Abstract
Objective.
To determine whether autophagy is involved in the degradation of misfolded HLA-B27 in experimental spondyloarthritis.
Methods.
Bone marrow derived macrophages from HLA-B27 and human β2m (hβ2m) transgenic rats were examined with and without IFNγ and proteasome or autophagy inhibitors. Immunoprecipitation, western blotting, and immunofluorescence were used to measure HLA-B27 heavy chains and autophagy. Autophagy was induced using rapamycin. HLA-B7/hβ2m transgenic and wild type rat macrophages were used as controls.
Results.
HLA-B27-expressing macrophages showed similar LC3B-II levels compared to both controls, before and after manipulating autophagy. Blocking autophagic flux with bafilomycin resulted in the accumulation of misfolded HLA-B27 dimers and oligomers as well as monomers, comparable to blocking endoplasmic reticulum-associated degradation (ERAD) with the proteasome inhibitor bortezomib. HLA-B7 monomers also accumulated after blocking each degradation pathway. The proportion of ubiquitinated heavy chains was ~3-fold lower for HLA-B27 compared to HLA-B7. Activation of autophagy with rapamycin rapidly eliminated ~50% of misfolded HLA-B27, while folded HLA-B27 or HLA-B7 monomeric heavy chains were minimally affected.
Conclusion.
These results demonstrate for the first time that both autophagy and ERAD play a role in the elimination of excess HLA class I heavy chains expressed in transgenic rats. We found no evidence that HLA-B27 expression modulated the autophagy pathway. Our results suggest that impaired ubiquitination of HLA-B27 may play a role in the accumulation of misfolded disulfide-linked dimers, whose elimination can be enhanced by activation of autophagy. Manipulation of the autophagy pathway should be further investigated as a potential therapeutic avenue in spondyloarthritis.
Keywords: HLA-B27, spondyloarthritis, ER quality control, ER-associated degradation, autophagy
INTRODUCTION
HLA-B27 is strongly associated with the development of spondyloarthritis (SpA), and in particular axial disease that progresses to ankylosing spondylitis (1). While the pathogenic mechanisms remain incompletely understood, abnormal properties of the HLA-B27 heavy chain have been implicated (2, 3). HLA-B27 exhibits a propensity to misfold and dimerize in the endoplasmic reticulum (ER) (4–6), and to form β2m-free homodimers on the cell surface (2). HLA-B27 has also been implicated in dendritic cell dysfunction (7, 8). Each of these properties has been mechanistically linked to the IL-23/IL-17 axis where several additional risk genes are located (9), supporting a central role for this axis in pathogenesis (10). Indeed, when HLA-B27 is expressed in rats along with human β2m (hβ2m), a SpA-like disease that is dependent on gut microbiota develops spontaneously (11), with striking activation of the IL-23/IL-17 axis in the inflamed gut and involved joints (12, 13). Interleukin-17A (IL-17A) has recently been established as a therapeutic target in ankylosing spondylitis (14), and there is evidence that blocking IL-23/IL-12 may be beneficial (15), underscoring the importance of this axis in human disease, and the relevance of this animal model for dissecting mechanisms.
The abnormal behavior of HLA-B27 in the endoplasmic reticulum (ER) involves the formation of disulfide-linked dimers by newly synthesized heavy chains (5, 16–18). These misfolded heavy chains accumulate bound to the ER chaperone BiP (also known as Grp78; encoded by Hspa5), and under certain circumstances can activate an ER stress response that alters immune signaling pathways (17, 19). There are ER quality control pathways that orchestrate the elimination of misfolded proteins, usually mitigating the threat they pose to ER homeostasis (20). HLA-B27 has been shown to undergo ER associated degradation (ERAD) (4, 21, 22), which involves dislocation of target proteins from the ER membrane, with retrotranslocation and ubiquitination in the cytosol where they are degraded by proteasomes (23). While the machinery differs for different proteins, ERAD of HLA class I and HLA-B27 is dependent on the E3 ubiquitin ligase, HRD1 (3-hydroxy-3-methylglutaryl-coenzyme A reductase degradation protein 1) (also known as synoviolin; encoded by SYVN1), and the E2 ubiquitin conjugating enzyme UBE2J1 (21, 22). Depletion of HRD1 as well as other components of the ERAD pathway (e.g. XBP1, derlin-1, derlin-2) promotes the accumulation of misfolded HLA-B27 heavy chain dimers in HeLa cells. Moreover, forced overexpression of EDEM1 (ER degradation enhancing α-mannosidase-like protein 1), which targets unsalvageable ER glycoproteins for ERAD, or upregulation of the ERAD pathway, enhances the elimination of HLA-B27 dimers (22). Despite ERAD serving a role in ER quality control (ERQC) for HLA-B27, it is insufficient to prevent the accumulation of misfolded dimers in cells expressing this HLA class I molecule (5, 17).
Autophagy delivers cytoplasmic organelles and macromolecules to lysosomes for destruction (24), and is important for cellular survival during short-term starvation, recycling of damaged organelles, differentiation of several cell types, and for the clearance of intracellular microbes (24–27). In macroautophagy (generally referred to as autophagy), a double membrane is formed around targeted organelles or aggregated proteins creating an autophagosome, that subsequently fuses with lysosomes (28). Autophagy is important for the elimination of terminally misfolded cytosolic and ER luminal proteins that are resistant to proteasomal degradation (29, 30), and more recently was shown to degrade non-aggregated membrane proteins that are resistant to ERAD (31). This prompted us to ask whether autophagy is also involved in the degradation of misfolded forms of HLA-B27.
Here, we show that autophagy as well as ERAD contributes to the elimination of misfolded disulfide-linked HLA-B27 heavy chains. Moreover, activation of autophagy with rapamycin can be used to promote degradation of misfolded HLA-B27. We also show that HLA-B27 dimers are poorly ubiquitinated relative to the control substrate HLA-B7, suggesting that the formation of disulfide linked dimers may be an impediment to efficient ERAD. These results raise the possibility that the autophagy pathway might be exploited for therapeutic benefit.
MATERIALS and METHODS
Animals.
Hemizygous HLA-B*27:05 and human β2m-transgenic (B27-Tg) Lewis rats carrying 55 copies of HLA-B27 and 66 copies of human β2m in the 33–3 locus on the Lewis background were used as described previously (32). The HLA-B*07:02 and human β2m-transgenic (B7-Tg) rat line (Lewis, 120–4 transgene locus) was kindly provided by Joel Taurog (University of Texas Southwestern Medical Center, Dallas, TX). Homozygous B7-Tg rats containing 52 copies of HLA-B7 and 10 copies of human β2m were used, as were wild type (WT) Lewis transgene negative littermates. Rats were bred and housed in Association for Assessment and Accreditation of Laboratory Animal Care-approved facilities on the Bethesda campus of the National Institutes of Health. All animal experiments were preapproved by the Animal Care and Use Committee at the National Institute of Arthritis and Musculoskeletal and Skin Disease.
Isolation of bone marrow-derived macrophages.
Bone marrow was obtained from the tibias and femurs of 8–12 week old Lewis rats following euthanasia. To derive macrophages, non-adherent bone marrow derived monocytes were incubated at 5 × 106/mL in high-glucose Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific, Waltham, MA) supplemented with 10% fetal calf serum (Thermo Fisher Scientific), 100 U/mL Penicillin, 100 ug/ml Streptomycin (Thermo Fisher Scientific), and 20 ng/mL macrophage colony-stimulating factor (M-CSF; Peprotech, Rocky Hill, NJ). Cells were cultured at 37°C in humified atmosphere containing 5% CO2. Media was changed every other day. Cells were used for experiments at day 7.
Reagents and antibodies.
IFNγ was used at 50 ng/ml for all experiments. For autophagy modulation the following reagents were used: Rapamycin (InvivoGen, San Diego, CA), Bafilomycin A1 (Bafilomycin) (InvivoGen) and Chloroquine diphosphate salt (Sigma-Aldrich, St. Louis, MO). Bortezomib (Cell Signaling, Danvers, MA) was used to block proteasomal degradation of proteins and hence ERAD. For western blotting and immunoprecipitation the following antibodies were used: rabbit anti-LC3B antibody (Sigma-Aldrich), rabbit anti-β Tubulin antibody (Abcam, Cambridge, UK), goat anti-Grp78/Hspa5 antibody (R&D Systems, Minneapolis, MN), mouse HC10 antibody (33) and rat 3B10.7 (5), recognizing HLA class I. For flow cytometry, the ME1 antibody was used. ME1 recognizes HLA-B27 and other B alleles, but not rat class I. As secondary antibodies for western blotting, horseradish peroxidase (HRP)-conjugated anti-rabbit, anti-goat, anti-mouse (R&D Systems) and anti-rat (SouthernBiotech, Birmingham, AL) were used. For immunofluorescence rabbit anti-LC3B antibody (Cell Signaling) and Alexa Fluor 488 conjugated donkey anti-rabbit secondary antibody (Life Technologies, Carlsbad, California) were used.
Immunofluorescence.
For immunofluorescence experiments, macrophages from WT, B7-Tg or B27-Tg rats were incubated with different autophagy modulators for 2 hours. Cells were then fixed and permeabilized with ice-cold methanol and incubated with 5% normal goat serum (Life Technologies) for one hour at room temperature, followed by overnight incubation at 4°C with the primary antibody (anti-LC3B, Cell Signaling). After washing with PBS, cells were incubated with the fluorescent-labeled secondary antibody (anti-rabbit Alexa 488) for 2 hours at room temperature. The nucleus was visualized with DAPI (Sigma-Aldrich). Immunofluorescence was evaluated by using the confocal fluorescence microscope Leica DMI6000 inverted microscope SP5 NLO (Leica Microsystems, Wetzlar, Germany) at 63x magnification.
Immunoprecipitation and immunoblotting.
To prevent spontaneous sulfhydryl bond formation and breakdown, at the end of each experiment and immediately prior to harvest, cells were treated for 20 min with 10 mM methyl methanethiosulfonate (MMTS, Thermo Scientific) in PBS on ice. Cells were then lysed with lysis buffer (0.025 M Tris, 0.15 M NaCl, 0.001 M EDTA, 1% NP-40, 5% glycerol; pH 7.4; Thermo Fisher Scientific) containing complete, EDTA-free proteinase inhibitor cocktail tablets (Roche, Indianapolis, IN) for 20 min on ice. After centrifugation at 13,000 g for 10 min to remove nuclei, supernatants were used for western blot analysis or immunoprecipitation. For western blotting, lysates were diluted with non-reducing 5x sample buffer (Thermo Fisher Scientific) and boiled for 5 min at 95°C. In order to reduce samples, 4x sample buffer containing dithiothreitol (Sigma-Aldrich) was used. Proteins were separated on a 4–20% Tris/Glycine gel (BioRad, Hercules, CA). After immunoblotting, the PVDF membrane was blocked with 5% milk in PBS containing 0.02% Tween (PBS-T) buffer for 1 hour and incubated overnight at 4°C in 5%BSA/PBS-T with the following first antibodies: 3B10.7 to detect HLA class I heavy chains, goat anti-BiP (R&D Systems), rabbit anti-LC3B (Sigma-Aldrich), rabbit anti-Ubiquitin (Cell Signaling) and rabbit anti-βTubulin (Abcam). After three washes the membrane was incubated with the respective HRP-conjugated the secondary antibody (R&D Systems) for 1 hour at room temperature. Proteins were visualized using the Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific). Prior to immunoprecipitation, sample protein concentration was measured using the BioRad DC Protein Assay (BioRad). Equal amounts of protein were used for immunoprecipitations. Immunoprecipitation with the HC10 antibody was performed using the Pierce Crosslink IP kit (Thermo Fisher Scientific) according to manufacturers guidelines. To immunoprecipitate polyubiquitin protein conjugates, the Pierce Ubiquitin Enrichment kit (Thermo Fisher Scientific) was used.
Flow cytometry.
To measure cell surface expression of HLA-B27, macrophages were washed and incubated with 5% BSA (Sigma-Aldrich) in PBS at 4oC for 30 min, and then incubated with allophycocyanin (APC)-labeled HC10 antibody or Pacific Blue-labeled ME1 antibody for 30 min at 4oC. Cells were washed and analyzed by FACS using a Beckman Coulter CyAn ADP (Beckman Coulter).
Statistical analysis.
Statistical analysis was performed using Student’s t test. Differences between means with p < 0.05 were considered statistically significant using the GraphPad Prism Version 6.0b (LaJolla, CA). Unless otherwise stated, the figures show three independent experiments.
RESULTS
Autophagy and ERAD contribute to the degradation of HLA-B27.
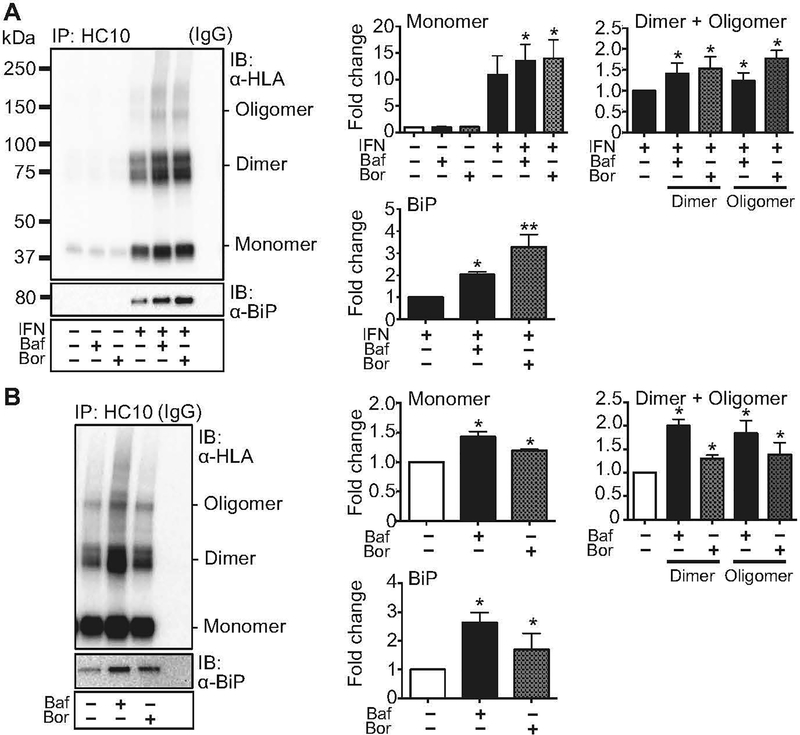
To determine whether autophagy plays a role in the degradation of HLA-B27 we assessed bone marrow-derived macrophages (BMM) from HLA-B27 Tg rats. BMM were derived in vitro in the absence of pro-inflammatory cytokines, and under these conditions express low levels of β2m-free HLA-B27 heavy chains (17, 34). To determine the relative contribution of autophagy and ERAD to the elimination of HLA-B27, BMM were incubated without or with IFNγ for 21 hours to upregulate HLA-B27 expression, and then bafilomycin A1 (bafilomycin) or bortezomib were added for the final 3 hours. Bafilomycin inhibits the vacuolar H+ ATPase, which prevents the maturation of autophagic vacuoles and autophagic degradation of proteins (35). Bortezomib binds to the active sites of the β5 and β1 subunits of the proteasome (36) and inhibits proteasomal degradation of proteins, which is essential for ERAD (37). Cells were lysed and HLA-B27 heavy chains immunoprecipitated (IP) from equal amounts of protein with the HC10 antibody, separated by non-reducing SDS-PAGE, and visualized by immunoblotting with the anti-HLA class I antibody 3B10.7 (Fig. 1). HC10 recognizes HLA-B (and HLA-C) heavy chains that are not associated with β2m (38), including unfolded and misfolded forms that exist as monomers, dimers, or oligomers (5). In rat cells that lack other HLA-B/C alleles, only HLA class I expressed from the transgene is recognized. Co-precipitating BiP bound to HLA-B27 heavy chain was also visualized by immunoblotting after running an aliquot of the same HC10 IP on a separate gel.
Figure 1.
Accumulation of HLA-B27 with inhibition of autophagy or ERAD. (A) BM-derived macrophages were left untreated or stimulated with IFNγ (IFN) for 21 hours, then treated for 3 hours with bafilomycin (100 nM) (Baf) to block the autophagy pathway, or with bortezomib (10 nM) (Bor) to block ERAD. Untreated cells and cells only treated with bafilomycin or bortezomib were used as controls. Immunoprecipitation (IP) and immunoblotting (IB) with HC10 and 3B10.7 (α-HLA) or α-BiP, were as indicated and described in Materials and Methods. Dimers and oligomers are visualized on non-reduced samples electrophoresed on non-reducing gels, while BiP is analyzed under reducing conditions. For quantification, samples were normalized to untreated or IFNγ-treated controls, as indicated, and expressed as fold change. For all figures, representative immunoblots are shown. Quantitative data represent mean ± SEM of five independent experiments. (*, p < 0.05) (B) HLA-B27 expressing C1R cells were stimulated for 3 hours with bafilomycin (Baf) or bortezomib (Bor), prior to immunoprecipitation and immunoblotting. Quantitative data represent mean ± SEM of six experiments with the same cell line. (*, p < 0.05). IgG lanes show background when IP was performed with non-specific IgG alone.
In the absence of IFNγ, relatively little β2m-free HLA-B27 heavy chain is detected in IPs from macrophages (Fig. 1A). HLA-B27 is strongly upregulated by IFNγ resulting in the accumulation of heterogeneous disulfide-linked dimers and oligomers of HLA-B27 (also referred to as misfolded forms), as well as monomers (also referred to as free heavy chains) (Fig. 1A). Proteasome inhibition with bortezomib (Bor) resulted in a 1.5 to 1.7-fold increase in dimeric and oligomeric forms of HLA-B27, respectively. This confirms previous results demonstrating that misfolded HLA-B27 undergoes ERAD in human cells (4, 21, 22), and extends these findings to HLA-B27 Tg rats. Blockade of autophagic flux with bafilomycin (Baf) induced a similar (1.3 to 1.4-fold) accumulation of HLA-B27 dimers and oligomers, with effects of proteasome inhibition being slightly more prominent. Bafilomycin and bortezomib also led to a 1.2–1.3-fold increase in the accumulation of monomers. Inhibition of autophagy and ERAD also resulted in the accumulation of BiP in HLA-B27 heavy chain immunoprecipitates (2 to 3-fold). BiP accumulates bound to misfolded HLA-B27 in the ER (17), and has been shown to preferentially accumulate with HLA-B27 dimers (16). Treatment with each inhibitor alone in cells not treated with IFNγ did not lead to accumulation of HLA-B27, indicating that neither autophagy nor ERAD are contributing significantly to rapid heavy chain degradation at baseline. Similar experiments were performed with wortmannin and LY294002, both of which inhibit autophagosome formation by blocking phosphatidylinositol 3-kinase. We obtained results similar to bafilomycin, supporting involvement of the autophagy pathway (unpublished observations).
We next asked whether transfected cells that constitutively express HLA-B27 (e.g. C1R.B27) in the absence of pro-inflammatory cytokines also use autophagy to degrade misfolded heavy chains. We have shown previously that C1R.B27 cells eliminate a proportion of HLA-B27 through the ERAD pathway, with deglycosylated heavy chains accumulating in the cytosol after 3 hours of proteasome inhibition (4). We treated C1R.B27 cells for 3 hours with bortezomib or bafilomycin, then assessed accumulation of various forms of HLA-B27 heavy chain (Fig. 1B). Both bafilomycin and bortezomib increased the accumulation of HLA-B27, with the effect on dimers being most prominent, followed by oligomers and monomers. Co-precipitating BiP also increased. Bafilomycin was more effective than bortezomib at inhibiting HLA-B27 degradation and causing accumulation of heavy chains bound to BiP in C1R.B27 cells. To demonstrate that both inhibitors were blocking the destruction of proteins targeted for degradation, we examined ubiquitinated complexes in whole cell lysates. In both rat BMM and C1R cells, incubation with bafilomycin or bortezomib for 3 hours led to accumulation of ubiquitinated proteins compared to untreated cells, or cells treated with IFNγ alone (Suppl. Fig. S1). Only bafilomycin caused an increase in LC3B-II, a marker of autophagy (35, 39), indicating that bortezomib was not affecting autophagic flux under these conditions (Suppl. Fig. S1). Taken together these results show that in addition to ERAD, rat BMM as well as C1R cells use the autophagy pathway to degrade misfolded dimers and oligomers, as well as some monomers of HLA-B27.
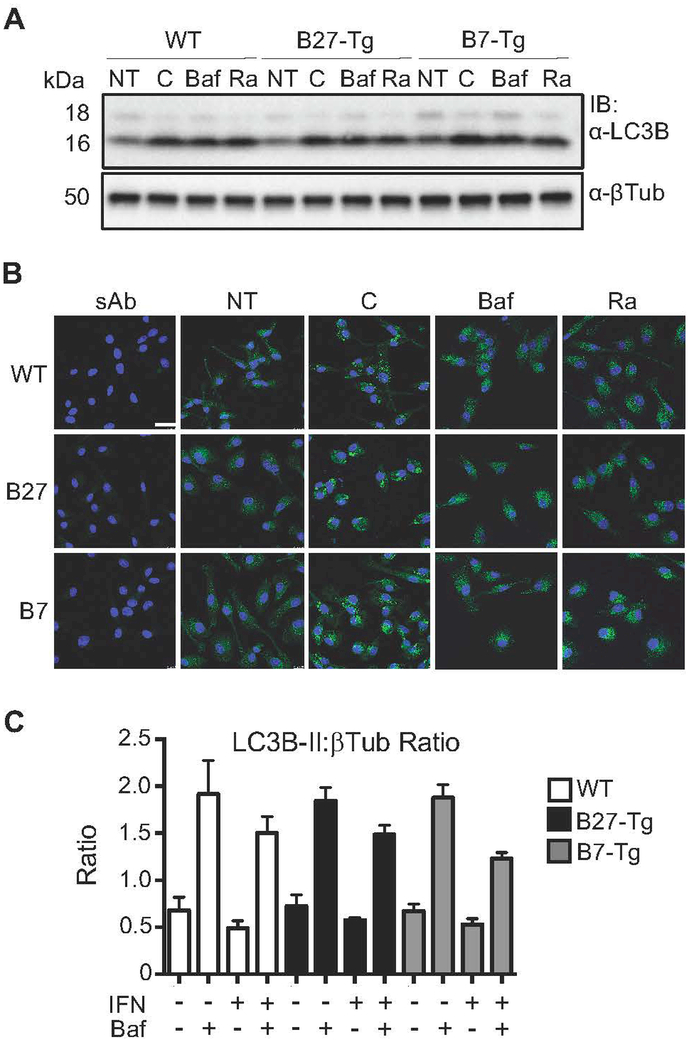
Autophagy is not altered by HLA-B27 expression in rat bone marrow macrophages.
The involvement of autophagy in HLA-B27 degradation raised the possibility that this pathway might be altered in HLA-B27-expressing rat cells. To test this, BMM from WT, B27-Tg and B7-Tg rats were incubated with bafilomycin or chloroquine to block autophagic flux, or with rapamycin, which activates autophagy by inhibiting mTOR. Changes in LC3B-II level were monitored as a marker for autophagic flux (39). The LC3B protein (encoded by Map1lc3b) is cleaved at the C-terminus forming LC3B-I, which is localized in the cytosol. During the initiation of autophagy, LC3B-I is conjugated to phosphatidylethanolamine (PE) to form LC3B-II, which becomes embedded in the autophagosome membrane. Hence, an increase in LC3B-II is widely used as a measure of autophagy as it reflects the number of autophagosomes. The antibody used for western blotting reveals two bands representing LC3B-I (upper) and LC3B-II (lower) (e.g. Fig. 2A; LC3B-II migrates faster due to the PE moiety). Increased accumulation of LC3B-II can reflect activation of autophagy (until autophagosomes are destroyed and steady state is reached), or inhibition of later stages of autophagy, which leads to accumulation of autophagosomes. Therefore, inhibitors (chloroquine and bafilomycin) as well as activators of autophagy (rapamycin) both lead to increases in LC3B-II under these conditions (13). No differences in LC3B-II levels were detected between untreated WT, B27-Tg, and B7-Tg macrophages by immunoblotting (Fig. 2A, NT lanes). Within each group, chloroquine, bafilomycin and rapamycin all increased LC3B-II accumulation as expected (Fig. 2A, lanes C, Baf, and Ra), but there were no differences between genotypes, indicating that the autophagy pathway is not altered by baseline (unstimulated) HLA-B27 or HLA-B7 expression. This was further confirmed by immunofluorescence staining of LC3B-II in macrophages. While chloroquine, bafilomycin, and rapamycin all increase vesicular staining for LC3B-II, there were no apparent differences between genotypes (Fig. 2B). We also asked whether treatment of cells with IFNγ would expose differences between genotypes due to either HLA-B27 or HLA-B7 upregulation. Again, there were no differences between WT and HLA class I-expressing cells incubated with bafilomycin alone, IFNγ alone, or both (Fig. 2C). It should be noted that activating autophagy in macrophages requires high concentrations of rapamycin that could be toxic (40). Therefore, we confirmed that the dose we used (25 μM) did not affect macrophage viability after 2–4 hours of incubation (unpublished observations). These data indicate that the autophagy pathway is not grossly altered by HLA-B27 or HLA-B7 expression in rat macrophages.
Figure 2.
Autophagy in rat macrophages expressing HLA class I transgenes. (A) BM macrophages from WT, B27-Tg, and B7-Tg rats were left untreated or stimulated with rapamycin (25 μM) (Ra) to induce autophagy, chloroquine (50 μM) (C) or bafilomycin (400 nM) (Baf) to block autophagic flux, and then collected 2 hours later and lysed for immunoblotting using anti-LC3B (α-LC3B), with anti-βTubulin (α-βTub) used as a loading control. Immunoblot shown is representative of three independent experiments. (B) Macrophages treated as described in (A) (NT, untreated; sAb, secondary Ab only) were analyzed by immunofluorescence with anti-LC3B (green), which has a stronger affinity for LC3B-II. Nuclei are visualized by DAPI staining (blue). Immunofluorescence was evaluated using confocal imaging with results shown at 63x magnification. Images are representative of three independent experiments. (C) BM macrophages from WT, B27-Tg, and B7-Tg rats were incubated without or with IFNγ (50 ng/ml) for 24 hours, followed by another 24 hours with bafilomycin (10 nM). LC3B-II expression was determined by immunoblotting (as for experiment shown in A), and is expressed as a ratio of LC3B-II:βTub. Results are representative of three independent experiments.
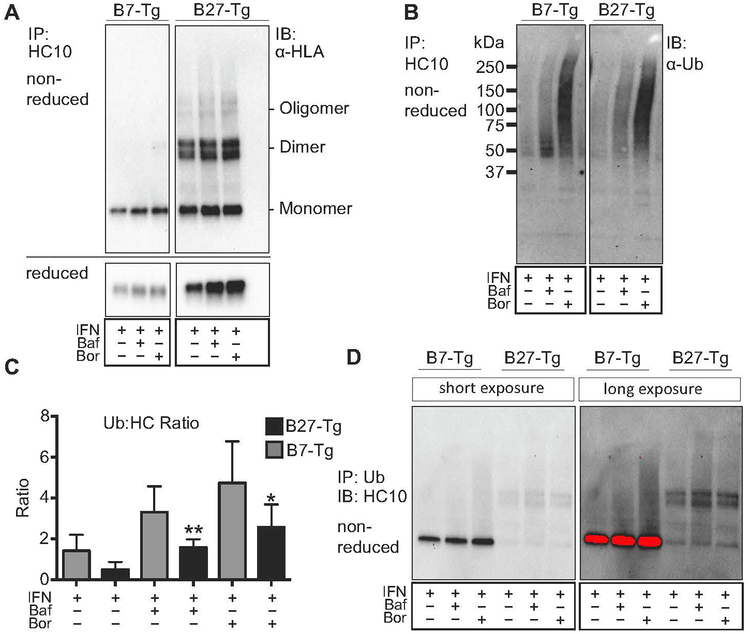
HLA-B27 heavy chains are poorly ubiquitinated relative to HLA-B7.
Many proteins that misfold in the ER are efficiently degraded by ERAD, while others (e.g. HLA-B27) can accumulate and result in cellular dysfunction (30, 41). Resistance to ERAD is incompletely understood, but one contributing factor is the formation of aberrant intermolecular disulfide bonds that result in dimerization or oligomerization (42). Since ERAD requires ubiquitination, we asked whether HLA-B27 heavy chains were ubiquitinated using HLA-B7 for comparison. Homozygous HLA-B7 Tg rats were used so that comparable amounts of HLA class I heavy chain (to HLA-B27) would be expressed and upregulated in response to IFNγ (34). In experiments similar to those shown in Figure 1, we noted that HLA-B7 heavy chain monomers accumulate in IFNγ-treated cells when autophagy or ERAD are inhibited (Fig. 3A), indicating that excess HLA-B7 heavy chains are also being eliminated by quality control pathways. Next, aliquots of the same HC10 immunoprecipitates shown in Fig. 3A were subjected to SDS-PAGE and probed for ubiquitin. Fig. 3B shows that highly ubiquitinated HLA class I heavy chains accumulate when ERAD is inhibited, and to a lesser extent when autophagy is blocked. We calculated the ratio of ubiquitinated material in the HC10 immunoprecipitates (e.g. Fig 3B) to total heavy chain in the same immunoprecipitates (e.g. Fig. 3A, reduced band) from five experiments, and plotted the ubiquitin to heavy chain ratio (Ub:HC) for both HLA class I molecules (Fig. 3C). These quantitative data reveal several important points. First, the Ub:HC ratio is 2–3 fold greater for HLA-B7 than HLA-B27. This must reflect a greater number of ubiquitinated heavy chains, since the alternative of more ubiquitin per heavy chain, is not reflected in the patterns shown in Fig. 3B (i.e. no shift toward higher molecular weight complexes for HLA-B7). Second, the Ub:HC ratio increases with blockade of autophagy or ERAD, but is most pronounced when ERAD is inhibited. This is consistent with ERAD being the primary pathway for degradation of ubiquitinated heavy chains. Third, even without blocking degradation (i.e. IFNγ upregulation alone) ubiquitinated heavy chains are more abundant for HLA-B7 than HLA-B27 (Fig. 3B,C IFNγ lanes). Note that the lower recovery of β2m-free HLA-B7 heavy chains compared to HLA-B27 (Fig. 3A, reduced panel) is due to more efficient folding and loss of HC10-reactivity as described previously (34).
Figure 3.
Differential ubiquitination of HLA-B7 and HLA-B27 heavy chains. (A) BM macrophages expressing HLA-B27 and HLA-B7 were treated for 21 hours with IFNγ, and then an additional 3 hours without or with bafilomycin (100 nM) (Baf) or bortezomib (10 nM) (Bor). HLA class I heavy chains immunoprecipitated with HC10 and analyzed under non-reducing or reducing conditions as indicated, using 3B10.7. (B) Samples from experiment described in (A) were immunoblotted with anti-ubiquitin antibody (α-Ub). (C) The ratio of ubiquitinated material (B) (from entire lane) to immunoprecipitated total heavy chain quantitated under non-reducing conditions (A, reduced) is plotted for HLA-B27 and HLA-B7 under each experimental condition. Results represent mean ± SEM (error bars) from five experiments. (*, p < 0.05) (D) Cells treated as described in (A) were used to immunoprecipitate all ubiquitinated material, and then blotted with HC10 to visualize heavy chains. In the long exposure image the red color indicates saturation.
To gain more insight into the predominant form of heavy chain that is ubiquitinated, we used an anti-ubiquitin antibody to perform immunoprecipitations, and then blotted for HLA class I heavy chains with HC10. Significantly more HLA-B7 is recovered than HLA-B27 (Fig. 3D), consistent with the increased Ub:HC ratio for HLA-B7 (Fig. 3C). Furthermore, the predominant form of HLA-B27 that is recovered is dimers, whereas a single band is recovered for HLA-B7. Taken together, these data indicate that there are major differences in how HLA-B7 and HLA-B27 heavy chains are handled by quality control pathways in rat BMM. Notably, HLA-B27 heavy chain dimers (and monomers) are inefficiently ubiquitinated compared to HLA-B7.
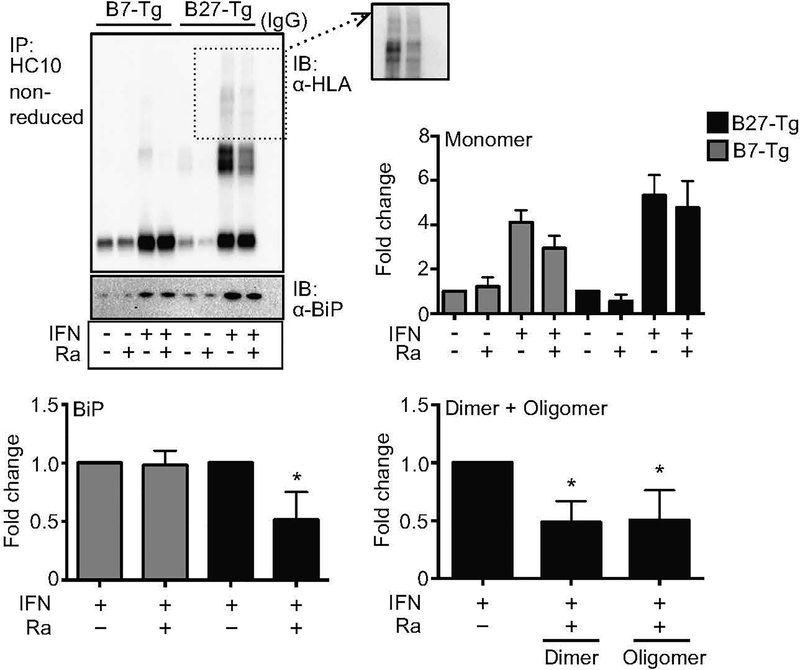
Activation of autophagy eliminates misfolded and unfolded forms of HLA-B27.
We next asked whether activating autophagy would enhance the degradation of HLA-B27. BMM were treated with IFNγ for 6 hours to upregulate class I expression, washed to remove IFNγ, and then incubated for an additional 2 hours without or with rapamycin to activate autophagy (Suppl. Fig. S2). HLA-B27 heavy chains were then immunoprecipitated with HC10, and visualized by immunoblotting. Oligomers and dimers of HLA-B27 that accumulate with IFNγ treatment were reduced at least 50%, whereas monomers were not significantly changed (Fig. 4). Rapamycin also reduced about 40% of BiP-bound HLA-B27 heavy chains (Fig. 4). Rapamycin is also effective at reducing dimers and oligomers of HLA-B27 in cells treated with IFNγ for 20–24 hours (unpublished observations).
Figure 4.
Activation of autophagy by rapamycin reduces misfolded HLA-B27. (A) BM macrophages were incubated without or with IFNγ (50 ng/ml) for 6 hours, washed, and treated without or with rapamycin (25 μM) (Ra) for 2 hours. HC10 was used to immunoprecipitate unfolded/misfolded heavy chains, followed by immunoblotting with 3B10.7 (α-HLA). Co-precipitating BiP was measured using an aliquot of the immunoprecipitate run on a separate reducing gel. Immunoblots depict representative results. Quantitative results are mean ± SEM from six independent experiments, showing monomers, dimers, oligomers, and co-precipitating BiP. Inset shows longer exposure of oligomers. Dimers, oligomers, and BiP were quantitated only in IFNγ-treated samples. (*, p < 0.05)
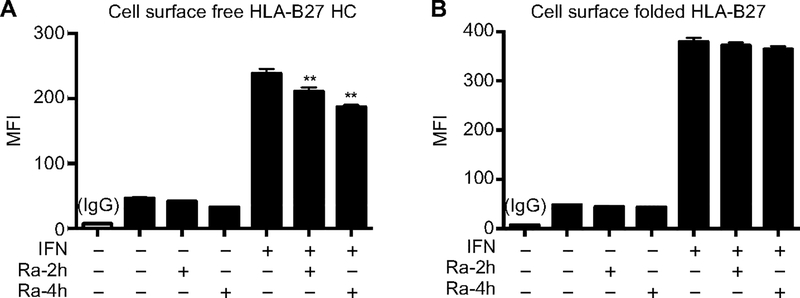
Cell surface β2m-free HLA-B27 heavy chains have been implicated in triggering killer immunoglobulin receptors (KIRs) on CD4+ Th17 T cells (2). To assess the effect of autophagy on cell surface HLA-B27, macrophages were incubated with IFNγ for 24 hours, with rapamycin added for the final 2–4 hours to activate autophagy. Cells were then washed and stained with either HC10 to detect free HLA-B27 heavy chains, or ME1 to measure folded HLA-B27 complexes, and analyzed by flow cytometry. Rapamycin treatment led to a rapid, time-dependent reduction in free HLA-B27 heavy chains on the cell surface of IFNγ-treated cells (Fig. 5A). In contrast, there was no significant effect on folded HLA-B27 complexes (Fig. 5B). These data indicate that activating autophagy can promote selective elimination of dimeric and oligomeric HLA-B27 heavy chains that accumulate with IFNγ treatment, and indicate that activating this pathway might be a beneficial strategy to ameliorate effects of HLA-B27 misfolding.
Figure 5.
Activation of autophagy by rapamycin reduces free HLA-B27 heavy chains on the cell surface. BM macrophages were incubated without or with IFNγ (50 ng/ml) for 24 hours, and treated without or with rapamycin (25 μM) for the last 2 (Ra-2h) or 4 hours (Ra-4h). (A) Cells were then stained with APC-conjugated HC10 to detect free heavy chains by flow cytometry. (B) For the detection of folded HLA-B27, cells were stained with Pacific Blue-conjugated ME1. Quantitative results are mean ± SEM from four independent experiments (**, p < 0.005).
DISCUSSION
Here, we investigated whether autophagy plays a role in the degradation of misfolded HLA-B27 heavy chains. Our results show that blocking autophagy for 3 hours led to accumulation of disulfide-linked HLA-B27 dimers and oligomers, as well as monomers, in primary cells from HLA-B27-Tg rats after HLA-B27 upregulation, and in transfected cells (C1R.B27) that continually express HLA-B27 heavy chains. Furthermore, a substantial proportion of accumulating HLA-B27 heavy chains are bound to the ER chaperone BiP, which has been shown to accumulate primarily with dimers of HLA-B27 (16). Importantly, we showed that activating autophagy with rapamycin led to a substantial and rapid reduction (50% in 2 hours) in the amount of misfolded HLA-B27 dimers and oligomers, including BiP-bound HLA-B27 heavy chains. Activation imposed by the exogenous stimulus (rapamycin) was more selective for the degradation of misfolded HLA-B27 (dimers and oligomers) than for monomers (Fig. 4). However, it should be noted that in some experiments, we have seen a reduction in HC10-reactive monomers along with more striking decreases (>50%) in dimers and oligomers, suggesting that autophagy is not entirely selective for misfolded forms (unpublished observations). Experimental variation in the strength of autophagy activation may account for these differences. Taken together, these results implicate autophagy as an ER quality control (ERQC) pathway involved in the elimination of misfolded and BiP-bound forms of HLA-B27. Rapamycin also reduced free HLA-B27 heavy chains on the cell surface (Fig. 5), which is not expected to be a consequence of ERQC. However, free class I heavy chains recycle through endosomes (2), and recycling endosomes contribute to autophagosome formation (43), suggesting a plausible mechanism for enhanced removal of free HLA-B27 heavy chains from the cell surface.
ERAD of HLA class I was first demonstrated for heavy chains synthesized when β2m or peptide is limited (44). HLA-B27 heavy chains were shown to undergo ERAD even in the presence of β2m and peptide, providing evidence for misfolding under normal physiologic conditions (4). HLA-B27 misfolding is a consequence of delayed folding imparted by polymorphic amino acids that also reduce its affinity for peptides and promote dimerization in the ER (4–6). Since these early studies, much more has been learned about ERAD of HLA class I heavy chains in general, and HLA-B27 in particular. EDEM1, together with the E3 ubiquitin ligase HRD1, and the ubiquitin conjugating enzyme UBE2J1, orchestrate the recognition, dislocation and ubiquitination of misfolded heavy chains including HLA-B27 dimers (21, 22). Furthermore, manipulation of the ERAD pathway, either by overexpressing EDEM1, or by pre-conditioning the ER with agents that activate the unfolded protein response, enhances the disposal of HLA-B27 dimers (22). Our studies confirm and extend previous results from transfected cells, by showing that ERAD plays a role in quality control of HLA-B27 after upregulation in primary rat macrophages. In rat macrophages blocking ERAD was slightly more effective at forcing the accumulation of HLA-B27 heavy chains than blocking autophagy (Fig. 1A). However, the opposite was seen in C1R cells expressing HLA-B27 (Fig. 1B). Barring differences in the sensitivity of primary macrophages and C1R (B lymphoblastoid) cells to these inhibitors, this suggests that there is a difference in the trafficking of misfolded HLA-B27 through these two pathways. This could be due to a number of factors, including cell type-specific differences in ERQC components, or perhaps differences in baseline expression of HLA-B27. The C1R.B27 cells continually express a protein that misfolds, which can be detrimental to survival. In contrast, the rat macrophages are derived from bone marrow progenitors in vitro in the absence of exogenous stimuli that upregulate HLA class I, and thus HLA-B27 expression is relatively low until the cells exposed to IFNγ. It will be of interest to compare the relative contributions of ERAD and autophagy to ERQC of HLA-B27 in other cells derived from HLA-B27 Tg rats.
HLA-B27 dimers form immediately after heavy chains are synthesized in the ER (5), exhibit a half-life exceeding 8 hours (22), and can be found in many cells at steady state (5, 18, 22) suggesting they are not efficiently eliminated and may be resistant to ERAD. This is in sharp contrast to many proteins that misfold in the ER and are efficiently degraded by ERAD, including MHC class I heavy chains produced in the absence of β2m or peptide (21, 44). This raises the question of why HLA-B27 heavy chain dimers and oligomers are not efficiently degraded. By comparing how rat cells handle HLA-B7 heavy chains, our results demonstrate that dimers of HLA-B27 are poorly ubiquitinated relative to HLA-B7 (monomers) (Fig. 3). This is even apparent in IFNγ-treated cells in the absence of autophagy or ERAD inhibitors (Fig. 3D). There is precedent for this as mutations in the T-cell receptor-α (TCRα) that increase the formation of disulfide-linked homo-oligomers also prevent it from being degraded by ERAD (45). Thus, we hypothesize rapid dimerization of HLA-B27 heavy chains is an impediment to efficient ERAD, and contributes to accumulation of misfolded forms.
Autophagy has been implicated in ER quality control of several proteins, including mutant α1-antitrypsin (AT) Z (ATZ). AT is normally produced and secreted in large amounts from the ER of hepatocytes, and functions as a serine protease inhibitor that limits proteolytic damage to tissues particularly during inflammation (30). A single point mutation in ATZ causes it to misfold and polymerize, forming insoluble aggregates in the extended ER. These insoluble aggregates are resistant to dislocation from the ER and thus escape ERAD, but are degraded by autophagy. A second ERQC autophagy pathway has been described for non-aggregated, ERAD-resistant membrane proteins (31), with gonadotropin-releasing hormone receptor type I (GnRHR) mutants as model substrates. Two different GnRHR mutants are subjected to ERQC, but degradation of one is blocked by proteasome inhibitors while the other is blocked by inhibitors of autophagy. Misfolded proteins that cannot be cleared by the proteosomal pathway can activate the autophagy pathway for degradation (46).
In summary, our studies implicate autophagy as well as ERAD in ERQC of misfolded HLA-B27. Nevertheless, neither the autophagy nor the proteasome pathway was sufficient under these conditions to prevent misfolded HLA-B27 from accumulating. In contrast to the proteasome pathway, the autophagy pathway can be further stimulated by starvation or the use of chemicals that inhibit the mTOR pathway (28). Several studies used successfully the mTOR inhibitor rapamycin in neurodegenerative diseases like Huntington or Alzheimer disease to partially resolve the misfolding proteins like tau by further activating autophagy (29, 47, 48), and it is widely used as an immunosuppressant to prevent transplant rejection. Hence, we treated cells with rapamycin for 2 h during upregulation of HLA-B27 or HLA-B7. Interestingly, mainly dimers and oligomers of HLA-B27 were reduced by activating autophagy, including heavy chains bound to BiP. Moreover, rapamycin was able to eliminate a significant proportion of cell surface free HLA-B27 heavy chains, which represent aberrant forms of HLA-B27 implicated in triggering CD4+ Th17 T cells to produce IL-17 (2). Therefore, these results raise the possibility that rapamycin could be tested therapeutically, although this remains a challenge as autophagy is involved in multiple homeostatics pathways inside and outside of the immune system.
Supplementary Material
ACKNOWLEDGEMENTS
Special thanks to Kristina Zaal and Evelyn Ralston (Light Imaging Section, NIAMS) for their help with the confocal immunofluorescence microscopy, and Nina Raben (NIAMS) for helpful discussions about autophagy.
Support: This work was supported by the NIAMS Intramural Research Program, Z01 AR041184.
Footnotes
Disclosure: The authors have no financial relationships to disclose.
REFERENCES
- 1.Taurog JD, Chhabra A, Colbert RA. Ankylosing Spondylitis and Axial Spondyloarthritis. The New England journal of medicine. 2016;374(26):2563–74. [DOI] [PubMed] [Google Scholar]
- 2.Bowness P HLA-B27. Annual review of immunology. 2015;33:29–48. [DOI] [PubMed] [Google Scholar]
- 3.Loll B, Fabian H, Huser H, Hee CS, Ziegler A, Uchanska-Ziegler B, et al. Increased Conformational Flexibility of HLA-B*27 Subtypes Associated With Ankylosing Spondylitis. Arthritis & rheumatology (Hoboken, NJ). 2016;68(5):1172–82. [DOI] [PubMed] [Google Scholar]
- 4.Mear JP, Schreiber KL, Munz C, Zhu X, Stevanovic S, Rammensee HG, et al. Misfolding of HLA-B27 as a result of its B pocket suggests a novel mechanism for its role in susceptibility to spondyloarthropathies. Journal of immunology (Baltimore, Md : 1950). 1999;163(12):6665–70. [PubMed] [Google Scholar]
- 5.Dangoria NS, DeLay ML, Kingsbury DJ, Mear JP, Uchanska-Ziegler B, Ziegler A, et al. HLA-B27 misfolding is associated with aberrant intermolecular disulfide bond formation (dimerization) in the endoplasmic reticulum. The Journal of biological chemistry. 2002;277(26):23459–68. [DOI] [PubMed] [Google Scholar]
- 6.Antoniou AN, Ford S, Taurog JD, Butcher GW, Powis SJ. Formation of HLA-B27 homodimers and their relationship to assembly kinetics. The Journal of biological chemistry. 2004;279(10):8895–902. [DOI] [PubMed] [Google Scholar]
- 7.Dhaenens M, Fert I, Glatigny S, Haerinck S, Poulain C, Donnadieu E, et al. Dendritic cells from spondylarthritis-prone HLA-B27-transgenic rats display altered cytoskeletal dynamics, class II major histocompatibility complex expression, and viability. Arthritis and rheumatism. 2009;60(9):2622–32. [DOI] [PubMed] [Google Scholar]
- 8.Fert I, Cagnard N, Glatigny S, Letourneur F, Jacques S, Smith JA, et al. Reverse interferon signature is characteristic of antigen-presenting cells in human and rat spondyloarthritis. Arthritis & rheumatology (Hoboken, NJ). 2014;66(4):841–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cortes A, Hadler J, Pointon JP, Robinson PC, Karaderi T, Leo P, et al. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nature genetics. 2013;45(7):730–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith JA, Colbert RA. Review: The interleukin-23/interleukin-17 axis in spondyloarthritis pathogenesis: Th17 and beyond. Arthritis & rheumatology (Hoboken, NJ). 2014;66(2):231–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taurog JD, Maika SD, Satumtira N, Dorris ML, McLean IL, Yanagisawa H, et al. Inflammatory disease in HLA-B27 transgenic rats. Immunological reviews. 1999;169:209–23. [DOI] [PubMed] [Google Scholar]
- 12.DeLay ML, Turner MJ, Klenk EI, Smith JA, Sowders DP, Colbert RA. HLA-B27 misfolding and the unfolded protein response augment interleukin-23 production and are associated with Th17 activation in transgenic rats. Arthritis and rheumatism. 2009;60(9):2633–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glatigny S, Fert I, Blaton MA, Lories RJ, Araujo LM, Chiocchia G, et al. Proinflammatory Th17 cells are expanded and induced by dendritic cells in spondylarthritis-prone HLA-B27-transgenic rats. Arthritis and rheumatism. 2012;64(1):110–20. [DOI] [PubMed] [Google Scholar]
- 14.Baeten D, Sieper J, Braun J, Baraliakos X, Dougados M, Emery P, et al. Secukinumab, an Interleukin-17A Inhibitor, in Ankylosing Spondylitis. The New England journal of medicine. 2015;373(26):2534–48. [DOI] [PubMed] [Google Scholar]
- 15.Braun J, Kiltz U, Heldmann F, Baraliakos X. Emerging drugs for the treatment of axial and peripheral spondyloarthritis. Expert opinion on emerging drugs. 2015;20(1):1–14. [DOI] [PubMed] [Google Scholar]
- 16.Tran TM, Satumtira N, Dorris ML, May E, Wang A, Furuta E, et al. HLA-B27 in transgenic rats forms disulfide-linked heavy chain oligomers and multimers that bind to the chaperone BiP. Journal of immunology (Baltimore, Md : 1950). 2004;172(8):5110–9. [DOI] [PubMed] [Google Scholar]
- 17.Turner MJ, Sowders DP, DeLay ML, Mohapatra R, Bai S, Smith JA, et al. HLA-B27 misfolding in transgenic rats is associated with activation of the unfolded protein response. Journal of immunology (Baltimore, Md : 1950). 2005;175(4):2438–48. [DOI] [PubMed] [Google Scholar]
- 18.Turner MJ, Delay ML, Bai S, Klenk E, Colbert RA. HLA-B27 up-regulation causes accumulation of misfolded heavy chains and correlates with the magnitude of the unfolded protein response in transgenic rats: Implications for the pathogenesis of spondylarthritis-like disease. Arthritis and rheumatism. 2007;56(1):215–23. [DOI] [PubMed] [Google Scholar]
- 19.Navid F, Colbert RA. Causes and consequences of endoplasmic reticulum stress in rheumatic disease. Nature reviews Rheumatology. 2017;13(1):25–40. [DOI] [PubMed] [Google Scholar]
- 20.Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nature reviews Molecular cell biology. 2008;9(12):944–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burr ML, Cano F, Svobodova S, Boyle LH, Boname JM, Lehner PJ. HRD1 and UBE2J1 target misfolded MHC class I heavy chains for endoplasmic reticulum-associated degradation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(5):2034–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guiliano DB, Fussell H, Lenart I, Tsao E, Nesbeth D, Fletcher AJ, et al. Endoplasmic reticulum degradation-enhancing alpha-mannosidase-like protein 1 targets isfolded HLA-B27 dimers for endoplasmic reticulum-associated degradation. Arthritis & rheumatology (Hoboken, NJ). 2014;66(11):2976–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christianson JC, Ye Y. Cleaning up in the endoplasmic reticulum: ubiquitin in charge. Nature structural & molecular biology. 2014;21(4):325–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469(7330):323–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432(7020):1032–6. [DOI] [PubMed] [Google Scholar]
- 26.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nature cell biology. 2010;12(9):814–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nature cell biology. 2010;12(9):823–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nature reviews Drug discovery. 2012;11(9):709–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. Alpha-Synuclein is degraded by both autophagy and the proteasome. The Journal of biological chemistry. 2003;278(27):25009–13. [DOI] [PubMed] [Google Scholar]
- 30.Perlmutter DH. Alpha-1-antitrypsin deficiency: importance of proteasomal and autophagic degradative pathways in disposal of liver disease-associated protein aggregates. Annual review of medicine. 2011;62:333–45. [DOI] [PubMed] [Google Scholar]
- 31.Houck SA, Ren HY, Madden VJ, Bonner JN, Conlin MP, Janovick JA, et al. Quality control autophagy degrades soluble ERAD-resistant conformers of the misfolded membrane protein GnRHR. Molecular cell. 2014;54(1):166–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Layh-Schmitt G, Yang EY, Kwon G, Colbert RA. HLA-B27 alters the response to tumor necrosis factor alpha and promotes osteoclastogenesis in bone marrow monocytes from HLA-B27-transgenic rats. Arthritis and rheumatism. 2013;65(8):2123–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stam NJ, Spits H, Ploegh HL. Monoclonal antibodies raised against denatured HLA-B locus heavy chains permit biochemical characterization of certain HLA-C locus products. Journal of immunology (Baltimore, Md : 1950). 1986;137(7):2299–306. [PubMed] [Google Scholar]
- 34.Colbert RA, DeLay ML, Klenk EI, Layh-Schmitt G. From HLA-B27 to spondyloarthritis: a journey through the ER. Immunological reviews. 2010;233(1):181–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell structure and function. 1998;23(1):33–42. [DOI] [PubMed] [Google Scholar]
- 36.Groll M, Berkers CR, Ploegh HL, Ovaa H. Crystal structure of the boronic acid-based proteasome inhibitor bortezomib in complex with the yeast 20S proteasome. Structure (London, England : 1993). 2006;14(3):451–6. [DOI] [PubMed] [Google Scholar]
- 37.Bonvini P, Zorzi E, Basso G, Rosolen A. Bortezomib-mediated 26S proteasome inhibition causes cell-cycle arrest and induces apoptosis in CD-30+ anaplastic large cell lymphoma. Leukemia. 2007;21(4):838–42. [DOI] [PubMed] [Google Scholar]
- 38.Stam NJ, Vroom TM, Peters PJ, Pastoors EB, Ploegh HL. HLA-A- and HLA-B-specific monoclonal antibodies reactive with free heavy chains in western blots, in formalin-fixed, paraffin-embedded tissue sections and in cryo-immuno-electron microscopy. International immunology. 1990;2(2):113–25. [DOI] [PubMed] [Google Scholar]
- 39.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12(1):1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harris J, Hanrahan O, De Haro SA. Measuring autophagy in macrophages Curr Protocol Immunol. 2009;Chapter14:Unit14.14. [DOI] [PubMed] [Google Scholar]
- 41.Bridges JP, Xu Y, Na CL, Wong HR, Weaver TE. Adaptation and increased susceptibility to infection associated with constitutive expression of misfolded SP-C. The Journal of cell biology. 2006;172(3):395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki Y, Schmitt MJ. Redox diversity in ERAD-mediated protein retrotranslocation from the endoplasmic reticulum: a complex puzzle. Biological chemistry. 2015;396(5):539–54. [DOI] [PubMed] [Google Scholar]
- 43.Longatti A, Tooze SA. Recycling endosomes contribute to autophagosome formation. Autophagy 2012;8:11, 1682–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hughes EA, Hammond C, Cresswell P. Misfolded major histocompatibility complex class I heavy chains are translocated into the cytoplasm and degraded by the proteasome. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(5):1896–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soetandyo N, Wang Q, Ye Y, Li L. Role of intramembrane charged residues in the quality control of unassembled T-cell receptor alpha-chains at the endoplasmic reticulum. Journal of cell science. 2010;123(Pt 7):1031–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cha-Molstad H, Sung KS, Hwang J, Kim KA, Yu JE, Yoo YD, et al. Amino-terminal arginylation targets endoplasmic reticulum chaperone BiP for autophagy through p62 binding. Nature cell biology. 2015;17(7):917–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berger Z, Ravikumar B, Menzies FM, Oroz LG, Underwood BR, Pangalos MN, et al. Rapamycin alleviates toxicity of different aggregate-prone proteins. Human molecular genetics. 2006;15(3):433–42. [DOI] [PubMed] [Google Scholar]
- 48.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nature genetics. 2004;36(6):585–95. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.