Abstract
Various fungi of the genera Aspergillus, Penicillium, and Malbranchea produce prenylated indole alkaloids possessing a bicyclo[2.2.2]diazaoctane ring system. After the discovery of distinct enantiomers of the natural alkaloids stephacidin A and notoamide B, from A. protuberus MF297-2 and A. amoenus NRRL 35660, another fungi, A. taichungensis, was found to produce their diastereomers, 6-epi-stephacidin A and versicolamide B, as major metabolites. Distinct enantiomers of stephacidin A and 6-epi-stephacidin A may be derived from a common precursor, notoamide S, by enzymes that form a bicyclo[2.2.2] diazaoctane core via a putative intramolecular hetero-Diels–Alder cycloaddition. This review provides our current understanding of the structural and stereochemical homologies and disparities of these alkaloids. Through the deployment of biomimetic syntheses, whole-genome sequencing, and biochemical studies, a unified biogenesis of both the dioxopiperazine and the monooxopiperazine families of prenylated indole alkaloids constituted of bicyclo[2.2.2]diazaoctane ring systems is presented.
1. Introduction
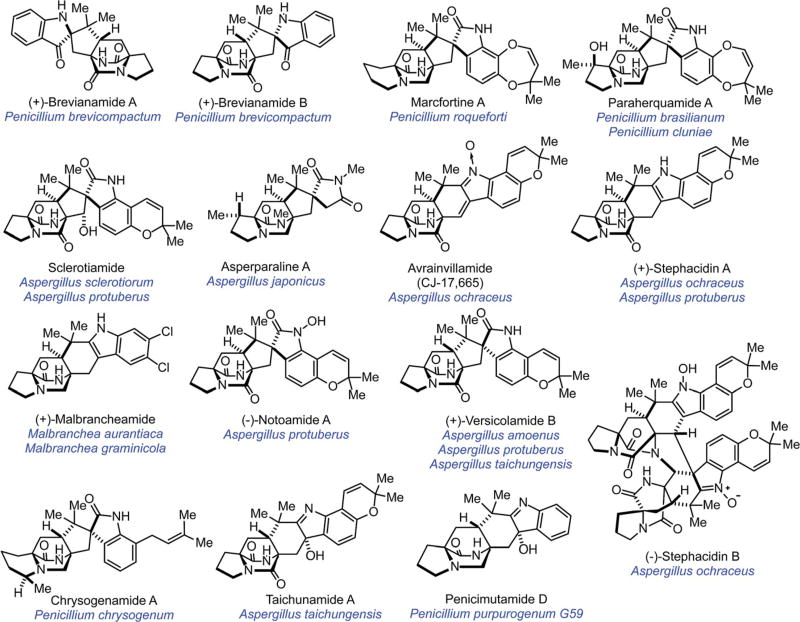
The isolation of natural products from marine and terrestrial-derived fungal species of the Aspergillus, Penicillium and related genera has allowed for a widely studied diverse class of secondary metabolites. In particular, various prenylated indole alkaloids have been discovered, including the brevianamides,1–7 marcfortines,8,9 paraherquamides,10–14 penicimutamides,15 sclerotiamide,16 asperparalines,17 avrainvillamide,18,19 stephacidins,20 malbrancheamides,21–23 notoamides,24–27 versicolamide B,28 chrysogenamide,29 and recently the taichunamides (Fig. 1).30 The presence of a bicyclo[2.2.2]diazaoctane ring system and densely functionalized indole-derived units form the bases for the structural allure of these naturally occurring metabolites. Many of these alkaloids have been found to show a variety of biological activity including insecticidal, cytotoxic, anthelmintic, calmodulin inhibition,31 osteoclast inhibition, and antibacterial properties.1–3,5,8–30,32–34 Reviews in this area have included the studies on the discovery of unexpected enantiomeric and diastereomeric relationships (Fig. 2) (see Section 2) as well as developments in the biosynthesis of these compounds (see Section 3), and biomimetic total syntheses (see Section 4).35–41 Herein, this review will focus on the biogenesis of the notoamides and their congeners, and provide the current understanding of the structural and stereochemical homologies and disparities that are particularly intriguing from a biosynthetic perspective.
Fig. 1.
Structures of representative prenylated indole alkaloids isolated from fungi of the genera Aspergillus, Penicillium, and Malbranchea.
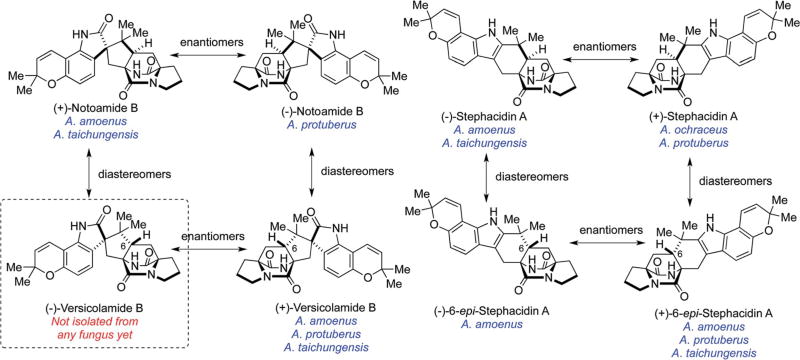
Fig. 2.
Structures of several prenylated indole alkaloids isolated from fungi and their stereochemical relationships.
Elucidation of the biogenesis of these compounds became of particular interest upon the isolation of antipodal metabolites from different species of the Aspergillus genus from both marine and terrestrial sources. Initial studies from the Williams laboratory42 were based on seminal work first reported by Sammes43 and Birch,44 and demonstrated that these natural metabolites arise from L-tryptophan, one or two isoprene units, and L-proline, β-methylproline, or pipecolic acid.
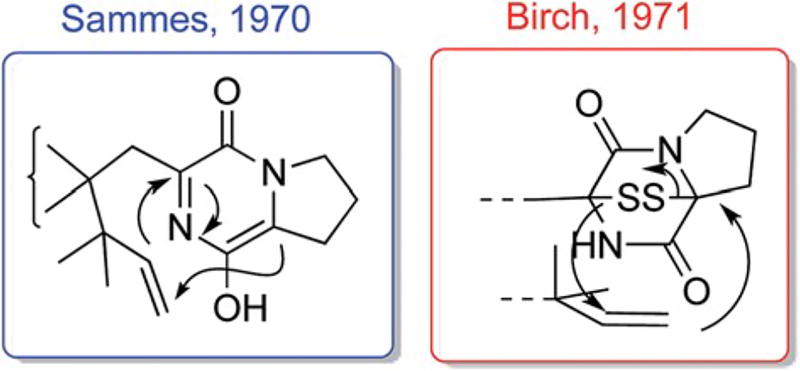
The first proposal concerning the biogenesis of the bicyclo [2.2.2]diazaoctane ring system was that of Sammes in 1970,43 wherein the intermediacy of an intramolecular hetero Diels–Alder construction through the agency of an azadiene species was proposed (left, Fig. 3). Shortly thereafter, Birch inferred in 1971 (ref. 45) the intermediacy of an epidithiapiperazinedione species, with net extrusion of S2 to form this ring system (right, Fig. 3), although the nature of the C–C bond formations as being heterolytic, radical, or stepwise were not further discussed.
Fig. 3.
Early suggestions of Sammes43 and Birch45 concerning the biogenesis of the bicyclo[2.2.2]diazaoctane ring system in the brevianamides.
At the time of the Sammes and Birch publications in the early 1970's, the absolute configuration of brevianamide B had not been rigorously assigned, but was thought to be of the same absolute sense as that of brevianamide A and being diastereomeric at the spiro-indoxyl stereogenic center. The Williams laboratory, in 1988,46 completed the first synthesis of (−)-brevianamide B and re-assigned the absolute configuration of natural (+)-brevianamide B, as depicted in Fig. 1.47,48 This revealed that brevianamides A and B have a pseudo-enantiomeric relationship (that is, the tricyclic portion constituted with the bicyclo[2.2.2]diazaoctane ring system are enantiomeric, but brevianamides A and B are technically diastereomers) which immediately raised provocative questions regarding the biogenesis of these natural substances in Penicillium brevicompactum. Williams suggested a modification to the intramolecular hetero Diels–Alder proposal, originally postulated by Sammes,43 as shown in Scheme 1.39,49,50 The differences between the Sammes and Williams biogenetic constructions involves the timing of the indole oxidation relative to the IMDA reaction. The Sammes proposal invokes a spiro-5 IMDA construction whereas the Williams alternative requires a spiro-6 IMDA. The Williams lab interrogated their alternative hypothesis through the synthesis and attempted precursor incorporation of DL-1. This potential metabolite was not incorporated into either brevianamide A nor brevianamide B in Penicillium brevicompactum,51,52 leaving the Sammes proposal as the most plausible. However, advances in confirming the assembly details of these fungal indole alkaloids were only made possible with identification of the notoamide gene cluster (see Section 3) as well as through labeled precursor incorporation studies of anticipated intermediates (see Section 2).
Scheme 1.
Alternative brevianamide biogenesis proposed by Williams.49
A deeper understanding of the enantio- and diastereo-divergence of the biosynthetic pathways exhibited by these closely related fungi, has led to the proposal of the functional presence of enzymes putatively mediating a Diels–Alder-type construction, an enzymatic function that is now receiving considerable attention from a number of laboratories investigating natural products biosynthetic pathways.53 Theoretical54,55 and experimental56–61 studies from our laboratories have provided corroborating evidence that an enzyme-mediated IMDA reaction is operative in the biosynthesis of the bicyclo [2.2.2]diazaoctane ring systems.38,53
2. Fungal metabolites and biosynthetic studies of prenylated bicyclo[2.2.2]diazaoctane indole alkaloids
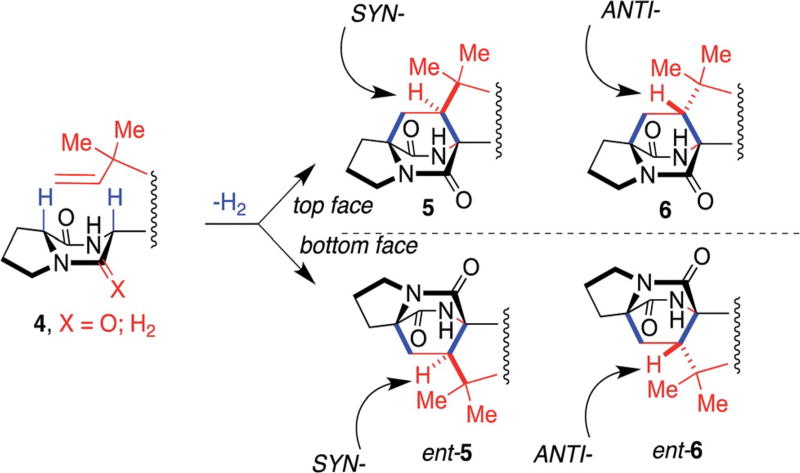
As to be discussed in detail below, there exists striking stereochemical similarities and differences between various members of this family of alkaloids for which biogenetic constructions need to accommodate. With respect to the construction of the bicyclo[2.2.2]diazaoctane ring system, the progenitor dioxopiperazine 4 (X = O) (or, for the paraherquamides, marcfortines, asperparalines, and malbrancheamides, the progenitor monooxopiperazine, 4, X = H2) suffers a net two-electron oxidation allowing the pendant isoprene-derived vinyl moiety to form the two new C–C bonds of the bicyclo[2.2.2]diazaoctane ring system. Four distinct diastereomeric transition states for this process can be operative resulting in the syn- (5) or anti-(6) diastereomeric relationships and a more subtle outcome being the manifestation of top-face or bottom-face cycloaddition as generically summarized in Fig. 4.
Fig. 4.
Stereochemical outcomes of the oxidative isoprene cyclization across the two amino acid a-carbons in the construction of the bicyclo [2.2.2]diazaoctane.62
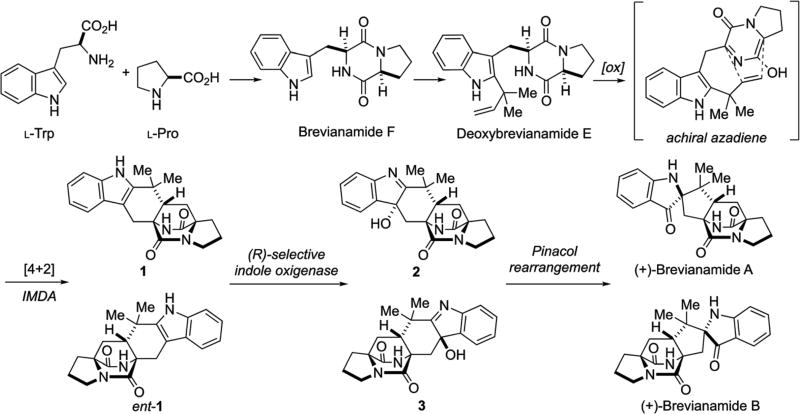
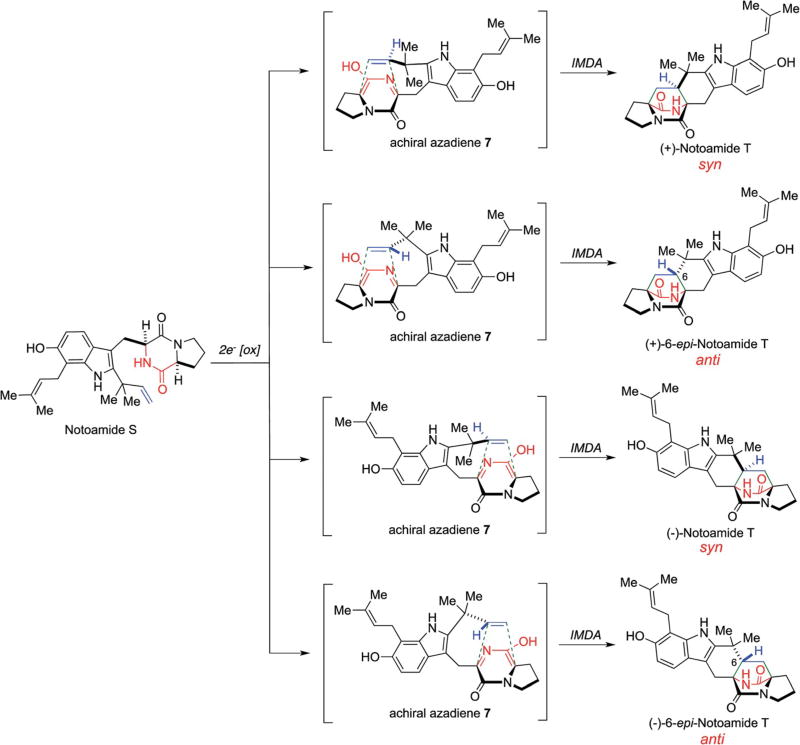
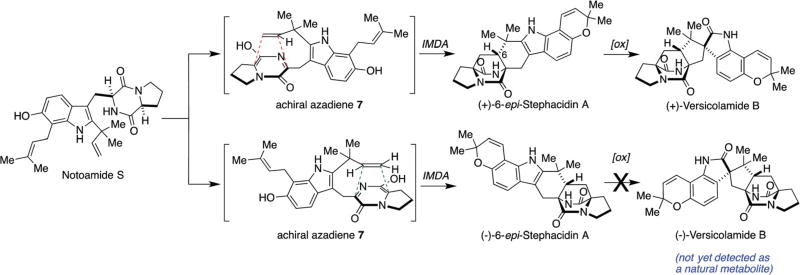
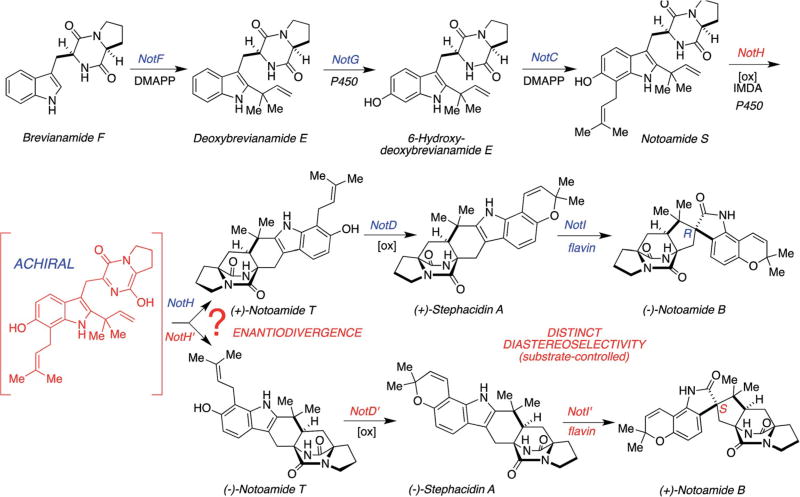
These stereochemical outcomes can be specifically identified in the known metabolic fates of notoamide S as illustrated in Scheme 2.61,63 Various metabolites have been identified, which were biosynthesized through both enantiomers of notoamide T and 6-epi-notoamide T.27,30,61,63–65 It has been suggested that all four of these substances arise via the intermediacy of common achiral azadiene species 7 (Scheme 2).
Scheme 2.
Putative IMDA reactions in the oxidative enzymatic transformation from notoamide S.61
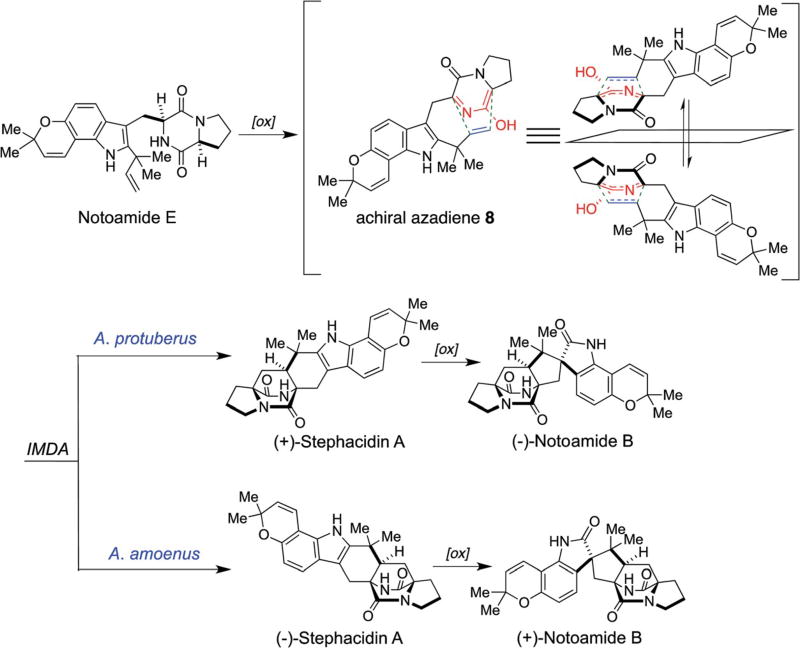
2.1 Isolation of prenylated indole alkaloids from the marine- derived fungus A. protuberus MF297-2 and the first proposal of the biogenesis of these alkaloids
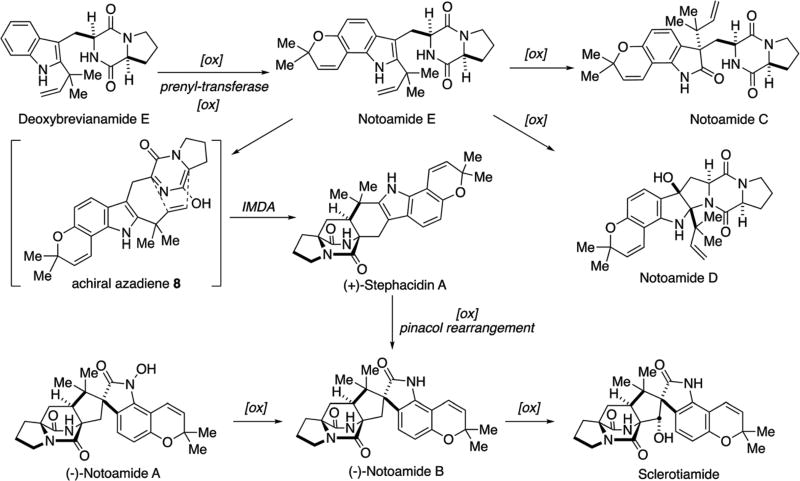
The marine-derived fungus A. protuberus MF297-2 was isolated from the mussel Mytilus edulis galloprovincialis, which was collected from the Noto peninsula (Japan) by the Tsukamoto laboratory. The fungal culture showed the activity of cell cycle inhibition in HeLa cells and the bioassay-guided purification afforded a fungal toxin, sterigmatocystin, as a cell cycle inhibitor along with seven prenylated indole alkaloids. Structure elucidation revealed that four of these were new alkaloids, notoamides A–D, and three were known alkaloids, sclerotiamide, stephacidin A, and deoxybrevianamide E.1,24 The structural relationship of these alkaloids led to the first proposal of the biosynthetic pathway illustrated in Scheme 3.66,67 Notoamide E was suggested to be a key precursor for this family, although it had not yet been isolated from the culture. The compounds containing a bicyclo [2.2.2]diazaoctane nucleus, namely stephacidin A, notoamides A and B, and sclerotiamide, were hypothesized to be produced from notoamide E through an achiral azadiene intermediate 8 by the IMDA reaction.
Scheme 3.
Structures of prenylated indole alkaloids isolated from A. protuberus MF297-2 and initial proposal regarding their possible biogenetic relationships.24,41,63,66–70
2.2 Precursor incorporation studies of notoamide E and structurally novel metabolites from A. protuberus MF297-2
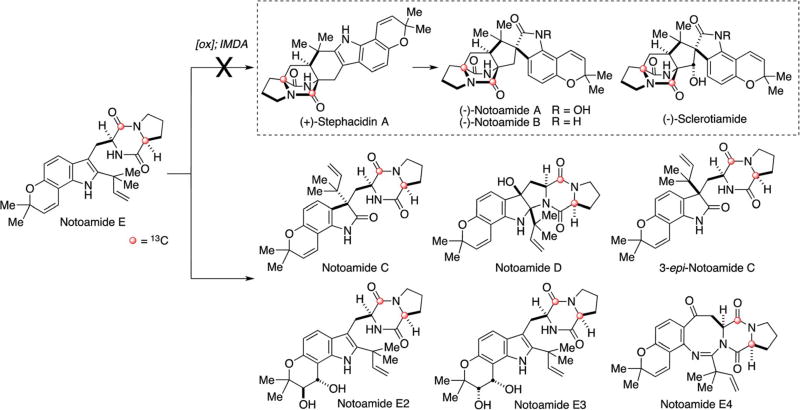
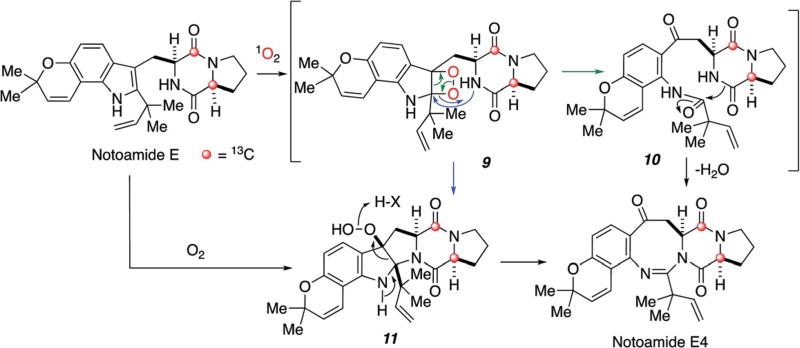
Since notoamide E was identified as a key intermediate in the biosynthesis of prenylated indole alkaloids in A. protuberus, Tsukamoto, et al. searched for notoamide E in the culture and found that it existed only in the fifth day of culture growth and immediately disappeared the next day.68,69 These results clearly indicated that notoamide E was produced by the fungus in the early phase of growth and was presumably rapidly converted to other downstream metabolites. Subsequently, precursor incorporation experiments with synthetic, [13C]2–notoamide E were carried out.68,71 Surprisingly, it was converted only to notoamides C and D as well as four new alkaloids, 3-epi-notoamides C and notoamides E2–E4, but not to the alkaloids containing a bicyclo[2.2.2]diazaoctane core, such as stephacidin A, notoamides A and B, nor sclerotiamide (Fig. 5).
Fig. 5.
Precursor incorporation results of [13C]2–notoamide E in A. protuberus MF297-2.68–71
Furthermore, in a precursor incorporation experiment, [13C]2–(+)-stephacidin A was, as expected, converted to (−)-notoamide B and (−)-sclerotiamide (Scheme 3).70 The four new alkaloids (3-epi-notoamide C and notoamides E2–E4) were not detected as metabolites from this organism in the normal nutrient-rich medium. Among these, the structure of notoamide E4 is particularly interesting, and two possible biosynthetic pathways from notoamide E can reasonably accommodate the biogenesis of these species (Scheme 4).68 One of the possible pathways would proceed via dioxetane intermediate 9, followed by fragmentation to the corresponding kynurenine derivative 10. Then, cyclization of 10 would afford the eight-membered ring derivative, notoamide E4. An alternative pathway is via peroxide species 11, which would be produced by proton-assisted nucleophilic attack of the indole 3-position on molecular oxygen. The results of this precursor incorporation experiment reveals that the addition of excess [13C]2–notoamide E (an endogenous metabolite) above the concentration levels of this metabolite normally produced, appears to alter the secondary metabolite profile of this organism. This provocative result warrants further investigation into how notoamide E, at elevated concentrations, must trigger the expression of dormant tailoring genes present that are not expressed under normal growth conditions.
Scheme 4.
Possible biosynthetic pathways to notoamide E4 from notoamide E.68
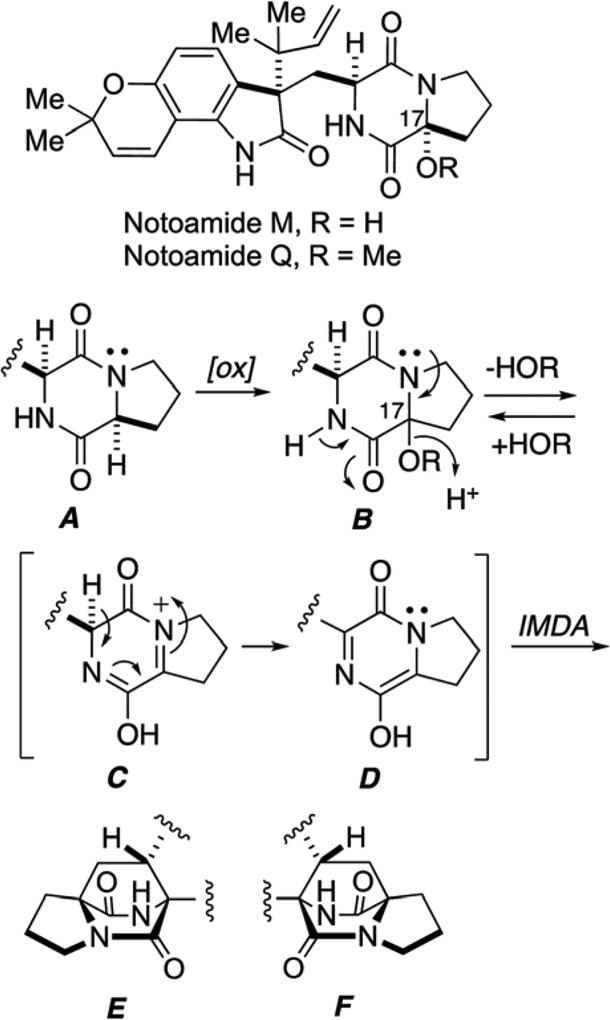
Tsukamoto, et al. succeeded in the isolation of notoamides M26 and Q,27 which have hydroxy and methoxyl groups at C-17 positions, respectively (Scheme 5). These metabolites represent the oxidation state of the putative azadiene species that have been strongly implicated in the construction of the bicyclo [2.2.2]diazaoctane ring system. Thus, it is possible that formation of azadiene intermediate (D), proceeds through oxidation at C-17 of dioxopiperazine A to B, followed by loss of water or methanol and tautomerization to D (Scheme 5). Alternatively, notoamides M and Q, might be artifacts of hydration/capture of azadiene species (C) and experiments to interrogate this possibility are being investigated.
Scheme 5.
Structures of notoamides M and Q and possible formation of the bicyclo[2.2.2]diazaoctane ring system, theoretically through notoamides M and Q.26,27,65
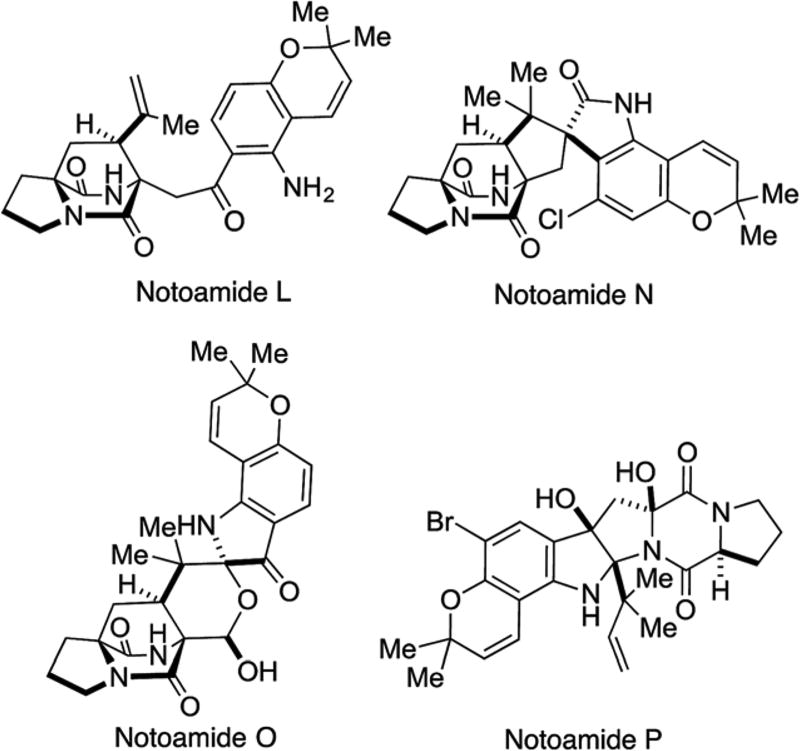
Tsukamoto, et al. isolated eighteen new prenylated indole alkaloids, notoamides A–R,24–27 from the marine fungus A. protuberus. Among these, carbon frameworks of notoamides L26 and O27 were novel in the family of prenylated indole alkaloids and notoamides N26 and P27 contained chlorine and bromine atoms, respectively (Fig. 6). Notoamide L is the first metabolite containing twenty-five carbons in this family, while all other alkaloids possess twenty-six carbons. In the structure of notoamide L, the C-2 carbon derived from tryptophan has been removed.
Fig. 6.
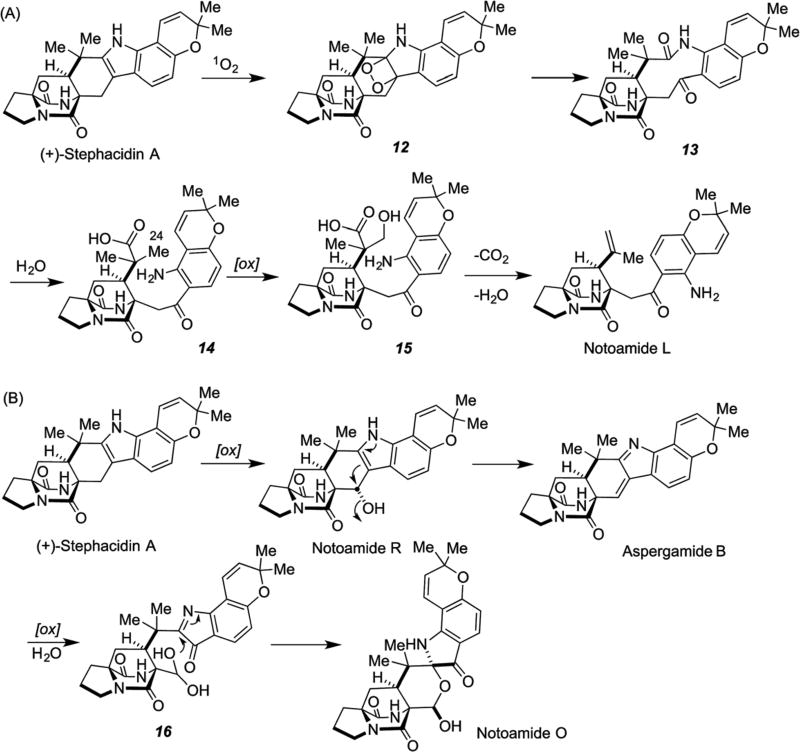
Notoamide O possesses a novel hemiacetal/hemiaminal ether functionality hitherto unknown among this family. Possibly, notoamides L and O may be derived from (+)-stephacidin A (Scheme 6). In the biogenesis of notoamide L, singlet oxygen reaction at the indole 2,3-position of (+)-stephacidin A would afford a dioxetane intermediate 12 followed by fragmentation to kynurenine 13. After hydrolysis of the amide bond of 13, oxidation at C-24 followed by decarboxylative dehydration would afford notoamide L.26 On the other hand, stephacidin A may be converted to aspergamide B72 via notoamide R. Oxidative cleavage of the tri-substituted alkene in aspergamide B would generate aldehyde hydrate 16; simple ring closure would afford notoamide O.27 Notoamide N is one of the rare chlorinated members of the family of prenylated indole alkaloids and this is the third chlorinated derivative following malbrancheamide,21 malbrancheamide B22 and spiromalbramide.23 On the other hand notoamide P is the first brominated member of this family.27,65 The existence of a halogenase system in these fungi is of interest and the identification of the genes responsible for the halogenations merits further investigation.
Scheme 6.
Possible biosynthetic pathways to notoamides L (A) and O (B).27
2.3 Isolation of enantiomers of stephacidin A and notoamide B from the terrestrial fungus Aspergillus amoenus NRRL 35660
The Gloer laboratory originally isolated (−)-stephacidin A and (+)-notoamide B from the terrestrial fungus A. amoenus (formerly A. versicolor) NRRL 35660, which was isolated from a basidioma of Ganoderma australe collected in a Hawaiian forest, along with a new alkaloid, (+)-versicolamide B (Fig. 2).28 (+)-Stephacidin A was originally isolated from the terrestrial fungus A. ochraceus by Bristol-Myers Squibb20 and subsequently, (+)-stephacidin A and (−)-notoamide B were isolated from the marine-derived fungus A. protuberus MF297-2, by the Tsukamoto laboratory.24 These facts reveal that these closely related Aspergillus fungi biosynthesize the opposite enantiomers of stephacidin A and notoamide B and it has been confirmed that the optical purity of these metabolites are >99%. The initial hypothesis for rationalizing how these opposite enantiomers might arise, is illustrated in Scheme 7 and invokes the oxidation of notoamide E, to the achiral azadiene species 8, followed by an entirely enantio-selective cycloaddition from this common precursor.28 Although [13C]2–stephacidin A was converted to [13C]2–notoamide B,70 as discussed above, the incorporation of [13C]2–notoamide E into stephacidin A and notoamide B, however, was not observed in A. amoenus73 nor A. protuberus.68 These results suggested alternate timing of the pyran and bicyclo[2.2.2]diazaoctane ring constructions as discussed below.
Scheme 7.
Initial proposal for the enantiodivergent biogenesis of enantiomeric prenylated indole alkaloids with a bicyclo[2.2.2]diazaoctane core in A. protuberus MF297-2 and A. amoenus NRRL 35660.28,70
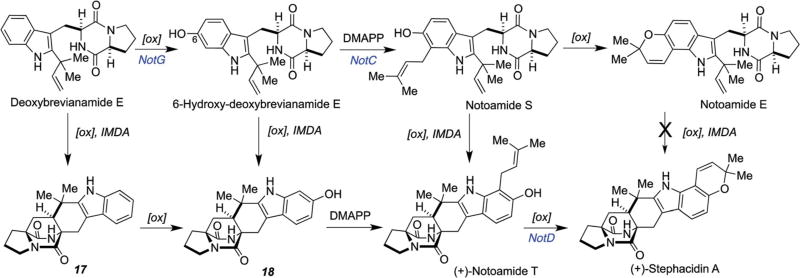
2.4 An alternative enantiodivergent biosynthetic pathway for stephacidin A biogenesis
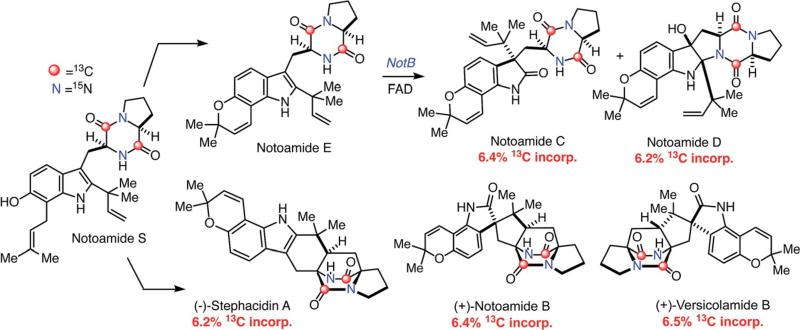
Since [13C]2–notoamide E was not incorporated into stephacidin A in either A. protuberus MF297-2 nor A. amoenus NRRL 35660, other testable hypotheses needed to be devised. The pathway from deoxybrevianamide E to stephacidin A contains four steps: the putative IMDA, an oxidation at C-6, prenylation, and final oxidation/cyclization to form a pyran ring (Scheme 8). From the results of the precursor incorporation experiments with notoamide E, three compounds, namely deoxybrevianamide E, 6-hydroxy-deoxybrevianamide E, and notoamide S, appeared to be potential precursors for IMDA reactions, although notoamide S had not yet been isolated from any species of the Aspergillus fungi.74 The incorporation experiments of synthetic [13C]2–[15N]–deoxybrevianamide E75 and [13C]2–[15N]–6-hydroxydeoxybrevianamide E75,76 afforded notoamides C and D but not stephacidin A nor notoamides A and B. Next, the administration of synthetic [13C]2–[15N]2–notoamide S in A. amoenus was examined and it was found to be incorporated into (−)-stephacidin A (6.2% incorporation), (+)-notoamide B (6.4% incorporation), and (+)-versicolamide B (6.5% incorporation), together with notoamides C (6.4% incorporation) and D (6.2% incorporation) (Fig. 7).77 These results clearly indicated that the construction of the bicyclo[2.2.2]diazaoctane ring system occurs prior to the formation of the pyran ring. Although notoamides S and T had not previously been found in the culture extract of A. amoenus, Kato, et al., have recently identified notoamide S as a minor metabolite of A. amoenus.61
Scheme 8.
Possible biosynthetic pathways from deoxybrevianamide E to stephacidin A and potential precursors for IMDA reactions.74–76
Fig. 7.
Results of the incorporation experiment of [13C]2–[15N]2–notoamide S in A. amoenus NRRL 35660.77
2.5 The incorporation of notoamide T and 6-epi-notoamide T
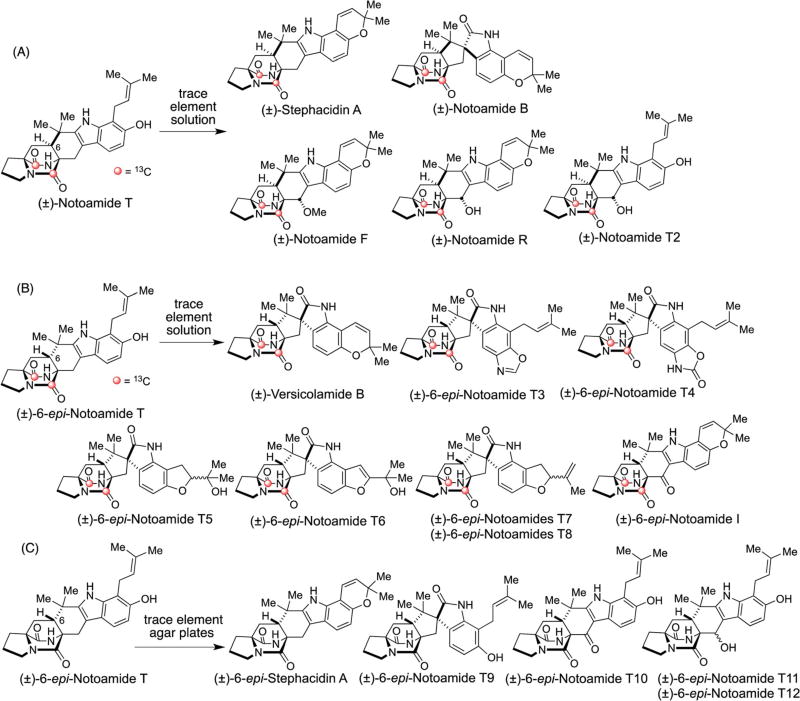
Since notoamide S was observed to be converted to compounds containing a bicyclo[2.2.2]diazaoctane core via notoamide T and 6-epi-notoamide T (Scheme 2), the precursor incorporation experiments of these two alkaloids were performed. Unexpectedly, the administration of synthetic [13C]2–(±)-notoamide T in A. protuberus aff orded racemic mixtures of stephacidin A, notoamide B, notoamide F, notoamide R, and a new compound, notoamide T2 (Fig. 8A).63 On the other hand, synthetic [13C]2–6-epi-(±)-notoamide T was converted to racemic mixtures of versicolamide B and eight new compounds, 6-epi-notoamides T3–T8 and 6-epi-notoamide I, in a trace element solution (Fig. 8B) and afforded racemic mixtures of 6-epi-stephacidin A and four new compounds, 6-epi-notoamides T9–T12, on trace element agar plates (Fig. 8C).64
Fig. 8.
Results of the incorporation experiments of [13C]2–(±)-notoamide T in a trace element solution (A), [13C]2–(±)-6-epi-notoamide T in a trace element solution (B), and (±)-6-epi-notoamide T on trace element agar plates (C) in A. protuberus MF297-2.63,64
Among them, notoamide T2, 6-epi-notoamides T3–T12, and 6-epi-notoamide I were not produced by the fungus under normal culture conditions to any detectable extent.63,64 These data suggest that the (presumably) endogenous, natural metabolites [13C]2–(±)-notoamide T and [13C]2–(±)-6-epi-notoamide T activated the expression of dormant tailoring genes that expands the metabolome of the producing organism. While notoamide T and 6-epi-notoamide T are presumed to be natural, endogenous metabolites in this organism, the precursor incorporation experiment with the easily accessible synthetic, racemic labeled substances was performed. In order to further interrogate this fascinating observation, sufficient quantities of the respective optically pure enantiomers of each will be required to query whether the metabolite profile alteration is triggered by the presence of the unnatural enantiomer for this producing organism.
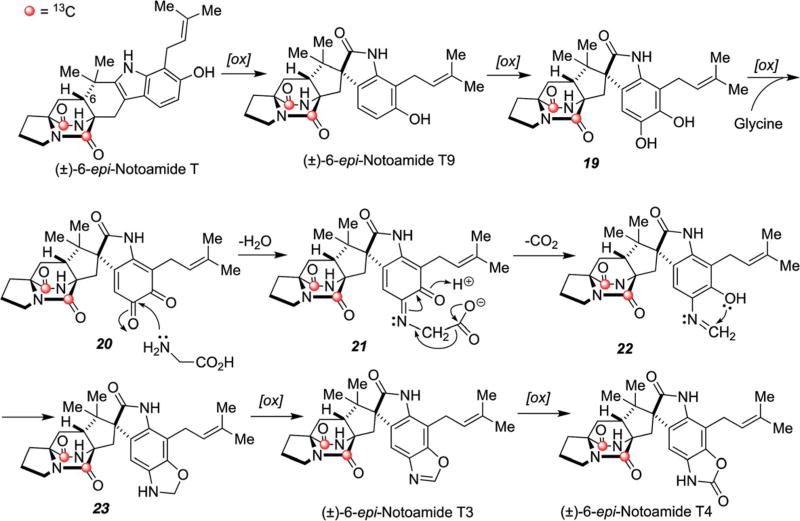
Of particular intrigue, 6-epi-notoamides T3 and T4 contain unprecedented structures wherein the aromatic benzenoid ring of the tryptophan moiety has suffered oxidative amination, culminating in the formation of the new fused oxazole and 2-oxazolone moieties, respectively. One possible mechanism for the oxidative amination of the tryptophan moiety is suggested in Scheme 9. Oxidation and subsequent pinacol rearrangement of [13C]2–(±)-6-epi-notoamide T would form the spiro-oxindole, [13C]2–(±)-6-epi-notoamide T9. Successive oxidation and incorporation of glycine into 20 to form imine 21, followed by decarboxylation, provides 22 that must cyclize (to 23) and then be oxidized to the oxazoline moiety of T3. Oxidation of the oxazoline of T3 would produce oxazolidinone in 6-epi-notoamide T4.64 It is reasonable to assume that the initial oxidation of the 2,3-disubstituted indole of 6-epi-notoamide T is conducted by NotI that Li, et al., previously predicted this oxidation and spiro-rearrangement.62 The downstream transformations from 6-epi-notoamide T9, requires a minimum of six enzyme-catalyzed reactions: (1) aromatic hydroxylase of T9 to 19; (2) a catechol oxidase to 20; (3) glycine aminotransferase to 21; (4) decarboxylase/cyclase to 23; (5) oxazolidine oxidase to T3; and (6) oxazolidinone oxidase to T4. The complex array of transformations required to construct T3 and T4 from 6-epi-notoamide T mandates that multiple dormant tailoring genes extant in this organism were activated by the presence of one (or both) enantiomers of 6-epi-notoamide T. Significant effort will need to be devoted to elucidating the nature of these remarkable transformations that have significant implications for the plasticity of secondary metabolomes in Nature.
Scheme 9.
Possible biosynthetic pathways to 6-epi-notoamide T3 and 6-epi-notoamide T4 from 6-epi-notoamide T.62,64
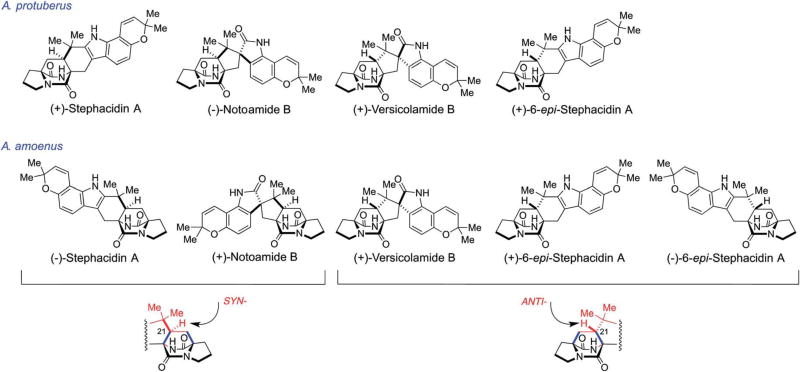
2.6 Isolation of an enantiomeric mixture of 6-epi- stephacidin A from A. amoenus NRRL 35660
The metabolic profiles of A. protuberus MF297-2 and A. amoenus. NRRL 35660 produced the opposite enantiomers of stephacidin A and notoamide B along with the same enantiomer of (+)-versicolamide B. (−)-Versicolamide B has yet to be detected as a natural metabolite in any of the fungi that have been examined to date and poses an enigma (Fig. 2). (+)-6-epi-Stephacidin A is the likely precursor of (+)-versicolamide B and was obtained from the culture of A. protuberus (Scheme 10).64 However, 6-epi-stephacidin A, which was isolated from A. amoenus, was found to be an enantiomeric mixture enriched with the (−)-isomer (1 : 2.4).61 This result clearly suggested that, in A. amoenus, notoamide S was converted to both (+)- and (−)-6-epi-stephacidin A and subsequently, only the (+)-isomer was converted into (+)-versicolamide B (Scheme 10). This result mandates that A. amoenus contains a highly enantio-specific indole oxidase that exclusively transforms the (+)-enantiomer of 6-epi-stephacidin A to (+)-versicolamide B, but apparently does not bind or oxidize the (−)-enantiomer of 6-epi-stephacidin A. While the formation of an enantiomerically-enriched metabolite has not yet been observed in any of the other fungal species examined, this begs the question of why this occurs in A. amoenus. The lack of optical purity of 6-epi-stephacidin A obtained in A. amoenus also requires an explanation and it is to such questions, that our labs have recently started investigations.
Scheme 10.
Possible biosynthetic pathways of an enantiomeric mixture of 6-epi-stephacidin A and (+)-versicoclamide B in A. amoenus NRRL 35660.61,64
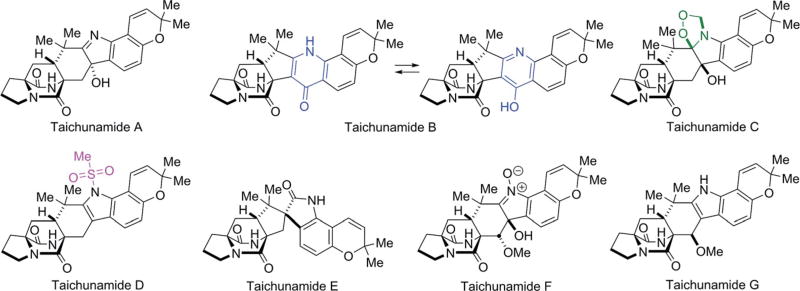
2.7 Isolation of taichunamides A–G from Aspergillus taichungensis IBT 19404: biogenetic considerations
The majority of the prenylated indole alkaloids thus far identified containing the bicyclo[2.2.2]diazaoctane ring system possess the syn-configuration (the syn- and anti-relationship is based on the relative configuration of H21 with respect to the bridging secondary amide; see Fig. 4 and 9). Distinct enantiomers of notoamide B and stephacidin A are the main metabolites present in A. protuberus and A. amoenus, and (+)-versicolamide B, a minor metabolite, was the first alkaloid identfied within this family with the corresponding anti-configuration. In 2013, Cai et al. isolated (+)-versicolamide B and (+)-6-epi-stephacidin A as major metabolites from A. taichungensis ZHN-7-07.78 Kagiyama, et al., have also investigated the isolation of alkaloids from A. taichungensis IBT 19404 collected from the soil in Taiwan and succeeded in isolating seven indole alkaloids with unprecedented structures and have named these substances taichunamides A–G (Fig. 10); all of these natural alkaloids possess the anti-bicyclo[2.2.2]diazaoctane core.30 Although the structure of taichunamide A was initially assigned to contain a very rare azetidine unit, recently it was revised as a 3-hydroxyindolenine.79 Taichunamide B was found to be constituted with a very rare 4-pyridone unit. In DMSO-d6 taichunamide B appears to be an equilibrium mixture of two tautomeric entities: a 4-pyridone and 4-pyridol, in a ratio of 3 : 1. This tautomeric equilibrium is highly solvent-dependent where it was observed a single keto or enol form was observed in CD3OD or acetone-d6, respectively. Taichunamides C and D contain endoperoxide and methylsulfonyl units, respectively.
Fig. 9.
Prenylated indole alkaloids illustrating the syn- and anti-diastereomeric relative configurations isolated from A. protuberus MF297-2 and A. amoenus NRL 35660.26,28,30,61
Fig. 10.
Structures of the taichunamides isolated from A. taichungensis IBT19404.30
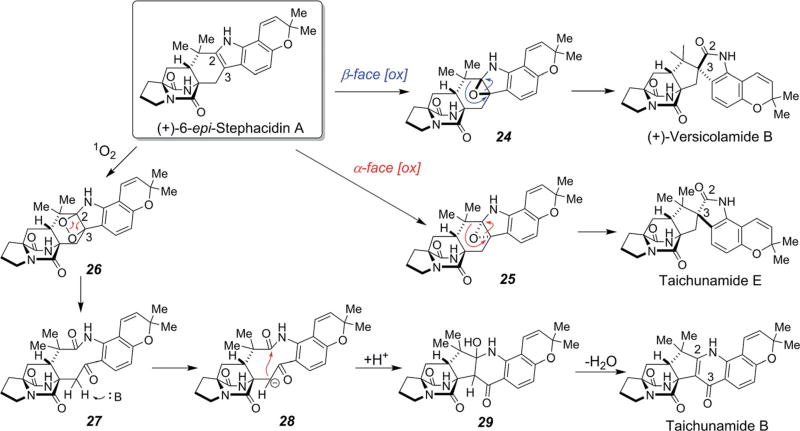
These new alkaloids, especially taichunamide B, constitute hitherto unknown structural arrays derived from tailoring of the tryptophan moiety and therefore must deploy a series of fascinating biochemical bond constructions. Biogenetic implications for taichunamides B and E as well as (+)-versicolamide B are represented in Scheme 11.30 β-Face oxidation (anti- to the bridged amide) of 6-epi-stephacidin A allows for the formation of a non-isolatable epoxide intermediate 24 which subsequently undergoes a pinacol rearrangement within the active site to form the spirooxindole in (+)-versicolamide B. Oxidation at the α-face (syn to the bridged amide) followed by pinacol rearrangement would allow for formation of taichunamide E. Compared with β-face oxidation, α-face oxidation is more sterically demanding with N19, as reflected by the metabolite ratios of (+)-versicolamide B (15.9 mg)/(+)-versicolamide C30,80 (a N1-OH derivative of versicolamide B)81 (240 mg) versus taichunamide E (0.43 mg). Although, in the biomimetic synthesis that deployed the IMDA reaction, taichunamide E was identified along with (+)-versicolamide B in a ratio of 1 : 1.4; this is the first report of its isolation from the fungal culture.30 Also, formation of peroxide 26 by oxidation of (+)-6-epi-stephacidin A would ultimately lead to the ring system found in taichunamide B, theoretically through compound 27.
Scheme 11.
Possible biogenesis of taichunamides B and E and versicolamide B from 6-epi-stephacidin A.30
A wide structural array of prenylated indole alkaloids have been isolated from the genera of Aspergillus and Penicillium to date, yet their respective carbon frameworks are very unique. The isolation of these compounds from A. taichungensis has greatly expanded the library of these secondary metabolites. Future biochemical studies and genome mining will aid in the identification of genes or gene clusters responsible for the evolution of these unprecedented metabolites.
2.8 Biogenetic implications of prenylated indole alkaloids in the Aspergillus genus
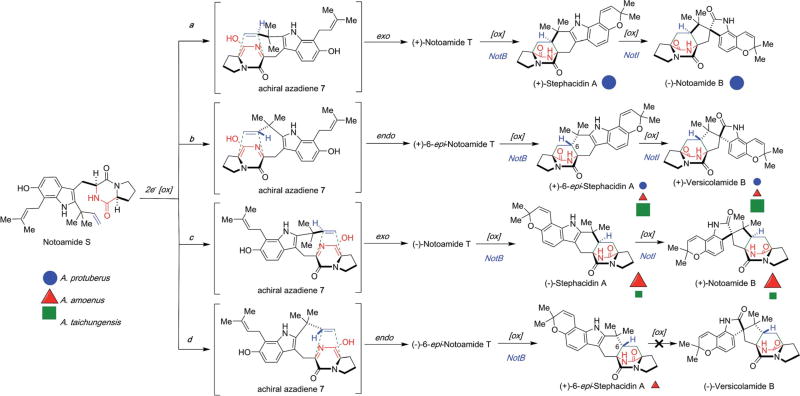
To date, numerous fungal species in the Aspergillus genus, including A. ochraceus, A. protuberus MF297-2, A. amoenus NRRL 35660, and A. taichungensis IBT 19404, described in this article that produce prenylated indole alkaloids, bearing the dioxopiperazine-based bicyclo[2.2.2]diazaoctane core most likely biosynthesized by the oxidative cycloaddition reaction of the common precursor, notoamide S (Scheme 12), have been identified. Each organism displays its own unique stereochemical signature wherein the absolute and relative stereochemistry within these molecular frameworks have been constructed are evident. In A. protuberus, the orientation of the dienophile to the diene in pathways a (main) and b (minor), respectively, leads to the construction of the corresponding exo-and endo-products namely, (+)-stephacidin A/(−)-notoamide B and (+)-6-epi-stephacidin A/(+)-versicolamide B, respectively. On the other hand, in A. taichungensis, the construction of the bicyclo[2.2.2]diazaoctane ring system favors the formation of the C-6-epi-diastereomeric via pathway b (main) and c (minor).30 Efforts to clarify the genetic and biochemical bases and relationships between the respective biosynthetic gene clusters are currently under intense investigation in our laboratories.
Scheme 12.
Proposed facial specificities of IMDA reactions for metabolites in A. protuberus MF297-2 (blue circles), A. amoenus NRRL 35660 (red triangles), and A. taichungensis IBT 19404 (green squares). Major and minor metabolites in each fungus are represented with large and small symbols, respectively.30
3. Genomic insights
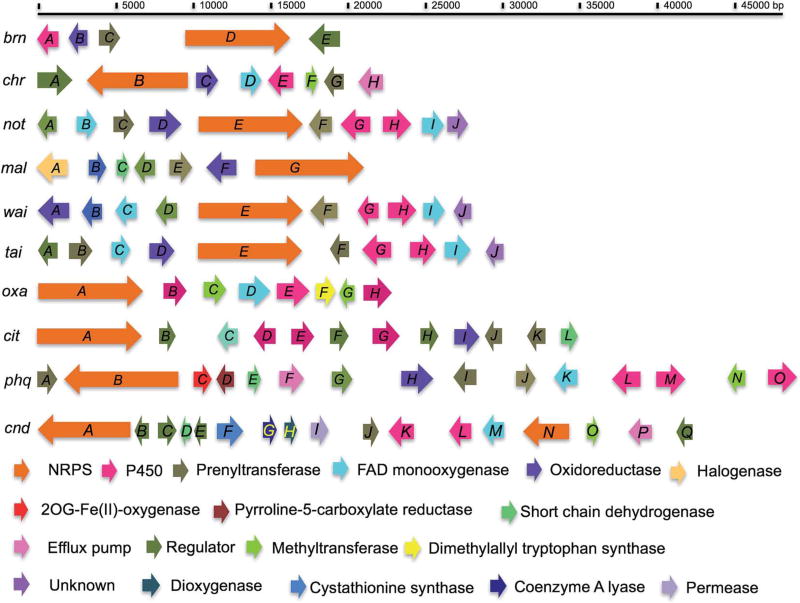
In order to obtain a deeper understanding of the biogeneses of these structurally fascinating natural alkaloids, and in particular, to obtain a higher level elucidation of the enantioselectivity and diastereoselectivity evident in the biogenesis of the stephacidins and notoamides, the genomes of ten fungi known to produce these alkaloids have been sequenced and the identification of relevant biosynthetic gene clusters (Fig. 11) has been realized. The mining of ftmA, a NRPS gene from the fumitremorgin Aspergillus species, A. fumigatus, and production of brevianamide F by heterologous gene expression in 2006 by Turner et al.,82 allowed Ding, et al., to identify the notoamide gene cluster in A. protuberus.83 Due to the crucial role brevianamide F plays in the early biogenesis of an enormous range of prenylated indole alkaloids within the Aspergillus genus, it was originally assumed that ftmA, or a comparable homolog, would also participate in biosynthetic transformations of A. protuberus. Using Roche 454FLX technology and ftmA to probe for homologous genes, notE was identified as the NRPS module analogous to ftmA, sharing a 47% nucleotide identity, in the A. protuberus species.83 Fig. 12 represents the (+)- and (−)-notoamide-producing gene clusters identified by genome sequencing and bioinformatics.83
Fig. 11.
Mining and annotation of biosynthetic gene clusters of fungal indole alkaloids. Brevianamide (Aspergillus versicolor MF030, brn); chrysogenamide (Penicillium. alligativum, chr); notoamide A (A. protuberus MF297-2, not); malbrancheamide (Malbranchea aurantiaca RRC1813, mal); waikialoid A (Aspergillus sp., wai); 3-epi-notoamide C (A. taichungensis IBT 19404, tai); oxaline (P. oxalicum F30, oxa); citrinalin (P. citrinum F53, cit); parahequamide A (P. fellutanum ATCC20841, phq); citrinadins (P. citrinum IBT 29821, cnd).62,84
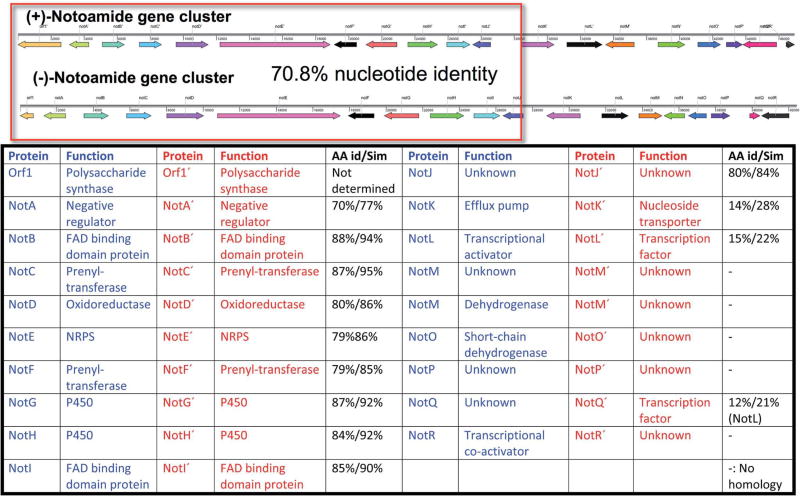
Fig. 12.
Comparison of the biosynthetic gene clusters of the (+)-notoamide (Aspergillus amoenus) and ( )-notoamide (Aspergillus protuberus) producing fungi.83
Bioinformatics analysis83 and deep annotation, has revealed a number of important facts and relationships between A. protuberus and A. amoenus that produce the opposite enantiomers of stephacidin A and notoamide B. First, it is clear from the structure and directionality of the open-reading frames of genes within the gene clusters, that the gene clusters and thus the organisms are orthologous (ancestrally related through a speciation event that occurred at some time during their evolutionary history). The high level of nucleotide identity overall between the gene clusters is almost 71% and within individual genes, the homology with respect to amino acid identity or similarity, is as high as 95% (for example NotC). Functions have been assigned to a number of genes within these orthologous gene clusters and are identified in Fig. 12, Scheme 13, and Table 1 which reflects our current understanding and hypotheses regarding the biogenesis of the enantiomeric pairs.63,83
Scheme 13.
Current understanding of (+)- and (−)-notoamide biogenesis in Aspergillus sp.63
Table 1.
Select notoamide biosynthetic enzymes
| Enzyme | bp/aa | Heterologous host | Function | Substrate |
|---|---|---|---|---|
|
| ||||
| NotB | 1563/456 | E. coli | FAD binding domain protein | Notoamide E |
| NotB´ | 1344/455 | - | FAD binding domain protein | Notoamide E |
| NotC | 1350/427 | E. coli | Prenyl transferase | 6-Hydroxydeoxybrevianamide E |
| NotD | 2025/621 | A. oryzae | Oxidoreductase | (+)-Notoamide T |
| NotD´ | 2025/612 | - | Oxidoreductase | (−)-Notoamide T |
| NotE | 6723/2241 | - | NRPS module | Pro + Trp |
| NotF | 1431/453 | E. coli | Reverse prenyl transferase | Brevianamide F |
| NotG | 1901/544 | - | Cytochrome P450 | Deoxybrevianamide E |
| NotH | 1836/502 | - | Cytochrome P450 | Notoamide S |
| NotH´ | 1836/499 | - | Cytochrome P450 | Notoamide S |
| NotI | 1423/434 | E. coli | FAD binding domain protein | (+)-Stephacidin A |
| NotI´ | 1423/433 | E. coli | FAD binding domain protein | (−)-Stephacidin A |
Li, et al., have successfully been able to clone and functionally express the following enzymes: NotF, NotG, NotC, NotD, NotD´, NotI and NotI´.62,83 Table 1 provides a summary of the relevant data on each individual gene and the heterologous host in which the protein was functionally expressed.
Li, et al., have tentatively assigned roles to other genes in the notoamide gene clusters as follows. NotG appears to be a cytochrome P450 oxidase responsible for the transformation of deoxybrevianamide E into 6-hydroxydeoxybrevianamide E, which was deduced through a process of elimination by comparative analysis with several fungal gene clusters.62,83 The other cytochrome P450, NotH is the current most plausible candidate for the oxidative transformation of notoamide S into notoamide T and 6-epi-notoamide T.83 Much to our chagrin, NotH and NotH´, which is currently the focus of where the key enantiodivergence occurs, is a membrane-associated cytochrome P450 that has proven recalcitrant to functional expression in a heterologous host and constitutes the focus of our current efforts.
Biochemical characterization, along with isotopically labeled precursor incorporation studies (vide supra), has determined the roles of NotF and NotC as aromatic prenyltransferases.83 Michaelis–Menten kinetics and isotopically labeled precursor incorporation studies have successfully demonstrated the substrate selectivity of the NotF and NotC enzymes. The reverse-prenylation of brevianamide F into deoxybrevianamide E, the first committed step of the post-NRPS tailoring cascade, is catalyzed by NotF. The subsequent normal prenylation of 6-hydroxydeoxybrevianamide E into notoamide S is catalyzed specifically by NotC.83
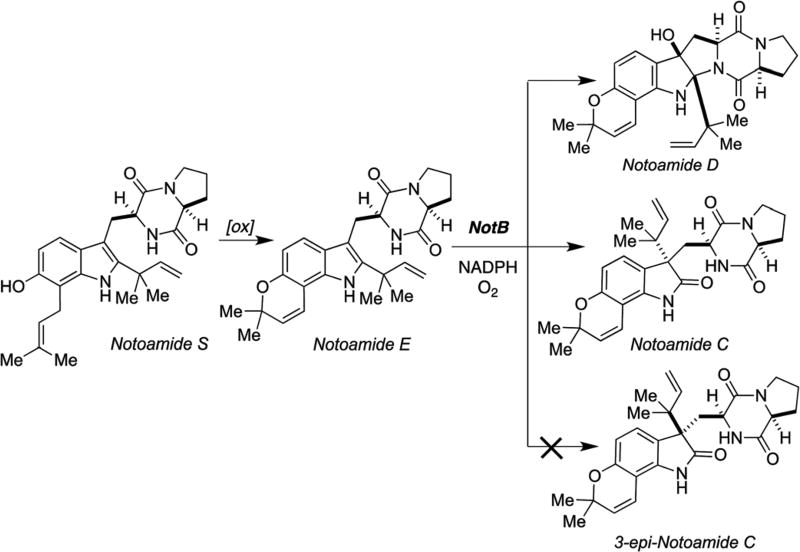
NotB and NotI are flavin-dependent monooxygenases (FMO) Biochemical characterization of NotB has determined its role as an FAD-dependent oxidase for converting notoamide E into notoamides C and D (Scheme 14).77 Curiously, 3-epi-notoamide C is not observed as a product when notoamide E is exposed to recombinant NotB. 3-epi-Notoamide C has been previously isolated from a precursor incorporation experiment with notoamide E as an added substrate, but has not been directly isolated from fermentation extracts of Aspergillus protuberus.68 NotI and NotI´ which are also flavin-dependent oxygenases, are predicted to catalyze the oxidative spiro-transformation of stephacidin A into notoamide B.83 The details of how NotI and NotI´ discriminate between the respective enantiomers of stephacidin A and the conversion into the corresponding enantiomers of notoamide B is of great interest, but has not yet been reported.
Scheme 14.
Role of NotB in the conversion of notoamide E into notoamides C and D.77
4. Biomimetic total syntheses
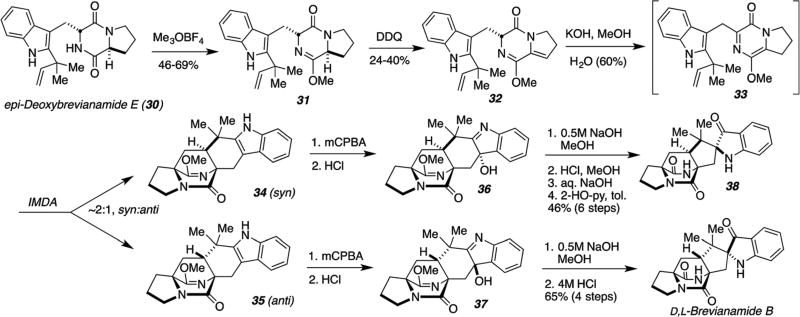
With the objective of interrogating the validity of the proposed biosynthetic intramolecular hetero Diels–Alder construction of the characteristic bicyclo[2.2.2]diazaoctane ring nucleus, Williams, et al., have investigated the laboratory version of the IMDA construction which successfully enabled biomimetic syntheses of numerous members of these natural alkaloids. These investigations provided additional insights as to the intrinsic facial bias of this cycloaddition with respect to the syn-/anti-diastereoselectivity.56–60 Theoretical calculations on these azadiene IMDA reactions published by our collaborator Domingo et al.,54,55 provide an important backdrop to the laboratory and putative biosynthetic constructions. The strategies for generating the key azadiene species evolved to ever more efficient protocols over the years and are shown here chronologically. Our first successful biomimetic synthesis was that of D,L-brevianamide B, as shown in Scheme 15, where DDQ oxidation of lactam ether 31 was employed to provide the unsaturated proline derivative 32.85 Base-mediated tautomerization generated the desired azadiene species, that proved stable enough to observe by 1H NMR, but spontaneously suffered IMDA cycloaddition to provide an approximately 2 : 1 syn : anti mixture of cycloadducts 34 and 35, respectively. Peracid oxidation to the corresponding diastereochemically pure 3-hydroxyindolenines (36 and 37) followed by base-mediated pinacol-type spiro-rearrangement and final acidic removal of the lactim ether furnished D,L-brevianamide B from 37 and the corresponding syn-diastereomer 38.85
Scheme 15.
First-generation biomimetic total synthesis of D,L-brevianamide B.85
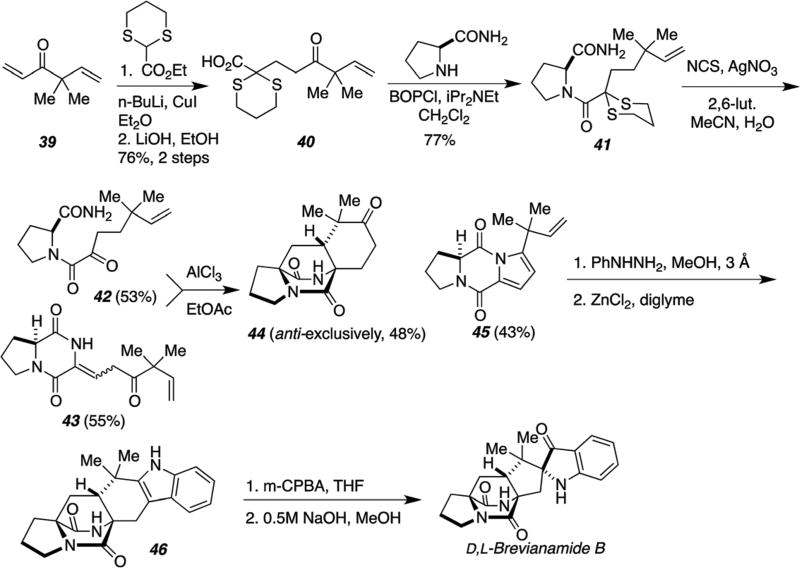
A second-generation synthesis, detailed in Scheme 16, generated the azadiene directly from an α-ketoamide proline amide species (i.e., 42).86 The IMDA reaction, carried out under strongly Lewis acidic conditions, interestingly provided the desired anti-cycloadduct 44, exclusively. Fisher indole synthesis to 46 and final peracid oxidation to the indoxyl provided D,L-brevianamide B.86
Scheme 16.
Second generation biomimetic total synthesis of D,L-brevianamide B.86
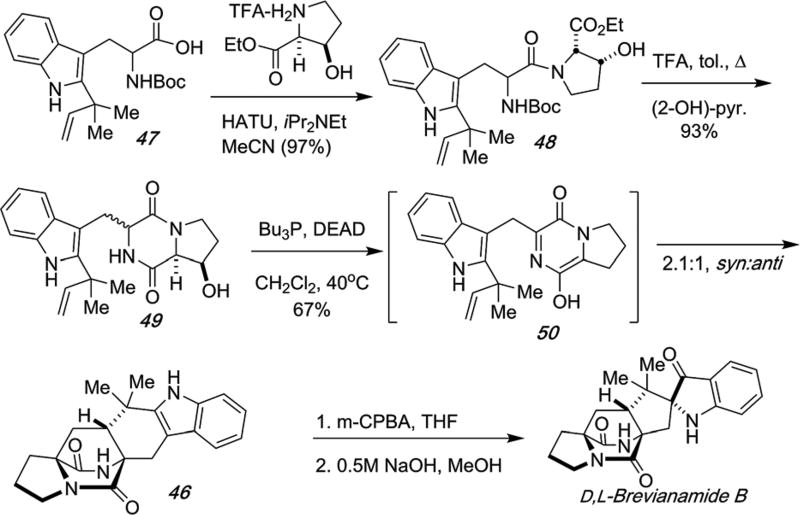
The most efficient biomimetic synthesis of brevianamide B that has been developed and reported to date, relied on using β-hydroxyproline as a surrogate representing the oxidation state of the azadiene, as shown in Scheme 17.60,85 Reverse-prenylated tryptophan species 47 was coupled with β-hydroxyproline ethyl ester to give dipeptide 48 that was deprotected and cyclized to 49. Mitsunobu dehydration of 49 in dichloromethane at 40 °C led directly to the production of meta-stable azadiene species 50 that spontaneously underwent IMDA cycloaddition to give a 2.1 : 1, syn : anti diastereomeric mixture that were readily separated by chromatography. The minor anti-cycloadduct 46 was subjected to peracid oxidation as above and rearranged under basic conditions furnishing D,L-brevianamide B. It is important to note that the intrinsic facial bias of the IMDA cycloadditions in all cases, was about 2 : 1 favoring the syn-diastereomer and in good agreement with theoretical predictions published by our collaborator Prof. Domingo (see below). The intrinsic facial bias of these IMDA reactions, have been interrogated by Domingo, et al., quantum chemically at the B3LYP/6-31G* level.54 He has examined both the dioxopiperazine-based azadienes (51b; 54b) relevant to the stephacidin and notoamide families, as well as the corresponding monooxopiperazine-based azadienes (51a; 54a) relevant to the paraherquamide, malbrancheamide and related family members.
Scheme 17.
Third-generation biomimetic total synthesis of D,L-brevianamide B.60,85
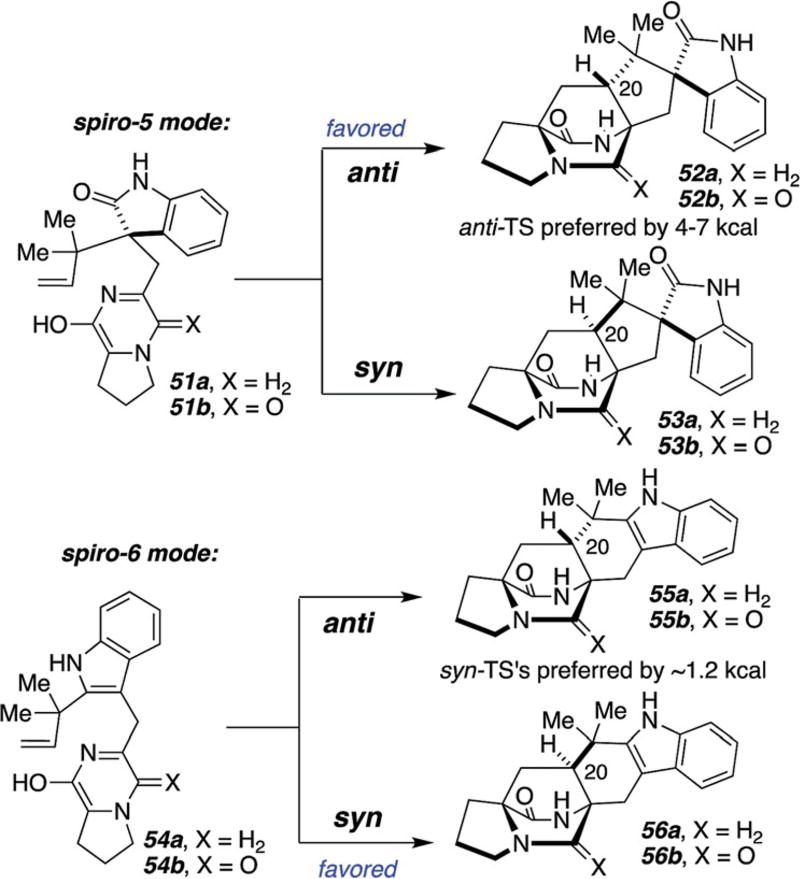
As depicted in Scheme 18, two main substrate platforms were evaluated: the spiro-5 mode, wherein the indole has been oxidized to the corresponding oxindole species and the spiro-6 mode, wherein the indole has yet to suffer oxidative tailoring.54 Strikingly, it was found that the spiro-5 mode greatly favors the anti-diastereomer (52) over the corresponding syn-diastereomer (53) by 4–7 kcal mol−1. In contradistinction, the corresponding spiro-6 mode from 54, revealed a modest preference for the syn- diastereomers 56 by about 1 kcal mol−1.54 These theoretical predictions have tracked quite well with experiment as can be seen from the modest syn : anti diastereoselectivities for the laboratory spiro-6 systems described above and below, where the syn : anti ratio is typically around 2.5 : 1. On the other hand, the experimentally observed spiro-5 cycloadditions have to date, yielded exclusively the anti-diastereomers, which will be detailed below.
Scheme 18.
Theoretical calculations predicting transition state energies and predicted product diastereoselectivities at the B3LYP/6-31G* level.54
The biomimetic syntheses developed above for brevianamide B, set the stage for deploying this technology to prepare more complex members of this family. The Williams lab has been able to successfully devise a biomimetic total synthesis of stephacidin A and its C-6-epimer, and more recently demonstrated C6-epi-stephacidin A to be a natural metabolite and, as noted above, these stephacidin A congeners are the direct biosynthetic precursors to notoamide B and versicolamide B, respectively. Subsequent to the biomimetic stephacidin A synthesis, these workers have been able to readily adopt this approach for successful biomimetic total syntheses of premalbrancheamide,87 malbrancheamide,88 spiromalbramide,89 marcfortine C,90 notoamide T and its C-6-epimer,63 also now known to be a natural metabolite as described above.
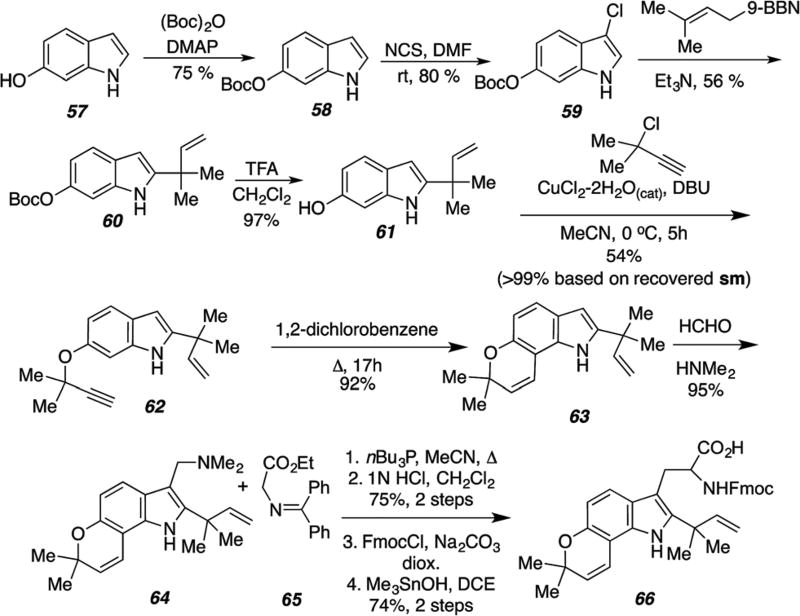
As shown in Scheme 19, the reverse-prenylated tryptophan derivative 66 was prepared on multi-gram scale from commercially available 6-hydroxyindole.63,83,91 Key steps included a Danishefsky 9-BBN reverse-prenylation of 3-chloroindole species 59 (ref. 92 and 100) and Claisen-based rearrangement of the propargyl ether 62 to the requisite pyran 63. Somei–Kametani93–96 gramine-based construction of the amino acid and protecting group adjustment furnished the key tryptophan derivative 66 that has been deployed in numerous total syntheses of the stephacidins and notoamides.
Scheme 19.
Synthesis of the reverse-prenylated tryptophan moiety.63,83,91–95
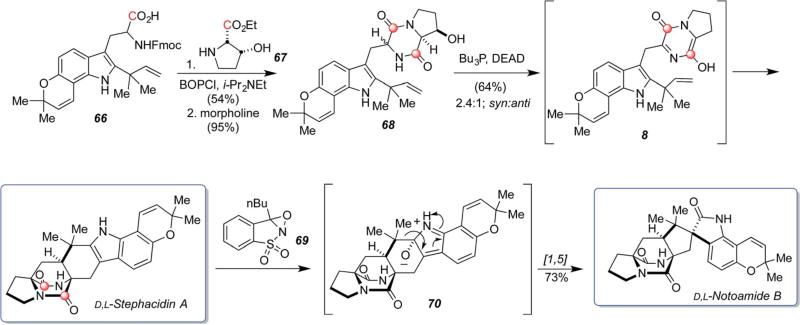
As described in Scheme 20, β-hydroxyproline ethyl ester 67 was coupled with tryptophan species 66 employing peptide coupling reagent BOPCl. This provided the incipient dipeptide, which was then subjected to removal of the N-Fmoc residue allowing spontaneous cyclization to dioxopiperazine 68. Mitsunobu dehydration directly led to the tautomerized azadiene species 8,97 that was stable enough to be isolated by PTLC and observable by 1H NMR, but spontaneously underwent IMDA cycloaddition to give stephacidin A and C6-epi-stephacidin A as a 2.4 : 1, syn : anti mixture in 64% combined yield.67
Scheme 20.
Biomimetic total synthesis of stephacidin A and notoamide B.67,97
Oxidation of synthetic stephacidin A with the Davis saccharin-derived oxaziridine (69)98 directly provided the spiro-oxindole product, notoamide B.67 Asymmetric syntheses of stephacidins A, B, and notoamide B were reported in 2007, utilizing a facially-controlled intramolecular SN2´ reaction.99
Notably, (−)-stephacidin A was achieved in 17 steps with a 6% overall yield. (+)-Stephacidin B is accessed from (−)-stephacidin A totaling a synthesis of 19 steps with a 1% overall yield. Additionally, (+)-notoamide B can be synthesized from the (−)-enantiomer of stephacidin A via the single oxidative pinacol step in 65% yield.99
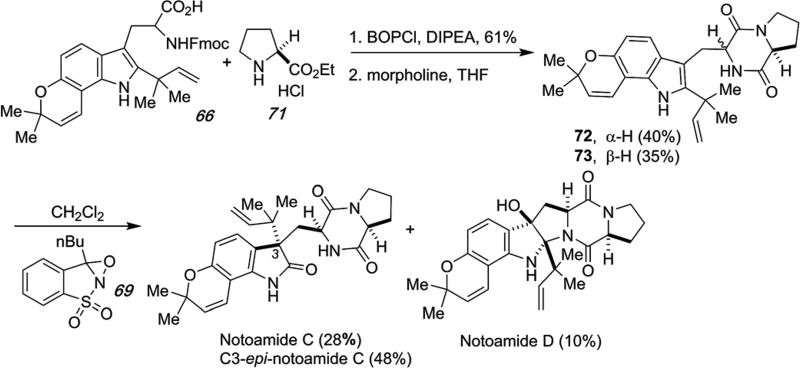
Grubbs, et al., have also been able to conscript the oxaziridine oxidation to a short, biomimetic synthesis of notoamide C, C3-epi-notoamide C and notoamide D as shown in Scheme 21.66 Using the same reverse-prenylated tryptophan species 66, coupling with (S)-proline and cyclization provided a diastereomeric mixture of dioxopiperazines 72 and 73 that were separated and oxidized with the Davis oxaziridine to provide notoamide C (28%), C3-epi-notoamide C (48%) and a small amount of notoamide D (together with the corresponding 2,3-epimer in 10% combined yield).66
Scheme 21.
Biomimetic total syntheses of notoamide C, C3-epi-notoamide C and notoamide D.66
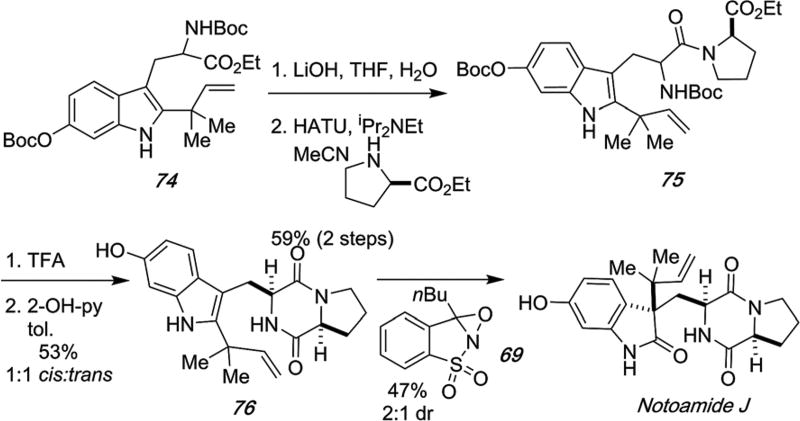
Similarly, notoamide J was prepared as detailed in Scheme 22 deploying the same Davis oxaziridine oxidation of indole 76 to provide notoamide J as a 2 : 1 diastereomeric mixture.92,100
Scheme 22.
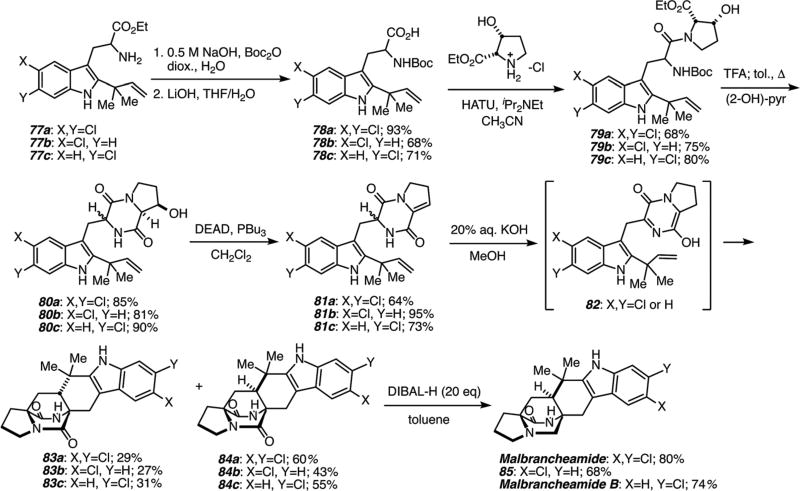
As detailed in Scheme 23, biomimetic total syntheses of the malbrancheamides have been successfully realized, deploying the same approach. In contrast to the stephacidin substrate, the Mitsunobu dehydration stops at the stage of dehydroproline derivative 81 that has to be treated with aqueous KOH in methanol at room temperature or below to effect tautomerization to the requisite azadiene that then suffers IMDA cycloaddition to give the cycloadducts 83 and 84 in a ~1 : 2 anti : syn diastereomeric ratio in good overall yields.88 Reduction of the tertiary amide in the presence of the secondary amide was readily achieved using di-isobutyl aluminum hydride.88
Scheme 23.
Biomimetic total synthesis of the malbrancheamides.88
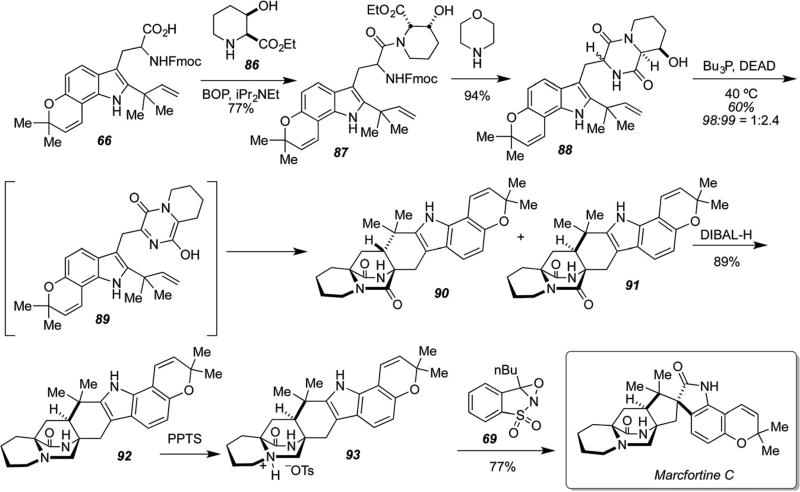
This same approach was also successfully deployed to effect a concise, biomimetic total synthesis of marcfortine C as illustrated in Scheme 24.90 Readily available β-hydroxypipecolic acid was coupled to reverse-prenylated tryptophan derivative 66 and carried through the analogous sequence deployed for the stephacidin A synthesis. The final spiro-oxindole oxidation required “protecting” the tertiary amine as the corresponding tosylate salt, which permitted smooth and diastereoselective oxidation and spiro-rearrangement to D,L-marcfortine C. This synthesis required a total of fifteen steps from commercially available 6-hydroxyindole with an overall yield of 3.7%.90
Scheme 24.
Biomimetic total synthesis of D,L-marcfortine C.90
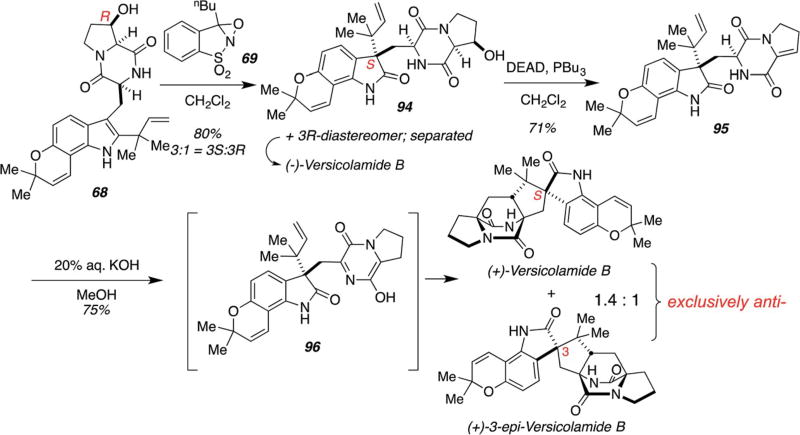
The asymmetric biomimetic total synthesis of versicolamide B has also been achieved and is described in Scheme 25.101,102 Similar to the previous discussion, pinacol rearrangement of reverse prenylated indole 68 allows for formation of a set of spiro-oxindole diastereomers. Upon separation, they can consequently be converted into both enantiomers of versicolamide B via IMDA cycloaddition. The C3-epimer of (+)-versicolamide B, recently isolated by the Tsukamoto laboratories from A. taichungensis and now denoted as ent-Taichunamide E,30 has thus also been achieved via this synthetic route (Scheme 25).
Scheme 25.
Biomimetic total synthesis of (+)-versicolamide B and (−)-versicolamide B.101,102
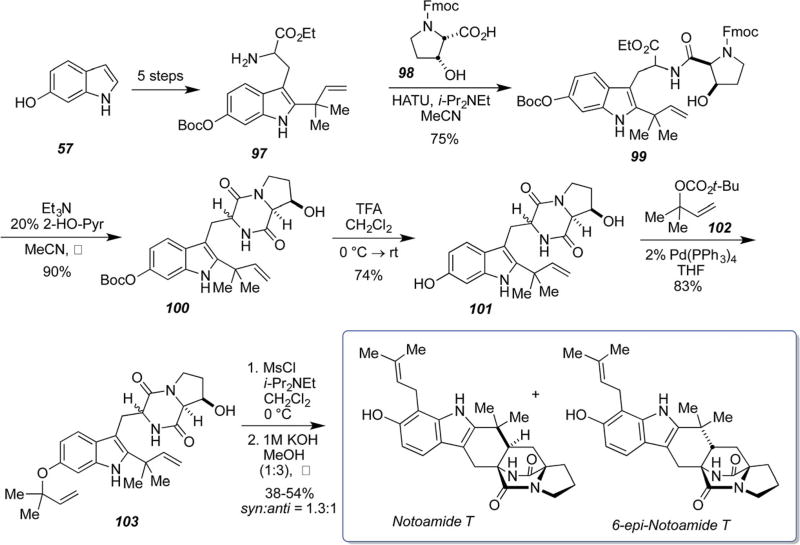
Synthetic, isotopically labeled samples of notoamide T and 6-epi-notoamide T were prepared by the biomimetic IMDA strategy as shown in Scheme 26.63 The doubly 13C-labeled samples of notoamide T and 6-epi-notoamide T were deployed in the precursor incorporation experiments described above (Scheme 9, Fig. 8) revealing that 6-epi-notoamide T is the biosynthetic precursor to versicolamide B and 6-epi-stephacidin A and provided the substrates from which the hitherto unobserved new metabolites were constructed from dormant tailoring genes.62–64 Efforts are underway to prepare optically pure samples of the individual enantiomers of 6-epi-notoamide T to more deeply interrogate the triggering mechanism for the expression of the dormant tailoring genes responsible for the biosynthesis of these new structures which were only produced in the presence of synthetic, racemic 6-epi-notoamide T.
Scheme 26.
Biomimetic total synthesis of D,L-notoamide T and D,L-6-epi-notoamide T.63
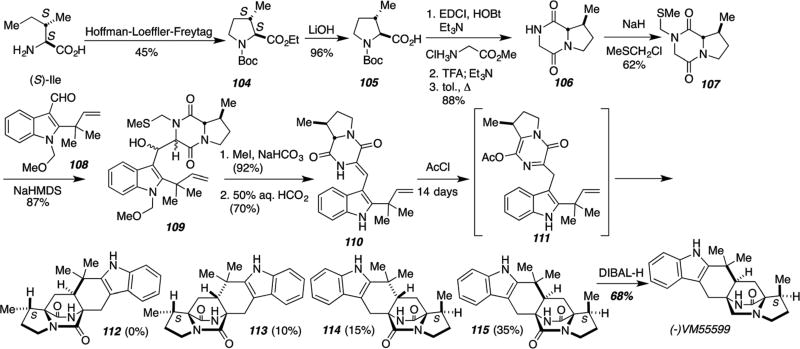
For studies relevant to the biosynthesis of the paraherquamides, the IMDA strategy has also been applied to complete the total syntheses of both racemic and optically pure VM55599 and diastereomeric congeners. As shown in Scheme 27, optically pure VM55599, a metabolite co-produced with paraherquamide A, was prepared by the IMDA cycloaddition of O-acyl azadiene species 111 generated under conditions reported by Liebscher.57
Scheme 27.
Biomimetic total synthesis of (−)-VM55599.57
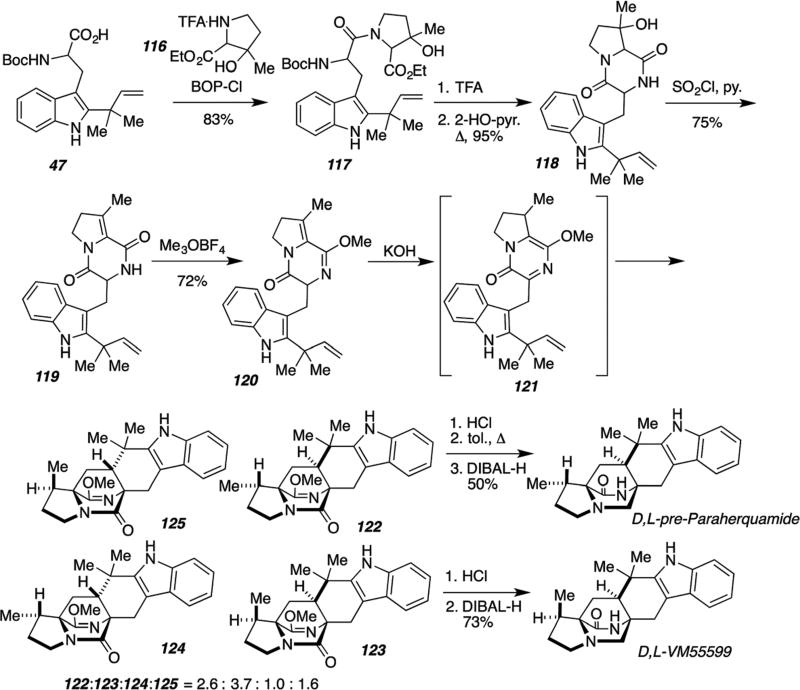
This synthesis enabled the confirmation of the absolute configuration of (−)-VM55599 as the original isolation of this natural, trace metabolite only assigned the relative configuration. As Stocking, et al. had demonstrated by labeled precursor incorporation experiments that the biogenetic source of β-methylproline in paraherquamide A was (S)-isoleucine,103 they had tentatively assigned the absolute stereostructure shown (Scheme 27).104–106 Culturing Penicillium fellutanum in a large format enabled the isolation of about 0.5 mg of natural (−)-VM55599.107 The synthetic material obtained, which was also derived from (S)-isoleucine via the Hoffman–Loeffler–Freytag protocol, exactly matched the natural species.57 Of additional significance, diastereomer 112 was not detected as a reaction product from this laboratory IMDA cycloaddition, revealing that the intrinsic facial bias of this particular IMDA construction favors the VM55599 stereochemistry and not the paraherquamide108–111 stereochemistry. This provides corroborating, indirect evidence that the enzyme responsible for the key cycloaddition that constructs the bicyclo[2.2.2]diazaoctane core in paraherquamide biosynthesis, must organize the conformation of the putative azadiene (presumably the unacylated congener of 111) to favor the paraherquamide stereochemistry in both a highly diastereoselective and enantioselective fashion. Stocking, et al., have also developed a racemic, biomimetic synthesis of VM55599 and the same group of diastereomeric congeners corresponding to 112–114 as illustrated in Scheme 28.58,112 In this instance, all four diastereomers were produced in the IMDA cycloaddition via 121, which permitted the preparation of an authentic specimen of pre-paraherquamide by dii-sobutyl aluminum hydride reduction of 122. As both amino acid subunits 47 and 116 were prepared from glycine, this synthesis was adopted to prepare double 13C-labeled versions of 122–125, VM55599 and pre-paraherquamide that were used in precursor incorporation experiments.112
Scheme 28.
Biomimetic total synthesis of D,L-VM55599 and D,L-pre-paraherquamide.58,112
5. The dioxopiperazine/monooxopiperazine quandary
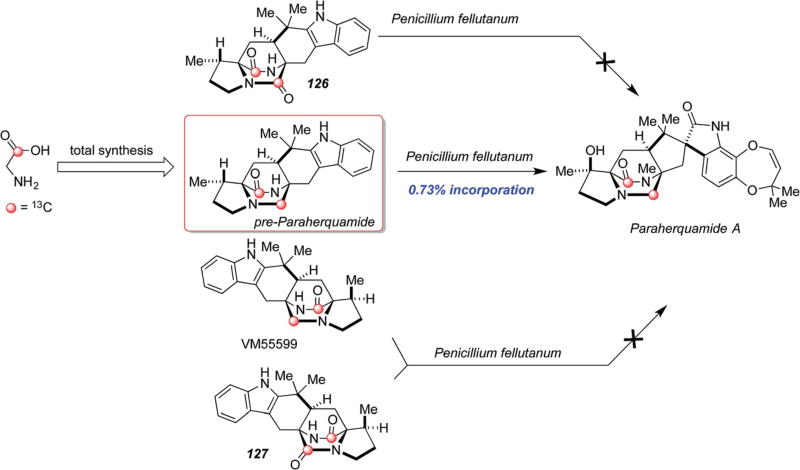
It was initially assumed that the bicyclo[2.2.2]diazaoctane ring systems that are common to the monooxopiperazine natural alkaloids, that includes the paraherquamides, marcfortines, asperparalines,113,114 and malbrancheamides, proceeded via biogenetic paths analogous to that of the dioxopiperazine families described in detail above, wherein a late-stage reduction of the tryptophan-derived carbonyl group was presumed to occur after the IMDA construction. This assumption has proven to be incorrect based on the following observations. As shown in Scheme 29, precursor incorporation studies with racemic, doubly 13C-labeled 126, 127, VM55599 and pre-paraherquamide were conducted in cultures of Penicillium fellutanum, and only pre-paraherquamide incorporated into the paraherquamide A isolated from this experiment (0.73% incorporation).112 In particular, the observed lack of incorporation of 126 was puzzling, which was presumed as the biosynthetic precursor to pre-paraherquamide.
Scheme 29.
Results of precursor incorporation studies in Penicillium fellutanum with isotopically labeled dioxopiperazine and monooxopiperazine substrates.112
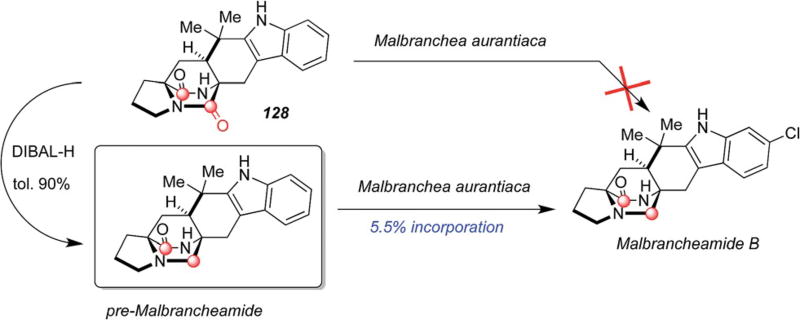
Ding, et al., observed exactly the same phenomena in malbrancheamide biosynthesis as shown in Scheme 30. Again here, the biomimetic synthesis of pre-malbrancheamide to prepare doubly 13C-labeled isotopomers of 128 and premalbrancheamide was adopted.115 When both substrates were presented to cultures of Malbranchea aurantiaca, only premalbrancheamide incorporated into malbrancheamide B (5.5% incorporation). The lack of incorporation of either dioxopiperazine species 126 and 128 begged the question as to when and how the tryptophan-derived carbonyl group is reduced by a net four-electrons? The answer to this quandary came in the most elegant way.
Scheme 30.
Results of precursor incorporation studies in Malbranchea aurantiaca with isotopically labeled dioxopiperazine and monooxopiperazine substrates.115
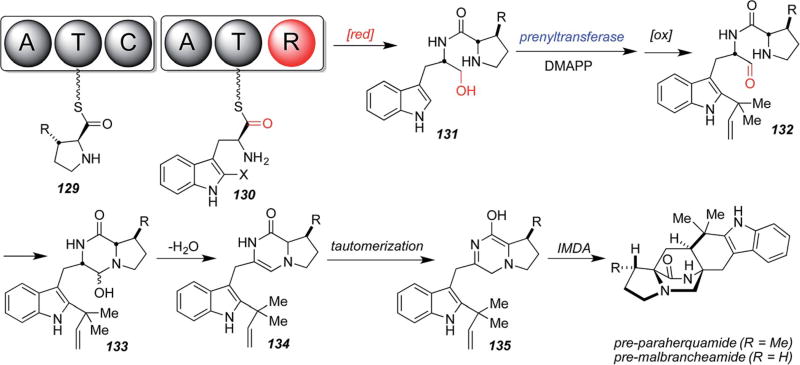
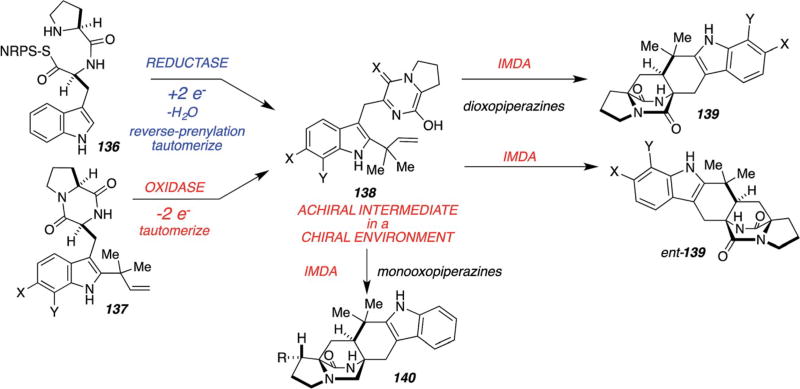
When the NRPS modules from the paraherquamide and malbrancheamide biosynthetic gene clusters were sequenced, it was revealed that the tryptophan module contains a terminal reductase domain, as opposed to the condensation domains evident in both amino acid-loading NRPS modules in notoamide biosynthesis. This is illustrated in Scheme 31.62 Thus, Li, et al., have devised a new biogenetic hypothesis that envisions reductive release of the pro–trp dipeptide from the NRPS module, by either a two-electron or four-electron process (the four-electron reduction is shown) providing species 131 (or, the corresponding aldehyde if the reductive release is a two-electron process) which is then reverse-prenylated and oxidized (for 131; or just reverse-prenylated for the two-electron process) to give key amino-aldehyde intermediate 132 that should spontaneously cyclize to carbinolamine 133. Subsequent dehydration to 134 and tautomerization to azadiene species 135 would then allow for IMDA cycloaddition, directly providing pre-paraherquamide (R = Me) and pre-malbrancheamide (R = H), respectively.62 This proposed biogenetic pathway explains why the dioxopiperazine species 126 and 128 did not incorporate as they are not pathway intermediates!
Scheme 31.
Molecular architecture of the monooxopiperazine NRPS module and a hypothetical biogenetic pathway to pre-paraherquamide and pre-malbrancheamide.62
The gene clusters for the post-NRPS module tailoring enzymes that construct species 132 merits additional scrutiny as the timing of the events leading from the NRPS module to the cycloadducts remains to be resolved.
6. Fungal producers of prenylated indole alkaloids
A large number of species of Penicillium and Aspergillus, both presently with approximately 400 described and accepted species116–119 are able to produce prenylated indole alkaloids, including penitrems, cyclopiazonic acid, aflavinins, paspalines fumigaclavines, janthitrems/shearinins, thiersindoles, fumitremorgins, roquefortines, rugulovasines etc.120,121 In Penicillium, these secondary metabolites are predominantly produced by species in subgenus Penicillium, but several producers are also found in subgenus Aspergilloides. While penitrems are restricted to subgenus Penicillium, the related shearinins are produced by species in both subgenera. Several species in Penicillium produce roquefortine C as the biosynthetic end-product, including P. expansum, P. griseofulvum, P. crustosum, P. hordei, P. robsamsonii, P. samsonianum, P. roqueforti, P. carneum and P. paneum, while other species accumulate meleagrin and oxalines, derived from roquefortine C, for example P. chrysogenum, P. rubens, P. oxalicum, P. compactum, P. concentricum, P. coprobium, P. glandicola, P. crystallinum and P. malodoratum.117,120,122 Interestingly, neoxaline is also produced by several species in Aspergillus section Nigri, but only in the uniseriate species.123,124 None of these secondary metabolites have been found outside the genera Aspergillus and Penicillium, except that few species in Talaromyces can produce rugulovasines.116,125
The bicyclo[2.2.2]diazaoctane ring system containing prenylated indole alkaloids are more restricted in their distribution, but again most of them are produced by species of Penicillium and Aspergillus. The known exception is the malbrancheamides produced by Malbranchea species,22 a species belonging to the Myxotrichaceae and unrelated to the Trichocomaceae. Since this species already produces the Penicillium and Aspergillus metabolite penicillic acid,126 there is a possibility that the fungus was indeed correctly identified as a Penicillium or Aspergillus. Most of these unique prenylated indole alkaloids are produced by both Aspergillus and Penicillium species except that the mangrovamides,127 brevianamides, chrysogenamide, paraherquamides and marcfortines have only been reported from Penicillium species, while the asperparalines, sclerotiamides, notoamides, versicolamides, avrainvillamides, stephacidins, and taichunamides have been recorded from Aspergillus species. However, chemotaxonomic investigations have shown that the mangrovamides and okaramines have been found in both genera (Frisvad, unpublished data). In Penicillium, the brevianamides are produced only by two species in subgenus Penicillium section Fasciculata (P. viridicatum) and subgenus Penicillium section Brevicompacta (P. brevicompactum).120,122 The marcfortines have until now only been found in Penicillium paneum in subgenus Penicillium section Roquefortorum.120,122 The paraherquamides have been found in Penicillium paraherquei10 (Penicillium section Lanata-Divaricata) and P. canescens (Penicillium section Canescentia) (originally determined as P. charlesii).11 Chrysogenamide was reported from P. chrysogenum,29 but the identity of the producing organisms has not been confirmed.
The producers of the related asperparalines (aspergillimides), avrainvillamides, notoamides, sclerotiamides, stephacidins, taichunamides, and versicolamides are produced by species in Aspergillus subgenus Circumdati sections Candidi, Circumdati, and Nigri, and in subgenus Nidulantes section Versicolores. Species producing those metabolites in section Candidi include A. subalbidus and A. taichungensis.128,129 Species in section Nigri producing aspergillamides include A. brunneoviolaceus (A. jiensis).124 In section Circumdati the species A. affinis, A. auricomus, A. bridgeri, A. melleus, A. ochraceus, A. ostianus, A. pallidofulvus, A. persii, A. salwaensis, A. sclerotiorum, A. sesamicola, A. subramanianii, A. westerdijkiae and A. westlandensis produce “Aspergamides”, which is a mixture of notoamides, avrainvillamides, stephacidins, and sclerotiamide.116 Finally, Aspergillus subgenus Nidulantes section Versicolores contains producers of “Aspergamides”: A. amoenus and A. protuberus,35 but “Aspergamides” could not be found in any species in section Nidulantes.130 Further studies will show whether the producing species in these sections of Aspergilli all have stereochemical diversity in their production of these compounds.
7. Conclusions
The biogenesis of prenylated indole alkaloid families containing the unique bicyclo[2.2.2]diazaoctane core, obtained from various genera of marine and terrestrial fungi has revealed a number of fascinating aspects of secondary metabolite biosynthesis. First, it is clear that the biosynthetic gene clusters responsible for directing the biosyntheses of these structurally alluring natural compounds are widely distributed in the biosphere. In particular, the orthologous pairs of marine and terrestrial fungi responsible for stephacidin and notoamide biosyntheses, have extensively interrogated the stereochemistry of these molecular frameworks in both a relative and absolute sense. A vast repertoire of tailoring genes is in evidence, and the experiments described in Scheme 9 and Fig. 8 reveal that many dormant tailoring genes likely lie in reserve in the genomes of these complex organisms. It is particularly intriguing to ponder the speciation events that occurred at some time during the evolutionary history of these fungi that resulted in enantiomerically distinct secondary metabolite profiles. Very little is known about the adaptive advantage with which these secondary metabolites embellish the producing fungi as the biological activities of these natural alkaloids have been only superficially examined and demand further elucidation.
It is also now clear that Nature has, perhaps, “accidentally” created the bicyclo[2.2.2]diazaoctane ring system through the generation of reactive, achiral azadiene species (138, Scheme 32). The achiral intermediates are conformationally restricted in the chiral environment of the enzyme binding sites that generate these species, and thus orchestrates these reactive azadiene species to undergo highly enantio- and diastereoselective cycloadditions culminating in the formation of the natural substances constituted with the bicyclo[2.2.2]diazaoctane core. Our biomimetic laboratory IMDA reactions, which proceed smoothly at or below room temperature in organic or aqueous solvents, are very facile transformations that in the biochemical setting, only require pre-organization of the pre-transition state conformations.
Scheme 32.
Unified biogenetic hypothesis for prenylated indole alkaloids containing the bicyclo[2.2.2]diazaoctane nucleus.53
It is further noteworthy that, in the monooxopiperazine families, the predominant relative stereochemistry is syn- (all known paraherquamide alkaloids in this family are syn-; see Fig. 1 and 4) and only a single absolute configuration is currently in evidence (with the exception of VM55599, a shunt metabolite of paraherquamide biosynthesis). To date, the only monooxopiperazine-producing fungi that construct the corresponding anti-diastereomeric series are Penicillium purpurogenum G59 (penicimutamides D and E; P. purpurogenum G59 has been mischaracterized and is a Penicillium citrinum),15 Penicillium chrysogenum (chrysogenamide A)29 and Penicillium citrinum F53 (the recently described citrinalin C).146 In stark contradistinction, the dioxopiperazine families display a broader range of relative and absolute stereochemistry, where a myriad of both syn- and anti-metabolites have been isolated in both enantiomeric forms. Most intriguing from an evolutionary perspective, the enantio-divergence evident in both marine- and terrestrial-producing fungi, provides for a very rich and unique opportunity to study molecular evolution of these organisms.
The application of whole-genome sequencing, bioinformatics analysis, cloning and functional expression of biosynthetic enzymes, synthesis of labeled pathway metabolites, precursor incorporation studies, structure determination of new metabolites and total syntheses of both new/undiscovered and known metabolites has been deployed to gain a deeper understanding of the secondary metabolome of marine and terrestrial fungi that produce these families of alkaloids. It should be noted that many laboratories in the synthetic community have published elegant, yet non-biomimetic total syntheses of numerous natural products in this family of alkaloids.41,46,47,57,58,66,67,85,86,88,90,95,97,99,102,108,115,131–146 These efforts have been reviewed by Miller and Williams in 2009 (ref. 40) and additional total syntheses have appeared in the literature since 2009.138–146 Complete knowledge of the structural and stereochemical homologies and disparities among these compounds continues to motivate research into the synthesis, chemistry, biochemistry, genetics and biology of natural fungal metabolites. It is anticipated that exciting new chapters of these fascinating natural alkaloids remain to be written.
Acknowledgments
This work was financially supported in part by Grants-in-Aid for Scientific Research (No. 23108518 and 25108719 to S. T. and 24710252 to H.K.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan and also by grants (S. T.) from the Naito Foundation, the Nagase Science and Technology Foundation, and the Yamada Science Foundation. Financial support from the National Institutes of Health (R01 CA070375 to R. M. W. and D. H. S.) is gratefully acknowledged. The authors wish to dedicate this manuscript to Professor Yoshito Kishi of Harvard University celebrating his 80th birthday.
Footnotes
Conflicts of interest
There are no conflicts to declare.
References
- 1.Birch AJ, Wright JJ. Tetrahedron Lett. 1970;26:2329–2344. doi: 10.1016/s0040-4020(01)92812-1. [DOI] [PubMed] [Google Scholar]
- 2.Birch AJ, Russel RA. Tetrahedron Lett. 1972;28:2999–3008. [Google Scholar]
- 3.Steyn PS. Tetrahedron Lett. 1973;29:107–120. [Google Scholar]
- 4.Birch AJ, Wright JJ. Chem. Commun. 1969:644–645. [Google Scholar]
- 5.Paterson RRM, Hawksworth DL. Trans. Br. Mycol. Soc. 1985;85:95–100. [Google Scholar]
- 6.Robbers JE, Straus JW. Lloydia. 1974;38:355–356. [PubMed] [Google Scholar]
- 7.Wilson BJ, Yang DTC, Harris TM. Appl. Microbiol. 1973;26:633–635. doi: 10.1128/am.26.4.633-635.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polonsky J, Merrien M-A, Prangé T, Pascard C, Moreau S. J. Chem. Soc., Chem. Commun. 1980:601–602. [Google Scholar]
- 9.Prangé T, Billion M-A, Vuilhorgne M, Pascard C, Polonsky J. Tetrahedron Lett. 1981;22:1977–1980. [Google Scholar]
- 10.Yamazaki M, Okuyama E, Kobayashi M, Inoue H. Tetrahedron Lett. 1981;22:135–136. [Google Scholar]
- 11.Ondeyka JG, Goegelman RT, Schaeffer JM, Kelemen L, Zitano L. J. Antibiot. 1990;43:1375–1379. doi: 10.7164/antibiotics.43.1375. [DOI] [PubMed] [Google Scholar]
- 12.Liesch JM, Wichmann CF. J. Antibiot. 1990;43:1380–1386. doi: 10.7164/antibiotics.43.1380. [DOI] [PubMed] [Google Scholar]
- 13.Blanchflower SE, Banks RM, Everet JR, Manger BR, Reading C. J. Antibiot. 1991;44:492–497. doi: 10.7164/antibiotics.44.492. [DOI] [PubMed] [Google Scholar]
- 14.Blanchflower SE, Banks RM, Everet JR, Manger BR, Reading C. J. Antibiot. 1993;46:1355–1363. doi: 10.7164/antibiotics.46.1355. [DOI] [PubMed] [Google Scholar]
- 15.Wu C-J, Li C-W, Gao H, Huang X-J, Cui C-B. RSC Adv. 2017;7:24718–24722. [Google Scholar]
- 16.Authrine C, Gloer JB. J. Nat. Prod. 1996;59:1093–1095. doi: 10.1021/np960607m. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi H, Nishimoto Y, Nozaki H. Tetrahedron Lett. 1997;38:5655–5658. [Google Scholar]
- 18.Fenical W, Jensen PR, Cheng XC. 6,066,635. U.S. Pat. 2000
- 19.Sugie Y, Hirai H, Inagaki T, Ishiguro M, Kim Y-J, Kojima Y, Sakakibara T, Sakemi S, Sugiura A, Suzuki Y, Brennan L, Duignan J, Huang LH, Sutcliffe J, Kojima N. J. Antibiot. 2001;54:911–916. doi: 10.7164/antibiotics.54.911. [DOI] [PubMed] [Google Scholar]
- 20.Qian-Cutrone J, Huang S, Shu Y-Z, Vyas D, Fairchild C, Menendez A, Krampitz K, Dalterio R, Klohr SE, Gao Q. J. Am. Chem. Soc. 2002;124:14556–14557. doi: 10.1021/ja028538n. [DOI] [PubMed] [Google Scholar]
- 21.Martínez-Luis S, Rodriguez R, Acevedo L, González MC, Lira-Rocha A, Mata R. Tetrahedron. 2006;62:1817–1822. [Google Scholar]
- 22.Figueroa M, González MC, Mata R. Nat. Prod. Res. 2008;22:709–714. doi: 10.1080/14786410802012361. [DOI] [PubMed] [Google Scholar]
- 23.Watts KR, Loveridge ST, Tenney K, Media J, Valeriote FA, Crews P. J. Org. Chem. 2011;76:6201–6208. doi: 10.1021/jo2009593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato H, Yoshida T, Tokue T, Nojiri Y, Hirota H, Ohta T, Williams RM, Tsukamoto S. Angew. Chem., Int. Ed. 2007;46:2254–2256. doi: 10.1002/anie.200604381. [DOI] [PubMed] [Google Scholar]
- 25.Tsukamoto S, Kato H, Samizo M, Nojiri Y, Onuki H, Hirota H, Ohta T. J. Nat. Prod. 2008;71:2064–2067. doi: 10.1021/np800471y. [DOI] [PubMed] [Google Scholar]
- 26.Tsukamoto S, Kawabata T, Kato H, Greshock TJ, Hirota H, Ohta T, Williams RM. Org. Lett. 2009;11:1297–1300. doi: 10.1021/ol900071c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsukamoto S, Umaoka H, Yoshikawa K, Ikeda T, Hirota H. J. Nat. Prod. 2010;73:1438–1440. doi: 10.1021/np1002498. [DOI] [PubMed] [Google Scholar]
- 28.Greshock TJ, Grubbs AW, Jiao P, Wicklow DT, Gloer JB, Williams RM. Angew. Chem., Int. Ed. 2008;47:3573–3577. doi: 10.1002/anie.200800106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Z, Wen J, Zhu T, Fang Y, Gu Q, Zhu W. J. Antibiot. 2008;61:81–85. doi: 10.1038/ja.2008.114. [DOI] [PubMed] [Google Scholar]
- 30.Kagiyama I, Kato H, Nehira T, Frisvad JC, Sherman DH, Williams RM, Tsukamoto S. Angew. Chem., Int. Ed. 2016;55:1128–1132. doi: 10.1002/anie.201509462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller KA, Figueroa M, Valente MWN, Greshock TJ, Mata R, Williams RM. Bioorg. Med. Chem. Lett. 2008;18:6479–6481. doi: 10.1016/j.bmcl.2008.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paterson RRM, Simmonds MJS, Kemmelmeier C, Blaney WM. Mycol. Res. 1990;94:538–542. [Google Scholar]
- 33.Paterson RRM, Simmonds MSJ, Blaney WM. J. Invertebr. Pathol. 1987;50:124–133. [Google Scholar]
- 34.Doerfler DL, Nulton CP, Bartman CD, Gottlieb FJ, Campbell IM. Can. J. Microbiol. 1978;24:1490–1500. doi: 10.1139/m78-239. [DOI] [PubMed] [Google Scholar]
- 35.Finefield JM, Frisvad JC, Sherman DH, Williams RM. J. Nat. Prod. 2012;75:812–833. doi: 10.1021/np200954v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finefield JM, Sherman DH, Kreitman M, Williams RM. Angew. Chem., Int. Ed. 2012;51:4802–4836. doi: 10.1002/anie.201107204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams RM. Chem. Pharm. Bull. 2002;50:711–740. doi: 10.1248/cpb.50.711. [DOI] [PubMed] [Google Scholar]
- 38.Stocking EM, Williams RM. Angew. Chem., Int. Ed. 2003;42:3078–3115. doi: 10.1002/anie.200200534. [DOI] [PubMed] [Google Scholar]
- 39.Williams RM, Cox RJ. Acc. Chem. Res. 2003;36:127–139. doi: 10.1021/ar020229e. [DOI] [PubMed] [Google Scholar]
- 40.Miller KA, Williams RM. Chem. Soc. Rev. 2009;38:3160–3174. doi: 10.1039/b816705m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sunderhaus JD, Sherman DH, Williams RM. Isr. J. Chem. 2011;51:442–452. doi: 10.1002/ijch.201100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams RM, Stocking EM, Sanz-Cervera JF. Top. Curr. Chem. 2000;209:97–173. [Google Scholar]
- 43.Porter AEA, Sammes PG. Chem. Commun. 1970;1103 [Google Scholar]
- 44.Baldas J, Birch AJ, Russell RA. J. Chem. Soc., Perkin Trans. 1. 1974;1:50. [Google Scholar]
- 45.Birch AJ. J. Agric. Food Chem. 1971;19:1088–1092. [Google Scholar]
- 46.Williams RM, Glinka T, Kwast E. J. Am. Chem. Soc. 1988;110:5927–5929. [Google Scholar]
- 47.Williams RM, Glinka T, Kwast E, Coffman H, Stille JK. J. Am. Chem. Soc. 1990;112:808–821. [Google Scholar]
- 48.Williams RM, Glinka T. Tetrahedron Lett. 1986;27:3581–3584. [Google Scholar]
- 49.Williams RM, Kwast E, Coffman H, Glinka T. J. Am. Chem. Soc. 1989;111:3064–3065. [Google Scholar]
- 50.Stocking EM, Williams RM, Sanz-Cervera JF. J. Am. Chem. Soc. 2000;122:9089–9098. [Google Scholar]
- 51.Sanz-Cervera JF, Glinka T, Williams RM. J. Am. Chem. Soc. 1993;115:347–348. [Google Scholar]
- 52.Sanz-Cervera JF, Glinka T, Williams RM. Tetrahedron. 1993;49:8471–8482. [Google Scholar]
- 53.Klas K, Tsukamoto S, Sherman DH, Williams RM. J. Org. Chem. 2015;80:11672–11685. doi: 10.1021/acs.joc.5b01951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Domingo LR, Zaragozá RJ, Williams RM. J. Org. Chem. 2003;68:2895–2902. doi: 10.1021/jo020564g. [DOI] [PubMed] [Google Scholar]
- 55.Domingo LR, Sanz-Cervera JF, Williams RM, Picher MT, Marco JA. J. Org. Chem. 1997;62:1662–1667. [Google Scholar]
- 56.Adams LA, Gray CR, Williams RM. Tetrahedron Lett. 2004;45:4489–4493. [Google Scholar]
- 57.Sanz-Cervera JF, Williams RM. J. Am. Chem. Soc. 2002;124:2556–2559. doi: 10.1021/ja017425l. [DOI] [PubMed] [Google Scholar]
- 58.Stocking EM, Sanz-Cervera JF, Williams RM. J. Am. Chem. Soc. 2000;122:1675–1683. [Google Scholar]
- 59.Sanz-Cervera JF, Williams RM, Marco JA, López-Sánchez JM, González F, Martínez ME, Sancenón F. Tetrahedron. 2000;56:6345–6358. [Google Scholar]
- 60.Williams RM, Sanz-Cervera JF, Sancenón F, Marco JA, Halligan KM. Bioorg. Med. Chem. 1998;6:1233–1241. doi: 10.1016/s0968-0896(98)00102-3. [DOI] [PubMed] [Google Scholar]
- 61.Kato H, Nakahara T, Sugimoto K, Matsuo K, Kagiyama I, Frisvad JC, Sherman DH, Williams RM, Tsukamoto S. Org. Lett. 2015;17:700–703. doi: 10.1021/ol5037198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li S, Srinivasan K, Tran H, Yu F, Finefield JM, Sunderhaus JD, McAfoos TJ, Tsukamoto S, Williams RM, Sherman DH. Med. Chem. Commun. 2012;3:987–996. doi: 10.1039/C2MD20029E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sunderhaus JD, McAfoos TJ, Finefield JM, Kato H, Li S, Tsukamoto S, Sherman DH, Williams RM. Org. Lett. 2013;15:22–25. doi: 10.1021/ol302901p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kato H, Nakahara T, Yamaguchi M, Kagiyama I, Finefield JM, Sunderhaus JD, Sherman DH, Williams M, Tsukamoto S. Tetrahedron Lett. 2015;56:247–251. doi: 10.1016/j.tetlet.2014.11.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsukamoto S, Umaoka H, Yoshikawa K, Ikeda T, Hirota H. J. Nat. Prod. 2013;76:1232–1232. doi: 10.1021/np1002498. [DOI] [PubMed] [Google Scholar]
- 66.Grubbs AW, Artman GD, III, Tsukamoto S, Williams RM. Angew. Chem., Int. Ed. 2007;46:2257–2261. doi: 10.1002/anie.200604377. [DOI] [PubMed] [Google Scholar]
- 67.Greshock TJ, Grubbs AW, Tsukamoto S, Williams RM. Angew. Chem., Int. Ed. 2007;46:2262–2265. doi: 10.1002/anie.200604378. [DOI] [PubMed] [Google Scholar]
- 68.Tsukamoto S, Kato H, Greshock TJ, Hirota H, Ohta T, Williams RM. J. Am. Chem. Soc. 2009;131:3834–3835. doi: 10.1021/ja810029b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsukamoto S, Kato H, Greshock TJ, Hirota H, Ohta T, Williams RM. J. Am. Chem. Soc. 2013;135:10878–10878. doi: 10.1021/ja810029b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Finefield JM, Kato H, Greshock TJ, Sherman DH, Tsukamoto S, Williams RM. Org. Lett. 2011;13:3802–3805. doi: 10.1021/ol201284y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cox RJ, Williams RM. Tetrahedron Lett. 2002;43:2149–2152. [Google Scholar]
- 72.Fuchser J. Ph. D. Dissertation. University of Gottingen; 1995. [Google Scholar]
- 73.Finefield JM, Greshock TJ, Sherman DH, Tsukamoto S, Williams RM. Tetrahedron Lett. 2011;52:1987–1989. doi: 10.1016/j.tetlet.2011.02.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McAfoos TJ, Li S, Tsukamoto S, Sherman DH, Williams RM. Heterocycles. 2010;82:461–472. doi: 10.3987/COM-10-S(E)19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kato H, Nakamura Y, Finefield JM, Umaoka H, Nakahara T, Williams RM, Tsukamoto S. Tetrahedron Lett. 2011;52:6923–6926. doi: 10.1016/j.tetlet.2011.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Finefield JM, Sherman DH, Tsukamoto S, Williams RM. J. Org. Chem. 2011;76:5954–5958. doi: 10.1021/jo200218a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li S, Finefield JM, Sunderhaus JD, McAfoos TJ, Williams RM, Sherman DH. J. Am. Chem. Soc. 2012;134:788–791. doi: 10.1021/ja2093212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cai S, Luan Y, Kong X, Zhu T, Gu Q, Li D. Org. Lett. 2013;15:2168–2171. doi: 10.1021/ol400694h. [DOI] [PubMed] [Google Scholar]
- 79.Li F, Zhang Z, Zhang G, Che Q, Zhu T, Gu Q, Li D. Org. Lett. 2018;20:1138–1141. doi: 10.1021/acs.orglett.8b00061. [DOI] [PubMed] [Google Scholar]
- 80.Cai S, Luan Y, Kong X, Zhu T, Gu Q, Li D. Org. Lett. 2013;15:2168–2171. doi: 10.1021/ol400694h. [DOI] [PubMed] [Google Scholar]
- 81.See ref. 24 for the isolation and structural elucidation of the corresponding N1–OH diastereomer of versicolamide C, notoamide A.
- 82.Maiya S, Grundmann A, Li S-M, Turner G. Chem. Bio. Chem. 2006;7:1062–1069. doi: 10.1002/cbic.200600003. [DOI] [PubMed] [Google Scholar]
- 83.Ding Y, deWet JR, Cavalcoli J, Li S, Greshock TJ, Miller KA, Finefield JM, Sunderhaus JD, McAfoos TJ, Tsukamoto S, Williams RM, Sherman DH. J. Am. Chem. Soc. 2010;132:12733–12740. doi: 10.1021/ja1049302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Newmister SA, Gober CM, Romminger S, Yu F, Tripathi A, Parra LLL, Williams RM, Berlinck RGS, Joullié MM, Sherman DH. J. Am. Chem. Soc. 2016;138:11176–11184. doi: 10.1021/jacs.6b04915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Williams RM, Sanz-Cervera JF, Sancenón F, Marco JA, Halligan K. J. Am. Chem. Soc. 1998;120:1090–1091. doi: 10.1016/s0968-0896(98)00102-3. [DOI] [PubMed] [Google Scholar]
- 86.Adams LA, Valente MWN, Williams RM. Tetrahedron. 2006;62:5195–5200. [Google Scholar]
- 87.Robins JG, Kim KJ, Chinn AJ, Woo JS, Scheerer JR. J. Org. Chem. 2016;81:2293–2301. doi: 10.1021/acs.joc.5b02744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Miller KA, Welch TR, Greshock TJ, Ding Y, Sherman DH, Williams RM. J. Org. Chem. 2008;73:3116–3119. doi: 10.1021/jo800116y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sherman DH, Williams RM, et al. unpublished results. [Google Scholar]
- 90.Greshock TJ, Grubbs AW, Williams RM. Tetrahedron. 2007;63:6124–6130. doi: 10.1016/j.tet.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Grubbs AW, Artman GD, III, Williams RM. Tetrahedron Lett. 2005;46:9013–9016. [Google Scholar]
- 92.Finefield JM, Williams RM. J. Org. Chem. 2010;75:2785–2789. doi: 10.1021/jo100332c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Williams RM, Cao J, Tsujishima H. Angew. Chem., Int. Ed. 2000;39:2540–2544. doi: 10.1002/1521-3773(20000717)39:14<2540::aid-anie2540>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 94.Cushing TD, Sanz-Cervera JF, Williams RM. J. Am. Chem. Soc. 1996;118:557–579. [Google Scholar]
- 95.Cushing TD, Sanz-Cervera JF, Williams RM. J. Am. Chem. Soc. 1993;115:9323–9324. [Google Scholar]
- 96.(a) Somei M, Karasawa Y, Kaneko C. Heterocycles. 1981;16:941–949. [Google Scholar]; (b) Kametani T, Kanaya N, Ihara M. J. Chem. Soc., Perkin Trans. 1. 1981;3:959. [Google Scholar]
- 97.Greshock TJ, Williams RM. Org. Lett. 2007;9:4255–4258. doi: 10.1021/ol701845t. [DOI] [PubMed] [Google Scholar]
- 98.Davis FA, Towson JC, Vashi DB, ThimmaReddy R, McCauley JP, Harakal ME, Gosciniak DJ. J. Org. Chem. 1990;55:1254–1261. [Google Scholar]
- 99.Artman GD, III, Grubbs AW, Williams RM. J. Am. Chem. Soc. 2007;129:6336–6342. doi: 10.1021/ja070259i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Finefield JM, Williams RM. J. Org. Chem. 2013;78:8214. [Google Scholar]
- 101.Miller KA, Tsukamoto S, Williams RM. Nat. Chem. 2009;1:165–165. doi: 10.1038/nchem.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Miller KA, Tsukamoto S, Williams RM. Nat. Chem. 2009;1:63–68. doi: 10.1038/nchem.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stocking EM, Sanz-Cervera JF, Unkefer CJ, Williams RM. Tetrahedron. 2001;57:5303–5320. [Google Scholar]
- 104.Stocking EM, Sanz-Cervera JF, Williams RM. Angew. Chem., Int. Ed. 2001;40:1296–1298. doi: 10.1002/1521-3773(20010401)40:7<1296::aid-anie1296>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 105.Stocking EM, Martinez RA, Silks LA, Sanz-Cervera JF, Williams RM. J. Am. Chem. Soc. 2001;123:3391–3392. doi: 10.1021/ja005655e. [DOI] [PubMed] [Google Scholar]
- 106.Stocking EM, Sanz-Cervera JF, Williams RM, Unkefer CJ. J. Am. Chem. Soc. 1996;118:7008–7009. [Google Scholar]
- 107.Ding Y, Gruschow S, Greshock TJ, Finefield JM, Sherman DH, Williams RM. J. Nat. Prod. 2008;71:1574–1578. doi: 10.1021/np800292n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Williams RM, Cao J, Tsujishima H, Cox RJ. J. Am. Chem. Soc. 2003;125:12172–12178. doi: 10.1021/ja036713+. [DOI] [PubMed] [Google Scholar]
- 109.Williams RM, Cao J. Tetrahedron Lett. 1996;37:5441–5444. [Google Scholar]
- 110.Williams RM, Cushing TD. Tetrahedron Lett. 1990;31:6325–6328. [Google Scholar]
- 111.Williams RM, Glinka T, Kwast E. Tetrahedron Lett. 1989;30:5575–5578. [Google Scholar]
- 112.Sommer K, Williams RM. Tetrahedron. 2009;65:3246–3260. doi: 10.1016/j.tet.2008.08.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gray CR, Sanz-Cervera JF, Silks LA, Williams RM. J. Am. Chem. Soc. 2003;125:14692–14693. doi: 10.1021/ja037687i. [DOI] [PubMed] [Google Scholar]
- 114.Gonzalez F, Sanz-Cervera JF, Williams RM. Tetrahedron Lett. 1999;40:4519–4522. [Google Scholar]
- 115.(a) Ding Y, Greshock TJ, Miller KA, Sherman DH, Williams RM. Org. Lett. 2008;10:4863–4866. doi: 10.1021/ol8019633. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Fraley AE, Garcia-Borras M, Tripathi A, Khare D, Mercado-Marin EV, Tran H, Dan Q, Webb GP, Watts KR, Crews P, Sarpong R, Williams RM, Smith JL, Houk KN, Sherman DH. J. Am. Chem. Soc. 2017;139:12060–12068. doi: 10.1021/jacs.7b06773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Visagie CM, Varga J, Houbraken J, Meijer M, Kocsubé S, Yilmaz N, Fotedar R, Seifert KA, Frisvad JC, Samson RA. Stud. Mycol. 2014;78:1–61. doi: 10.1016/j.simyco.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Visagie CM, Houbraken J, Frisvad JC, Hong SB, Klaassen CHW, Perrone G, Seifert KA, Varga J, Yaguchi T, Samson RA. Stud. Mycol. 2014;78:343–371. doi: 10.1016/j.simyco.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Samson RA, Visagie CM, Houbraken J, Hong S-B, Hubka V, Klaassen CHW, Perrone G, Seifert KA, Susca A, Tanney JB, Varga J, Kocsubé S, Szigeti G, Yaguchi T, Frisvad JC. Stud. Mycol. 2014;78:141–173. doi: 10.1016/j.simyco.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Frisvad JC. Front Microbiol. 2015;5:773. doi: 10.3389/fmicb.2014.00773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Frisvad JC, Smedsgaard J, Larsen TO, Samson RA. Stud. Mycol. 2004;49:201–241. [Google Scholar]
- 121.Frisvad JC, Larsen TO. Appl. Microbiol. Biotechnol. 2015;99:7859–7877. doi: 10.1007/s00253-015-6839-z. [DOI] [PubMed] [Google Scholar]
- 122.Houbraken J, Wang L, Lee HB, Frisvad JC. Persoonia. 2016;36:299–314. doi: 10.3767/003158516X692040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Samson RA, Noonim P, Meijer M, Houbraken J, Frisvad JC, Varga J. Stud. Mycol. 2007;59:129–145. doi: 10.3114/sim.2007.59.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Varga J, Frisvad JC, Kocsubé S, Brankovics B, Tóth B, Szigeti G, Samson RA. Stud. Mycol. 2011;69:1–17. doi: 10.3114/sim.2011.69.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Samson RA, Yilmaz N, Houbraken J, Spierenburg H, Seifert KA, Peterson SW, Varga J, Frisvad JC. Stud. Mycol. 2011;70:159–183. doi: 10.3114/sim.2011.70.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Martínez-Luis S, González MC, Ulloa M, Mata R. Phytochemistry. 2005;66:1012–1016. doi: 10.1016/j.phytochem.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 127.Yang B, Dong J, Lin X, Zhou X, Zhang Y, Liu Y. Tetrahedron. 2014;70:3859–3863. [Google Scholar]
- 128.Varga J, Frisvad JC, Samson RA. Stud. Mycol. 2007;59:75–88. doi: 10.3114/sim.2007.59.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hubka V, Lyskova P, Frisvad JC, Peterson SW, Skorepova M, Kolarik M. Med. Mycol. 2014;52:565–576. doi: 10.1093/mmy/myu022. [DOI] [PubMed] [Google Scholar]
- 130.Chen AJ, Frisvad JC, Sun BD, Varga J, Kocsubé S, Dijksterhuis J, Kim DH, Hong S-B, Houbraken J, Samson RA. Stud. Mycol. 2016;84:1–118. doi: 10.1016/j.simyco.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Baran PS, Guerrero CA, Ambhaikar NB, Hafensteiner BD. Angew. Chem., Int. Ed. 2005;44:606–609. doi: 10.1002/anie.200461864. [DOI] [PubMed] [Google Scholar]
- 132.Baran PS, Guerrero CA, Hafensteiner BD, Ambhaikar NB. Angew. Chem., Int. Ed. 2005;44:3892–3895. doi: 10.1002/anie.200500655. [DOI] [PubMed] [Google Scholar]
- 133.Baran PS, Hafensteiner BD, Ambhaikar NB, Guerrero CA, Gallagher JD. J. Am. Chem. Soc. 2006;128:8678–8693. doi: 10.1021/ja061660s. [DOI] [PubMed] [Google Scholar]
- 134.Trost BM, Cramer N, Bernsmann H. J. Am. Chem. Soc. 2007;129:3086–3087. doi: 10.1021/ja070142u. [DOI] [PubMed] [Google Scholar]
- 135.Greshock TJ, Grubbs AW, Jiao P, Wicklow DT, Gloer JB, Williams RM. Angew. Chem., Int. Ed. 2008;47:3573–3577. doi: 10.1002/anie.200800106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pichowicz M, Simpkins NS, Blake AJ, Wilson C. Tetrahedron. 2008;64:3713–3735. [Google Scholar]
- 137.Frebault F, Simpkins NS, Fenwick A. J. Am. Chem. Soc. 2009;131:4214–4215. doi: 10.1021/ja900688y. [DOI] [PubMed] [Google Scholar]
- 138.Frebault FC, Simpkins NS. Tetrahedron. 2010;66:6585–6596. [Google Scholar]
- 139.McAfoos TJ, Li S, Tsukamoto S, Sherman DH, Williams RM. Heterocycles. 2010;82:461–472. doi: 10.3987/COM-10-S(E)19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Laws SW, Scheerer JR. J. Org. Chem. 2013;78:2422–2429. doi: 10.1021/jo3026059. [DOI] [PubMed] [Google Scholar]
- 141.Simpkins NS, Pavlakos I, Weller MD, Male L. Org. Biomol. Chem. 2013;11:4957–4970. doi: 10.1039/c3ob40979a. [DOI] [PubMed] [Google Scholar]
- 142.Sunderhaus JD, McAfoos TJ, Finfield JM, Kato H, Li S, Tsukamoto S, Sherman DH, Williams RM. Org. Lett. 2013;15:22–25. doi: 10.1021/ol302901p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Trost BM, Bringley DA, Zhang T, Cramer N. J. Am. Chem. Soc. 2013;135:16720–16735. doi: 10.1021/ja409013m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mercado-Marin EV, Sarpong R. Chem. Sci. 2015;6:5048–5052. doi: 10.1039/c5sc01977j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhang B, Zheng W, Wang X, Sun D, Li C. Angew. Chem., Int. Ed. 2016;55:10435–10438. doi: 10.1002/anie.201604754. [DOI] [PubMed] [Google Scholar]
- 146.Mukai K, de Sant'Ana DP, Hirooka Y, Mercado-Marin EV, Stephens DE, Kou KGM, Richter SC, Kelley N, Sarpong R. Nat. Chem. 2018;10:38–44. doi: 10.1038/nchem.2862. [DOI] [PMC free article] [PubMed] [Google Scholar]