Abstract
Chronic kidney disease (CKD) affects millions of people and constitutes a major health and financial burden worldwide. People of African descent are at an increased risk of developing kidney disease, which is mostly explained by two variants in the Apolipoprotein L1 (APOL1) gene that are found only in people of West African origin. It is hypothesized that these variants were genetically selected due to the protection they afford against African sleeping sickness, caused by the parasite Trypanosome brucei. Targeting mutant APOL1 could have substantial therapeutic potential for treating kidney disease. In this review, we will describe the intriguing interplay between microbiology, genetics, and kidney disease as revealed in APOL1-associated kidney disease, and discuss APOL1-induced cytotoxicity and its therapeutic implications.
Keywords: Apolipoprotein 1, chronic kidney disease, genetics
APOL1 in chronic kidney disease- a game changer
Chronic kidney disease (CKD) and end stage renal disease (ESRD) affect millions of people of African ancestry throughout the world. African-Americans, while only 13% of the general population in the US, constitute 32% of dialysis patients [1, 2]. Much of this increased risk is attributed to two risk variants (named G1 and G2) that were recently identified in the Apolipoprotein L1 (APOL1) gene [3]. About 40% of all African-American dialysis patients carry these variants [4] and it is estimated that these variants explain about 70% of the excess ESRD risk in this population.
Variants in APOL1 are thought to have arisen as a result of positive genetic selection, as they confer resistance against Trypanosome brucei rhodesiense, a parasite that causes African sleeping sickness [5]. In sub-Saharan Africa, sleeping sickness threatens millions of people. During the most recent epidemic the prevalence reached 50% in some regions. While having one risk variant (G1 or G2) imparts this crucial resistance against sleeping sickness, having two risk alleles significantly increases the risk of developing kidney disease [6]. It is estimated that 20% of people with two risk alleles will eventually develop kidney disease [7]. Genetic variants increase CKD risk with an odds ratio of 2–89, depending on other environmental factors such as HIV infection. Understanding the mechanism by which these risk variants cause kidney disease will be important for future therapeutic targeting.
A tale of two mutations: The interplay between the human Apolipoprotein L1 and the Trypanosome
The first studies that analyzed the potential contribution of genetic factors to CKD/ESRD development in the African-American population used admixture linkage disequilibrium (MALD) methods. These studies identified a genomic region on chromosome 22 that showed an increase in African ancestry marks compared to healthy controls [8, 9]. The initial analysis proposed that the risk variant is located on the intronic region of MYH9. In 2010, using denser genotyping data from the 1,000 genome project, two groups reported that the association can be explained by coding variants in the Apolipoprotein L1 gene [10, 11]. After controlling for the APOL1 variants, the MYH9 locus was no longer associated with kidney disease [10]. G1 represents two missense mutations that are in near-complete linkage disequilibrium, and G2 results in deletion of two amino acids. Both variants are in the C-terminal domain on the gene. The G1 and G2 alleles are common, with 50% of African-Americans carrying one or two risk alleles and 10–15% carrying two risk alleles (G1/G1, G2/G2 or G1/G2) [12, 13]. Having two of the risk alleles significantly and reproducibly increased the risk for a wide range of kidney diseases including focal segmental glomerulosclerosis (FSGS), lupus nephritis, sickle cell disease, hypertensive renal disease, and HIV associated nephropathy (HIVAN) [14–20].
APOL1 is a recently evolved gene. It is present only in humans and a few primates [21, 22]. It circulates as part of the high density lipoprotein (HDL) complex and imparts resistance to infection with Trypanosoma Brucei through lysosomal swelling and lysis of the parasite [23–25]. Several Trypanosome subspecies (T b rhodesiense and T b gambiense), developed mechanisms to avoid APOL1-mediated lysis and cause African sleeping sickness in humans, a potentially lethal disease [26, 27]. T. b. rhodesiense escapes from APOL1-induced toxicity by expressing a protein called serum resistance-associated protein (SRA), that binds to and neutralizes APOL1 [28]. It is hypothesized that genetic variants in APOL1 emerged as a result of positive genetic selection [29]. The selective sweep originated in West Africa and mostly involved the G1 allele, although evidence of a selective sweep has also been shown for the G2 allele [3]. Long-range haplotype tests based on extended haplotype homozygosity suggest that the selective sweep occurred within the last 10,000 years [30]. Recently, studies have added important insight into the evolutionary genetics of APOL1 risk variants. G1’s and G2’s protective properties against T b rhodesiense and T b gambiense were demonstrated in an epidemiological study in West and East Africa [31]. G2 was shown to protect against T b rhodesiense in East Africa, thus explaining the high allele frequency and its association to this region. In a study in West Africa, G1 did not protect against T b gambiense but was rather associated with a latent asymptomatic infection. On the other hand, a different study in Central Africa failed to identify a protective effect of G2 against T b rhodesiense [32]. These results suggest that the relationship between the evolution of APOL1 risk variants and the trypanosome might actually be more complex than previously imagined.
APOL1 genetic variants: Proving causality
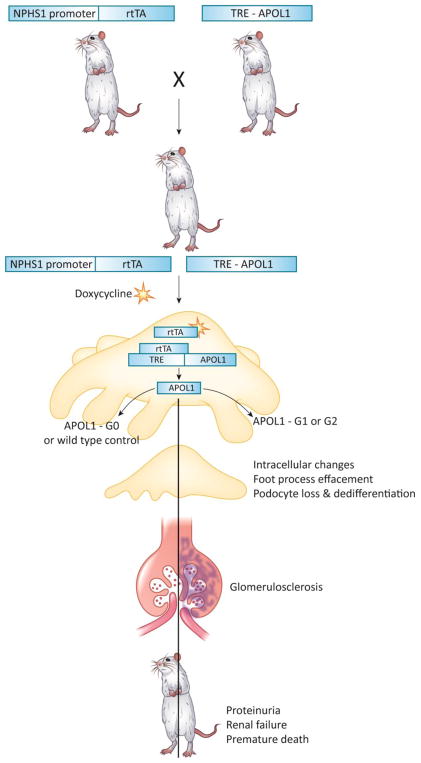
The properties of the G1 and G2 APOL1 genetic variants are highly unusual. The allele frequencies of the G1 and G2 APOL1 risk variants are high [13], and the odds of disease development with either allele are also unusually high for a complex trait variant [7]. In addition, while most complex trait variants are non-coding, G1 and G2 are in the coding region of the gene, resulting in alterations in the translated protein. Animal models are currently the most effective tool to establish causality between genetic variants and disease. Such models can aid the understanding of disease mechanisms and drug development. However, APOL1 is a recently evolved gene that exists only in the genome of humans and some primates, making the construction of animal models challenging. To avoid this problem, several groups have targeted the entire APOL1 locus when designing animal models. While the genetic knock-in of the human locus should be a powerful method, it is not necessarily expected to lead to phenotype development as only 20% of people with two risk alleles develop kidney disease [33]. Furthermore, neither the regulation of APOL1 nor the disease causing cell type is well understood [34, 35]. To this end, our lab has generated a conditionally inducible G0, G1 or G2 APOL1 mouse model, where the transgene can be induced by doxycycline administration [36]. By mating this mouse model with cell-type specific inducer mouse strains, cell-type specific APOL1 expression can be achieved. In our mouse model, expression of G1 or G2 APOL1 variants in podocytes induced albuminuria. Leakage of serum albumin into the urine is one of the key phenotypic manifestations seen in patients with high-risk APOL1 genotypes. The albuminuria was most likely the result of structural damage to the glomerular filter—including podocyte foot process effacement and solidified glomerulosclerosis—observed in the animal model, as well as in patients with APOL1-associated kidney disease [36]. As a result of the severe functional and structural damage, APOL1 transgenic animals developed azotemia and died of renal failure. (Figure 1) Molecular studies, using RNA sequencing of podocyte-specific APOL1 transgenic animals, indicated similarities in the mouse model to the transcriptomes of human FSGS patients with APOL1 high risk genotype, further supporting that the mouse model recapitulates the human disease [37].
Figure 1. Transgenic expression of podocyte specific APOL1 induces kidney disease in mice.
The doxycycline inducible (rtTA) system was used to generate mice with podocyte specific conditional inducible APOL1 expression. Transgene expression was controlled by doxycycline and the nephrin (NPHS1) rtTA promoter was used to drive podocyte specific expression. Either APOL1 reference (G0) or risk allele (G1 or G2) was expressed (TRE-APOL1). When exposed to doxycycline, transgenic mice expressing risk allele APOL1 developed kidney disease including functional (proteinuria and renal failure) and structural (glomerulosclerosis and foot process effacement) changes, similar to the human kidney disease. Transgenic mice expressing the reference allele did not develop disease.
A previous study described a mouse model with podocyte-specific constitutive expression of G0 or G2 APOL1 that did not develop glomerulosclerosis or proteinuria. In this model, APOL1 expression was significantly lower in the G2 mice compared to G0 mice. The constitutive expression of APOL1 potentially caused a selective pressure and decreased toxicity. Yet, this mouse model developed podocyte loss over time and a phenotype similar to preeclampsia, suggesting that even under constitutive and lower expression of APOL1, podocyte loss can be demonstrated, supporting a causal role [33].
Using the conditionally inducible transgenic approach, expression of APOL1 in kidney tubule cells did not result in phenotype development in our study, indicating cell type specific toxicity [36]. Furthermore, studies using mice with hydrodynamic gene transfer-mediated expression of APOL1 in the circulation also failed to induce renal phenotype development [30]. Animal model studies are consistent with human observational studies, indicating that kidney-specific expression of risk variant APOL1 is critical for phenotype development. Studies from the transplant literature indicate that the kidney genotype, rather than the recipient genotype, correlates with inferior outcome after kidney transplantation. High-risk allele donor allografts are associated with decreased graft survival, independent of the recipient genotype [38–42] (see Clinician’s Corner, Box 1).
In humans, the majority of APOL1 circulates in the plasma [23]. Most APOL1 is secreted by the liver, as evidenced by mass spectrometry of liver transplant patients [43]. On the other hand, transplanting low or high risk variant APOL1 livers had no effect on liver graft survival [44]. There is also no correlation between circulating APOL1 concentrations and renal phenotype (GFR or proteinuria) in a large population-based cohort [45]. On the other hand, our group found a significant negative correlation between glomerular APOL1 transcript level and kidney function in patients [36]. The clinical observations are consistent with in vitro and animal model results. Deletion of the APOL1 signal peptide did not abolish toxicity in human embryonic kidney (HEK) cell line 293T transfected with risk allele APOL1, indicating that the endogenous (i.e. cell autonomous) and not the secreted (circulating) APOL1 is important for disease development [46]. A recent study suggested that APOL1 may form a complex with soluble urokinase-type plasminogen activator receptor (suPAR) and integrins on the podocyte plasma membrane [47]. This complex was identified using surface plasma resonance (SPR) studies, and confirmed in vitro in transfected HEK cells and in human cultured podocytes using immunoprecipitation and integrin activation studies, respectively. Furthermore, expression of risk variant APOL1 in mice using gene delivery technique induced proteinuria as well as integrin activation; no proteinuria was observed following the expression of APOL1 in suPAR-deficient mice. However, the origin of the APOL1, either exogenous or endogenous that is secreted from the podocyte and re-internalized through the suPAR-integrin complex, was not determined. Taken together, these data suggest that G1 and G2 variants are pathogenic mutations and that cell autonomous expression of APOL1 in podocytes plays a key role in kidney disease development.
The second hit: environmental hits are necessary for disease development
While the risk of chronic kidney disease development for people with G1 and G2 APOL1 variants is significant, close to 80% of people with two risk alleles will still not develop disease [7]. Characterizing the role of environmental factors as second hits is crucial to the understanding of APOL1 associated kidney disease. Such studies indicate that the high-risk APOL1 genotype confers an odds ratio of 30–90 for HIV associated nephropathy (HIVAN). The lifetime risk for HIVAN in untreated HIV-infected individuals carrying two risk alleles is 50% [7, 19, 20, 48, 49]. Indeed, 60–70% of patients who present with classic HIVAN (collapsing FSGS pathology) have the high-risk APOL1 genotype. Similarly, a subset of patients with the high-risk APOL1 genotypes who received interferon injections for other disease conditions developed proteinuria and histological lesions such as FSGS [50]. These studies strongly support that viral infections, most likely via an interferon-mediated mechanism, present an important second hit for kidney disease development [51, 52].
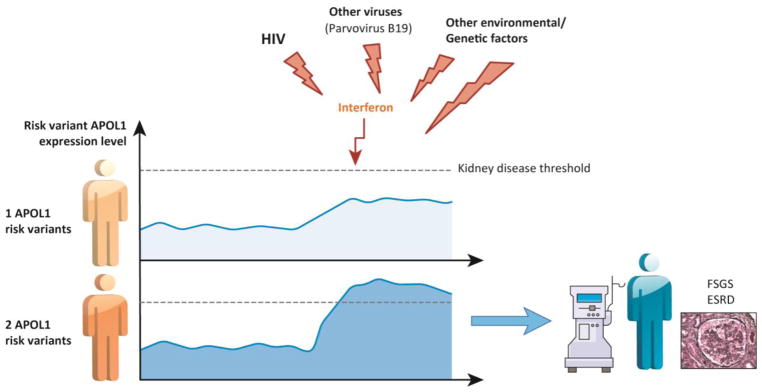
In cultured human podocytes and endothelial cells, interferon is one of the strongest inducers of APOL1 RNA and protein expression, as APOL1 has multiple STAT-binding sites on its regulatory regions [36, 53]. Thus, a relatively simple model has been proposed, in which environmental factors induce kidney disease development by increasing risk variant APOL1 expression in podocytes. (Figure 2) Multiple observations support this hypothesis. In animal models and cell culture systems, APOL1-induced toxicity not only depends on risk variant APOL1, but it is also highly dose dependent [30, 54–56]. Increasing APOL1 mRNA or protein expression is associated with higher toxicity [36]. Furthermore, transcriptomic studies of a large collection of human kidney glomerular samples indicated that APOL1 transcript expression is higher in patients with kidney disease and the amount linearly correlates with kidney function [36]. Taken together, this data suggests a possible mechanism in which environmental triggers, such as viral infections, induce interferon expression, which then increases APOL1 concentration above a certain critical threshold that leads to podocyte injury and progressive CKD. (Figure 2)
Figure 2. Environmental hits are necessary for disease development.
Most people with two APOL1 risk alleles will not develop disease. A second hit is required to induce kidney disease in high risk genotype people. Environmental factors serve as second hits. Viral infections, most likely via an interferon mediated mechanism, present an important second hit for kidney disease development. HIV is a well-established second hit. Multiple studies suggest that environmental factors induce kidney disease by increasing risk variant APOL1 transcript and protein expression in podocytes. Environmental triggers such as viral infections induce interferon expression, which then increases APOL1 protein expression above a certain critical threshold that leads to progressive CKD. People with one risk allele have a lower expression of risk variant that are less likely to reach this threshold.
Several other factors have been proposed as second hits [57]. For example, it was published that serum suPAR is a good predictive biomarker for kidney disease development among people with the high-risk APOL1 genotype [47]. suPAR can potentially bind to podocyte integrins and APOL1, triggering the disease. Further studies are needed to identify factors that serve as second hits and the mechanism by which these factors cause disease.
Gained or lost? APOL1 recessive mode of inheritance and gain of function mutation: are they truly contradictory?
Studies indicate that APOL1 variants bind Trypanosomal SRA less efficiently than wild type APOL1 [18]. The variant therefore represents a gain-of-function variation, whereby the Trypanosome is unable to escape the lytic activity of APOL1. However, most studies have been unable to detect a statistically significant increased risk for kidney disease in patients carrying only one risk variant [11, 15]. The recessive mode of inheritance usually suggests a loss of function mechanism. Whether the G1 and G2 variants induce kidney disease by loss or gain of function has been a matter of extensive discussion [58].
Studies indicate that knock-down of the APOL gene cluster in zebrafish results in renal phenotype development, thus supporting a loss-of-function mechanism which is consistent with the mode of inheritance [59]. However, the APOL1 gene is not conserved in most species and it is unclear whether the phenotype is APOL1 related [21]. Moreover, an individual with homozygous loss of function APOL1 has been identified in India [60]. This individual presented with repeated episodes of Trypanosomiasis, but no obvious renal abnormalities were detected. These observations indicate that APOL1 is a non-essential gene and therefore the renal phenotype is unlikely to be caused by a loss-of-function.
There are several possible explanations for the need of two risk alleles for phenotype development. One possibility is that APOL1 is present in the body as a multimer [61]. The multimerization model proposes that APOL1 toxicity is antagonized by an unknown factor that interacts with wild-type APOL1, and that this antagonism is attenuated by binding to G1 and G2. When one risk allele APOL1 is present, there is sufficient APOL1 multimer, that carry at least one wild-type APOL1, thus the toxicity-blocking binding of the factor is retained. When two risk alleles are present, multimers lacking wild-type APOL1 predominate, resulting in toxicity. The ability of APOL1 to multimerize in vitro has been reported, yet there is no formal proof of this model [61]. Another alternative could be that the effect of a single risk allele is small and larger studies are needed to identify a significant association. A recent study indicates that in Sub-Saharan Africa, where both APOL1 risk variants and HIV incidence is much higher than in previously studied African American populations, even heterozygous individuals have a significantly increased risk of developing CKD [49]. In addition, because the APOL1 risk variant associated toxicity is dose dependent, it is possible that having two risk alleles increases the likelihood of reaching the critical threshold to induce kidney disease. (Figure 2)
APOL1-mediated Trypanosome lysis and renal injury: Similar mechanisms?
APOL1 is a 398 amino acid protein. Five functional domains have been defined in the protein: the S domain is a secretory signal, the MAD (membrane-addressing domain) is a pH sensor and regulator of cell death, the BH3 domain is associated with programmed cell death, the PFD (pore forming domain) and SRA (serum resistance-associated) binding domains confer resistance to Trypanosoma brucei [62–64]. The G1 and G2 variants alter the SRA binding domain of the protein, therefore specific attention has been paid to this domain. The structure of SRA was recently determined through crystallography and the APOL1-binding site was identified [65]. This finding highlighted flaws in the current SRA-APOL1 interaction model, and may help identify SRA-like candidates for APOL1 binding in human cells. The C-terminal coiled coil domain of APOL1, which directly interacts with SRA, has also been recently characterized through nuclear magnetic resonance (NMR) and molecular dynamics simulations [66]. The model predicts a helical hairpin, comprised of two antiparallel helices, that has a more compact and stable fold in G0 than G1 or G2, affecting SRA binding, as well as potentially human protein targets.
APOL1 circulates in the human blood as part of the high density lipoprotein complex. A model based on studies of APOL1 in planar lipid bilayers measuring pH-dependent ion conductance was recently described. The model stipulates that APOL1 is endocytosed by the parasite and inserts into the vesicular membrane when exposed to the low lysosomal pH [67]. Later, upon APOL1 recycling to the plasma membrane and exposure to neutral pH, it creates cation channels that transport sodium, with secondary chloride and potassium fluxes, thus inducing osmotic swelling and death of the parasite [68]. This model suggests that SRA prevents APOL1 from inserting into endocytic membranes, so when recycled to the plasma membrane, it is released from the parasite without creating lethal channels. The resemblance of the APOL1 pore-forming domain to the bacterial pore forming colicinA is the structural basis of this hypothesis.
Multiple publications suggest that APOL1 forms cation channels in membranes; either sodium or potassium [69–71]. Yet, other studies indicate that APOL1 functions as an anion channel that is activated in low pH, transporting chloride across lysosomal membranes, thus inducing lysosomal swelling and cell death [54, 72–74]. A recent study offers an inclusive model that reconciles APOL1 anion and cation channel activity, by its pH-switchable permeability properties [75]. The study used purified recombinant APOL1 in a vesicle-based approach. They found that APOL1 confers chloride selective permeability when allowed to insert into vesicles and tested at low pH, and calcium dependent potassium permeability when inserted at low pH but tested at neutral pH. This model indicates that APOL1 is taken up by endocytosis and the endosomal acidification triggers APOL1 membrane insertion, creating anion channels that affect the lysosomal pH gradient. When APOL1 is targeted to other membranes, such as the plasma membrane, it is exposed to neutral pH and its channel activity is switched from anion to cation permeability [75].
A limitation of these studies is that they were all based on an artificial lipid bilayer or liposome systems, acknowledging that in intact cells, ion conductance, especially secondary ion currents, is likely to be more complex. Apart from the lysosomal and plasma membrane, a recent work indicated that APOL1 can also insert into the mitochondrial membrane, inducing the release of factors that further enhance trypanolysis [73]. Understanding the mechanism by which APOL1 creates channels to mediate trypanolysis may help to understand its pathogenic mechanism in kidney disease.
Indeed, studies by the Pollak group indicate the role of potassium efflux in mammalian cytotoxicity as well [76, 77]. Exogenous expression of APOL1 using APOL1 cRNA injections in Xenopus oocytes increased ion permeability and induced morphological toxicity [78]. The currents were mainly potassium and calcium fluxes with a secondary chloride flux and were influenced by extracellular pH. Elevated extracellular potassium concentration diminished APOL1-associated morphological toxicity. Other reports, including a recent paper by the same group, also found an abnormal potassium efflux in HEK cells transfected with risk variant APOL1, which preceded APOL1-induced cell death [76, 79]. Increasing extracellular potassium, again abrogated these cellular effects, including cytotoxicity. According to this model, APOL1 pore activity plays an important role in initiating a cascade of events leading both to trypanosome death and to human kidney cytotoxicity. The mechanisms by which the channels are created, the membranes involved and the precise intracellular events that follow will require further investigations.
Endocytosis: APOL1 intracellular voyage
APOL1 undergoes a complex endo-lysosomal vesicle trafficking in the Trypanosome. Several lines of evidence suggest that APOL1 risk variants interfere with normal trafficking and these alterations are important for toxicity [54, 70]. Indeed, SRA and APOL1 colocalize in the endosome of Trypanosome brucei, as SRA traffics to the flagellar pocket of the parasite [27].
Unbiased screen studies in yeast, using the risk allele APOL1, indicated that APOL1 risk variant-associated toxicity was increased in mutant strains with defective endosomal trafficking [80]. The interaction screen indicated a role for VPS36. VPS36 protein is a subunit of endosomal sorting complex required for transport II (ESCRT-II). This protein plays a complex role in the sorting of ubiquitinated membrane proteins during endocytosis. In Drosophila, variant APOL1 expression lead to disturbances in the endocytic function and acidification of the nephrocyte; the Drosophila podocyte [80, 81].
Structural homology searches to identify proteins similar to the APOL1-interacting domain of trypanosomal SRA, identified VAMP8 as a potential binding partner of APOL1 [82]. VAMP8 is a SNARE protein involved in autophagy through the direct control of autophagosome and lysososome fusion [83]. Experimental data suggest that G1 and G2 variants could alter VAMP8 binding of APOL1, thus affecting vesicular trafficking and membrane fusion [82].
In human cultured podocytes, we showed that APOL1 is associated with intracellular vesicles, mainly the late endosome, also called multivesicular body compartment [36]. Transfection of HEK cell line with APOL1 risk variants resulted in a massive increase in the late endosome compartment. On the molecular level, we showed poor acidification of the endocytic vesicle in risk variant APOL1, which was associated with a defect in fusion of endocytic vesicles with lysosomes. This fusion step is mediated by the STX17/VAMP8 interaction [36]. Since APOL1 can bind to VAMP8, this model could potentially define the molecular defect induced by risk variant APOL1. We did not identify a defect in the lysosomal compartment in our studies using human podocytes and transfected HEK cells [36]. However, another study did suggest an increase in lysosomal permeability in human podocytes transduced with risk variant APOL1 lentivirus [54]. In summary, several unbiased and targeted experiments indicate that APOL1 might play a role in the fusion of endocytic vesicles with the lysosome.
A dynamic interplay exists between endocytosis and autophagy [84, 85], and several lines of evidence suggest that risk variant APOL1 interferes with autophagy. BH3 proteins, such as APOL1, play critical roles in regulating autophagy; specifically autophagosome formation [86, 87]. Earlier research suggested that wild type APOL1 protein expression induced the formation of autophagic vacuoles and activated translocation of LC3II (a marker of autophagosomes) to the vacuoles. APOL1 expression in autophagy deficient mouse embryonic fibroblast cells did not induce cell death, indicating the role of APOL1-induced autophagy in cell death [86, 87]. Binding of APOL1 to PI3P, a derivative of membrane phospholipids that plays a role in autophagosome formation, was thought to mediate this effect. Deletion or replacement of APOL1 BH3 domain has been shown to reduce APOL1-associated toxicity in transfected HEK cells[46].
Studies testing the effect of risk variants on autophagy have shown seemingly conflicting results. APOL1 risk variant was associated with an increase in LC3II abundance (a marker of autophagosomes) in HEK and hepatoma cell lines and in primary human hepatocytes [55, 88]. Our group examined autophagy in risk variant APOL1 transfected HEK and HeLa cells. We found that while there was an increase in LC3II at baseline, treatment with chroloquine, a lysosomal blocker, was associated with lower LC3II protein expression indicating a block in autophagy flux secondary to a defective fusion of autophagosomes with lysosomes. The block in autophagy flux was confirmed by other methods such as electron microscopy analysis and immunofluorescence staining of autophagic vesicles in human podocytes and transfected HEK cells [36]. Other studies were mostly based on simply quantifying LC3II protein expression without analyzing the flux and therefore were unable to detect significant differences in autophagy flux when G0 and G1, G2 variants were compared [89]. In summary, model organisms and cell biology data indicate that risk variant APOL1 induced toxicity is mediated by a defect in late endosome-lysosome fusion (a known VAMP8-mediated process) and a downstream defect in autophagy flux.
Risk allele APOL1 cytotoxicity: How do podocytes die?
Most investigators agree that exogenous expression of APOL1 in podocytes, transfected HEK293 cells and transfected hepatoma cells is associated with increased cell death [54–56]. Expression of the exogenous reference allele, G0 also seems to induce some toxicity in most in vitro systems. One study reported no significant differences in cytotoxicity between G0 and G1, G2 variants in transfected HEK cells [89], while most other groups identified a dose dependent toxicity induced by APOL1 in which G1 and G2 variants are more toxic than G0.
Transgenic expression of G1 or G2 APOL1 in podocytes, resulted in podocyte loss and dedifferentiation in mice [36]. In a Drosophila transgenic model system, G1 was more toxic to the fly nephrocyte than G0, manifested as nephrocyte hypertrophy and ultimately the death of the organism [81]. By contrast, in zebrafish, exogenous APOL1 expression caused only endothelial and podocyte damage, more pronounced in G1 and G2 compared to G0, with no significant kidney-related phenotype [90]. In summary, most data suggest risk variant APOL1 induce cell death in vivo.
Different signaling pathways have been suggested to be involved in APOL1-induced cytotoxicity (Figure 3). Most studies failed to observe caspase3 activation and apoptosis following APOL1 expression [36, 55]. Other mechanisms include the expression of stress kinases such as JNK/SAPK and p38 MAPK as well as mitochondrial damage and energy depletion [88, 91, 92]. While most studies described an increase in LC3II protein expression following exogenous APOL1 expression, they mostly failed to conclusively demonstrate that autophagy is the mechanism of cell death following APOL1 expression. Furthermore, our study indicated a decrease in autophagy flux rather than increased autophagy [36]. It appears that non-inflammatory cell death pathways were not consistently associated with APOL1 induced damage.
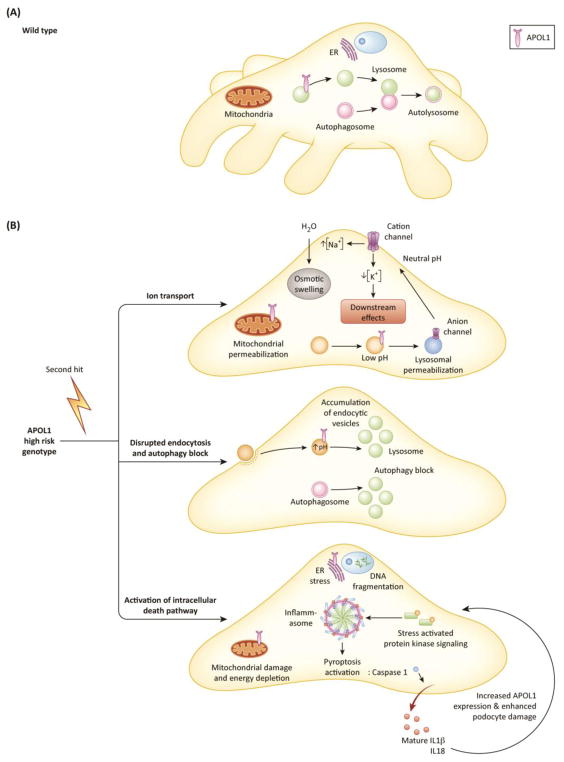
Figure 3. Intracellular mechanisms of APOL1-induced cytotoxicity.
(A) In wild type podocytes, reference allele APOL1 does not affect endocytosis and autophagy. Autophagy inhibits pyroptosis, and mitochondrial and ER functions are unperturbed. (B) Upon exposure to a second hit, a cascade of events is initiated in podocytes expressing two APOL1 risk alleles that culminate in podocyte loss and dedifferentiation. APOL1 inserts into membranes and creates anion (when exposed to low pH, e.g. in the lysosome) or cation channel (when exposed to neutral pH, e.g. in the plasma membrane). These channels mediate ion fluxes that induce osmotic swelling and further activate toxic intracellular pathways (top panel). Due to defective endo-lysosomal trafficking and impaired acidification of endocytic vacuoles, there is a defect in autophagosome maturation resulting in an autophagy block and accumulation of endocytic vacuoles (middle panel). Consequently, various intracellular pathways are activated; including stress activated protein kinase signaling, mitochondrial and endoplasmic reticulum damage and inflammasome activation, resulting in caspase1-mediated inflammatory cell death, pyroptosis. Pyroptosis involves the release of IL1 and IL18, DNA fragmentation, and by a positive feedback loop can increase APOL1 protein expression and could further enhance cytotoxicity (bottom panel).
Unbiased studies indicate the activation of inflammatory cell death pathways by APOL1 [55, 93–95]. Studies of HIV nephropathy in human podocytes and in kidneys of HIV-transgenic mice (Tg26) showed increased expression of caspase1, its upstream regulator NLRP3 and IL1β mRNA and proteins [93–94]. Caspase1 inhibitors decreased cytotoxicity in cultured podocytes and in APOL1 transfected hepatoma cells [55, 95]. We found an increase in activation and cleavage of caspase1 and downstream IL1 and IL18 expression. Inhibition of caspase1 or NLRP3 reduced APOL1-induced cytotoxicity in transfected HeLa cells [36]. These results are consistent with a pyroptosis-mediated cell death [96]. Blockage of non-inflammatory cell death mechanisms such as autophagy, results in inflammasome activation in other cellular systems, mainly in macrophages [97, 98]. Analysis of our podocyte specific G1 and G2 transgenic mice confirmed the activation of inflammatory cell death components including NLRP3 and caspase1.
Risk variant APOL1 induced kidney disease appears to be podocyte specific. There are several potential explanations for this observation. Podocytes are terminally differentiated cells with minimal, if any, capacity to renew. Podocytes therefore rely on multiple highly active mechanisms to eliminate defective proteins, such as a strong autophagic activity [99–101]. Podocyte specific autophagy defect resulted in glomerulosclerosis development in autophagy-deficient mice, mostly via the accumulation of defective proteins, causing dedifferentiation and cell death [102, 103]. Studies are consistent with the observation that APOL1 interferes with autophagic flux in podocytes [36]. The cytokine release associated with inflammatory cell death might fuel a positive feed-back loop by increasing APOL1 expression and enhancing toxicity. (Figure 3) Additional in vivo animal models and human studies are needed to confirm and define APOL1 induced cell death pathways. [104].
Concluding remarks
APOL1-associated kidney disease is another remarkable example of how evolutionary genetics is critical for human disease understanding. While the field has made several major important observations, even eight years after the discovery of APOL1 risk variants, key pieces are still missing to complete the puzzle. One major question that needs to be addressed is the cellular mechanism of podocyte toxicity, as it could directly lead to therapeutics development. (see Outstanding Questions). Cellular systems and animal models seem to indicate that inflammatory cell death pathways and alterations in endo-lysosomal function play a key role in the process, but confirmatory human studies are still missing. In the meantime, major advances have also been made. These include the characterization of the structure of SRA and the SRA-interacting domain of APOL1, the understanding of the complex protection APOL1 risk variants against the different Trypanosome subspecies in Africa, and defining the ion channel activities of APOL1. Additionally, better understanding of trypanosome biology, the distribution and epidemiology of one or two APOL1 risk allele, the full spectrum of the human phenotype and APOL1 mechanisms of injury outside the podocyte will broaden our understanding of APOL1-associated kidney disease. Furthering APOL1 kidney disease research will hopefully enable further development of therapies for a disease that affects millions of people around the world.
Outstanding questions box.
What makes the kidney and specifically, podocytes, vulnerable to APOL1 toxicity?
Are there differences in G1 and G2 risk allele induced cytotoxicity? Do they have different toxic effects, reflected in mechanism, phenotype, or magnitude?
What are the second hits, either environmental or genetic triggers, which induce kidney disease in APOL1 high-risk genotype people?
Which intracellular pathway, affected by APOL1 risk alleles, is the best target for therapeutic intervention?
Clinician’s corner box (box 1).
African-Americans have an increased risk of developing focal segmental glomerulosclerosis (FSGS), HIV associated nephropathy (HIVAN) and end stage renal disease (ESRD) compared with Americans of European descent
Much of this increased risk is explained by coding variants in apolipoprotein L1 (APOL1), and two risk alleles, present in about 13% of African-American population confer this risk.
In kidney transplant patients, a kidney from a high-risk genotype donor increases the risk for graft loss, while recipient genotype does not affect graft survival. Questions remain over whether scoring and kidney allocation should be formally changed to include APOL1 genotype
As specific treatments are developed, it will become more important to genotype people of recent African ancestry to identify subjects with high risk genotypes for early specific treatment and follow up
Highlights.
Nucleotide variants in the Apolipoprotein L1 (APOL1) gene contribute to the increased risk of kidney disease in people of African ancestry.
It is believed that the high allele frequency of these variants is due to the resistance they confer against the disease-causing Trypanosome species, which is implicated in Human African sleeping sickness.
The mechanism by which APOL1 causes kidney disease is not fully understood. Experiments suggest that mutant APOL1 proteins create ion channels in membranes, disrupt the endo-lysosomal function, alter autophagy, and induce inflammatory cell death and podocyte damage.
Concise glossary
- African trypanosomes
African trypanosomiasis, also known as sleeping sickness, is a disease spread by an infected tsetse fly, found in rural areas of sub-Saharan Africa. Sleeping sickness is caused by two different parasite subspecies, Trypanosoma brucei rhodesiense and Trypanosoma brucei gambiense and is fatal if left untreated
- Chronic kidney disease (CKD) and End stage renal disease (ESRD)
CKD is the gradual loss of kidney function, as manifested by decrease in glomerular filtration rate (GFR). CKD is divided into 5 stages, based on GFR. CKD can result from a variety of causes. Treatment is based on slowing the progression of the kidney damage, usually by controlling the underlying cause, if possible. CKD can progress to ESRD, which is fatal without artificial filtering (dialysis) or a kidney transplant
- Podocytes
The kidney’s functional unit is the nephron, which consists of a size selective filter (glomerulus) and a long tubule system. Podocytes are specialized epithelial cells in the glomerulus. Leakage of albumin and other proteins (proteinuria or albuminuria) is caused by damage to this filtration barrier. Genetic mutations in podocyte proteins lead to proteinuria.
- Tubular cells
epithelial cells that line the tubules, the segment of the nephron that processes the primary filtrate
- SNARE protein
a large protein family that mediates vesicle fusion with their target membrane bound compartment
- VAMP8
(vesicle associated membrane protein 8) is a SNARE involved in autophagy that mediates the fusion of autophagosome membrane with the lysosome membrane
- Pyroptosis
a form of inflammatory programmed cell death pathway, activated mainly by caspase1 and involves the activation of the inflammasome. Pyroptosis is mainly used by the host to control infection by inducing inflammation. Pathologically persistent pyroptosis has been implicated in autoimmune diseases
Footnotes
Competing interests: The Susztak lab receives research support from Biogen, Boehringer Ingelheim, Celgene, GSK, Merck, Regeneron and ONO Pharma for work not related to this manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Centers for Disease Control and Prevention. Chronic Kidney Disease Surveillence System-United States. [Google Scholar]
- 2.Hsu CY, et al. Racial differences in the progression from chronic renal insufficiency to end-stage renal disease in the United States. J Am Soc Nephrol. 2003;14(11):2902–7. doi: 10.1097/01.asn.0000091586.46532.b4. [DOI] [PubMed] [Google Scholar]
- 3.Limou S, et al. Sequencing rare and common APOL1 coding variants to determine kidney disease risk. Kidney Int. 2015;88(4):754–63. doi: 10.1038/ki.2015.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freedman BI, et al. Differential effects of MYH9 and APOL1 risk variants on FRMD3 Association with Diabetic ESRD in African Americans. PLoS Genet. 2011;7(6):e1002150. doi: 10.1371/journal.pgen.1002150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kruzel-Davila E, Wasser WG, Skorecki K. APOL1 Nephropathy: A Population Genetics and Evolutionary Medicine Detective Story. Semin Nephrol. 2017;37(6):490–507. doi: 10.1016/j.semnephrol.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Tin A, et al. Patterns of Kidney Function Decline Associated with APOL1 Genotypes: Results from AASK. Clin J Am Soc Nephrol. 2016;11(8):1353–9. doi: 10.2215/CJN.12221115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dummer PD, et al. APOL1 Kidney Disease Risk Variants: An Evolving Landscape. Semin Nephrol. 2015;35(3):222–36. doi: 10.1016/j.semnephrol.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kopp JB, et al. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat Genet. 2008;40(10):1175–84. doi: 10.1038/ng.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kao WH, et al. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat Genet. 2008;40(10):1185–92. doi: 10.1038/ng.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Genovese G, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329(5993):841–5. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tzur S, et al. Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum Genet. 2010;128(3):345–50. doi: 10.1007/s00439-010-0861-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foster MC, et al. APOL1 variants associate with increased risk of CKD among African Americans. J Am Soc Nephrol. 2013;24(9):1484–91. doi: 10.1681/ASN.2013010113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman DJ, et al. Population-based risk assessment of APOL1 on renal disease. J Am Soc Nephrol. 2011;22(11):2098–105. doi: 10.1681/ASN.2011050519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedman DJ, Pollak MR. Apolipoprotein L1 and Kidney Disease in African Americans. Trends Endocrinol Metab. 2016;27(4):204–215. doi: 10.1016/j.tem.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freedman BI, et al. End-stage renal disease in African Americans with lupus nephritis is associated with APOL1. Arthritis Rheumatol. 2014;66(2):390–6. doi: 10.1002/art.38220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larsen CP, et al. Apolipoprotein L1 risk variants associate with systemic lupus erythematosus-associated collapsing glomerulopathy. J Am Soc Nephrol. 2013;24(5):722–5. doi: 10.1681/ASN.2012121180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kopp JB, et al. Clinical Features and Histology of Apolipoprotein L1-Associated Nephropathy in the FSGS Clinical Trial. J Am Soc Nephrol. 2015;26(6):1443–8. doi: 10.1681/ASN.2013111242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipkowitz MS, et al. Apolipoprotein L1 gene variants associate with hypertension-attributed nephropathy and the rate of kidney function decline in African Americans. Kidney Int. 2013;83(1):114–20. doi: 10.1038/ki.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenberg AZ, et al. HIV-associated nephropathies: epidemiology, pathology, mechanisms and treatment. Nat Rev Nephrol. 2015;11(3):150–60. doi: 10.1038/nrneph.2015.9. [DOI] [PubMed] [Google Scholar]
- 20.Papeta N, et al. APOL1 variants increase risk for FSGS and HIVAN but not IgA nephropathy. J Am Soc Nephrol. 2011;22(11):1991–6. doi: 10.1681/ASN.2011040434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith EE, Malik HS. The apolipoprotein L family of programmed cell death and immunity genes rapidly evolved in primates at discrete sites of host-pathogen interactions. Genome Res. 2009;19(5):850–8. doi: 10.1101/gr.085647.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedman DJ, Pollak MR. Genetics of kidney failure and the evolving story of APOL1. J Clin Invest. 2011;121(9):3367–74. doi: 10.1172/JCI46263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weckerle A, et al. Characterization of circulating APOL1 protein complexes in African Americans. J Lipid Res. 2016;57(1):120–30. doi: 10.1194/jlr.M063453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrington JM, Howell S, Hajduk SL. Membrane permeabilization by trypanosome lytic factor, a cytolytic human high density lipoprotein. J Biol Chem. 2009;284(20):13505–12. doi: 10.1074/jbc.M900151200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanhamme L, et al. Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature. 2003;422(6927):83–7. doi: 10.1038/nature01461. [DOI] [PubMed] [Google Scholar]
- 26.Bart JM, et al. Localization of serum resistance-associated protein in Trypanosoma brucei rhodesiense and transgenic Trypanosoma brucei brucei. Cell Microbiol. 2015;17(10):1523–35. doi: 10.1111/cmi.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephens NA, Hajduk SL. Endosomal localization of the serum resistance-associated protein in African trypanosomes confers human infectivity. Eukaryot Cell. 2011;10(8):1023–33. doi: 10.1128/EC.05112-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibson WC. The SRA gene: the key to understanding the nature of Trypanosoma brucei rhodesiense. Parasitology. 2005;131(Pt 2):143–50. doi: 10.1017/s0031182005007560. [DOI] [PubMed] [Google Scholar]
- 29.Pays E, et al. The molecular arms race between African trypanosomes and humans. Nat Rev Microbiol. 2014;12(8):575–84. doi: 10.1038/nrmicro3298. [DOI] [PubMed] [Google Scholar]
- 30.Thomson R, et al. Evolution of the primate trypanolytic factor APOL1. Proc Natl Acad Sci U S A. 2014;111(20):E2130–9. doi: 10.1073/pnas.1400699111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cooper A, et al. APOL1 renal risk variants have contrasting resistance and susceptibility associations with African trypanosomiasis. Elife. 2017:6. doi: 10.7554/eLife.25461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimuda MP, et al. No evidence for association between APOL1 kidney disease risk alleles and Human African Trypanosomiasis in two Ugandan populations. PLoS Negl Trop Dis. 2018;12(2):e0006300. doi: 10.1371/journal.pntd.0006300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bruggeman LA, et al. APOL1-G0 or APOL1-G2 Transgenic Models Develop Preeclampsia but Not Kidney Disease. J Am Soc Nephrol. 2016;27(12):3600–3610. doi: 10.1681/ASN.2015111220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma L, et al. Localization of APOL1 protein and mRNA in the human kidney: nondiseased tissue, primary cells, and immortalized cell lines. J Am Soc Nephrol. 2015;26(2):339–48. doi: 10.1681/ASN.2013091017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madhavan SM, et al. APOL1 localization in normal kidney and nondiabetic kidney disease. J Am Soc Nephrol. 2011;22(11):2119–28. doi: 10.1681/ASN.2011010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beckerman P, et al. Transgenic expression of human APOL1 risk variants in podocytes induces kidney disease in mice. Nat Med. 2017;23(4):429–438. doi: 10.1038/nm.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sampson MG, et al. Integrative Genomics Identifies Novel Associations with APOL1 Risk Genotypes in Black NEPTUNE Subjects. J Am Soc Nephrol. 2016;27(3):814–23. doi: 10.1681/ASN.2014111131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee BT, et al. The APOL1 genotype of African American kidney transplant recipients does not impact 5-year allograft survival. Am J Transplant. 2012;12(7):1924–8. doi: 10.1111/j.1600-6143.2012.04033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reeves-Daniel AM, et al. The APOL1 gene and allograft survival after kidney transplantation. Am J Transplant. 2011;11(5):1025–30. doi: 10.1111/j.1600-6143.2011.03513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freedman BI, et al. APOL1 Genotype and Kidney Transplantation Outcomes From Deceased African American Donors. Transplantation. 2016;100(1):194–202. doi: 10.1097/TP.0000000000000969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riella LV, Sheridan AM. Testing for High-Risk APOL1 Alleles in Potential Living Kidney Donors. Am J Kidney Dis. 2015;66(3):396–401. doi: 10.1053/j.ajkd.2015.04.046. [DOI] [PubMed] [Google Scholar]
- 42.Freedman BI, et al. Apolipoprotein L1 gene variants in deceased organ donors are associated with renal allograft failure. Am J Transplant. 2015;15(6):1615–22. doi: 10.1111/ajt.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shukha K, et al. Most ApoL1 Is Secreted by the Liver. J Am Soc Nephrol. 2017;28(4):1079–1083. doi: 10.1681/ASN.2016040441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dorr CR, et al. Deceased-Donor Apolipoprotein L1 Renal-Risk Variants Have Minimal Effects on Liver Transplant Outcomes. PLoS One. 2016;11(4):e0152775. doi: 10.1371/journal.pone.0152775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kozlitina J, et al. Plasma Levels of Risk-Variant APOL1 Do Not Associate with Renal Disease in a Population-Based Cohort. J Am Soc Nephrol. 2016;27(10):3204–3219. doi: 10.1681/ASN.2015101121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lan X, et al. Protein domains of APOL1 and its risk variants. Exp Mol Pathol. 2015;99(1):139–44. doi: 10.1016/j.yexmp.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hayek SS, et al. A tripartite complex of suPAR, APOL1 risk variants and alphavbeta3 integrin on podocytes mediates chronic kidney disease. Nat Med. 2017;23(8):945–953. doi: 10.1038/nm.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kopp JB, et al. APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol. 2011;22(11):2129–37. doi: 10.1681/ASN.2011040388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kasembeli AN, et al. APOL1 Risk Variants Are Strongly Associated with HIV-Associated Nephropathy in Black South Africans. J Am Soc Nephrol. 2015;26(11):2882–90. doi: 10.1681/ASN.2014050469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Markowitz GS, et al. Treatment with IFN-{alpha}, -{beta}, or -{gamma} is associated with collapsing focal segmental glomerulosclerosis. Clin J Am Soc Nephrol. 2010;5(4):607–15. doi: 10.2215/CJN.07311009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moudgil A, et al. Association of parvovirus B19 infection with idiopathic collapsing glomerulopathy. Kidney Int. 2001;59(6):2126–33. doi: 10.1046/j.1523-1755.2001.00727.x. [DOI] [PubMed] [Google Scholar]
- 52.Tanawattanacharoen S, et al. Parvovirus B19 DNA in kidney tissue of patients with focal segmental glomerulosclerosis. Am J Kidney Dis. 2000;35(6):1166–74. doi: 10.1016/s0272-6386(00)70055-2. [DOI] [PubMed] [Google Scholar]
- 53.Nichols B, et al. Innate immunity pathways regulate the nephropathy gene Apolipoprotein L1. Kidney Int. 2015;87(2):332–42. doi: 10.1038/ki.2014.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lan X, et al. APOL1 risk variants enhance podocyte necrosis through compromising lysosomal membrane permeability. Am J Physiol Renal Physiol. 2014;307(3):F326–36. doi: 10.1152/ajprenal.00647.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng D, et al. Biogenesis and cytotoxicity of APOL1 renal risk variant proteins in hepatocytes and hepatoma cells. J Lipid Res. 2015;56(8):1583–93. doi: 10.1194/jlr.M059733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khatua AK, et al. Exon 4-encoded sequence is a major determinant of cytotoxicity of apolipoprotein L1. Am J Physiol Cell Physiol. 2015;309(1):C22–37. doi: 10.1152/ajpcell.00384.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen TK, et al. Examination of Potential Modifiers of the Association of APOL1 Alleles with CKD Progression. Clin J Am Soc Nephrol. 2015;10(12):2128–35. doi: 10.2215/CJN.05220515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anderson BR, et al. In vivo Modeling Implicates APOL1 in Nephropathy: Evidence for Dominant Negative Effects and Epistasis under Anemic Stress. PLoS Genet. 2015;11(7):e1005349. doi: 10.1371/journal.pgen.1005349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kotb AM, et al. Knockdown of ApoL1 in Zebrafish Larvae Affects the Glomerular Filtration Barrier and the Expression of Nephrin. PLoS One. 2016;11(5):e0153768. doi: 10.1371/journal.pone.0153768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johnstone DB, et al. APOL1 null alleles from a rural village in India do not correlate with glomerulosclerosis. PLoS One. 2012;7(12):e51546. doi: 10.1371/journal.pone.0051546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Limou S, et al. APOL1 toxin, innate immunity, and kidney injury. Kidney Int. 2015;88(1):28–34. doi: 10.1038/ki.2015.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Duchateau PN, et al. Apolipoprotein L gene family: tissue-specific expression, splicing, promoter regions; discovery of a new gene. J Lipid Res. 2001;42(4):620–30. [PubMed] [Google Scholar]
- 63.Duchateau PN, et al. Apolipoprotein L, a new human high density lipoprotein apolipoprotein expressed by the pancreas. Identification, cloning, characterization, and plasma distribution of apolipoprotein L. J Biol Chem. 1997;272(41):25576–82. doi: 10.1074/jbc.272.41.25576. [DOI] [PubMed] [Google Scholar]
- 64.Currier RB, et al. Decoding the network of Trypanosoma brucei proteins that determines sensitivity to apolipoprotein-L1. PLoS Pathog. 2018;14(1):e1006855. doi: 10.1371/journal.ppat.1006855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zoll S, et al. The structure of serum resistance-associated protein and its implications for human African trypanosomiasis. Nat Microbiol. 2018;3(3):295–301. doi: 10.1038/s41564-017-0085-3. [DOI] [PubMed] [Google Scholar]
- 66.Sharma AK, et al. Structural characterization of the C-terminal coiled-coil domains of wild-type and kidney disease-associated mutants of apolipoprotein L1. FEBS J. 2016;283(10):1846–62. doi: 10.1111/febs.13706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thomson R, Finkelstein A. Human trypanolytic factor APOL1 forms pH-gated cation-selective channels in planar lipid bilayers: relevance to trypanosome lysis. Proc Natl Acad Sci U S A. 2015;112(9):2894–9. doi: 10.1073/pnas.1421953112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Greene AS, Hajduk SL. Trypanosome Lytic Factor-1 Initiates Oxidation-stimulated Osmotic Lysis of Trypanosoma brucei brucei. J Biol Chem. 2016;291(6):3063–75. doi: 10.1074/jbc.M115.680371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rifkin MR. Trypanosoma brucei: biochemical and morphological studies of cytotoxicity caused by normal human serum. Exp Parasitol. 1984;58(1):81–93. doi: 10.1016/0014-4894(84)90023-7. [DOI] [PubMed] [Google Scholar]
- 70.del Molina-Portela MP, et al. Trypanosome lytic factor, a subclass of high-density lipoprotein, forms cation-selective pores in membranes. Mol Biochem Parasitol. 2005;144(2):218–26. doi: 10.1016/j.molbiopara.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 71.O’Toole JF, et al. The Cell Biology of APOL1. Semin Nephrol. 2017;37(6):538–545. doi: 10.1016/j.semnephrol.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Perez-Morga D, et al. Apolipoprotein L-I promotes trypanosome lysis by forming pores in lysosomal membranes. Science. 2005;309(5733):469–72. doi: 10.1126/science.1114566. [DOI] [PubMed] [Google Scholar]
- 73.Vanwalleghem G, et al. Coupling of lysosomal and mitochondrial membrane permeabilization in trypanolysis by APOL1. Nat Commun. 2015;6:8078. doi: 10.1038/ncomms9078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hager KM, et al. Endocytosis of a cytotoxic human high density lipoprotein results in disruption of acidic intracellular vesicles and subsequent killing of African trypanosomes. J Cell Biol. 1994;126(1):155–67. doi: 10.1083/jcb.126.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bruno J, et al. Apolipoprotein L1 confers pH-switchable ion permeability to phospholipid vesicles. J Biol Chem. 2017;292(44):18344–18353. doi: 10.1074/jbc.M117.813444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Olabisi OA, et al. APOL1 kidney disease risk variants cause cytotoxicity by depleting cellular potassium and inducing stress-activated protein kinases. Proc Natl Acad Sci U S A. 2016;113(4):830–7. doi: 10.1073/pnas.1522913113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Olabisi OA, Heneghan JF. APOL1 Nephrotoxicity: What Does Ion Transport Have to Do With It? Semin Nephrol. 2017;37(6):546–551. doi: 10.1016/j.semnephrol.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Heneghan JF, et al. BH3 domain-independent apolipoprotein L1 toxicity rescued by BCL2 prosurvival proteins. Am J Physiol Cell Physiol. 2015;309(5):C332–47. doi: 10.1152/ajpcell.00142.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Edwards JK. Chronic kidney disease: Potassium efflux in APOL1 nephropathy. Nat Rev Nephrol. 2016;12(3):124. doi: 10.1038/nrneph.2016.4. [DOI] [PubMed] [Google Scholar]
- 80.Kruzel-Davila E, et al. APOL1-Mediated Cell Injury Involves Disruption of Conserved Trafficking Processes. J Am Soc Nephrol. 2017;28(4):1117–1130. doi: 10.1681/ASN.2016050546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fu Y, et al. APOL1-G1 in Nephrocytes Induces Hypertrophy and Accelerates Cell Death. J Am Soc Nephrol. 2017;28(4):1106–1116. doi: 10.1681/ASN.2016050550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Madhavan SM, et al. APOL1 variants change C-terminal conformational dynamics and binding to SNARE protein VAMP8. JCI Insight. 2017;2(14) doi: 10.1172/jci.insight.92581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Minton K. Membrane dynamics: How lysosomes SNARE autophagosomes. Nat Rev Mol Cell Biol. 2013;14(2):65. doi: 10.1038/nrm3506. [DOI] [PubMed] [Google Scholar]
- 84.Lamb CA, Dooley HC, Tooze SA. Endocytosis and autophagy: Shared machinery for degradation. Bioessays. 2013;35(1):34–45. doi: 10.1002/bies.201200130. [DOI] [PubMed] [Google Scholar]
- 85.Tooze SA, Abada A, Elazar Z. Endocytosis and autophagy: exploitation or cooperation? Cold Spring Harb Perspect Biol. 2014;6(5):a018358. doi: 10.1101/cshperspect.a018358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wan G, et al. Apolipoprotein L1, a novel Bcl-2 homology domain 3-only lipid-binding protein, induces autophagic cell death. J Biol Chem. 2008;283(31):21540–9. doi: 10.1074/jbc.M800214200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhaorigetu S, et al. ApoL1, a BH3-only lipid-binding protein, induces autophagic cell death. Autophagy. 2008;4(8):1079–82. doi: 10.4161/auto.7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Granado D, et al. Intracellular APOL1 Risk Variants Cause Cytotoxicity Accompanied by Energy Depletion. J Am Soc Nephrol. 2017;28(11):3227–3238. doi: 10.1681/ASN.2016111220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.O’Toole JF, et al. ApoL1 Overexpression Drives Variant-Independent Cytotoxicity. J Am Soc Nephrol. 2018;29(3):869–879. doi: 10.1681/ASN.2016121322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Olabisi O, et al. From man to fish: What can Zebrafish tell us about ApoL1 nephropathy? Clin Nephrol. 2016;86(13):114–118. doi: 10.5414/CNP86S116. [DOI] [PubMed] [Google Scholar]
- 91.Ma L, et al. APOL1 Renal-Risk Variants Induce Mitochondrial Dysfunction. J Am Soc Nephrol. 2017;28(4):1093–1105. doi: 10.1681/ASN.2016050567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kloft N, et al. Pore-forming toxins activate MAPK p38 by causing loss of cellular potassium. Biochem Biophys Res Commun. 2009;385(4):503–6. doi: 10.1016/j.bbrc.2009.05.121. [DOI] [PubMed] [Google Scholar]
- 93.Doitsh G, et al. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature. 2014;505(7484):509–14. doi: 10.1038/nature12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Haque S, et al. HIV Promotes NLRP3 Inflammasome Complex Activation in Murine HIV-Associated Nephropathy. Am J Pathol. 2016;186(2):347–58. doi: 10.1016/j.ajpath.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mikulak J, et al. Impact of APOL1 polymorphism and IL-1beta priming in the entry and persistence of HIV-1 in human podocytes. Retrovirology. 2016;13(1):63. doi: 10.1186/s12977-016-0296-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gabay C, Lamacchia C, Palmer G. IL-1 pathways in inflammation and human diseases. Nat Rev Rheumatol. 2010;6(4):232–41. doi: 10.1038/nrrheum.2010.4. [DOI] [PubMed] [Google Scholar]
- 97.Byrne BG, et al. Inflammasome components coordinate autophagy and pyroptosis as macrophage responses to infection. MBio. 2013;4(1):e00620–12. doi: 10.1128/mBio.00620-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Turner CM, et al. Is the inflammasome a potential therapeutic target in renal disease? BMC Nephrol. 2014;15:21. doi: 10.1186/1471-2369-15-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tagawa A, et al. Impaired Podocyte Autophagy Exacerbates Proteinuria in Diabetic Nephropathy. Diabetes. 2016;65(3):755–67. doi: 10.2337/db15-0473. [DOI] [PubMed] [Google Scholar]
- 100.Kume S, et al. Autophagy: emerging therapeutic target for diabetic nephropathy. Semin Nephrol. 2014;34(1):9–16. doi: 10.1016/j.semnephrol.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 101.Hartleben B, Wanner N, Huber TB. Autophagy in glomerular health and disease. Semin Nephrol. 2014;34(1):42–52. doi: 10.1016/j.semnephrol.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 102.Kawakami T, et al. Deficient Autophagy Results in Mitochondrial Dysfunction and FSGS. J Am Soc Nephrol. 2015;26(5):1040–52. doi: 10.1681/ASN.2013111202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hartleben B, et al. Autophagy influences glomerular disease susceptibility and maintains podocyte homeostasis in aging mice. J Clin Invest. 2010;120(4):1084–96. doi: 10.1172/JCI39492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Larsen CP, Freedman BI. Apolipoprotein L1-associated nephropathy and the future of renal diagnostics. J Am Soc Nephrol. 2015;26(6):1232–5. doi: 10.1681/ASN.2014101052. [DOI] [PMC free article] [PubMed] [Google Scholar]