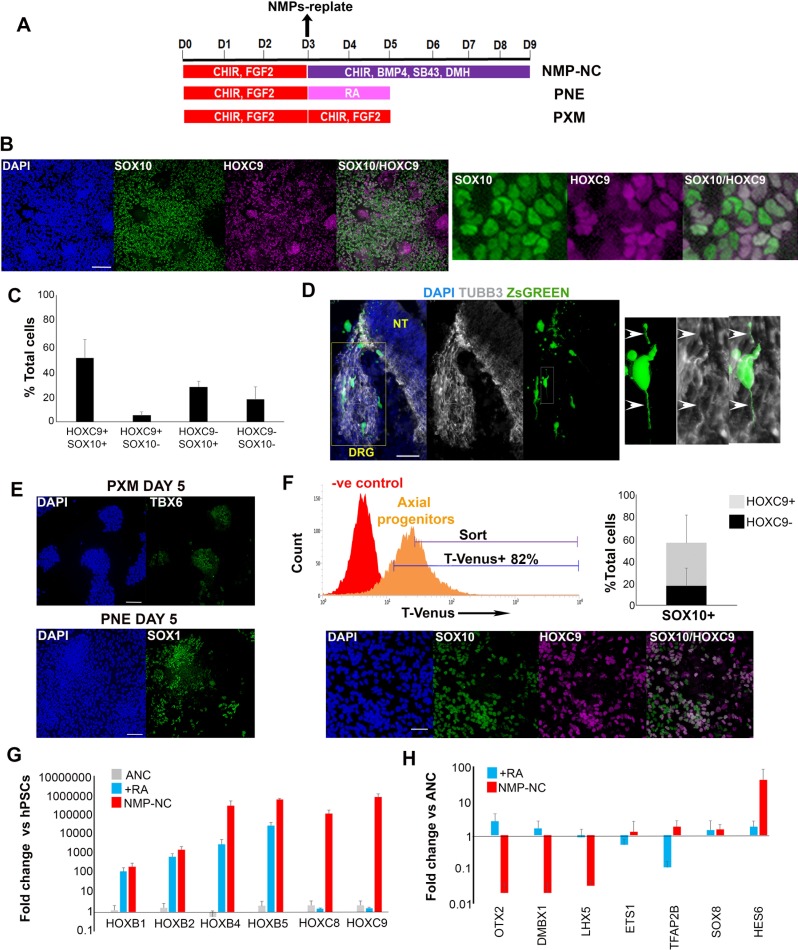
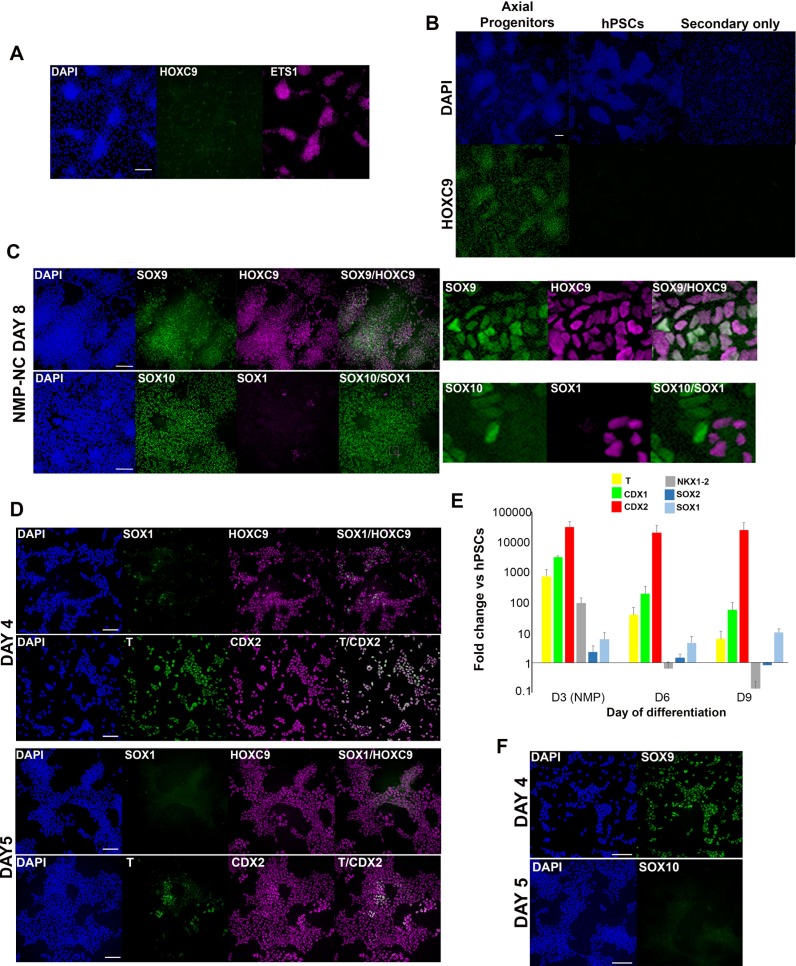
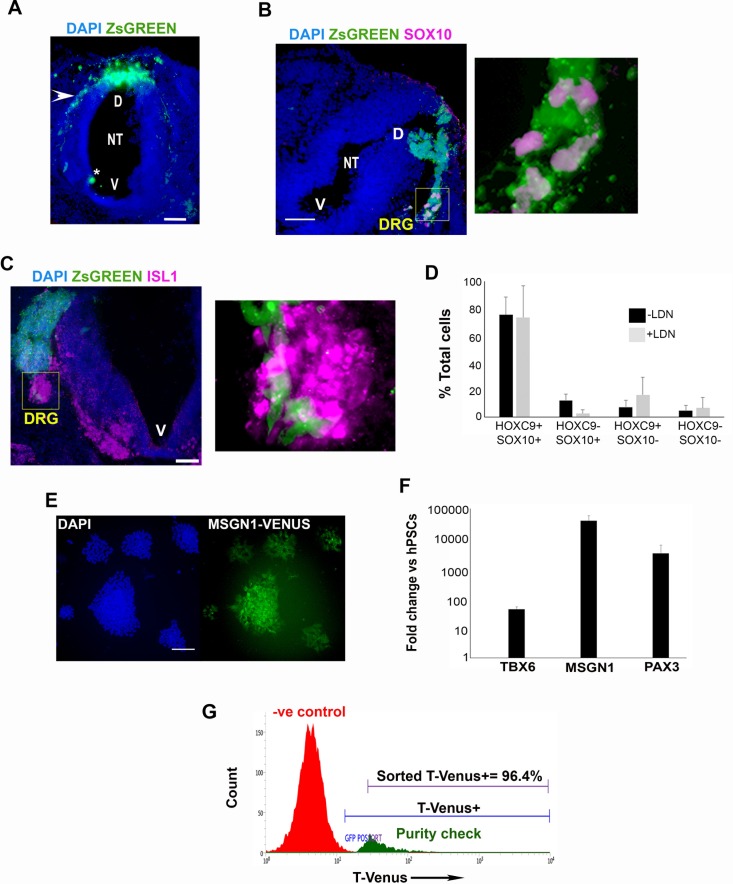
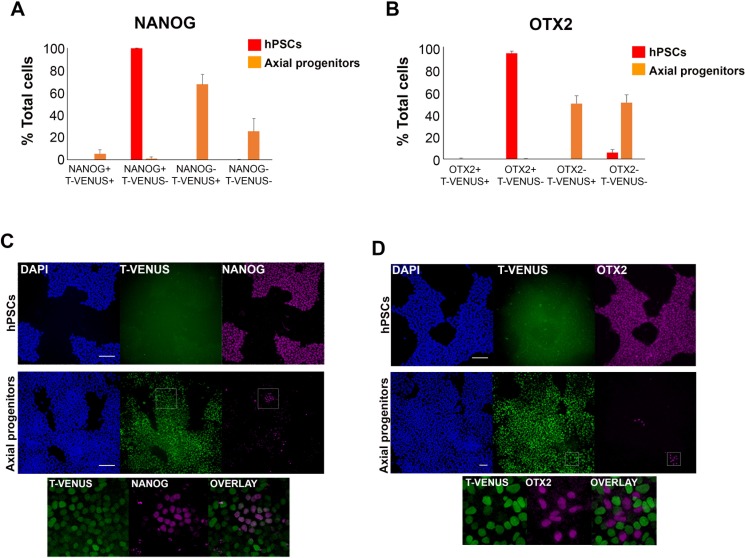
Figure 3. In vitro-derived axial progenitors generate trunk neural crest efficiently.
(A) Diagram depicting the culture conditions employed to direct trunk NC, posterior neurectoderm (PNE) and paraxial mesoderm (PXM) differentiation from hPSC-derived axial progenitors. (B) Immunofluorescence analysis of the expression of the definitive NC marker SOX10 and the thoracic/trunk marker HOXC9 in trunk NC cells derived from axial progenitors after 8 days of differentiation (NMP-NC, see Figure 3A). A magnified region corresponding to the inset is also shown. Scale bar = 100 µm. (C) Quantification of cells marked by different combinations of HOXC9 and SOX10 expression in day eight trunk NC cultures derived from axial progenitors following image analysis. The data in the graph were obtained after scoring three random fields per experiment (two independent replicates) that is a total of 6 fields for two experiments and the error bars/standard deviation represent the variation across all 6 fields and two experiments. Total number of cells scored = 5366, average number of cells/field = 894, error bars = s.d. (D) Immunofluorescence analysis of ZsGREEN and TUBB3 expression in a section of a chick embryo grafted with ZsGREEN+ human axial progenitor-derived trunk NC cells. The DRG region is marked by a yellow box. The images on the right are magnifications of the region marked by the white inset within the DRG region. Arrowheads mark co-localisation of the ZsGREEN and TUBB3 proteins in a donor cell derived, DRG-localised neurite. V, ventral neural tube. Scale bar = 100 µm. (E) Immunofluorescence analysis of TBX6 (left) or SOX1 (right) expression in axial progenitors treated with CHIR-FGF2 (pro-PXM conditions) and RA (pro-PNE conditions) respectively. Scale bar = 100 µm. (F) Top left: Representative FACS histogram indicating the gated T-VENUS +hPSC derived axial progenitors as well as its flow-sorted fraction (‘sort’) which was subsequently plated in NC-inducing conditions. Top right: Average percentage of SOX10+ cells (in relation to HOXC9 expression) following 5 day differentiation of sorted T-VENUS+ axial progenitors in NC-inducing conditions, immunostaining and image analysis. The data in the graph were obtained after scoring 8–10 random fields per experiment (N = 5). The error bars/standard deviation represent the variation across all fields and five experiments. Error bars = s.d. Bottom: A representative field depicting immunofluorescence analysis of SOX10 and HOXC9 expression in NC cells derived from sorted T-VENUS+ axial progenitors. Scale bar = 100 µm. (G) qPCR expression analysis of indicated HOX genes in hPSC-derived anterior cranial (ANC), retinoic acid (RA)-treated NC (+RA), and axial progenitor-derived NC cells (NMP-NC) relative to hPSCs. Error bars = S.E.M. (n = 3). (H) qPCR expression analysis of indicated NC markers in +RA and axial progenitor-derived NC cells relative to untreated anterior cranial NC cells. Error bars = S.E.M. (n = 3).