Abstract
It is generally accepted that cytokinin oxidases, which oxidatively remove cytokinin side chains to produce adenine and the corresponding isopentenyl aldehyde, play a major role in regulating cytokinin levels in planta. Partially purified fractions of cytokinin oxidase from various species have been studied for many years, but have yet to clearly reveal the properties of the enzyme or to define its biological significance. Details of the genomic organization of the recently isolated maize (Zea mays) cytokinin oxidase gene (ckx1) and some of its Arabidopsis homologs are now presented. Expression of an intronless ckx1 in Pichia pastoris allowed production of large amounts of recombinant cytokinin oxidase and facilitated detailed kinetic and cofactor analysis and comparison with the native enzyme. The enzyme is a flavoprotein containing covalently bound flavin adenine dinucleotide, but no detectable heavy metals. Expression of the oxidase in maize tissues is described.
Regulation of active cytokinin levels in planta may occur through control of synthesis, through reversible conjugation to inactive glucosides, or through irreversible inactivation (Jameson, 1994). Although the plant cytokinin biosynthetic gene remains to be isolated, progress has been made in identifying catabolic genes responsible for conversion of active cytokinins into inactive glucosyl or xylosyl conjugates, ZOG1 and ZOX1 (Martin et al., 1999a, 1999b), or for oxidative side chain removal, cytokinin oxidase, ckx1 (Houba-Herin et al., 1999; Morris et al., 1999). Further genetic and biochemical characterization of these genes should provide insight into their roles in regulating the reversible and irreversible inactivation of cytokinins in plants.
In many plant tissues oxidative cytokinin degradation appears to be the major pathway for cytokinin inactivation (for review, see Armstrong, 1994). Maize (Zea mays) tissues have historically been shown to have potent cytokinin degrading activities, and cytokinin oxidase activity has been partially purified from maize kernels and seedlings (Whitty and Hall, 1974; McGaw and Horgan, 1983; Burch and Horgan, 1989; Horgan et al., 1990). Characterization of the substrate specificity and other biochemical properties of these oxidases has been limited by the relative impurity of the enzyme preparations and the lack of accurate and rapid assays. Nevertheless, previous reports found that the maize cytokinin oxidase had a substrate preference for isopentenyladenine, although the reported physical and kinetic properties of the enzyme varied between studies (for review, see Armstrong, 1994; Hare and Van Staden, 1994a).
With the cloning of the maize ckx1 gene, rigorous molecular and biochemical studies are now possible and may lead to experiments that will identify the role played by cytokinin catabolism in plant development. In this report data are presented on the genomic organization of maize ckx1 and seven homologs from Arabidopsis, on the substrate specificity of the native and recombinant maize enzymes, on the nature of the inhibition of the enzyme by phenylurea derivatives, on the nature of the flavin cofactor, and on the expression pattern of the oxidase in different maize tissues. A preliminary report has already appeared (Morris et al., 1999).
RESULTS
Cytokinin Oxidase Gene Organization in Maize and Arabidopsis
In a previous paper we described the isolation of a glycosylated cytokinin oxidase from maize and its use to identify the corresponding structural gene, ckx1 (Morris et al., 1999). The genomic organization of ckx1 is shown schematically in Figure 1. It is predicted to consist of three exons separated by two small introns, 412 and 94 bp, respectively. The location and sizes of these introns were confirmed by sequencing reverse transcriptase (RT)-PCR products of maize kernel cytokinin oxidase mRNA (data not shown) and by intron removal and expression of the resulting cDNA in Pichia pastoris to give active, recombinant cytokinin oxidase.
Figure 1.
Genomic organization of the maize and Arabidopsis cytokinin oxidase genes. Restriction map and gene structure of maize ckx1 and gene structures of seven Arabidopsis cytokinin oxidase homologs. Exons (numbered rectangles) and introns (solid and dashed lines, not to scale) are indicated. Restriction enzymes: N, NotI; H, HindIII; and B, BamHI. Amino acid identity is for all exons of each gene compared with maize ckx1 exons. Oxidase activity is that exhibited by P. pastoris supernatants expressing the appropriate cDNA. ND, Gene activity not determined. *, All introns were predicted; **, first intron was predicted.
The 1,602-bp coding region of ckx1 predicts a polypeptide with a molecular mass of 57,141 D (57 kD). As reported previously, features of the oxidase include a predicted signal peptide, eight possible N-glycosylation sites, a domain homologous to flavoproteins, and spectroscopic properties expected for a flavin enzyme (Morris et al., 1999). Predicted glycosylation of the enzyme is consistent with its affinity for the lectin concanavalin A (Chatfield and Armstrong, 1988).
There are several uncharacterized sequences in GenBank that show identity to maize CKX1 at the amino acid level. To date there are seven such homologs in Arabidopsis (Fig. 1): two on chromosome 2 (designated AtCKX1 and AtCKX2), two on chromosome 5 (designated AtCKX3 and AtCKX5), one on chromosome 4 (designated AtCKX4), one on chromosome 1 (designated AtCKX6), and one on chromosome 3 (designated AtCKX7). The amino acid identity of these putative proteins with maize CKX1 ranges from 39% to 47%, and intron locations are significantly conserved. Upon RT-PCR cloning of a subset of the Arabidopsis homologs and expression of the resulting cDNAs in P. pastoris (Morris et al., 1999), proteins encoded by AtCKX2, AtCKX3, and AtCKX4 were found to be secreted and to have cytokinin oxidase activity (Fig. 1). The AtCKX1 homolog was not secreted from P. pastoris, nor was it active, possibly due to differences in signal peptide recognition. It remains to be determined whether this gene actually encodes a functional cytokinin oxidase. The other Arabidopsis homologs have not yet been characterized. We have also cloned and expressed an active cytokinin oxidase from oilseed rape (K. Bilyeu, J. Laskey, and R.O. Morris, unpublished data). Full details of the properties of these Arabidopsis and oilseed rape cytokinin oxidases will be described elsewhere. In addition, more than 20 other expressed sequence tag and genome survey sequences have been identified in GenBank that have identity to regions of CKX1.
Physical and Enzymatic Properties of Maize Cytokinin Oxidase
Because the yield of native enzyme from maize was low and the protein was difficult to purify, determination of complete kinetic parameters for the native enzyme was not feasible. However, expression of the ckx1 structural gene in the yeast P. pastoris (Despreaux and Manning, 1993; Paifer et al., 1994) resulted in the secretion of active glycosylated recombinant enzyme (Morris et al., 1999) in amounts sufficient for physical and enzymatic studies. The endogenous ckx1 signal peptide directed secretion of substantial cytokinin oxidase activity into P. pastoris supernatants compared with the modest amounts of active enzyme that can be isolated from maize tissues (Morris et al., 1999); Replacement of the putative native signal peptide, which was predicted by a method based on trained neural networks (Nielsen et al., 1997), with the Saccharomyces cerevisiae α-factor propeptide (Cregg et al., 1993; Scorer et al., 1993) resulted in a dramatic increase in cytokinin oxidase secretion (Fig. 2A). We can presume that the yeast signal peptide directs more efficient extracellular targeting and secretion in P. pastoris than does the native maize signal peptide. A single-step purification of the recombinant cytokinin oxidase from P. pastoris culture supernatants by size exclusion chromatography resulted in a cytokinin oxidase preparation that was better than 95% homogeneous by SDS-PAGE.
Figure 2.
Expression of recombinant maize cytokinin oxidase. A, Time course of expression of maize ckx1 in P. pastoris. Black bars, Cytokinin oxidase activity in P. pastoris supernatants expressing maize ckx1 (introns removed) with its native signal peptide (MAVVYYLLLA GLIACSHA-LAA); cross-hatched bars, cytokinin oxidase activity in supernatants of P. pastoris in which the native signal peptide of CKX1 was replaced by the yeast α-factor propeptide signal peptide (MRFPSIFTAV LFAASSALAA PVNTTTEDET AQIPAEAVIG YSDLEGDFAV AVLPFSNSTN NGLLFINTTI ASIAAKEEGV SLEKR-LAA). B, Western analysis of recombinant CKX1 produced with the α-factor propeptide and native CKX1 before and after chemical deglycosylation. Proteins were separated by SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membranes, and probed with anti-CKX1 monoclonal antibodies (19F4). Lane 1, Recombinant glycosylated CKX1 produced in P. pastoris (5 ng); lane 2, partially purified glycosylated native maize cytokinin oxidase (3 μg); lane 3, mixture of native and recombinant enzymes; lane 4, chemically deglycosylated recombinant oxidase (3 ng); lane 5, chemically deglycosylated partially purified native maize cytokinin oxidase (14 μg); lane 6, mixture of deglycosylated native and recombinant enzymes. The position and size of protein molecular mass markers is indicated to the left of the blot.
For purposes of comparison the native maize cytokinin oxidase was also partially purified from cornmeal using a protocol abridged from one previously described for immature maize kernels (Morris et al., 1999). Following a final chromatographic purification on concanavalin A, active fractions were pooled, dialyzed, and used for kinetic analysis. The total amount of active enzyme obtained was adequate for Km determination, but was not sufficiently homogeneous for accurate kcat determination.
Western analysis (Fig. 2B) indicated that monoclonal antibodies to recombinant CKX1 expressed in P. pastoris reacted equally well with the native maize cytokinin oxidase and with the recombinant cytokinin oxidase produced with the α-factor propeptide, but that there was an apparent size difference. However, western analysis of the chemically deglycosylated native and recombinant enzymes (Fig. 2B) indicated that they were the same size. The larger apparent molecular mass of the recombinant cytokinin oxidase is therefore solely due to differential glycosylation between the P. pastoris and maize enzymes (Grinna and Tschopp, 1989). Because recombinant expression in P. pastoris produced a highly active enzyme, it is likely that the properties of the recombinant enzyme accurately reflect those of the native oxidase, although differences in enzyme stability or substrate access due to steric interference from the yeast glycosylation are possible in theory.
Substrate Specificity and Inhibitors for Cytokinin Oxidase
The kinetic parameters of a number of partially purified cytokinin oxidases have been reported from a variety of sources (for review, see Armstrong, 1994; Hare and Van Staden, 1994a). Most of these earlier studies used a fixed-time radiometric assay, which is not particularly amenable to generation of precise data. The availability of a continuous spectrophotometric assay based on the transfer of reducing equivalents to the dye 2,6-dichlorophenolindophenol (DCPIP; Morris et al., 1999) allowed rigorous kinetic and inhibitory studies.
Table I lists the Km values for several cytokinins for the recombinant and native cytokinin oxidase enzymes using this new assay. The kcat values are for the recombinant enzyme only and are based on accurate rate data and the known molar extinction coefficient of the enzyme. The Km values for the recombinant and native cytokinin oxidases were quite similar, with zeatin, isopentenyladenine, and isopentenyladenosine serving as good substrates for both enzymes. Cleavage of zeatin riboside and cis-zeatin was detected with large amounts of enzyme, although the Km values were higher and kcat values were lower than for the other substrates. Dihydrozeatin, kinetin, and benzylaminopurine were not substrates. It is surprising that zeatin-9-glucoside was not a substrate, in contrast to an earlier report (McGaw and Horgan, 1983). Based on kcat/Km values, isopentenyladenine is the preferred substrate with a kcat/Km value of 45 μM−1 s−1, with zeatin a close second at 9 μM−1 s−1. It should be noted that DCPIP is a far better electron acceptor than is oxygen (J. Laskey, K. Bilyeu, P. Patterson, and R.O. Morris, unpublished data); observed rates of reaction are far greater than reported previously (McGaw and Horgan, 1983; Laloue and Fox, 1989; Houba-Herin et al., 1999).
Table I.
Substrate specificity of native and recombinant cytokinin oxidases
| Substrate | Native (Km) | Recombinant
|
||
|---|---|---|---|---|
| Km | kcat | kcat/Km | ||
| μm | s−1 | μm−1 s−1 | ||
| Isopentenyladenine | 2.8 | 1.5 | 67 | 44.6 |
| Trans-zeatin | 11 | 14 | 126 | 9.0 |
| Isopentenyladenosine | 15 | 11 | 48 | 4.4 |
| Cis-zeatin | n.d.a | 46 | 17 | 0.37 |
| Trans-zeatin riboside | n.d. | 54 | 18 | 0.34 |
Substrate oxidation was assayed by following A600 after addition of enzyme to assay mixtures containing buffer, DCPIP, and substrates at various concentrations. Km and rate values were calculated based on Lineweaver-Burk plots of the data. Cis-zeatin riboside, zeatin-9-glucoside, dihydrozeatin, benzylaminopurine, and kinetin were not substrates (kcat < 8 s−1).
n.d., Activity not determined.
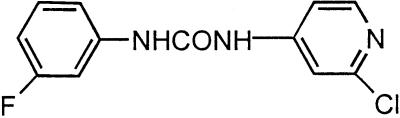
It has long been known that many phenylureas have cytokinin-like effects in cytokinin bioassays and tissue culture systems (for review, see Shudo, 1994). The question remains as to whether these ureas have innate cytokinin activity, or whether they exert their effects by inhibition of endogenous cytokinin oxidases. Earlier data (Burch and Horgan, 1989) indicated maize cytokinin oxidase was inhibited by N-(2-chloro-4-pyridyl)-N′-phenylurea (CPPU) in a non-competitive manner, and N-phenyl-N′-1,2,3-thidiazol-5-urea (thidiazuron) inhibited a soybean callus cytokinin oxidase in a predominantly uncompetitive manner (Hare and Van Staden, 1994b). To shed more light on the nature of the inhibition, we tested phenylureas with a range of biological activities. Figure 3 shows Lineweaver-Burk plots for zeatin as a substrate and CPPU as inhibitor. It is clear that inhibition is competitive. All other inhibitors tested were also competitive. Their structures and Ki values are listed in Table II. N-(2-chloro-4-pyridyl-N′-5-fluoro-phenylurea) (F-PU) was the strongest with a Ki of 0.1 μM, but N-(2-methoxy-4-pyridyl)-N′-phenylurea (methoxy-PU), thidiazuron, and 6-(N-isopent-2-enyl-N-methylamino) purine (Me-iP; Wang and Letham, 1995) were also effective. It is interesting that the good correlation observed (Shudo, 1994) between the high biological activity of a urea in vivo and low Ki for oxidase inhibition in vitro does appear to hold.
Figure 3.
Inhibition of cytokinin oxidase activity by CPPU. No CPPU (●), 1.5 μm CPPU (▴), and 3 μM CPPU (▪). Linear regressions through each data set intersect on the ordinate, indicating competitive inhibition.
Table II.
Structure and K1 values for cytokinin oxidase inhibitors
Inhibition of activity was determined as in Figure 3. All inhibition was competitive and allowed calculation of Ki values using three concentrations of each inhibitor.
Identification of the Cytokinin Oxidase Cofactor
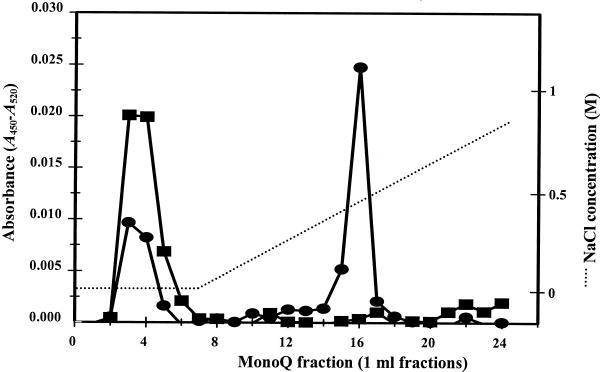
As indicated previously (Morris et al., 1999), concentrated solutions of the recombinant enzyme are yellow and have absorption spectra that are characteristic of flavoproteins. However, the absorption spectrum of the enzyme did not allow one to determine whether the cofactor was FAD or flavin mononucleotide (FMN). To determine the identity of the flavin and the nature of the flavin-protein interaction, the enzyme was precipitated with trichloroacetic acid or boiled in 70% (v/v) ethanol. Under both conditions, the flavin failed to separate from the protein, indicating that the association was covalent. To determine whether the flavin was FMN or FAD, denatured cytokinin oxidase was digested proteolytically and the flavin-containing peptide was purified and then treated with nucleotide pyrophosphatase. FAD and FMN peptides have previously been shown to have distinct chromatographic properties (Cook et al., 1984). Figure 4 illustrates the chromatographic properties of the flavin peptide on Mono Q before and after digestion with nucleotide pyrophosphatase. Prior to digestion the majority of the flavinated peptide eluted in fraction 16, although some of the peptide had reduced binding to the column, possibly due to cleavage of FAD during sample manipulation. After digestion the binding of the flavopeptide to the column was reduced significantly, consistent with the conversion of the dinucleotide FAD to the mononucleotide FMN. It is apparent that cytokinin oxidase contains covalently bound FAD.
Figure 4.
Identification of flavin cofactor. A flavinated peptide was isolated from a tryptic digest of homogeneous recombinant CKX1. It was fractionated on MonoQ before (●) or after (▪) nucleotide pyrophosphatase treatment. The difference between A450 and A520 is plotted for each sample.
Cytokinin oxidases have also been thought to be copper-containing proteins (Burch and Horgan, 1989; Hare and Van Staden, 1994). However, we found no evidence to support this. No heavy metals (Cu, Zn, Fe, and Mn) were detected by atomic absorption analysis and the copper chelator diethyldithiocarbamic acid had no inhibitory effect on the enzyme's activity (data not shown).
Cytokinin Oxidase Distribution in Maize Tissues
To determine the distribution of the cytokinin oxidase in maize, western analysis was performed on a number of maize tissues and compared with oxidase activity in the same tissues. Preliminary experiments indicated that rabbit polyclonal antibodies raised against the recombinant oxidase were able to immunoprecipitate native cytokinin oxidase activity from cornmeal and could detect cytokinin oxidase as a 70-kD band in samples of glycosylated maize proteins in westerns. Therefore, maize glycosylated proteins from different tissues were extracted, separated by SDS-PAGE, and subjected to western analysis. As shown in Figure 5A, most immunological cross-reactivity was seen in immature maize kernels (17 d after pollination) and cornmeal; unpollinated cobs and roots had some cross-reactivity in westerns after overexposure and other tissues had none.
Figure 5.
Distribution of glycosylated cytokinin oxidase in maize. A, Western analysis of the distribution of CKX1 protein in different maize tissues. Total glycosylated proteins were isolated from each tissue, normalized for fresh weight (each lane represents 25 mg of starting material), separated by SDS-PAGE, transferred to PVDF membranes, and probed with anti-CKX1 polyclonal rabbit antibodies. The position and size of protein molecular mass markers is indicated to the right of the blot. B, Glycosylated cytokinin oxidase activity in different maize tissues. Glycosylated protein samples were normalized for fresh weight. Oxidase activity was assayed by following conversion of [3H]zeatin into [3H]adenine by thin-layer chromatography.
Figure 5B shows the corresponding cytokinin oxidase activity present in glycosylated protein preparations from the same set of tissues. To account for all possible cytokinin oxidase activities present in the tissues, assays were carried out by measuring the conversion of [3H]zeatin to [3H]adenine. The highest cytokinin oxidase activity was detected in immature maize kernels (17 d after pollination) and in cornmeal. Significant activity was also present in unpollinated cobs. Other tissues tested had very little cytokinin oxidase activity. It is interesting that the activity in unpollinated cobs was higher than would be expected based on the intensity of the western band. The possibility of the presence of immunologically unrelated cytokinin oxidase activities in this latter tissue must be considered.
DISCUSSION
Cytokinins and cytokinin oxidase activity have been implicated in a number of plant developmental events, including the control of apical dominance, leaf senescence, grain fill potential, and seed germination (for review, see Mok, 1994). However, progress has been limited by the absence of a rapid and convenient cytokinin oxidase assay and by a lack of information on the cognate structural genes. The recent cloning of the maize cytokinin oxidase gene ckx1 (Houba-Herin et al., 1999; Morris et al., 1999) has reversed this situation and opened the way for studies of biological function.
The significant homology of ckx1 to seven sequences from Arabidopsis that show conserved gene organization suggested that these genes might also encode active cytokinin oxidases. This proved to be the case for at least three of the four homologs tested. Although Arabidopsis deduced peptide sequences are only 39% to 47% identical to those of maize ckx1, three of the homologs (AtCKX2, AtCKX3, and AtCKX4) have cytokinin oxidase activity when appropriately expressed in P. pastoris. Conserved regions that might be necessary for cytokinin oxidase function are beginning to be revealed. The number of species that have sequenced ckx1 homologs is growing, and it will soon be possible to determine which amino acid residues are functionally conserved in the active genes. The lack of activity in the case of AtCKX1 is puzzling, but may perhaps be attributed to the fact that the gene has a very different predicted signal peptide or, more likely, because it has a longer C-terminal sequence than the active homologs. It is known (Fraaije et al., 1998) that the C terminus of other FAD oxidases participates in correct folding of the FAD-binding domain. Misfolding of the C terminus may inactivate AtCKX1.
The discovery of the ability of semi-purified cytokinin oxidase to efficiently transfer reducing equivalents to the synthetic dye DCPIP led to the development of a rapid and specific assay, which in turn allowed the purification of the maize kernel oxidase to homogeneity and identification of the structural gene ckx1. Full details of this assay will be published elsewhere (J. Laskey, K. Bilyeu, P. Patterson, and R.O. Morris, unpublished data). The ability of cytokinin oxidase to react more effectively with DCPIP than with oxygen in vitro raises the question as to which factors (oxygen, redox proteins, or other cofactors) ultimately accept the electrons in planta and whether or not these other factors contribute to the overall rate of degradation of cytokinins in vivo. An immediate benefit of the DCPIP assay was easy characterization of the substrate preference of maize cytokinin oxidase. The preferred substrates generally agree with previous reports (Whitty and Hall, 1974; McGaw and Horgan, 1983) using partially purified cytokinin oxidase fractions, but the Km and kcat values can now be considered accurate due to the availability of the homogeneous recombinant enzyme. It will be interesting to compare the substrate specificity of the maize oxidase with that of the dicot cytokinin oxidases.
The ability of phenylurea derivatives to have cytokinin-like effects in bioassays (for review, see Shudo, 1994), and the finding that these compounds competitively inhibit cytokinin oxidase activity in vitro has raised questions in the past about the nature of their activity in vivo. If their activity is dependent on raising endogenous cytokinin levels by blocking cytokinin oxidase activity, then an experiment to address the issue would be to bioassay phenylurea cytokinins using tissue in which ckx1 and other cytokinin oxidases had been inactivated genetically.
The majority of cytokinin oxidase activity was detected in maize reproductive tissue. A previous study established a temporal correlation between decreasing cytokinin levels and increasing cytokinin oxidase activity in developing maize kernels (Dietrich et al., 1995). One function of cytokinin oxidase might be to destroy endogenous cytokinins at the appropriate time during kernel development. Identifying the subcellular and intracellular location of expression of cytokinin oxidase will be of great interest. Phenotypic analysis of plants with repressed oxidase activity will also be very important in determining the function of cytokinin oxidase in the kernel and during other stages of plant development. One alternative role for maize oxidase activity may be the protection of the kernel from pathogen invasion, because maize fungal pathogens are known to be capable of producing cytokinins (Mills and Van Staden, 1978; Angra et al., 1990).
Is ckx1 the only maize oxidase? It is not likely because there is evidence for the presence of homologous maize genes in the GenBank and other enzyme activities have been detected in crude maize kernel extracts (Morris et al., 1999). In addition, the presence of multiple active cytokinin oxidase homologs in Arabidopsis suggests that there will be multiple cytokinin oxidase genes found in maize and in other plant species. Substrate specificity might differ for each enzyme or the proteins could be physically separated by subcellular distribution or tissue-specific expression and have distinct functions. The available promoter sequences for maize ckx1 and the Arabidopsis homologs will allow detailed studies of expression patterns.
MATERIALS AND METHODS
Isolation of Arabidopsis Cytokinin Oxidase Homologs
cDNAs encoding Arabidopsis cytokinin oxidases were obtained by RT-PCR of whole plant mRNA using gene-specific primers. Arabidopsis CKX4 is derived from base numbers 76,187 through 78,968 of GenBank accession number AL079344; Arabidopsis CKX3 is derived from the reverse complement of base numbers 32,716 through 29,379 of GenBank accession number AB024035; ArabidopsisCKX2 is derived from base numbers 39,050 through 42,040 of GenBank accession number AC005917; Arabidopsis CKX1 is derived from base numbers 15,517 through 17,752 of GenBank accession number AC002510; Arabidopsis CKX7 is derived from base numbers 46,630 through 48,565 of GenBank accession number AL163818 (introns were predicted); and Arabidopsis CKX6 is derived from the reverse complement of base numbers 56,922 through 54,018 of GenBank accession number AC023754 (introns were predicted). Arabidopsis CKX5 contains a Kazusa bacterial artificial chromosome end clone (CIC 7E11 1), and a full-length cDNA, as well as partial genomic sequences were cloned (the first intron has not yet been confirmed).
Oxidase Isolation
A recombinant version of the maize cytokinin oxidase lacking the introns, but maintaining the native signal peptide, was constructed from the genomic sequence as described previously (Morris et al., 1999). The recombinant gene was further modified by the removal of the predicted maize signal (MAVVYYLLAGLIACSHA-LA) and its replacement by the Saccharomyces cerevisiae α-factor propeptide (Cregg et al., 1993; Scorer et al., 1993) in frame with the remaining coding sequence. Transformation of the modified ckx1 into Pichia pastoris, selection, growth, and methanol induction were conducted as described previously (Morris et al., 1999). At harvest, P. pastoris cultures were centrifuged, the supernatants were further cleared by filtration (0.22 micron), and the cultures were concentrated with buffer exchange into Tris-EDTA (TE) buffer (10 mm Tris-Cl, pH 8.0, and 1 mm EDTA) in a stirred-cell filtration apparatus. Purification of recombinant, active cytokinin oxidase to better than 95% electrophoretic homogeneity was achieved by size exclusion chromatography on Superose 12 (Amersham Pharmacia Biotech, Piscataway, NJ).
Native oxidase was partially purified from 10 kg of commercial degerminated cornmeal (Quaker Oats, Chicago), which was blended with 16 L of a buffer containing 50 mm Tris-Cl, 5 mm EDTA, 0.4% (w/v) ascorbic acid, and 10 mm β-mercaptoethanol, pH 8.5. After settling and filtration through Miracloth (Calbiochem, La Jolla, CA), a 45% to 60% (w/v) ammonium sulfate fraction was collected. The pellet was dissolved in and dialyzed against TE, pH 8.5, and separated into four 200-mL aliquots before preparative scale diethylaminoethyl-cellulose fractionation (5 × 22 cm of DE52, Whatman, Maidstone, UK) in TE with a linear gradient (5 mL/min for 100 min) of KCl to 0.2 m. Active fractions were pooled and dialyzed against a buffer containing 20 mm Tris-Cl, pH 7.0, 0.5 m NaCl, 1 mm CaCl2, and 1 mm MnCl2. The sample was divided in half and each was applied to a concanavalin A-agarose (Sigma, St. Louis) column (15 mL) and eluted with a step gradient (29 mL) of α-d-methylmannoside (1 m). The resulting glycosylated fractions were pooled and dialyzed against TE.
An adaptation of this procedure was developed for rapid extraction of oxidase activity from small tissue samples. Tissues (2.5 g) were powdered in liquid nitrogen and homogenized in extraction buffer (20 mL) containing 100 mm Tris-Cl, pH 7.5, 0.2 m KCl, 0.4% (w/v) ascorbic acid, and 10 mm β-mercaptoethanol. Diethylaminoethyl-cellulose suspension was added (1 g wet weight in extraction buffer) and then pelleted by centrifugation. Supernatants were adjusted to 2 mm CaCl2 and MnCl2, concanavalin A-agarose (0.375 g wet weight) was added, and samples were gently shaken for 20 min at 37°C. Non-glycosylated proteins were removed by washing twice with a buffer containing 20 mm Tris-Cl, pH 7.5, 0.5 m NaCl, 1 mm CaCl2, and 1 mm MnCl2 and glycosylated proteins were eluted by incubation at 37°C for 1 h with 1.5 mL of the buffer containing α-d-methylmannoside (1 m).
Western Analysis
Proteins were separated by SDS-PAGE (Laemmli, 1970), electrotransferred to PVDF membranes (NEN, Boston), and the membranes were then blocked for 1 to 18 h with bovine serum albumin (2%, w/v). Tissue culture supernatants (designated 19F4) containing monoclonal antibodies against recombinant cytokinin oxidase were purified over Protein A (HiTrap, Amersham Pharmacia Biotech). Rabbit polyclonal antibodies were raised against recombinant oxidase and purified over Protein A. Membranes were sequentially incubated with primary antibodies diluted 1:100 (monoclonal-mouse) or 1:2,000 (polyclonal-rabbit) in Tris-buffered saline plus Tween 20 (TBST; 10 mm Tris-Cl, pH 8, 0.9% [w/v] NaCl, and 0.05% [v/v] Tween 20) for 45 to 60 min at room temperature; with TBST, for three 5-min washes; with secondary antibodies (anti-mouse IgG-alkaline phosphatase conjugate [Bio-Rad, Hercules, CA] diluted 1:3,000 or anti-rabbit IgG-alkaline phosphatase conjugate [Sigma] diluted 1:10,000) for 25 min; and with TBST, for three 5-minute washes. After a 2-min equilibration in development buffer (Tris-Cl, 100 mm, pH 9.5, 100 mm NaCl, and 50 mm MgCl2), colorimetric detection was initiated by addition of 0.225 mg mL−1 4-nitroblue tetrazolium chloride and 0.35 mg mL−1 5-bromo-4-chloro-3-indolyl-phosphate solution (Roche Molecular Biochemicals, Indianapolis) in this buffer. Reactions were stopped by rinsing membranes in water, then in TE, and then air drying.
Kinetic Analysis
Kinetic data were acquired on an HP 8453 diode array spectrophotometer and reduced using ChemStation software Rev A. 05.02 (Hewlett-Packard, Palo Alto, CA). Unless otherwise stated, cytokinin oxidase activity was measured by the continuous DCPIP reduction assay described previously (Morris et al., 1999). Reactions were carried out in 3 mL of total volume at 30°C and contained sodium phosphate (100 mm, pH 7.0), EDTA (1 mm), DCPIP (0.05 mm), bovine serum albumin or ovalbumin (1 mg mL−1), and appropriate amounts of substrate. Initial rates of reduction of DCPIP were measured at 600 nm at 5-s intervals upon addition of oxidase and were corrected for background drift. Three repetitions were performed for each substrate concentration. Inhibitors were dissolved in the minimum amount of dimethyl sulfoxide and diluted into ovalbumin (1 mg mL−1) just prior to use. Inhibitor studies were carried out by addition of substrate and inhibitor prior to the addition of enzyme. The absolute amount of enzyme present was determined from A280 using a calculated molar absorbance of 9.1 × 104 m −1 cm −1 (based on amino acid composition).
In some experiments oxidase activity was followed by measurement of the rate of conversion of [3H]zeatin into [3H]adenine. Reaction mixtures (50 μL) contained TE, pH 8.0, zeatin (10 μM, including 4.37 × 10 4 Bq [3H]zeatin), and DCPIP (100 μM) and were incubated for up to 2 h at 37°C. Five microliters of each sample was applied to thin-layer chromatography plates (Silica Gel IB-F, J. T. Baker, Phillipsburg, NJ) preloaded with cold zeatin and adenine (8 nmol each) and separated chromatographically in methanol:chloroform:5 n sodium hydroxide (50:50:1). Radioactivity in each spot was determined by scintillation counting in ScintiSafe 30% (Fisher, Pittsburgh).
Flavin Identification
Recombinant cytokinin oxidase (4 mg in 1 mL TE) was denatured by boiling for 12 min. The enzyme was digested with trypsin and chymotrypsin (0.4 mg each) in the presence of 5 mm MgCl2 at 37°C for 19 h. The resulting peptides were applied to a Mono Q HR 5/5 column (Amersham Pharmacia Biotech) equilibrated with 10 mm 2-N-morpholinoethanesulfonic acid, pH 5.5, and 1 mm EDTA, washed for 7 min, and eluted with a 20-min (1 mL min−1) linear gradient of 0 to 1 m NaCl. Absorbance was measured at 210, 280, 350, and 450 nm. A single fraction containing the flavopeptide (maximum absorbance at 350 and 450 nm) was adjusted to 15 mm MgCl2 and divided in half for further analysis. One-half was reapplied to the Mono Q column and refractionated. The remainder was digested with 0.5 mg of nucleotide pyrophosphatase (Type II, Crotalus adamanteus, Sigma) at 37°C for 2 h, applied to the Mono Q column, and refractionated.
Chemical Deglycosylation
Chemical deglycosylation of native and recombinant cytokinin oxidase samples using anhydrous trifluoromethanesulfonic acid (Sojar and Bahl, 1987) was performed with the GlycoFree deglycosylation kit (Oxford GlycoSystems, Rosedale, NY) according to the manufacturer's directions.
ACKNOWLEDGMENTS
We thank Paige Patterson for skilled technical assistance, Kevin McGrory for help in the initial purification of the cornmeal enzyme, Dale Blevins and Tim Reinbott for maize kernel samples, and the Monsanto Corporation for providing Me-iP and the AtCKX1 cDNA clone. The phenylurea derivative compounds methoxy-PU and F-PU were generous gifts from Koichi Shudo.
Footnotes
This research was supported by grants from the Monsanto Corporation and the Illinois-Missouri Biotechnology Alliance.
LITERATURE CITED
- Angra R, Mandahar CL, Gulati A. The possible involvement of cytokinins in the pathogenicity of Helminthosporium maydis. Mycopathologia. 1990;109:177–182. [Google Scholar]
- Armstrong DJ. Cytokinin oxidase and the regulation of cytokinin degradation. In: Mok DWS, Mok MC, editors. Cytokinins: Chemistry, Action, and Function. Boca Raton, FL: CRC Press; 1994. pp. 139–154. [Google Scholar]
- Burch LR, Horgan R. The purification of cytokinin oxidase from Zea mays kernels. Phytochemistry. 1989;28:1313–1319. [Google Scholar]
- Chatfield JM, Armstrong DJ. Cytokinin oxidase from Phaseolus vulgaris callus cultures: affinity for concanavalin A. Plant Physiol. 1988;88:245–247. doi: 10.1104/pp.88.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook RJ, Kunio SM, Wagner C. Identification of the covalently bound flavin of dimethylglycine dehydrogenase and sarcosine dehydrogenase from rat liver mitochondria. J Biol Chem. 1984;259:12475–12480. [PubMed] [Google Scholar]
- Cregg JM, Vedvick TS, Raschke WC. Recent advances in the expression of foreign genes in Pichia pastoris. Bio/Technology. 1993;11:905–910. doi: 10.1038/nbt0893-905. [DOI] [PubMed] [Google Scholar]
- Despreaux CW, Manning RF. The dacA gene of Bacillus stearothermophilus coding for D-alanine carboxypeptidase: cloning, structure and expression in Escherichia coli and Pichia pastoris. Gene. 1993;131:35–41. doi: 10.1016/0378-1119(93)90666-q. [DOI] [PubMed] [Google Scholar]
- Dietrich JT, Kaminek M, Blevins DG, Reinbott TM, Morris RO. Changes in cytokinins and cytokinin oxidase activity in developing maize kernels and the effects of exogenous cytokinin on kernel development. Plant Physiol Biochem. 1995;33:327–336. [Google Scholar]
- Fraaije M, Van Berkel W, Benen J, Visser J, Mattevi A. A novel oxidoreductase family sharing a conserved FAD-binding domain. Trends Biochem Sci. 1998;23:206–207. doi: 10.1016/s0968-0004(98)01210-9. [DOI] [PubMed] [Google Scholar]
- Grinna LS, Tschopp JF. Size distribution and general structural features of N-linked oligosaccharides from the methylotrophic yeast, Pichia pastoris. Yeast. 1989;5:107–115. doi: 10.1002/yea.320050206. [DOI] [PubMed] [Google Scholar]
- Hare PD, Van Staden J. Cytokinin oxidase: biochemical features and physiological significance. Physiol Plant. 1994a;91:128–136. [Google Scholar]
- Hare PD, Van Staden J. Inhibitory effect of thidiazuron on the activity of cytokinin oxidase isolated from soybean callus. Plant Cell Physiol. 1994b;35:1121–1125. [Google Scholar]
- Horgan R, Burch LR, Palni LMS. Cytokinin oxidase and the degradative metabolism of cytokinins. In: Pharis RP, Rood SB, editors. Proceedings of the 13th International Conference on Plant Growth Substances. Berlin: Springer-Verlag; 1990. pp. 282–290. [Google Scholar]
- Houba-Herin N, Pethe C, d'Alayer J, Laloue M. Cytokinin oxidase from Zea mays: purification, cDNA cloning and expression in moss protoplasts. Plant J. 1999;17:615–626. doi: 10.1046/j.1365-313x.1999.00408.x. [DOI] [PubMed] [Google Scholar]
- Jameson PE. Cytokinin metabolism and compartmentation. In: Mok DWS, Mok MC, editors. Cytokinins: Chemistry, Activity, and Function. Boca Raton, FL: CRC Press; 1994. pp. 113–128. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laloue M, Fox JE. Cytokinin oxidase from wheat: partial purification and general properties. Plant Physiol. 1989;90:899–906. doi: 10.1104/pp.90.3.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RC, Mok MC, Mok DWS. A gene encoding the cytokinin enzyme zeatin O-xylosyltransferase of Phaseolus vulgaris. Plant Physiol. 1999a;120:553–557. doi: 10.1104/pp.120.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RC, Mok MC, Mok DWS. Isolation of a cytokinin gene, ZOG1, encoding zeatin O-glucosyltransferase from Phaseolus lunatus. Proc Natl Acad Sci USA. 1999b;96:284–289. doi: 10.1073/pnas.96.1.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaw BA, Horgan R. Cytokinin oxidase from Zea mays kernels and Vinca rosea crown-gall tissue. Planta. 1983;159:30–37. doi: 10.1007/BF00998811. [DOI] [PubMed] [Google Scholar]
- Mills LJ, Van Staden J. Extraction of cytokinins from maize, smut tumors of maize and Ustilago maydis cultures. Physiol Plant Pathol. 1978;13:73–80. [Google Scholar]
- Mok MC. Cytokinins and plant development: an overview. In: Mok DWS, Mok MC, editors. Cytokinins: Chemistry, Activity, and Function. Boca Raton, FL: CRC Press; 1994. pp. 155–166. [Google Scholar]
- Morris RO, Bilyeu KD, Laskey JG, Cheikh NN. Isolation of a gene encoding a glycosylated cytokinin oxidase from maize. Biochem Biophys Res Commun. 1999;255:328–333. doi: 10.1006/bbrc.1999.0199. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Paifer E, Margolles E, Cremata J, Montesino R, Herrera L, Delgado JM. Efficient expression and secretion of recombinant α amylase in Pichia pastoris using two different signal sequences. Yeast. 1994;10:1415–1419. doi: 10.1002/yea.320101104. [DOI] [PubMed] [Google Scholar]
- Scorer CA, Buckholz RG, Clare JJ, Romanos MA. The intracellular production and secretion of HIV-1 envelope protein in the methylotrophic yeast Pichia pastoris. Gene. 1993;136:111–119. doi: 10.1016/0378-1119(93)90454-b. [DOI] [PubMed] [Google Scholar]
- Shudo K. Chemistry of phenylurea cytokinins. In: Mok DWS, Mok MC, editors. Cytokinins: Chemistry, Activity, and Function. Boca Raton, FL: CRC Press; 1994. pp. 35–42. [Google Scholar]
- Sojar HT, Bahl OP. A chemical method for the deglycosylation of proteins. Arch Biochem Biophys. 1987;259:52–57. doi: 10.1016/0003-9861(87)90469-3. [DOI] [PubMed] [Google Scholar]
- Wang J, Letham DS. Cytokinin oxidase-purification by affinity chromatography and activation by caffeic acid. Plant Science. 1995;112:161–166. [Google Scholar]
- Whitty CD, Hall RH. A cytokinin oxidase in Zea mays. Can J Biochem. 1974;52:789–799. doi: 10.1139/o74-112. [DOI] [PubMed] [Google Scholar]