2017; doi:10.1007/s00294-017-0796-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Trauner A., Lougheed K.E.A., Bennett M.H., Hingley-Wilson S.M., Williams H.D.. The dormancy regulator DosR controls ribosome stability in hypoxic mycobacteria. J. Biol. Chem. 2012; 287:24053–24063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Voskuil M.I., Schnappinger D., Visconti K.C., Harrell M.I., Dolganov G.M., Sherman D.R., Schoolnik G.K.. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 2003; 198:705–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Polikanov Y.S., Blaha G.M., Steitz T.A.. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science. 2012; 336:915–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bunker R.D., Mandal K., Bashiri G., Chaston J.J., Pentelute B.L., Lott J.S., Kent S.B.H., Baker E.N.. A functional role of Rv1738 in Mycobacterium tuberculosis persistence suggested by racemic protein crystallography. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:4310–4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sherman D.R., Voskuil M., Schnappinger D., Liao R., Harrell M.I., Schoolnik G.K.. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding -crystallin. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:7534–7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Li X., Sun Q., Jiang C., Yang K., Hung L.W., Zhang J., Sacchettini J.C.. Structure of ribosomal silencing factor bound to Mycobacterium tuberculosis ribosome. Structure. 2015; 23:1858–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Andersson S.G.E., Sharp P.M.. Codon usage in the Mycobacterium tuberculosis complex. Microbiology. 1996; 142:915–925. [DOI] [PubMed] [Google Scholar]
- 74. Pan A., Dutta C., Das J.. Codon usage in highly expressed genes of Haemophillus influenzae and Mycobacterium tuberculosis: translational selection versus mutational bias. Gene. 1998; 215:405–413. [DOI] [PubMed] [Google Scholar]
- 75. Presnyak V., Alhusaini N., Chen Y.H., Martin S., Morris N., Kline N., Olson S., Weinberg D., Baker K.E., Graveley B.R. et al. Codon optimality is a major determinant of mRNA stability. Cell. 2015; 160:1111–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Leng T., Pan M., Xu X., Javid B.. Translational misreading in Mycobacterium smegmatis increases in stationary phase. Tuberculosis. 2015; 95:678–681. [DOI] [PubMed] [Google Scholar]
- 77. Bratulic S., Toll-Riera M., Wagner A.. Mistranslation can enhance fitness through purging of deleterious mutations. Nat. Commun. 2017; 8:15410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Javid B., Sorrentino F., Toosky M., Zheng W., Pinkham J.T., Jain N., Pan M., Deighan P., Rubin E.J.. Mycobacterial mistranslation is necessary and sufficient for rifampicin phenotypic resistance. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:1132–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Su H.W., Zhu J.H., Li H., Cai R.J., Ealand C., Wang X., Chen Y.X., Kayani M. ur R., Zhu T.F., Moradigaravand D. et al. The essential mycobacterial amidotransferase GatCAB is a modulator of specific translational fidelity. Nat. Microbiol. 2016; 1:16147. [DOI] [PubMed] [Google Scholar]
- 80. Lorenz C., Lünse C.E., Mörl M.. tRNA modifications: impact on structure and thermal adaptation. Biomolecules. 2017; 7:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Dedon P.C., Begley T.J.. A system of RNA modifications and biased codon use controls cellular stress response at the level of translation. Chem. Res. Toxicol. 2014; 27:330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Chionh Y.H., McBee M., Babu I.R., Hia F., Lin W., Zhao W., Cao J., Dziergowska A., Malkiewicz A., Begley T.J. et al. tRNA-mediated codon-biased translation in mycobacterial hypoxic persistence. Nat. Commun. 2016; 7:13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Maisonneuve E., Shakespeare L.J., Jorgensen M.G., Gerdes K.. Bacterial persistence by RNA endonucleases. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:13206–13211. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 84. Sala A., Bordes P., Genevaux P.. Multiple toxin-antitoxin systems in Mycobacterium tuberculosis. Toxins (Basel). 2014; 6:1002–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Keren I., Minami S., Rubin E., Lewis K.. Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. MBio. 2011; 2:e00100-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Gupta A., Venkataraman B., Vasudevan M., Gopinath Bankar K.. Co-expression network analysis of toxin-antitoxin loci in Mycobacterium tuberculosis reveals key modulators of cellular stress. Sci. Rep. 2017; 7:5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Jankevicius G., Ariza A., Ahel M., Ahel I.. The toxin-antitoxin system DarTG catalyzes reversible ADP-ribosylation of DNA. Mol. Cell. 2016; 64:1109–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Albrethsen J., Agner J., Piersma S.R., Højrup P., Pham T. V, Weldingh K., Jimenez C.R., Andersen P., Rosenkrands I.. Proteomic profiling of Mycobacterium tuberculosis Identifies Nutrient-starvation-responsive toxin–antitoxin systems. Mol. Cell. Proteomics. 2013; 12:1180–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Winther K.S., Brodersen D.E., Brown A.K., Gerdes K.. VapC20 of Mycobacterium tuberculosis cleaves the Sarcin–Ricin loop of 23S rRNA. Nat. Commun. 2013; 4:2796. [DOI] [PubMed] [Google Scholar]
- 90. Schifano J.M., Edifor R., Sharp J.D., Ouyang M., Konkimalla A., Husson R.N., Woychik N.A.. Mycobacterial toxin MazF-mt6 inhibits translation through cleavage of 23S rRNA at the ribosomal A site. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:8501–8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Schifano J.M., Woychik N.A.. 23S rRNA as an a-Maz-ing new bacterial toxin target. RNA Biol. 2014; 11:101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Schifano J.M., Vvedenskaya I.O., Knoblauch J.G., Ouyang M., Nickels B.E., Woychik N.A.. An RNA-seq method for defining endoribonuclease cleavage specificity identifies dual rRNA substrates for toxin MazF-mt3. Nat. Commun. 2014; 5:3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Schifano J.M., Cruz J.W., Vvedenskaya I.O., Edifor R., Ouyang M., Husson R.N., Nickels B.E., Woychik N.A.. TRNA is a new target for cleavage by a MazF toxin. Nucleic Acids Res. 2015; 44:1256–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Cole S.T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C.E. et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998; 393:537–544. [DOI] [PubMed] [Google Scholar]
- 95. Hwang J.Y., Buskirk A.R.. A ribosome profiling study of mRNA cleavage by the endonuclease RelE. Nucleic Acids Res. 2017; 45:D327–D336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Singh R., Barry C.E., Boshoff H.I.M.. The three RelE homologs of Mycobacterium tuberculosis have individual, drug-specific effects on bacterial antibiotic tolerance. J. Bacteriol. 2010; 192:1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Schuessler D.L., Cortes T., Fivian-Hughes A.S., Lougheed K.E.A., Harvey E., Buxton R.S., Davis E.O., Young D.B.. Induced ectopic expression of HigB toxin in Mycobacterium tuberculosis results in growth inhibition, reduced abundance of a subset of mRNAs and cleavage of tmRNA. Mol. Microbiol. 2013; 90:195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Matzov D., Aibara S., Basu A., Zimmerman E., Bashan A., Yap M.-N.F., Amunts A., Yonath A.E.. The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus. Nat. Commun. 2017; 8:723. [DOI] [PMC free article] [PubMed] [Google Scholar]