Abstract
Murine infection models are critical for understanding disease pathogenesis and testing the efficacy of novel therapeutics designed to combat causative pathogens. Infectious pneumonia is among the most common infections presented by patients in the clinic and thus warrants an appropriate in vivo model. Typical pneumonia models use intranasal inoculation, which deposits excessive organisms outside the lung, causing off-target complications and symptoms, such as sinusitis, gastritis, enteritis, physical trauma, or microparticle misting to mimic aerosol spread more typical of viral, tuberculous, or fungal pneumonia. These models do not accurately reflect the pathogenesis of typical community- or healthcare-acquired bacterial pneumonia. In contrast, this murine model of oropharyngeal aspiration pneumonia mimics the droplet route in healthcare-acquired pneumonia. Inoculating 50 µL of the bacteria suspension into the oropharynx of anesthetized mice causes reflexive aspiration, which results in pneumonia. With this model, one can examine the pathogenesis of pneumonia-causing pathogens and new treatments to combat these diseases.
Keywords: Medicine, Issue 136, Infectious diseases, murine model, pneumonia, oropharyngeal aspiration, ventilator-associated pneumonia, hospital-acquired pneumonia
Introduction
Lower respiratory infection is the world's deadliest communicable disease and the most common cause of death in developing countries1. Globally, these infections account for more than 3.2 million deaths1. In addition, nosocomial pneumonia is among the most common and deadly forms of healthcare acquired infections, and is caused by the most antibiotic-resistant pathogens2,3. The typical route of acquisition of bacterial pneumonia for both community-acquired and nosocomial pneumonia is the aspiration of oropharyngeal contents into the alveoli. Murine models used to study these diseases often use intranasal inoculation4, depositing much of the bacteria outside the lung, causing off-target complications and symptoms like sinusitis and physical trauma, which are incongruent with the disease progression in human that the models were designed to emulate. Other models may use inhalation chambers and micromisting devices, which more accurately mimic viral, tuberculous, and fungal pneumonias, but do not accurately recapitulate the normal route of acquisition for typical bacterial pneumonias.
The murine oropharyngeal aspiration pneumonia model may be utilized to simulate the natural route and pathogenesis of bacterial pneumonia. By inoculating 50 µL of the bacterial suspension into the oropharynx of anesthetized mice using a pipette, reflexive aspiration ensues, which results in infectious pneumonia. Using this model, one can examine the pathogenesis of pneumonia-causing pathogens and new treatments to combat these diseases with a higher fidelity model, more analogous to aspiration pneumonia infections observed in human. Additionally, unlike similar models that infect through the oral cavity5,6, this model ensures that the full inoculum reaches the lungs instead of the gut, where it can cause off-site inflammation and infections, such as gastritis and enteritis. Finally, unlike another published model that requires a laryngoscope and inoculates through the trachea7, this model does not obstruct the airway with a gavage needle and does not require injection for inoculum delivery. Instead, inoculation relies on the natural aspiration reflex of the mouse.
Protocol
All procedures involving animals must be approved by the researcher’s Institutional Animal Care and Use Committee (IACUC).
1. Preparation of Bacterial Inoculum
- Isolate bacterial colonies.
- Streak a bacterial strain (e.g., A. baumannii “HUMC1”) on appropriate sterile agar medium (e.g., Tryptic soy agar), being careful to generate isolated colonies.
- Incubate at appropriate conditions (e.g., overnight at 37 °C).
- Grow overnight culture.
- Select a representative, isolated colony from the agar plate with a sterile inoculating loop and use it to inoculate 10 mL of appropriate sterile broth medium (e.g., Tryptic soy broth).
- Allow the sample to reach the stationary phase at the proper conditions (e.g., 37 °C with shaking at 200 rpm overnight in a vented-cap, 50-mL conical vial).
- Grow the subculture.
- Transfer 100 μL of overnight culture to 10 mL of fresh sterile broth using a pipette.
- Allow to reach exponential/log phase at the proper conditions (e.g., 37 °C with shaking at 200 rpm for 3 h in a vented-cap, 50-mL conical vial).
- Wash the subculture.
- Remove the subculture from its incubating environment.
- Centrifuge at 4,000 × g for 5 min to pellet the bacteria.
- Aspirate and discard the supernatant.
- Add 10 mL of sterile phosphate-buffered saline (PBS) to the pellet and vortex vigorously until fully resuspended.
- Repeat steps 1.4.2 - 1.4.4 twice, for a total of 3 washes.
- Adjust the concentration of bacterial suspension.
- Using a spectrophotometer that measures optical density at 600 nm (OD600), measure the optical density of the bacterial suspension.
- Add sterile PBS to the bacterial suspension until the optical density of the inoculum drops to an OD600 of 0.5, using the equation Ci×Vi = Cf×Vf, where “C” is concentration, “V” is volume, “i” is initial, and “f” is final. *Cf should always be 0.5; Vf is the variable to solve.
- If the bacterial suspension drops below an OD600 of 0.5, centrifuge it at 4,000 × g for 5 min to pellet the bacteria and remove an appropriate amount of the supernatant to reach an OD600 of 0.5.
- Perform serial dilutions in sterile PBS (e.g., three 1:100 dilutions to achieve 1 × 10-6) and plate on sterile agar to determine the correlation coefficient that reveals the bacterial concentration in colony-forming units per mL (CFUs/mL) at an OD600 of 0.5. Note: The value of this correlation coefficient will vary for each strain of each species and must be determined prior to inoculum preparation for an infection.
- After determining the correlation coefficient, use it to create an inoculum of the desired concentration (e.g., 2 × 108- 1 × 109 CFUs/mL); if the desired inoculum is greater than the OD600 of 0.5, centrifuge the bacterial suspension at 4,000 × g for 5 min to pellet the bacteria and remove an appropriate amount of the supernatant to reach the desired concentration.
- Perform in vivo pilot studies to determine the virulence of each bacterial isolate in each strain of mouse as these values will vary between bacterial isolates of the same species and mice of different genetic backgrounds. For various Gram-negative species, the LD100 in mice is generally 1 - 5 × 108 CFUs/mouse, although certain strains are lethal at lower inocula.
- Freeze identical aliquots for future use as on-demand, precise inocula, as previously published8 (optional).
- Prepare 1 L of bacterial inoculum as described above, transfer to two 500-mL conical vials, and centrifuge at 4,000 × g for 5 min.
- Discard 450 mL of the supernatant from each conical vial and resuspend the pellets of bacteria in the remaining supernatant.
- Transfer the concentrated bacterial inoculum to a 250 mL beaker containing a magnetic stir bar and continuously mix on top of a stir plate at 300 rpm.
- Carefully transfer exactly 600 μL of concentrated bacterial inoculum (e.g., 1 × 1010 CFUs/mL) using a pipette to a 1.6-mL cryogenic vial containing exactly 300 μL of sterile H2O and 300 μL of sterile glycerol; mix and store at -80 °C. Note: The glycerol necessary for storage can confound the experimental results and cause excess mortality so it is necessary to wash it out.
- When ready for use, thaw the sample for 10 min at room temperature.
- Transfer 1 mL to a 50-mL conical vial, add 9 mL of sterile PBS, and centrifuge at 4,000 × g for 5 min to pellet the bacteria. Aspirate the supernatant and resuspend the pre-determined quantity of CFUs (1 mL × frozen stock concentration in CFUs/mL) in an appropriate volume of PBS to reach the desired concentration for the infectious inoculum.
2. Anesthetizing Mice
- Ketamine/Xylazine Note: Anesthesia with ketamine/xylazine has a long duration, which offers enough time for the researcher who is new at this technique to complete the inoculation procedure before the mouse awakens.
- Prepare 10 mL of injectable solution by combining 8.5 mL of pharmaceutifcal grade PBS, 1 mL of 100 mg/mL Ketamine stock solution (final concentration 10 mg/mL), and 0.5 mL of 20 mg/mL Xylazine stock solution (final concentration 10 mg/mL).
- Administer 10 μL/g by intraperitoneal (IP) injection (e.g., 250 μL for a 25-g mouse).
- Apply ophthalmic ointment to the eyes of the mice anesthetized with Ketamine/Xylazine because their eyes are prone to damage during extended periods of anesthesia.
- Place the mouse in a cage and monitor until it wakes up from anesthesia. Note: The animal should not be left unattended until it has regained sufficient consciousness to maintain sternal recumbency.
- Isoflurane Note: Mice quickly wake up from isoflurane-induced anesthesia after being removed from the induction chamber, so this method of anesthesia should only be used after becoming proficient at the inoculation procedure.
- Place the naïve mice in an appropriately sized induction chamber with oxygen flowing.
- After the chamber is sealed shut, anesthetize the mice using a precision vaporizer to add isoflurane at 4% (v/v) for at least 5 min; confirm the anesthesia by removing a mouse from the induction chamber and measuring how long it takes to wake up from anesthesia (~60 s). Note: Be aware that anesthesia times may vary based on institutional guidelines and some animals may require more than 5 min to become fully anesthetized.
- Maintain the anesthesia for up to 10 min by adjusting the isoflurane concentration to 2 - 3%; if total duration of anesthesia is >15 min, apply ophthalmic ointment to the eyes of anesthetized mice.
- Place the mouse in a cage and monitor until it wakes up from anesthesia, as in step 2.1.
3. Inoculating/Infecting Mice
Suspend an anesthetized mouse, hanging it by its top incisors on a strong, thin string (e.g., dental floss) secured to a fixed object at a height approximately twice the mouse’s body length (e.g., 20 cm) above the operating surface (e.g., laboratory bench).
Gently pull the mouse's tongue out of its mouth using sterile, blunt-ended forceps. Transfer the tongue to a sterile, gloved finger to prevent accidental crushing of the mouse’s tongue. Hold the tongue outside of the mouth, allowing access to the oropharynx and preventing swallowing of the inoculum to the side.
While holding the mouse’s tongue, place the 50-μL inoculum (bacterial suspension) into the oropharynx using a micropipette; the oropharynx is located at the junction of the oral cavity and the pharynx toward the back of the mouth. Note: It is very important to only deliver liquid by pipetting to the first stop on the micropipette. The mouse will stop breathing for a few seconds when the inoculum is placed in the oropharynx; eventually, reflexive aspiration will cause the mouse to inhale the bacterial suspension, indicated by the distinctive crackling noise of liquid entering the lungs, which results in pneumonia infection.
If the inoculum is not placed far back enough to block normal respiration, pinch the nares with forceps to force reflexive aspiration through the mouth and subsequent inhalation of the inoculum.
After inoculation, release the tongue, remove the mouse from the suspension string, place it in a cage, and monitor until it wakes up from anesthesia, as in step 2.1.
4. Monitoring Disease Progression
Note: Due to animal suffering, various indicators should be used to indicate when mice become moribund; euthanasia should be performed following this determination, in accordance with a previously approved IACUC protocol; various markers of moribundity include body temperature, weight loss, appearance, gait, and other biomarkers that can be obtained from blood (e.g., via iSTAT)9,10,11,12,13,14,15,16.
Euthanize the mice according to previously approved IACUC protocol (e.g., CO2 followed by cervical dislocation).
- Remove the lungs from euthanized mouse by cutting the trachea, pulmonary artery, and pulmonary vein close to the lungs.
- Microscopy
- Place the lungs (or portion) in a specimen mold.
- Fill the specimen mold with Optimal Cutting Temperature (O.C.T.) compound, completely submerging the tissue.
- Freeze the sample at -80 °C.
- Send the sample to a pathology lab to section and mount on slides for microscopy.
- Bacterial burden
- Weigh the lung tissue on an analytical balance.
- Transfer the lung tissue to 50-mL conical vial containing 2 - 5 mL of sterile PBS (depending on the size of the tissue).
- Homogenize the lung tissue with a tissue homogenizer.
- Perform serial dilutions of the homogenate in sterile PBS to achieve approximately 1,000 CFUs/mL, based on expected bacterial burden in lungs.
- For example, if 1 × 109 CFUs/mg is expected, perform three 1:100 dilutions: Transfer 100 μL of homogenate to 9.9 mL of PBS and vortex; this is the first 1:100 dilution. Transfer 100 μL of the first 1:100 dilution to 9.9 mL of PBS and vortex; this is the second 1:100 dilution. Transfer 100 μL of the second 1:100 dilution to 9.9 mL of PBS and vortex; this is the third and final 1:100 dilution.
- If bacterial burden in the lungs is unknown, perform a range of dilutions to capture the expected range.
- Plate dilution(s) on sterile agar.
- Prepare sterile Tryptic Soy Broth (TSB) with agar (TSA) plates: Combine 30 g of TSB, 15 g of agar, and 1 L of water, mix with a magnetic stirrer, and heat until boiling at ~100 °C for 5 min. Allow to cool and then autoclave on liquid cycle for 15 min. Allow to cool to ~55 °C, transfer 10 mL freshly autoclaved TSA to sterile Petri dishes, and allow to cool to room temperature. Let dry on benchtop for 2 - 5 d or uncovered under a biosafety cabinet for 10 min.
- Transfer 100 μL of the dilution to a Petri dish containing sterile TSA and spread across TSA using a spreader or glass beads.
- Store the Petri dish overnight at 37 °C in a humidified incubator (i.e., 37 °C incubator containing an uncovered 1-L beaker filled with water)
- Determine CFUs/mL lung homogenate based on agar plating (e.g., 100 CFUs on TSA plate with 100 μL of third and final dilution described above would be 100 CFUs/100 μL × 100dil#1 × 100dil#2 × 100dil#3 = 1×109 CFUs/mL).
- Calculate CFUs/mg lung tissue based by dividing CFUs/mL lung homogenate by mg lung tissue/mL homogenate (e.g., if sample described above used 100 mg of lung tissue homogenized in 5 mL of PBS, then 1 × 109 CFUs/mL ÷ 100 mg/5 mL = 5 × 107 CFUs/mg).
Representative Results
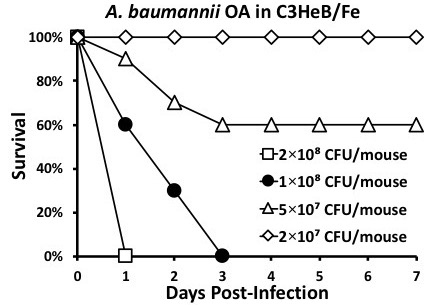
By carefully following the protocol, reproducible and robust data can be easily obtained. It is critical to strictly adhere to one's customized inoculum preparation protocol for experiments to be compared to one each other. It is also important to properly handle mice during the infection procedure. Be sure to place mice into an anesthesia chamber devoid of isoflurane. Mice will panic if they are placed into a chamber that has been pre-filled with isoflurane and may experience excess stress, which can possibly compromise experimental results. After securing the container lid, slowly introduce isoflurane by incrementally increasing its concentration from 0% to 4% (v/v); hastily administering isoflurane can also cause mice to panic. Once the mice become unconscious, reduce the isoflurane concentration to 2 - 3% (v/v) and allow them to remain in the chamber for an additional few minutes. At this point, mice are ready to be infected (Figure 1).
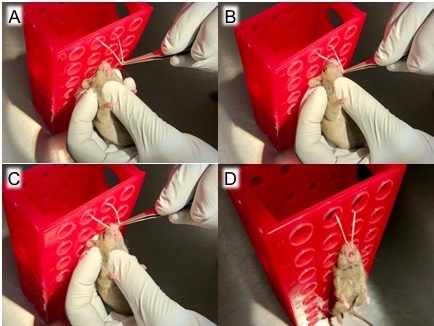
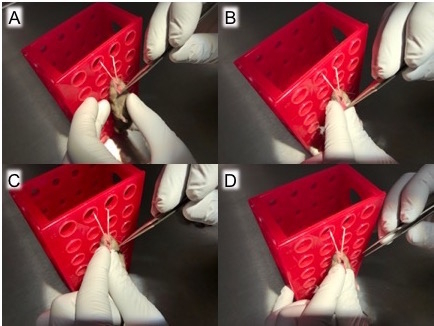
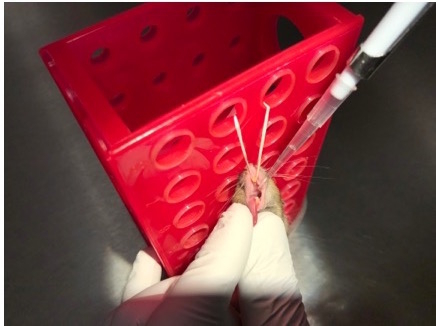
Remove a mouse from the anesthesia chamber and suspend it by its top incisors to allow access to the tongue (Figure 2). Use blunt-ended forceps to gently pull out the tongue (Figure 3) then transfer the forceps-held tongue to a sterile, gloved fingers (Figure 4) to prevent trauma to the mouse's tongue. With the tongue still outside the mouth, transfer the 50-µL inoculum to the oropharynx (Figure 5). Here, it is very important to only deliver liquid by pipetting to the first stop on the micropipette. Continuing to the second stop can introduce a large bubble into the inoculum that interferes with the infection.
Once the infection is complete, there are a myriad of options for testing mice. For pathogenesis studies, one can compare uninfected to infected tissue (Figure 6). If analyzing the effectiveness of novel therapeutics, one can remove the lungs and have them sectioned and stained. This can be done at one time point or at multiple time points to show disease progression (Figure 7).
Another option is to assess the bacterial burden in the lungs of infected mice. This is done by removing the lungs, transferring to a vial containing a known volume of sterile PBS, then homogenizing with a tissue homogenizer (rinsing between each sample to prevent cross contamination). Plating serial dilutions of the lung homogenate on nutrient-rich agar allows for calculating the CFUs/mL for each lung homogenate and subsequently CFUs/mg lung tissue (Figure 8). This, too can be done at one time point or at multiple time points to show disease progression.
There are countless other assays that can be performed, including cytokine analyses (Figure 9), sepsis biomarkers by iSTAT (Figure 10), flow cytometry to investigate cell surface markers, cellular profiling, RNAseq, etc.
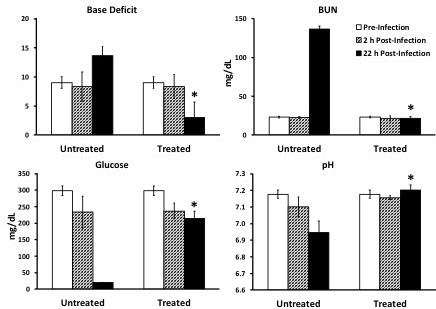
Figure 1: Determining LD100. It is necessary to infect mice with various inocula concentrations to determine the LD100. Here, the inoculum 2 × 108 CFUs/mouse is too high, 5 × 107 and 2 × 107 are too low, and 1 × 108 is just right.
Figure 2: Hang mice by top incisors. After the mice are anesthetized, remove one mouse from the induction chamber, (A) use forceps to pull out a loop of string secured 20 - 30 cm above the work surface, (B) move the string behind the mouse's top incisors, (C) ensure the string is secured behind top incisors, and (D) let the mouse hang by its top incisors on the loop of string.
Figure 3: Pull tongue out of mouth with forceps and transfer grip to gloved fingers. After hanging the mouse by its top incisors, (A) use forceps to gently grasp the mouse's tongue, (B) gently pull out the tongue, (C) carefully transfer the tongue from forceps to gloved fingers to prevent trauma to the mouse's tongue, and (D) hold the tongue in place with gloved fingers. Be sure to apply enough pressure to prevent the tongue from slipping back into the mouth but not so much pressure that trauma ensues. The tongue should be held outside of the moth and to the side to allow for micropipette access in the next step. At this point, the mouse is ready for infection.
Figure 4: Infect the mouse. Maintaining the tongue outside the mouth, use a micropipette to transfer the 50-µL inoculum to the oropharynx. Place the pipet tip inside the mouth at the back of the tongue and dispense the bacterial suspension, only going to the first top on the micropipette; do not go to the second stop as this can introduce a large bubble into the inoculum that can interfere with complete aspiration.
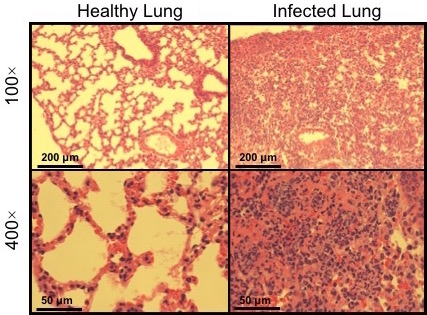
Figure 5: Micrograph of healthy (uninfected) vs infected lung tissue. Hematoxylin and eosin (H&E) staining of lung sections before and after oropharyngeal aspiration of A. baumannii, which recapitulates the route of ventilator-associated pneumonia (VAP), resulting in death typically within 1 - 3 days and substantial alveolar inflammation as seen here.
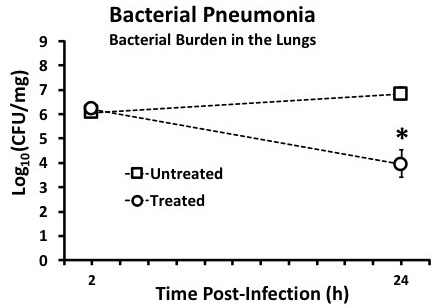
Figure 6: Assess bacterial burden; reprinted and modified with permission14. After infecting mice, remove lungs by dissection following successful euthanasia in compliance with the applicable IACUC protocol. Collect the mass of the lung tissue, transfer to a vial containing a known volume (2 - 5 mL) of sterile PBS, then homogenize with a tissue homogenizer. Be sure to rinse the homogenizer with ethanol and sterile PBS in between each sample to prevent cross contamination. Perform serial dilutions of the lung homogenate and plate various dilutions on nutrient-rich agar and incubate appropriately for the chosen pathogen. Calculate the CFUs/mL for each lung homogenate based on CFUs/plate × dilution, and then divide by the mg lung tissue/mL lung homogenate. This can be done at one time point or at multiple time points to show disease progression. Medians are displayed with error bars representing interquartile ranges. *p <0.05 Treated vs. Untreated group.
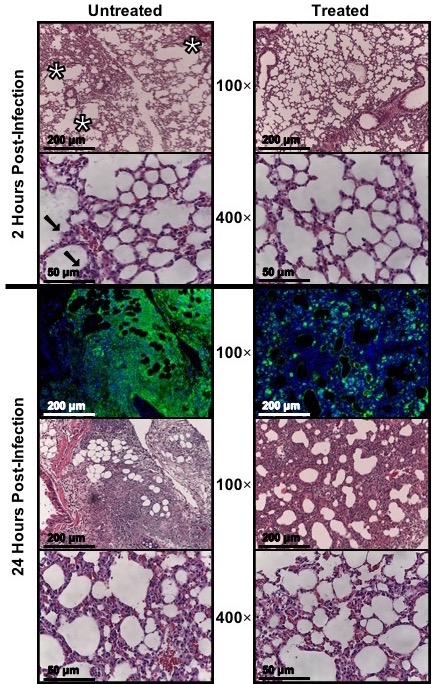
Figure 7: Micrograph of infected lung tissue, untreated vs treated; reprinted and modified with permission14. After infecting mice, remove lungs by dissection following successful euthanasia in compliance with the applicable IACUC protocol. Appropriately preserve the tissue (e.g., immersed in Optimal Cutting Temperature gel and frozen at -80 °C) then send to pathology lab or other proficient individual for sectioning and staining. This can be done at one time point or at multiple time points to show disease progression.
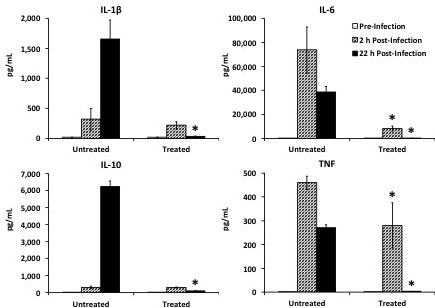
Figure 8: Cytokine analysis; reprinted and modified with permission14. To analyze circulating cytokines, procure sufficient blood (e.g., 50 µL via tail nicking), allow to coagulate for 30 min at room temperature, centrifuge at 1,000 × g for 10 min at 4 °C, and collect serum supernatant. The serum can then be analyzed by Luminex multiplex for cytokines. This can be done at one time point or at multiple time points to show disease progression. Medians are displayed with error bars representing interquartile ranges. *p <0.05 treated vs. untreated group at same time-point.
Figure 9: Sepsis biomarkers; reprinted and modified with permission14. To analyze sepsis biomarkers, procure 75 µL blood (e.g., via tail nicking) and quickly transfer to an iSTAT cartridge that tests for desired analytes. This can be done at one time point or at multiple time points to show disease progression. Medians are displayed with error bars representing interquartile ranges. *p <0.05 treated vs. untreated group at same time-point.
Discussion
To be sure, mice are not miniature humans. Results obtained from mouse models must be considered in context and subsequently interpreted for applicability to humans, based on differences and similarities between the two species6. It is also important to choose the appropriate mouse strain as certain are more susceptible to some infections than others; the same applies to the pathogen strain of choice16.
It is essential to perform infections in an exacting and highly reproducible way. Inocula can be lethal to mice at one value yet harmless at even 90% of that value. Thus, it is imperative that all conditions listed in this protocol are reproduced in the exact same way, particularly when preparing the inoculum and when infecting. Additionally, each pathogen will cause disease at a unique inoculum. Therefore, it is necessary to perform pilot experiments to determine the appropriate inoculum for each strain of pathogen and in each strain of mouse.
With Gram-negative bacteria, an inoculum of ≤5 × 108 CFUs/mouse is sufficient to cause death in virulent strains. It is advised to not use strains that are non-lethal at ≤1 × 109 CFUs/mouse because they suggest dubious relevance to pathogenesis based on sheer quantity of material being placed into the lungs16.
Compared to others, there are very few technical complications for the oropharyngeal aspiration pneumonia model and reproducibility is far greater. Only one minor complication of the model results when the surface tension around the 50-µL inoculum placed in the oropharynx allows for nasal respiration, precluding aspiration-the cause of infection. In this case, simply pinching the nares with forceps compels reflexive aspiration through the mouth and subsequent inhalation of the inoculum to induce infectious pneumonia. This is, however, a technical error by the technician, which is easily avoided with minimal practice.
In terms of its relevance to human disease, the one limitation of this model, like other models of Gram-negative bacterial infections, is the rapidity of the progression of the disease. Humans are rarely infected with a large bolus of a bacterial suspension, which is why mice progress to disease so rapidly. Nevertheless, all FDA-approved treatments undergo such translational research screenings in animal models to assess their therapeutic potential before moving on to clinical trials in humans. Ironically, the rapidity of the model is also an attractive feature, since it can provide clarity on therapeutic efficacy within a brief period of time.
No murine model is perfect, but this is a model with the ability to faithfully emulate the route of infection and pathogenesis of human diseases and assess the effectiveness of potential therapeutics, thereby allowing for the rapid translation of novel therapies that are desperately needed in the clinic.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
This work was supported by the National Institute of Allergy and Infectious Diseases at the National Institutes of Health [Grant Numbers R01 AI117211, R01 AI130060, R21 AI127954, and R42 AI106375 to BS] and US Food and Drug Administration [Contract HHSF223201710199C to BML].
References
- Media Centre WHO. World Health Organization. Fact Sheet - Top 10 Causes of Death. 2017. Available from: http://www.who.int/mediacentre/factsheets/fs310/en/
- Spellberg B, Talbot GH. Recommended Design Features of Future Clinical Trials of Antibacterial Agents for Hospital-Acquired Bacterial Pneumonia and Ventilator-Associated Bacterial Pneumonia. Clinical Infectious Diseases. 2010;51(S1):S150–S170. doi: 10.1086/653065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalil AC, et al. Management of Adults With Hospital-acquired and Ventilator-associated Pneumonia: 2016 Clinical Practice Guidelines by the Infectious Diseases Society of America and the American Thoracic Society. Clinical Infectious Diseases. 2016;63(5):e61–e111. doi: 10.1093/cid/ciw353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina E. Murine model of pneumococcal pneumonia. Methods in Molecular Biology. 2010. pp. 405–410. [DOI] [PubMed]
- Azoulay-Dupuis E, et al. In vivo efficacy of a new fluoroquinolone, sparfloxacin, against penicillin-susceptible and -resistant and multiresistant strains of Streptococcus pneumoniae in a mouse model of pneumonia. Antimicrobial Agents and Chemotherapy. 1992;36(12):2698–2703. doi: 10.1128/aac.36.12.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizgerd JP, Skerrett SJ. Animal models of human pneumonia. American Journal of Physiology-Lung Cellular and Molecular Physiology. 2008;294(3):L387–L398. doi: 10.1152/ajplung.00330.2007. [DOI] [PubMed] [Google Scholar]
- Rayamajhi M, et al. Non-surgical intratracheal instillation of mice with analysis of lungs and lung draining lymph nodes by flow cytometry. Journal of Visualized Experiments. 2011. [DOI] [PMC free article] [PubMed]
- Nielsen TB, Bruhn KW, Pantapalangkoor P, Junus JL, Spellberg B. Cryopreservation of virulent Acinetobacter baumannii to reduce variability of in vivo studies. BMC Microbiology. 2015;15:252. doi: 10.1186/s12866-015-0580-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trammell RA, Toth LA. Markers for predicting death as an outcome for mice used in infectious disease research. Comparative Medicine. 2011;61(6):492–498. [PMC free article] [PubMed] [Google Scholar]
- Bast DJ, et al. Novel murine model of pneumococcal pneumonia: use of temperature as a measure of disease severity to compare the efficacies of moxifloxacin and levofloxacin. Antimicrobial Agents and Chemotherapy. 2004;48(9):3343–3348. doi: 10.1128/AAC.48.9.3343-3348.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankenson FC, et al. Weight loss and reduced body temperature determine humane endpoints in a mouse model of ocular herpesvirus infection. Journal of the American Association for Laboratory Animal Science. 2013;52(3):277–285. [PMC free article] [PubMed] [Google Scholar]
- Adamson TW, Diaz-Arevalo D, Gonzalez TM, Liu X, Kalkum M. Hypothermic endpoint for an intranasal invasive pulmonary aspergillosis mouse model. Comparative Medicine. 2013;63(6):477–481. [PMC free article] [PubMed] [Google Scholar]
- Nielsen TB, et al. Diabetes Exacerbates Infection via Hyperinflammation by Signaling through TLR4 and RAGE. mBio. 2017;8(4) doi: 10.1128/mBio.00818-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen TB, et al. Monoclonal Antibody Protects Against Acinetobacter baumannii Infection by Enhancing Bacterial Clearance and Evading Sepsis. Journal of Infectious Diseases. 2017;216(4):489–501. doi: 10.1093/infdis/jix315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng BL, et al. Evaluation of serotypes 5 and 8 capsular polysaccharides in protection against Staphylococcus aureus in murine models of infection. Human Vaccine Immunotherapy. 2017;13(7):1609–1614. doi: 10.1080/21645515.2017.1304334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong D, et al. Clinical and Pathophysiological Overview of Acinetobacter Infections: a Century of Challenges. Clinical Microbiology Reviews. 2017;30(1):409–447. doi: 10.1128/CMR.00058-16. [DOI] [PMC free article] [PubMed] [Google Scholar]


