Abstract
Base editing is an exciting new genome engineering technology. C-to-T mutations in genomic DNA have been achieved using ribonucleoprotein complexes comprised of rat APOBEC1 single-stranded DNA deaminase, Cas9 nickase (Cas9n), uracil DNA glycosylase inhibitor (UGI), and guide (g)RNA. Here, we report the first real-time reporter system for quantification of APOBEC-mediated base editing activity in living mammalian cells. The reporter expresses eGFP constitutively as a marker for transfection or transduction, and editing restores functionality of an upstream mCherry cassette through the simultaneous processing of two gRNA binding regions that each contain an APOBEC-preferred 5′TCA target site. Using this system as both an episomal and a chromosomal editing reporter, we show that human APOBEC3A-Cas9n-UGI and APOBEC3B-Cas9n-UGI base editing complexes are more efficient than the original rat APOBEC1-Cas9n-UGI construct. We also demonstrate coincident enrichment of editing events at a heterologous chromosomal locus in reporter-edited, mCherry-positive cells. The mCherry reporter also quantifies the double-stranded DNA cleavage activity of Cas9, and may therefore be adaptable for use with many different CRISPR systems. The combination of a rapid, fluorescence-based editing reporter system and more efficient, structurally defined DNA editing enzymes broadens the versatility of the rapidly expanding toolbox of genome editing and engineering technologies.
INTRODUCTION
APOBEC enzymes are single-stranded (ss) polynucleotide cytosine deaminases. Human cells encode nine active family members with AID functioning in antibody DNA diversification, APOBEC1 in mRNA editing, and APOBEC3A-H in DNA virus and transposon restriction (1–4). APOBEC1 is also an efficient DNA mutator (5,6), and the rat enzyme was recently combined with Cas9 and guide (g)RNA to create ribonucleoprotein complexes capable of editing single cytosine nucleobases and making site-specific C-to-T mutations in genomic DNA (7). A construct comprised of rat APOBEC1, Cas9 nickase (Cas9n), and uracil DNA glycosylase inhibitor (UGI) has been shown to yield base editing frequencies ranging from 5 to 50% (BE3) (7–10). This editing complex has already been adopted by many labs and harnessed for biotechnology applications (10–17). Two orthologs, human AID and lamprey PmCDA1, have also been combined with Cas9n but with lower overall base editing efficiencies, likely due to lower intrinsic enzyme activities (18–22). PmCDA1 has also been used in plant genome engineering (20).
A significant impediment to optimizing base editing technologies and deployment in limitless cell types is a lack of an efficient, real-time, rapid, and quantitative editing assay (ideally one that is also transferable across species and, at least initially, independent of DNA sequencing to assess efficiencies). Here, we report a fluorescence-based reporter system for quantification of real-time editing in living mammalian cells. We refer to the system as ‘ACE’ because it monitors both APOBEC- and Cas9-mediated Editing in real-time. The reporter is a bicistronic construct with a mutated mCherry cassette and a downstream eGFP gene (a constitutive indicator of reporter abundance). The mCherry gene was rendered inactive through a 43 base-pair insertion that introduces a frame-shift, thus ablating fluorescence. Restoration of fluorescence can only occur through APOBEC–Cas9n-UGI-mediated editing of dual APOBEC-preferred trinucleotide motifs, 5′-TCA-to-TUA, within the 43-base-pair insertion. Editing at these motifs generates uracil lesions, which are substrates for uracil excision and ssDNA cleavage by canonical base excision repair enzymes (UNG2 and APE1, respectively) (23,24). Simultaneous cleavage of the opposing DNA strand by the Cas9 nickase then results in two DNA double-strand breaks that are most likely fused by non-homologous end joining (NHEJ) to restore mCherry fluorescence. The ratio of mCherry-positive to eGFP-positive cells thereby enables rapid quantification of DNA editing frequencies by fluorescence microscopy or flow cytometry. The ACE system was validated episomally in transient transfection experiments and chromosomally following stable integration of the reporter by lentivirus-mediated transduction. The ACE system was used to develop highly efficient base editing constructs based upon APOBEC3A and APOBEC3B (catalytic domain) that, like Cas9, are defined structurally. Additional utility of the ACE reporter system was shown by using it to enrich for cells with editing events at heterologous chromosomal sites. The success of these two applications demonstrates the power and utility of ACE as a rapid, fluorescence-based, DNA editing reporter system that can easily be adapted for applications in different systems.
MATERIALS AND METHODS
Cell lines and culture conditions
293T cells were maintained in DMEM (Hyclone) supplemented with 10% FBS (Gibco) and 0.5% penicillin/streptomycin (50 units). HeLa were maintained in RPMI (Hyclone) supplemented with 10% FBS (Gibco) and 0.5% penicillin/streptomycin (50 units). 293T and HeLa cells were transfected with TransIT-LT1 (Mirus) according to the manufacturer's protocol. SSM2c, CHO, and COS-7 cells were maintained in DMEM (Euroclone) supplemented with 10% FBS (Carlo Erba), 2 mM l-glutamine (Carlo Erba), and 1 mM penicillin/streptomycin (Carlo Erba). SSM2c were transfected with PEI (Sigma-Aldrich) according to the manufacturer's protocol. CHO and COS-7 cells were transfected with Lipofectamine LTX (Invitrogen) according to the manufacturer's protocol. Single time point episomal editing experiments were harvested 72 h post-transfection, and chromosomal editing experiments were harvested 96 h post-transfection.
APOBEC- and Cas9-mediated editing reporter construct
The ACE system was derived from HIV-1 NL4-3 by excising the gag-pol, vif, and vpr open reading frames using SwaI and SalI restriction sites and blunt end ligation. vpr and the first ∼1200 bp of env were removed using SacI and PsiI restriction sites and blunt end ligation to retain the Rev response element (RRE). A gBlock, synthesized by Integrated DNA Technologies (IDT) to introduce a CMV promoter with a 3′ AgeI restriction site, was cloned into the nef open reading frame using BamHI and KpnI restriction sites. mCherry was PCR amplified using Phusion high-fidelity DNA polymerase (NEB) from a pcDNA3.1 expression plasmid with primers that introduce a 3′ T2A self-cleaving peptide sequence (primers in Table 1) and cloned into a CloneJET PCR cloning vector (Thermo Fisher). eGFP was PCR amplified from a pcDNA3.1 expression plasmid with primers introducing scrambled nucleotide sequences at the 5′ and 3′ ends of the gene that retained the wild-type protein sequence (primers in Table 1). This was done to eliminate recombination during reverse-transcription of the viral reporter because the 5′ and 3′ ∼20 nt of mCherry and eGFP are identical. The eGFP PCR amplicon was cloned into the mCherry-T2A cloning vector using XhoI and KpnI restriction sites. Finally, the single mCherry-T2A-eGFP cassette was cloned into the modified NL4-3 vector using AgeI and KpnI restriction sites. 8 different mCherry mutants were created using site-directed mutagenesis with Phusion DNA polymerase (NEB) (primers in Table 1 and data not shown). Functional testing of several candidate mCherry L59S mutants identified one that reverted to mCherry positive with BE3. Subsequent DNA sequencing revealed a near triplication of the site-directed mutation oligonucleotide sequence, equating to a net insertion of 43 bp, likely created during the PCR amplification step of the construction. The full sequence of this region is shown in Figure 1A, which includes two flanking gRNA binding sites that each contain an APOBEC-preferred 5′-TCA deamination target. For comparison, another round of site-directed mutagenesis generated a sequence-confirmed L59S mCherry single amino acid substitution mutant, which retained wild-type mCherry fluorescence activity (Supplemental Figure S1 and discussed further below in the Results section).
Table 1.
Oligonucleotide sequences
| Primer | Sequence (5′ to 3′) |
|---|---|
| A3A Cloning Forward Primer | AGATCCGCGGCCGCGCCGCCACCATGATGGAAGCCAGCCCAGCATCCGGGC |
| A3A Cloning Reverse Primer | TGAGGTCCCGGGAGTCTCGCTGCCGCTTCCGTTTCCCTGATTCTGGAGAATG |
| A3Bctd Cloning Forward Primer | AGATCCGCGGCCGCGCCGCCACCATGGATCCAGACACATTCACTTTCAACT |
| A3Bctd Cloning Reverse Primer | TGAGGTCCCGGGAGTCTCGCTGCCGCTGTTTCCCTGATTCTGGAGAATGGCC |
| mCherry L59S SDM Forward Primer | AAGGGTGGCCCCTCACCCTTCGCCTGGG |
| mCherry L59S SDM Reverse Primer | CCCAGGCGAAGGGTGAGGGGCCACCCTT |
| Codon #59-directed mCherry gRNA Forward Primer | ACACCTGGCCCCTCACCCTTCGCCTG |
| Codon #59-directed mCherry gRNA Reverse Primer | AAAACAGGCGAAGGGTGAGGGGCCAG |
| NS gRNA Forward | ACACCGCACTACCAGAGCTAACTCAG |
| NS gRNA Reverse | AAAACTGAGTTAGCTCTGGTAGTGCG |
| T2A Cloning Forward Primer | CTGGCTACCGGTATGGTGAGCAAGGGCGAGG |
| T2A Cloning Reverse Primer | TTAAAGGTACCAGGGCCGGGATTCTCCTCCACGTCACCGCATGTTAGAAGACTTCCTCTGCCCTCCTTGTACTCGAGATCTGCACCGGGCTTGTACAGCTCGTCCATGCC |
| eGFP Cloning Forward Primer | GCAGATCTCGAGTACAAGGAGGGCAGAGGAAGTCTTCTAACATGCGGTGACGTGGAGGAGAATCCCGGCCCTCTGGTCAGTAAAGGTGAAGAACTGTTCACCG |
| eGFP Cloning Reverse Primer | CTTAAAGGTACCTTATTTATATAATTCATCCATACCGAGAG |
| mCherry Amplification Forward Primer | ATGGCCATCATCAAGGAGTT |
| mCherry Amplification Reverse Primer | CTCTGCCCTCCTTGTACTCG |
| CloneJET Sequencing Forward Primer | CGACTCACTATAGGGAGAGCGGC |
| CloneJET Sequencing Reverse Primer | AAGAACATCGATTTTCCATGGCAG |
| L1 Intron into BE3 Forward | NNNGAGCTCAGAGACTGGCCCAGTGGCTGTGGACCCCACATTGAGGTGAGTCCAGGAGA |
| L1 Intron into BE3 Reverse | NNNGAGCTCTCTCGGATCGAAGAATACCTCAAACTCATGGGGCTCGATCCGCCGTCTGT |
Figure 1.
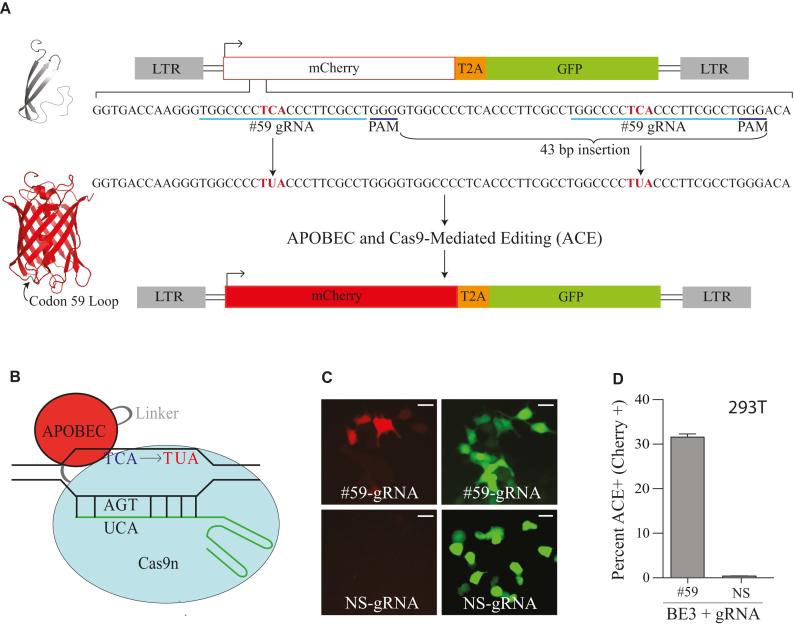
A real-time fluorescent reporter for APOBEC- and Cas9-mediated editing. (A) Schematic of the APOBEC- and Cas9-mediated editing (ACE) reporter in the context of a lentiviral construct with a CMV promoter that drives expression of a bicistronic message encoding mutant mCherry and wild-type eGFP. The sequence of the gRNA displaced DNA strand is shown below with flanking APOBEC 5′-TCA deamination hotspots (red), PAM sites, and 43 bp insertion labeled. See text for a description of editing, cleavage, and processing events required for reversion to mCherry-positive. Ribbon schematics of defective mCherry (gray) and functionally restored mCherry (red) with the flexible loop position of residue 59 shown (model based on pdb 2H5Q). (B) Schematic of an APOBEC–Cas9n/gRNA editosome engaging a DNA target. C-to-U editing occurs in the ssDNA loop displaced by gRNA annealing to target DNA. (C) Representative images of mCherry-positive 293T cells catalyzed by BE3 and mCherry codon 59-directed gRNA (#59-gRNA) but not with NS-gRNA (NS, non-specific; inset white bar = 30 μm). (D) Quantification of the base editing experiment in panel C (n = 3; average ± SD).
Base editing constructs
The rat APOBEC1-Cas9n-UGI-NLS construct (BE3) was provided by David Liu, Harvard University (7). A3A and A3Bctd cDNA sequences, each disrupted by an L1 intron to prevent toxicity in Escherichia coli (25), were amplified using primers in Table 1 and used to replace rat APOBEC1 in BE3 using a NotI site in the MCS and a XmaI site in the XTEN linker. gRNAs targeting mCherry or non-specific (NS) sequence as a control (Table 1) were cloned into MLM3636, obtained from J. Keith Joung, Harvard University, through Addgene (Plasmid #43860), using the accompanying Joung Lab gRNA cloning protocol. An L1 intron was amplified from the A3Ai construct using primers in Table 1 and cloned into the SacI site in the rat APOBEC1 region of BE3 to create the BE3i editing construct.
Episomal DNA editing experiments
Semi-confluent 293T, SSM2c, CHO and COS-7 cells in a six-well plate format were transfected with 200 ng gRNA, 400 ng ACE, and 600 ng of each base-editor [10 min, RT with 6 μl of TransIT-LT1 (Mirus) and 200 μl of serum-free DMEM (Hyclone)]. Cells were harvested at indicated time points for editing quantification by flow cytometry.
Chromosomal DNA editing experiments
A semi-confluent 10 cm plate of 293T cells was transfected with 8 μg of an HIV-1 Gag-Pol packaging plasmid, 1.5 μg of a VSV-G expression plasmid, and 3 μg of the ACE lentiviral reporter plasmid. Virus was harvested 48 h post-transfection, frozen at –80°C for 8 h, thawed, and used to transduce target cells (MOI = 1). Forty eight hours post-transduction, 600 ng APOBEC–Cas9n-UGI editor and 250 ng of targeting or NS-gRNA were transfected into a semi-confluent six-well plate of ACE-transduced cells. Cells were harvested 96 h post-transfection and editing was quantified by flow-cytometry.
In a subset of experiments, mCherry-positive cells were recovered by FACS, converted to genomic DNA (Gentra Puregene), and subjected to high-fidelity PCR using Phusion (NEB) to amplify mCherry target sequences (primers in Table 1). PCR products were gel-purified (GeneJET Gel Extraction Kit, Thermo Fisher Scientific) and cloned into a sequencing plasmid (CloneJET PCR Cloning Kit, Thermo Fisher Scientific). Sanger sequencing was done in 96-well format (Genewiz) using primers recommended with the CloneJET PCR Cloning Kit (Table 1).
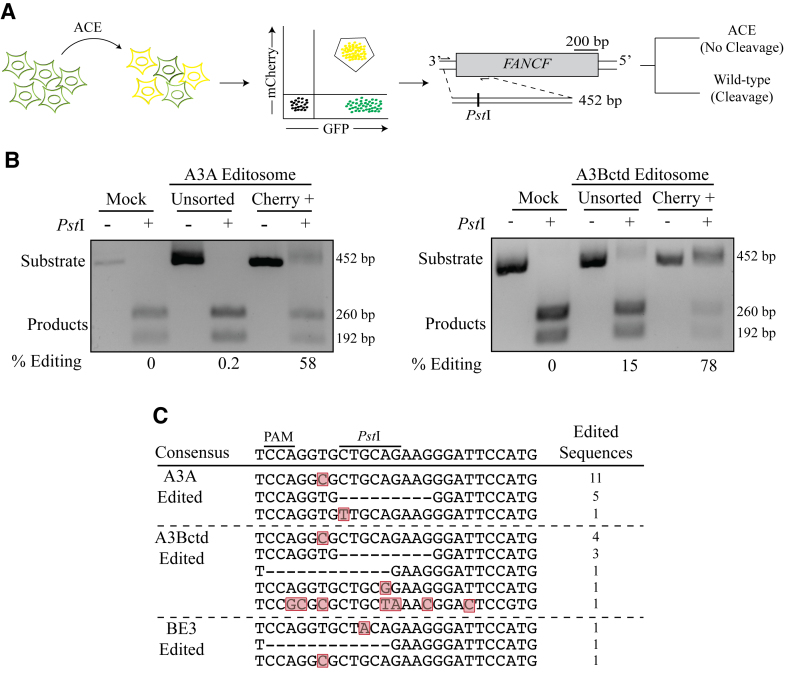
To carry out FANCF editing enrichment experiments, semi-confluent 293T cells transduced with ACE were co-transfected with 600 ng of A3Bctd-Cas9n-UGI and 200 ng of gRNA targeting both mCherry and FANCF in a six-well format. Seventy two hours post-transfection, cells were harvested and FACS was used to collect cells expressing mCherry. gDNA was harvested and a 452 bp fragment of FANCF was PCR amplified using nested primers shown in Table 1. A PstI-HF (New England Biolabs) digest was done, and products were fractionated on an agarose gel to quantify editing efficiencies.
Immunoblots
1 × 106 cells were lysed directly into 2.5x Laemmli sample buffer, separated by a 4–20% gradient SDS-PAGE gel, and transferred to PVDF-FL membranes (Millipore). Membranes were blocked in 5% milk in PBS and incubated with primary antibody diluted in 5% milk in PBS supplemented with 0.1% Tween20. Secondary antibodies were diluted in 5% milk in PBS supplemented with 0.1% Tween20 and 0.01% SDS. Membranes were imaged with a LICOR Odyssey instrument. Primary antibodies used in these experiments were rabbit anti-Cas9 (Abcam ab204448) and mouse anti-HSP90 (BD Transduction Laboratories 610418). Secondary antibodies used were goat anti-rabbit IRdye 800CW (LICOR 827-08365) and goat anti-mouse Alexa Fluor 680 (Molecular Probes A-21057).
RESULTS
Construction and initial validation of the APOBEC- and Cas9-mediated editing (ACE) reporter system
The APOBEC- and Cas9-mediated editing reporter (ACE) system utilizes a CMV driven dual fluorescence reporter cassette (mCherry-T2A-eGFP) to enable expression and quantification of real-time editing in living mammalian cells (reporter schematic in Figure 1A and APOBEC editosome schematic in Figure 1B). To maximize versatility, we used an HIV-based proviral vector as the backbone for the system, which enables use as a transient multi-copy plasmid-based episomal editing reporter or as a stable single-copy chromosomal DNA editing reporter. The eGFP fluorescence marker is used to quantify reporter delivery to target cells. The most important aspect of the ACE system is tight ‘off-to-on’ gain of function fluorescence activity in which mCherry mutational inactivation creates an APOBEC deamination hotspot 5′-TCA, and then APOBEC-catalyzed editing is able to restore mCherry function. Eight different APOBEC mutational hotspots in mCherry were tested and most failed to completely ablate fluorescence, were not located an appropriate distance from a gRNA anchoring motif (PAM), and/or did not become substrates for editing (e.g. Supplemental Figure S1 and data not shown).
One mutant mCherry construct proved robust with no background fluorescence and strong mCherry-positive signal upon transient co-expression of an appropriate mCherry-directed gRNA and the rat APOBEC1 editosome BE3 (fluorescence microscopy images in Figure 1C and quantification in Figure 1D; construct schematic in Figure 1A and details in methods). DNA editing and restoration of mCherry fluorescence requires codon 59 (#59) gRNA-mediated targeting of the APOBEC–Cas9 complex to the intended hotspot because a non-specific (NS) gRNA does not restore fluorescence activity (fluorescence microscopy images in Figure 1C and quantification in Figure 1D). This system is portable and capable of providing real-time read-outs of editing activity in a variety of different mammalian cell lines (e.g. 293T in Figure 1C–D; COS-7, CHO, and SSM2c in Supplemental Figure S2).
Surprisingly, DNA sequencing showed that the site-directed mutagenesis procedure used to generate the reporter had created a 43 bp insertion within mCherry, which shifted it out of frame for translation. The net result was generation of two codon 59 gRNA binding sites, each with an APOBEC-preferred editing hotspot 5′-TCA (the intervening region is also a potential gRNA binding site but it lacks a Cas9 PAM motif; Figure 1A). Combining the aforementioned genetic requirements and sequence results of positive editing events (detailed below), the most likely molecular mechanism for reversion from mCherry negative to positive is simultaneous APOBEC-mediated editing of codon 59 5′-TCA hotspots to 5′-TUA (and/or flanking C’s to U’s) followed by cleavage of this DNA strand by the concerted action of canonical base excision repair enzymes (uracil excision by UNG2 and DNA cleavage by APE1), cleavage of the opposing DNA strand by the nickase activity of Cas9n, and repair of the resulting double-stranded breaks by non-homologous end joining (NHEJ) (outcome depicted in Figure 1A and rationalized further below). Activation of the ACE reporter requires restoration of the full mCherry open reading frame, which at the structural level is due to fluorescence emission from a canonical β-barrel structure (Figure 1A). An additional notable feature of the ACE reporter (and a likely additional explanation for why most constructs initially tested negative) is the structural location of mCherry residue 59 (residue encoded by APOBEC hotspot) within a flexible loop region, which is more tolerant of amino acid substitution and flanking mutations including small insertions and deletions (e.g. a single L59S amino acid substitution retains wild-type mCherry activity; Supplemental Figure S1 and data not shown).
Application of the ACE system to create highly efficient next-generation base editing constructs based on human APOBEC3A and APOBEC3B
Optimization of base editing technologies is likely to require editosomes with the highest possible efficiencies and structural information to guide rational improvements such as single nucleobase specificity [APOBEC1 and PmCDA1 have yet to yield structures, and the crystalized form of AID is diverged from wild-type (26)]. We therefore tested human APOBEC3A (A3A) and APOBEC3B (A3B) catalytic domain for Cas9n-directed DNA editing. These enzymes are the most efficient ssDNA C-to-U deaminases in human cells (27–30), and high-resolution crystal structures of both apo- and ssDNA-bound forms have been determined (31–34). A3A-ssDNA and A3B C-terminal domain (A3Bctd)-ssDNA structures share a unique U-shaped bound ssDNA conformation and provide an atomic explanation for the intrinsic 5′-TC specificity of these enzymes (33,34). As a testament to the utility of this structural information, it informed a single amino acid change in a loop region adjacent to the active site of A3A that altered its intrinsic specificity from 5′-TC to 5′-CC (33). Additional enzyme customization may enable tailoring of these enzymes to other di-nucleotide contexts as well as extensions to tri-nucleotide contexts.
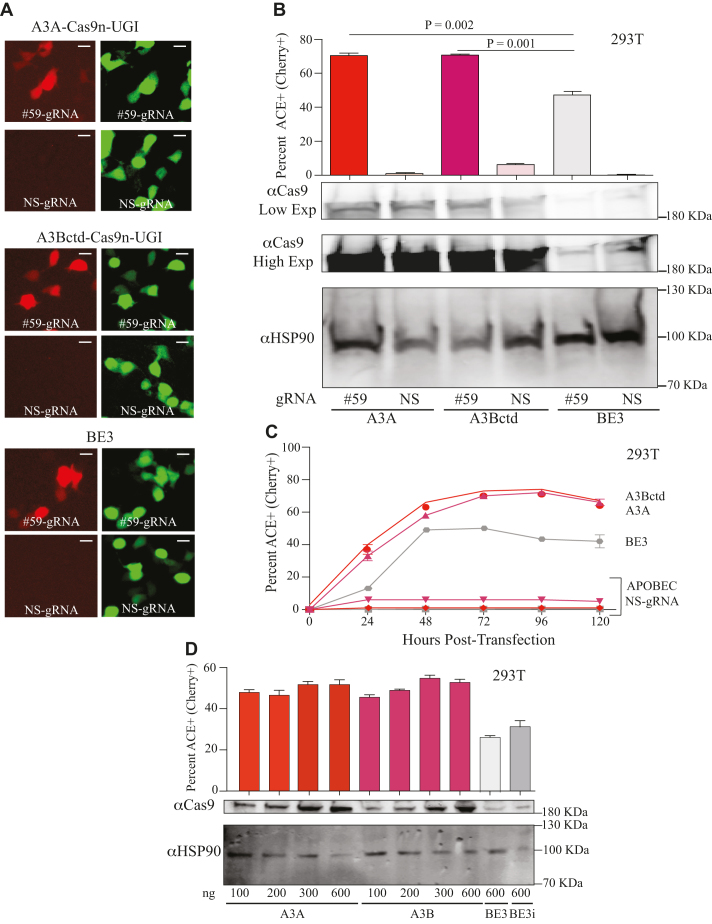
A3A-Cas9n-UGI and A3Bctd-Cas9n-UGI constructs were assembled and tested in parallel with BE3 to directly compare editing efficiencies. These constructs were co-transfected into 293T cells with ACE and a gRNA to direct editosomes to the insertion at mCherry codon 59 (#59) or a NS-gRNA (NS) as a negative control. In a single time point experiment, the rat APOBEC1 editosome yielded 47% mCherry-positive cells, and both A3A and A3Bctd achieved 70% mCherry-positive cells (representative fluorescence images in Figure 2A and quantification in Figure 2B). DNA deaminase activity was required because catalytic glutamate mutant constructs, A3A-E72A and A3Bctd-E255A, were defective in ACE reporter activation (data not shown). Higher editing efficiencies were also observed in time course studies in 293T cells, with both A3A and A3Bctd editosomes achieving nearly 40% mCherry fluorescence by 24 h and maximal fluorescence of 70% by 72 h before declining (as expected for transient transfection with non-replicating plasmids; Figure 2C). Anti-Cas9 immunoblots indicated that at least some of the improved editing efficiencies might be due to higher expression levels of the A3A- and A3Bctd-Cas9n-UGI editosomes (Figure 2B immunoblot images, Supplemental Figure S2B). A titration of A3A- and A3Bctd-Cas9n-UGI was performed to compare editing efficiencies of the newly developed editosomes to those of BE3 and, despite achieving similar expression levels (lanes 1, 5 and 9), our newly developed A3A- and A3Bctd-Cas9n-UGI editosomes still exhibited higher editing frequencies (Figure 2D). In addition, an intron was cloned into BE3 (identical to that in the A3A and A3Bctd editosome constructs), and the newly created BE3 intron (BE3i) construct was tested against the ACE reporter to rule out intron-associated differences in protein expression (Figure 2D). As for the original BE3, the intron-containing derivative still had expression and DNA editing levels lower than those of A3A and A3Bctd editosomes.
Figure 2.
High-efficiency editing by human A3A and A3Bctd editosomes. (A) Representative fluorescence microscopy images of ACE-activated, mCherry-positive 293T cells catalyzed by human A3A, human A3Bctd, or rat APOBEC1/BE3 editosomes (mCherry codon 59-directed gRNA versus NS-gRNA; inset white bar = 30 μm). (B) Quantification of the experiment in panel ‘A’ together with 2 independent parallel experiments (n = 3; average ± SD). The corresponding immunoblots of expressed APOBEC–Cas9n-UGI constructs are shown below (low and high exposures to help visualize BE3) with HSP90 as a loading control. (C) Time course of ACE activation in 293T cells catalyzed by human A3A, human A3Bctd, or rat APOBEC1/BE3 editosomes (mCherry codon 59-directed gRNA versus NS-gRNA; n = 3; mean ± SD; error bars smaller than symbols are not shown). (D) Titration data for 293T cells co-transfected with the ACE reporter, mCherry codon 59-directed gRNA, and different amounts of the indicated editosome constructs (100–600 ng; n = 3; mean ± SD). BE3i has an intron in the rat APOBEC1 portion of the construct, identical to the intron required for propagation of A3A and A3Bctd constructs in E. coli and for expression in mammalian cells.
Improved chromosomal DNA editing efficiencies using A3A and A3Bctd editosomes
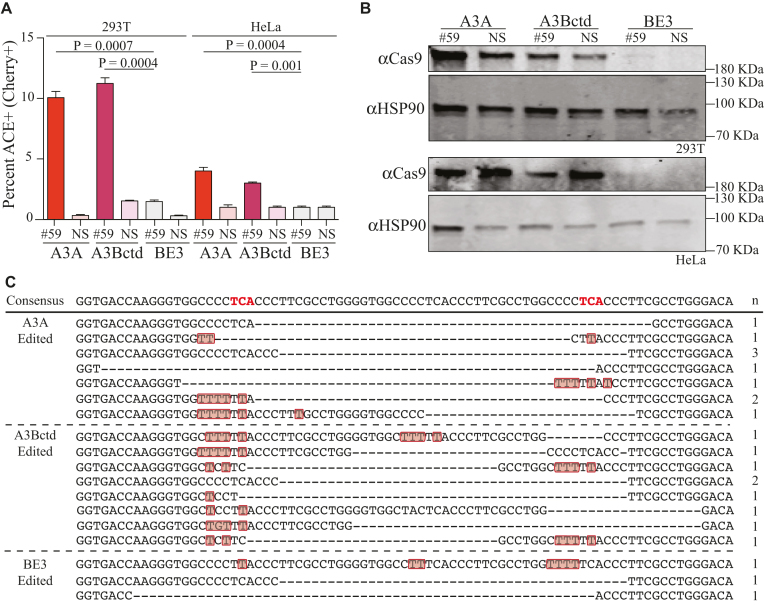
To further compare the efficiencies of these editosomes, ACE was pre-delivered to 293T and HeLa cells by lentiviral transduction (MOI of 1). Following stable introduction of the reporter, the resulting mCherry-negative/eGFP-positive pools were co-transfected with editosome constructs and either a gRNA directed to the insertion at mCherry codon 59 or a NS-gRNA as a negative control. As above, the A3A and A3Bctd editosomes performed better than the rat APOBEC1 editosome (Figure 3A and B). However, the single copy nature of the ACE system in the context of the chromosome caused a 10-fold reduction in the overall efficiency of each editosome. This result is to be expected because reversion of a single copy chromosomal reporter, which is chromatinized to varying degrees depending on integration position, will occur less frequently than editing of one of many episomal copies in a transient co-transfection experiment.
Figure 3.
Chromosomal editing by A3A and A3Bctd editosomes. (A) Editing of single copy genomic ACE reporter by the indicated editosomes in 293T and HeLa cells (n = 3, average ± SD). (B) Immunoblots corresponding to a representative experiment in panel A showing APOBEC–Cas9n-UGI expression levels and HSP90 as a loading control. (C) Sanger sequencing results for the gRNA-binding region of the ACE reporter recovered by high-fidelity PCR of mCherry-positive 293T. Mutated nucleotides are depicted in red and deleted nucleotides by hyphens. The number of times each sequence was recovered is indicated to the right.
To further investigate the mechanism of ACE reporter activation, DNA sequencing was used to ask whether editing events catalyzed by APOBEC editisomes are specific to the intended 5′-TCA motifs or distributed more broadly within the ssDNA loops created by gRNA base pairing to the duplicated target region. FACS was used to enrich for mCherry-positive cells with chromosomal editing events and, single high-fidelity PCR amplicons were cloned into a plasmid vector for Sanger sequencing (Figure 3C). As expected, almost all clones contained editing-associated deletions that caused mCherry to be shifted into the correct open reading frame, which is essential for fluorescence restoration. Interestingly, many of the sequences had C-to-T mutations in flanking regions displaced by annealing of the gRNA, but not in surrounding DNA regions that are presumably double-stranded and protected from the single-strand specific DNA deaminase activity of the APOBEC enzymes. Thus, in addition to enabling quantification of editing efficiencies in episomes and chromosomes, the ACE system unexpectedly reports both on-target and target-adjacent editing events. This helps to explain why several other tested sites in mCherry were not amenable to being developed into an editing reporter system.
Canonical Cas9 DNA cleavage quantification using the ACE reporter
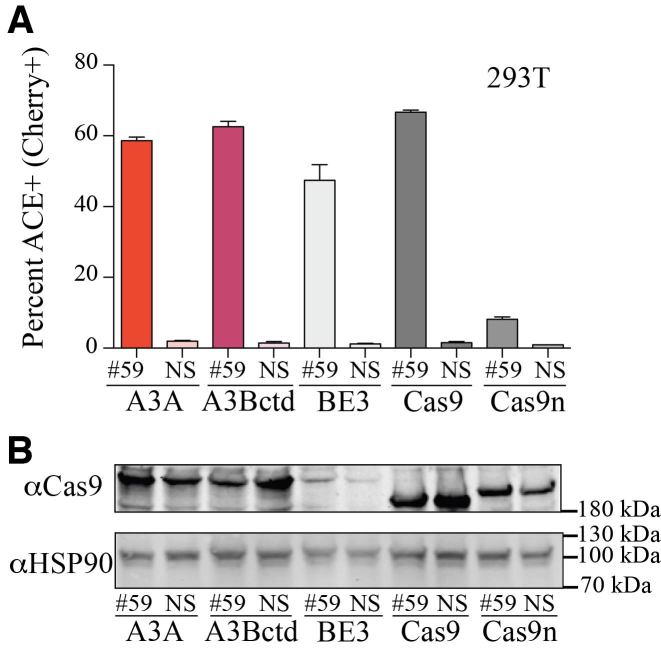
The tight coupling of editing and deletion mutagenesis led us to hypothesize that the ACE reporter would also be capable of quantifying the double-stranded DNA cleavage activity of Cas9. CRISPR/Cas9 is one of the most widely used new technologies in biology and medicine, and having a method to visualize its editing activity in real-time would be useful. To test this idea, the ACE system was simultaneously analyzed using A3A and A3Bctd editosomes, BE3, and Cas9 nuclease constructs (Figure 4). As already described, A3A and A3Bctd editosomes had editing levels higher than those of BE3. In addition, the Cas9 nuclease drove editing to levels comparable to A3A and A3Bctd editosomes, thus expanding the utility of the ACE reporter system. In comparison, the Cas9 nickase alone only elicited modest reporter activation.
Figure 4.
ACE reporter activation through Cas9 nucleolytic cleavage. (A) Quantification of ACE reporter activation in 293T cells 72 h after co-transfection of ACE reporter, mCherry codon 59 targeting gRNA or NS-gRNA, and A3A-Cas9n-UGI, A3Bctd-Cas9n-UGI, rat APOBEC1-Cas9n-UGI/BE3, Cas9, or Cas9n expression constructs (n = 3; mean ± SD). (B) Anti-Cas9 and anti-HSP90 immunoblots from a representative experiment reported in panel A.
Application of the ACE reporter system to enrich for editing events at heterologous chromosomal sites
We next asked if the ACE system could be used to enrich for chromosomal DNA editing events at an unlinked genetic locus with disease relevance. ACE-transduced eGFP-positive 293T cells were transfected with an A3A-, A3Bctd-, or rat APOBEC1-Cas9n-UGI base editing construct and gRNAs targeting mCherry codon 59 and FANCF codon 5. After 96 h incubation, mCherry-positive (ACE-edited) cells were purified by FACS and editing events at FANCF were assessed using a PCR and restriction enzyme-based assay (Figure 5A). Wild-type FANCF DNA amplicons are 452 bp, and restriction by PstI (5′-CTGCAG) results in two fragments, 192 and 260 bp, visible by agarose gel electrophoresis. APOBEC-mediated editing has the potential to destroy the PstI cleavage site and preserve the full-length fragment. The A3A and A3Bctd reactions yielded >10 000 mCherry-positive cells for this analysis, and unfortunately the rat APOBEC1 editosome yielded too few fluorescent cells for reliable purification (concordant with low frequency chromosomal editing data in Figure 3). Nevertheless, this restriction assay yielded clear results with FANCF editing events being highly enriched in sorted mCherry-positive cells in comparison to unsorted pools (Figure 5B; 290-fold and 5-fold for A3A and A3Bctd editosomes, respectively). Sequencing data showed that ACE-sorted cells are edited at the FANCF locus and nearly all edited sequences have mutations in the PstI cut site (Figure 5C). These data indicated that the ACE reporter system may be broadly useful for isolating subpopulations of cells with heterologous chromosomal editing events.
Figure 5.
ACE enriches for base-editing events at heterologous genomic loci. (A) Schematic of a co-transfection experiment resulting in ACE reporter activation (yellow shading represents mCherry and eGFP double-positive cells). FANCF and the PstI restriction assay used to quantify chromosomal base editing of this locus. Base editing events destroy the PstI cleavage site and block cleavage of the 452 bp amplicon into 260 and 192 bp products. (B) Representative agarose gels images showing the results of FANCF base editing by A3A and A3Bctd editsomes in 293T cells. The percentage of base editing was calculated by dividing the percentage of substrate band by the total of substrate and product bands following PstI cleavage for both unsorted and mCherry-positive cell populations. (C) Sanger sequencing results for the gRNA-binding region of the FANCF gene, which was recovered by high-fidelity PCR using genomic DNA from mCherry-positive 293T. Mutated nucleotides are highlighted in red and deleted nucleotides are indicated by hyphens. The number of times each sequence was recovered is indicated to the right.
DISCUSSION
We report the development of an APOBEC- and Cas9-mediated editing (ACE) reporter system for rapid, efficient, and quantitative fluorescent read-outs of DNA editing activity in living mammalian cells. The ACE reporter system is also, to our knowledge, the first to enable comparisons of the DNA editing efficiencies of the same isogenic reporter system in two different subcellular contexts - episomal high-copy conditions and chromosomal single-copy conditions. Standard molecular biology procedures may be used to adapt this system to other mammalian and non-mammalian cell types. The ACE reporter system may be used for a wide variety of applications, such as enrichment for heterologous editing events in reporter-activated cells. For example, as shown here for FANCF, transduction of the ACE reporter and subsequent transfection of an APOBEC editosome, along with gRNAs targeting mCherry codon 59 and FANCF, enabled FACS enrichment of mCherry-positive cells and enrichments for FANCF editing events.
Overall DNA editing efficiencies are also an important consideration. Here, we use the ACE system to validate new editosome complexes comprised of A3A and A3Bctd and show that these are more efficient than the previously described rat APOBEC1-based editosome BE3 (7) (Figures 2-4; Supplemental Figure S2). Immunoblots indicate that at least part of the increased efficiencies may be due to higher expression levels (Figures 2–4; Supplemental Figure S2). However, when titrations were carried out and similar protein expression levels were achieved, the A3A and A3Bctd editosomes still showed nearly twice as much editing (Figure 2D). Regardless of the full molecular explanation for the higher editing efficiencies demonstrated here, many applications such as site-directed mutation and anti-viral mutagenesis are likely to benefit from using the most efficient editosome complexes available.
Significant issues requiring further work are target-adjacent editing and local deletion outcomes, both recognized in the original BE3 study (7) and confirmed in subsequent work (8,9,11,15). These events appear to be confined to the DNA regions engaged by gRNA during the editing process as flanking double-stranded regions are not mutated. Target-adjacent editing events are likely due in part to the long dwell time of gRNA/Cas9 complexes and also to high APOBEC editing efficiencies. A recent report seems to have partly overcome this issue by using less efficient base editing complexes (16). Local deletions are most likely, as inferred here, due to simultaneous deamination of nearby cytosine bases, DNA strand breakage by the concerted activity of UNG2 and APE1, opposing DNA strand breakage by the nickase activity of Cas9, and error-prone resealing of the broken DNA ends by NHEJ. A variety of strategies have been reported to minimize deletion outcomes including adding more UGI (cis or trans), deleting UNG2, and using the DNA end-binding protein Gam (10,16). However, despite these advances, additional work is needed to completely eliminate undesirable outcomes and maximize desired on-target outcomes. The ACE system described here and/or future derivatives, together with further structural studies (ideally with full APOBEC editosome complexes), have the potential to facilitate the development of truly specific editing reactions and help advance base editing technologies toward clinical applications.
DATA AVAILABILITY
Key plasmids from our studies including the ACE reporter and the A3A and A3Bctd editing constructs are available through Addgene (ID 109425-109432).
Supplementary Material
ACKNOWLEDGEMENTS
We thank David Liu for providing BE3, Erik Toso and Michael Kyba for FACS, Hideki Aihara and Maria Montiel-González for helpful comments and Wendy Gordon for adapting the ACE system to solve additional problems.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Harris lab contributions were supported by NIGMS [R01 GM118000]; NIAID [R37 AI064046] NCI [R21 CA206309]; Conticello lab studies were supported by a grant from Ministero della Salute [PE-2013-02357669]; A.S. received salary support from NSF-GRFP 00039202, D.S. from NIH T90DE022732 and F.D. from AIRC [IG-17701]; R.S.H. is the Margaret Harvey Schering Land Grant Chair for Cancer Research, a Distinguished McKnight University Professor and an Investigator of the Howard Hughes Medical Institute. Funding for open access charge: HHMI.
Conflict of interest statement. R.S.H. is a co-founder, shareholder and consultant of ApoGen Biotechnologies Inc. The other authors declare no competing financial interests.
REFERENCES
- 1. Conticello S.G. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 2008; 9:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Harris R.S., Dudley J.P.. APOBECs and virus restriction. Virology. 2015; 479–480C:131–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nabel C.S., Manning S.A., Kohli R.M.. The curious chemical biology of cytosine: deamination, methylation, and oxidation as modulators of genomic potential. ACS Chem. Biol. 2012; 7:20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Simon V., Bloch N., Landau N.R.. Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat. Immunol. 2015; 16:546–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Harris R.S., Petersen-Mahrt S.K., Neuberger M.S.. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol. Cell. 2002; 10:1247–1253. [DOI] [PubMed] [Google Scholar]
- 6. Severi F., Chicca A., Conticello S.G.. Analysis of reptilian APOBEC1 suggests that RNA editing may not be its ancestral function. Mol. Biol. Evol. 2011; 28:1125–1129. [DOI] [PubMed] [Google Scholar]
- 7. Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R.. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016; 533:420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rees H.A., Komor A.C., Yeh W.H., Caetano-Lopes J., Warman M., Edge A.S.B., Liu D.R.. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat. Commun. 2017; 8:15790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim Y.B., Komor A.C., Levy J.M., Packer M.S., Zhao K.T., Liu D.R.. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol. 2017; 35:371–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang L., Xue W., Yan L., Li X., Wei J., Chen M., Wu J., Yang B., Yang L., Chen J.. Enhanced base editing by co-expression of free uracil DNA glycosylase inhibitor. Cell Res. 2017; 27:1289–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim D., Lim K., Kim S.T., Yoon S.H., Kim K., Ryu S.M., Kim J.S.. Genome-wide target specificities of CRISPR RNA-guided programmable deaminases. Nat. Biotechnol. 2017; 35:475–480. [DOI] [PubMed] [Google Scholar]
- 12. Kim K., Ryu S.M., Kim S.T., Baek G., Kim D., Lim K., Chung E., Kim S., Kim J.S.. Highly efficient RNA-guided base editing in mouse embryos. Nat. Biotechnol. 2017; 35:435–437. [DOI] [PubMed] [Google Scholar]
- 13. Li J., Sun Y., Du J., Zhao Y., Xia L.. Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol. Plant. 2017; 10:526–529. [DOI] [PubMed] [Google Scholar]
- 14. Lu Y., Zhu J.K.. Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system. Mol. Plant. 2017; 10:523–525. [DOI] [PubMed] [Google Scholar]
- 15. Zong Y., Wang Y., Li C., Zhang R., Chen K., Ran Y., Qiu J.L., Wang D., Gao C.. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 2017; 35:438–440. [DOI] [PubMed] [Google Scholar]
- 16. Komor A.C., Zhao K.T., Packer M.S., Gaudelli N.M., Waterbury A.L., Koblan L.W., Kim Y.B., Badran A.H., Liu D.R.. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci. Adv. 2017; 3:eaao4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lei L., Chen H., Xue W., Yang B., Hu B., Wei J., Wang L., Cui Y., Li W., Wang J. et al. . APOBEC3 induces mutations during repair of CRISPR-Cas9-generated DNA breaks. Nat. Struct. Mol. Biol. 2018; 25:45–52. [DOI] [PubMed] [Google Scholar]
- 18. Hess G.T., Fresard L., Han K., Lee C.H., Li A., Cimprich K.A., Montgomery S.B., Bassik M.C.. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat. Methods. 2016; 13:1036–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nishida K., Arazoe T., Yachie N., Banno S., Kakimoto M., Tabata M., Mochizuki M., Miyabe A., Araki M., Hara K.Y. et al. . Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016; 353:aaf8729. [DOI] [PubMed] [Google Scholar]
- 20. Shimatani Z., Kashojiya S., Takayama M., Terada R., Arazoe T., Ishii H., Teramura H., Yamamoto T., Komatsu H., Miura K. et al. . Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 2017; 35:441–443. [DOI] [PubMed] [Google Scholar]
- 21. Kuscu C., Adli M.. CRISPR-Cas9-AID base editor is a powerful gain-of-function screening tool. Nat. Methods. 2016; 13:983–984. [DOI] [PubMed] [Google Scholar]
- 22. Banno S., Nishida K., Arazoe T., Mitsunobu H., Kondo A.. Deaminase-mediated multiplex genome editing in Escherichia coli. Nat. Microbiol. 2018; 3:423–429. [DOI] [PubMed] [Google Scholar]
- 23. Lindahl T., Wood R.D.. Quality control by DNA repair. Science. 1999; 286:1897–1905. [DOI] [PubMed] [Google Scholar]
- 24. Krokan H.E., Drablos F., Slupphaug G.. Uracil in DNA–occurrence, consequences and repair. Oncogene. 2002; 21:8935–8948. [DOI] [PubMed] [Google Scholar]
- 25. Hultquist J.F., Lengyel J.A., Refsland E.W., LaRue R.S., Lackey L., Brown W.L., Harris R.S.. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J Virol. 2011; 85:11220–11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pham P., Afif S.A., Shimoda M., Maeda K., Sakaguchi N., Pedersen L.C., Goodman M.F.. Activation-induced deoxycytidine deaminase: Structural basis for favoring WRC hot motif specificities unique among APOBEC family members. DNA Repair. 2017; 54:8–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stenglein M.D., Burns M.B., Li M., Lengyel J., Harris R.S.. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat. Struct. Mol. Biol. 2010; 17:222–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Carpenter M.A., Li M., Rathore A., Lackey L., Law E.K., Land A.M., Leonard B., Shandilya S.M., Bohn M.F., Schiffer C.A. et al. . Methylcytosine and normal cytosine deamination by the foreign DNA restriction enzyme APOBEC3A. J. Biol. Chem. 2012; 287:34801–34808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Burns M.B., Lackey L., Carpenter M.A., Rathore A., Land A.M., Leonard B., Refsland E.W., Kotandeniya D., Tretyakova N., Nikas J.B. et al. . APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013; 494:366–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ito F., Fu Y., Kao S.A., Yang H., Chen X.S.. Family-wide comparative analysis of cytidine and methylcytidine deamination by eleven human APOBEC proteins. J. Mol. Biol. 2017; 429:1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bohn M.F., Shandilya S.M., Silvas T.V., Nalivaika E.A., Kouno T., Kelch B.A., Ryder S.P., Kurt-Yilmaz N., Somasundaran M., Schiffer C.A.. The ssDNA mutator APOBEC3A is regulated by cooperative dimerization. Structure. 2015; 23:903–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shi K., Carpenter M.A., Kurahashi K., Harris R.S., Aihara H.. Crystal structure of the DNA deaminase APOBEC3B catalytic domain. J. Biol. Chem. 2015; 290:28120–28130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shi K., Carpenter M.A., Banerjee S., Shaban N.M., Kurahashi K., Salamango D.J., McCann J.L., Starrett G.J., Duffy J.V., Demir O. et al. . Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B. Nat. Struct. Mol. Biol. 2017; 24:131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kouno T., Silvas T.V., Hilbert B.J., Shandilya S.M.D., Bohn M.F., Kelch B.A., Royer W.E., Somasundaran M., Kurt Yilmaz N., Matsuo H. et al. . Crystal structure of APOBEC3A bound to single-stranded DNA reveals structural basis for cytidine deamination and specificity. Nat. Commun. 2017; 8:15024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Key plasmids from our studies including the ACE reporter and the A3A and A3Bctd editing constructs are available through Addgene (ID 109425-109432).