Abstract
To determine the transcriptional profile of synovium during the molecular phase of post-traumatic osteoarthritis, anterior cruciate ligament transections (ACL) were performed in 36 Yucatan minipigs. Equal numbers were randomly assigned to no further treatment, ACL reconstruction or repair. Perimeniscal synovium for histopathology and RNA-sequencing was harvested at 1 and 4 weeks post-operatively and from 6 healthy control animals. Microscopic synovitis scores significantly worsened at 1 (p<0.001) and 4 weeks (p=0.003) post-surgery relative to controls, and were driven by intimal hyperplasia and increased stromal cellularity without inflammatory infiltrates. Synovitis scores were similar between no treatment, reconstruction and repair groups (p≥.668). Relative to no treatment at 1 week, 88 and 367 genes were differentially expressed in the reconstruction and repair groups, respectively (227 and 277 at 4 weeks). Relative to controls and with the treatment groups pooled, 1683 transcripts were concordantly differentially expressed throughout the post-surgery time-course. Affected pathways included, Proteolysis_Connective tissue degradation (including upregulations of protease-encoding MMP1, MMP13, and ADAMTS4), and Development_Cartilage development (including upregulations of ACAN, SOX9 and RUNX2), amongst others. Using linear regression, significant associations of post-surgery synovial expression levels of 20 genes with the articular cartilage glycosaminoglycan loss were identified. These genes were predominantly related to Embryonic skeletal system development and included RUNX2. In conclusion, this study confirmed an increased synovial expression of genes that may serve as targets to prevent cartilage degradation, including MMP1, MMP13, and ADAMTS4, in knees with microscopic synovitis and cartilage proteoglycan loss. Attractive novel targets include regulators of embryonic developmental processes in synovium.
Keywords: Osteoarthritis, Synovium, RNA-sequencing, Animal Model, ACL
Introduction
Synovitis has been linked to symptoms and structural severity of knee osteoarthritis (OA)1,2. The link between synovitis and OA has been established in-vitro3 and clinically2. From early organ culture experiments, Fell and Jubb concluded that “the synovium has a direct, presumably enzymatic action on the cartilage matrix, and an indirect effect mediated through the chondrocytes”3. While synovitis is a common feature of idiopathic OA4, it also occurs following acute knee injury5–8. Given the association between synovitis and cartilage loss, therapies that target synovitis soon after injury may potentially modulate symptoms and the structural progression of the disease9–11.
The matrix metalloproteinases (MMPs) are thought to play a significant role in cartilage breakdown in both early and late stage osteoarthritis. MMP-1, -8, -13 and -14 have been implicated, with MMP-13 thought to be the most critical player12–14. Studies have demonstrated that mice lacking MMP-13 were protected from cartilage damage following surgical induction of OA, likely due to reduced type II collagen proteolysis14. Aggrecan, another major structural protein of cartilage, is also cleaved by proteinases in the “A Disintegrin and Metalloproteinase with Thrombospondin motifs” (ADAMTS) family15. While there has been interest in MMP and ADAMTS expression in chondrocytes13; little is known about the expression of these genes in the synovium in early post-traumatic OA.
Using a porcine model, we recently reported the transcriptional and histologic response of chondrocytes and articular cartilage to the surgical induction of OA, in which microscopic cartilage damage was observed at 1 and 4 weeks after anterior cruciate ligament (ACL) surgery when compared to healthy cartilage from uninjured control animals16. From the same animals, we obtained synovium samples, and performed microscopic synovitis scoring and RNA-sequencing (RNA-seq) to define the response of the synovium to the surgical induction of OA. We hypothesized that microscopic synovitis would develop and that concomitant changes in synovial gene expression would be present. Combining these findings with our recently reported articular cartilage histology data16, we also hypothesized that we could identify expression of synovial genes that are associated with articular cartilage glycosaminoglycan loss.
Materials & Methods
Study design
A controlled, large animal experiment with cross-sectional assessments at two post-surgical time points was designed. Approval of the Institutional Animal Care and Use Committee was obtained prior to performing this study. Forty-two adolescent Yucatan minipigs (Sinclair BioResources, Columbia MO) were allocated to receive unilateral ACL transection surgery (n=36) or no surgery (INTACT, n=6). Animals receiving ACL transection surgery were allocated to euthanasia and assessment at 1 week (1W, n=18) or 4 weeks (4W, n=18) after surgery. Within each time-point, 6 of 18 animals were allocated to no treatment following ACL transection, 6 of 18 to immediate ACL reconstruction surgery, and 6 of 18 to immediate ACL repair surgery. A computer-based random permutation stratified for sex determined each animal's group allocation and side of unilateral surgery with an equal number of males and females in each group.
Microscopicinflammation of the perimeniscal medial synovium (semiquantitative scoring)17 and site-matched transcriptome-wide gene expression levels were the primary outcomes, which were compared between the ACL surgery and intact control groups. Pathway enrichment analysis was used to identify functional relations between the differentially expressed genes. Correlations between synovial gene expression levels and articular cartilage glycosaminoglycan (GAG) loss (loss of red saturation in Safranin-O staining) in surgical knees were performed. Secondary outcomes included the macroscopic synovitis score (Osteoarthritis Research Society International (OARSI) guidelines)18 and the individual parameters of the microscopic synovitis score17.
Animal model
While the animals were under anesthesia, a medial arthrotomy was performed and the fat pad partially resected to expose the ACL. The ACL was transected between the proximal and middle thirds of the ligament. A clinical exam was performed to verify ACL transection. In the animals assigned to receive ACL reconstruction surgery, a fresh-frozen bone-patellar-tendon-bone allograft harvested from an age-, weight-, and sex-matched donor, was implanted as previously described19. In the animals assigned to ACL repair surgery, an extracellular matrix scaffold in combination with autologous blood was implanted as previously described19. Knees were thoroughly irrigated with 500 mL of normal saline following ACL transection in the group that did not receive further surgical treatment or following bone tunnel placement in those that received ACL reconstruction or repair. All incisions were closed in layers. No postoperative immobilization was used. Detailed information regarding animal husbandry and pain management are available in the supplemental methods (see Supplementary Methods).
Synovium sample collection
Upon harvest, the medial meniscus of the surgical leg along with the attached synovium and capsule were excised. This tissue sample was divided according to the frontal plane through the center of the pars intermedia. The synovium posterior of this plane was harvested for immediate RNA extraction, as described below. The meniscus, along with the attached synovium and capsule anterior of this plane, was immediately immersed in 10% neutral buffered formalin and used subsequently used for histopathological analyses.
Gross synovium analysis
The gross characteristics of the perimeniscal synovium were scored according to the OARSI recommendations18 by one reader, who was blinded to the group and time allocation. The ordinal grades ranged from 0 (Normal, i.e. opal white, semitranslucent, smooth, with sparse well defined blood vessels) to 5 (Severe, i.e. diffuse involvement, severe discoloration, consistent and severe fibrillation, thickening to the point of fibrosis, severe proliferation and hypervascularity).
Sample processing for histopathology
The medial perimeniscal synovium samples described above were fixed in 10% neutral buffered formalin for 48 hours. Samples were then dehydrated in 70%, 95% and 100% Ethanol, 1:1 Ethanol/Xylene and 100% Xylene at room temperature for 24 hours each, immersed in paraffin at 60°C for 48 hours and then embedded. 6 μm sections were mounted on silanized microscope slides (Superfrost Plus, Thermo Scientific, Waltham, MA) and stained with hematoxylin and eosin. Staining and whole slide imaging were performed in one batch. The images were acquired using the 20× objective on a VS120-S6-W microscope system (Olympus, Tokyo, Japan).
Microscopic synovium analysis
Microscopic scoring of the perimeniscal synovium was performed by one reader on the whole slide images using the semiquantitative scoring system developed by Krenn et al.17. The group and time assignment for each slide was blinded during scoring and so were the corresponding gene expression and cartilage outcomes. The assessment included the degree of intimal hyperplasia, stromal cellularity and inflammatory infiltration (each parameter from 0-absent to 3-strong)17. The intimal hyperplasia parameter was scored based on the number of lining cell layers (ranging from 0-one layer to 3-more than five layers). The stromal cellularity parameter was scored based on the density of resident synovial cells, such as fibroblasts, fibrocytes, endothelial cells and macrophages (ranging from 0-normal cellularity to 3-greatly increased cellularity with presence of multinucleated giant cells). The inflammatory infiltration parameter was scored based on the presence of non-resident synovial cells, such as lymphocytes and plasma cells (ranging from 0-no inflammatory infiltrate to 3-dense band like inflammatory infiltrate or numerous large follicle-like aggregates). Lymphocytes, plasma cells and granulocytes can readily be identified and distinguished from resident synovial cells on H&E stained sections. The microscopic sum score represented the sum of all parameters and ranged from 0-9 (0-no synovitis, 9-highest degree of synovitis). Detailed instructions of the scoring system are provided in the original publication17.
In addition, two microscopic quantitative measures of synovitis were employed on the H&E stained sections, including the synovial cell count and the tissue area. The synovial cell count was calculated by using image analysis software (FIJI ImageJ 1.5 20) to determine the number of cell nuclei present in the synovial tissue in a 5.5 mm × 5.2 mm field of view centered on the base of the medial meniscus. One reader, blinded to group allocation and other outcome measures, manually selected the synovial tissue. The synovial tissue area is the area of the described selection in pixel and was determined using image analysis software (FIJI ImageJ 1.5 20).
Synovium RNA-seq
Synovium samples were immediately placed in 2 ml Lysing matrix S tubes (MP Biomedical) and homogenized in 1 ml TRIzol (Life Technologies) using a FastPrep-24 Instrument (MP Biomedical). Samples were then frozen in liquid nitrogen, transferred on dry ice and stored at -80°C until RNA isolation. For total RNA extraction, the homogenisates were brought to room temperature followed by phenol-chloroform separation and on-column purification using the PureLink™ RNA Mini Kit (Life technologies). RNA samples were treated with DNase I (PureLink DNase Set; Life Technologies) and assessed for purity with a NanoDrop (Thermo scientific) and for integrity with an Agilent 2100 Bioanalyzer. The mean 260/280 absorbance ratios were 1.98, 2.08, and 2.08 for the INTACT, 1W, and 4W groups, respectively; the mean 260/230 ratios were 1.67, 2.35, and 2.20, and mean RNA integrity numbers (RINs) were 7.5, 8.4, and 8.2, respectively (Table 1; see Supplementary Table 1 for detailed information about RNA purity and integrity information for all groups). The samples were then enriched for poly(A+) messenger RNA, reverse transcribed with random hexamers, ligated with indexed adapters, and amplified with 17 cycles of polymerase chain reaction using a TruSeq RNA Sample Preparation Kit version 2 (Illumina). Following the removal of primer dimers by magnetic bead–based purification, samples were pooled and sequenced with 10 or 11 libraries per lane on an Illumina HiSeq 2500 machine with 50-bp paired-end reads (Biopolymers Facility, Harvard Medical School, Boston, MA). An average of 17.4, 17.5, and 14.4 million reads per sample for the INTACT, 1W and 4W groups were obtained. Of those 83.1%, 79.5%, 81.2%, were uniquely mapped to the pig genome (susScr3 at the University of California, Santa Cruz Genome Browser [http://hgdownload.cse.ucsc.edu/downloads.html]) with the RNA-seq Unified Mapper (RUM) pipeline21 (Table 1; see Supplementary Table 1 for number of reads and mapping information for all groups). Reads uniquely aligned to the exons of each gene were counted with a custom R script22 that uses Rsamtools23 and GenomicFeatures24 packages. Reads per kilobase of transcript per million mapped reads (RPKM) were calculated for each transcript as a measure of transcript abundance in the cartilage samples. Differential expression and ontology term enrichment analyses were performed as described below. Transcriptome-wide gene expression data is available at ArrayExpress (E-MTAB-[will be added upon approval]).
Table 1.
Group-specific synovitis assessments, RNA and sequencing qualities, extent of synovial transcriptomes and quantitative cartilage assessment (surgical treatment groups pooled; see Supplementary Table 1 for the comparison of the surgical treatment groups; see Sieker et al. JOR 2017 for baseline characteristics including age, weight and sex).
| Scale | Intact (n=6) | 1W (n=18) | 4W (n=18) | 1W/Intact | 4W/Intact | 4W/1W | |
|---|---|---|---|---|---|---|---|
| Synovitis Assessments | Median (Range) | Median (Range) | Median (Range) | P value | P value | P value | |
| Macroscopic score (OARSI) | 0-5 | 1 (0, 2) | 2 (1, 3) | 2 (1, 3) | .125 | .140 | .753 |
| Microscopic sum (Krenn) | 0-9 | 0 (0, 1) | 4 (1, 8) | 3 (1, 8) | <.001 | .003 | .179 |
| Lining score | 0-3 | 0 (0, 1) | 2 (1, 3) | 1.5 (0, 3) | <.001 | .006 | .123 |
| Stromal cellularity score | 0-3 | 0 (0, 0) | 1 (0, 3) | 1 (0, 3) | .001 | .005 | .363 |
| Infiltration score | 0-3 | 0 (0, 0) | 0.5 (0, 2) | 0 (0, 2) | .097 | .307 | .313 |
| Mean (95% CI) | Mean (95% CI) | Mean (95% CI) | P value | P value | P value | ||
| Cellcount (in 1000 cells) | 0-∞ | 0.89 (-0.22, 2.00) | 8.80 (5.45, 12.16) | 7.15 (5.19, 9.11) | .002 | .002 | .860 |
| Tissue area (in million px) | 0-∞ | 0.36 (0.01, 0.71) | 3.77 (1.84, 5.70) | 2.43 (1.82, 3.03) | .002 | .002 | .892 |
| RNA and Sequencing quality | Mean (95% CI) | Mean (95% CI) | Mean (95% CI) | P value | P value | P value | |
| Concentration in ng/μl | |||||||
| Pooled | 0-∞ | 126.3 (66.8, 185.9) | 899.9 (712.3, 1087.6) | 1081.9 (867.7, 1296.2) | .005 | <.001 | .202 |
| ACLT only | 0-∞ | 903 (710, 1097) | 786 (560, 1010) | 0.239 | 0.431 | 1 | |
| RECON only | 0-∞ | 870 (538, 1203) | 1270 (764, 1776) | 0.239 | 0.0038 | 1 | |
| REPAIR only | 0-∞ | 926 (468, 1384) | 1191 (951, 1431) | 0.239 | 0.004 | 1 | |
| 260/280 absorbance ratio | |||||||
| Pooled | 0-∞ | 1.98 (1.81, 2.14) | 2.08 (2.06, 2.10) | 2.08 (2.06, 2.09) | .594 | .694 | .594 |
| ACLT only | 0-∞ | 2.1 (2.1, 2.1) | 2.1 (2.0, 2.1) | 1 | 1 | 1 | |
| RECON only | 0-∞ | 2.1 (2.0, 2.1) | 2.1 (2.0, 2.1) | 1 | 1 | 1 | |
| REPAIR only | 0-∞ | 2.1 (2.0, 2.1) | 2.1 (2.1, 2.1) | 1 | 1 | 1 | |
| 260/230 absorbance ratio | |||||||
| Pooled | 0-∞ | 1.67 (1.12, 2.23) | 2.35 (2.16, 2.53) | 2.20 (2.11, 2.30) | .081 | .209 | .407 |
| ACLT only | 0-∞ | 2.3 (2.3, 2.3) | 2.2 (2.1, 2.4) | 0.61 | 0.91 | 1 | |
| RECON only | 0-∞ | 2.2 (2.0, 2.4) | 2.3 (2.3, 2.3) | 1 | 1 | 1 | |
| REPAIR only | 0-∞ | 2.6 (2.0, 3.1) | 2.3 (2.3, 2.3) | 0.44 | 0.86 | 1 | |
| RNA integrity number (RIN) | |||||||
| Pooled | 7.5 (7.0, 7.9) | 8.4 (8.1, 8.8) | 8.2 (7.7, 8.6) | .027 | .065 | .509 | |
| ACLT only | 8.3 (7.6, 8.9) | 8.0 (7.2, 8.8) | 1 | 1 | 1 | ||
| RECON only | 8.4 (7.6, 9.2) | 8.2 (7.3, 9.1) | 0.955 | 1 | 1 | ||
| REPAIR only | 8.7 (8.2, 9.1) | 8.3 (7.8, 8.7) | 0.131 | 1 | 1 | ||
| Number of reads in millions | |||||||
| Pooled | 0-∞ | 17.4 (13.5, 21.3) | 17.5 (12.8, 22.2) | 14.4 (13.3, 15.4) | .635 | .635 | .635 |
| ACLT only | 0-∞ | 14.7 (12.0, 17.5) | 14.6 (12.8, 16.3) | 1 | 1 | 1 | |
| RECON only | 0-∞ | 15.9 (8.9, 22.8) | 15.3 (13.3, 17.3) | 1 | 1 | 1 | |
| REPAIR only | 0-∞ | 21.9 (9.8, 33.9) | 13.2 (11.6, 14.8) | 1 | 1 | 0.761 | |
| Reads uniquely mapped in % | |||||||
| Pooled | 0-100 | 83.1 (82.2, 84.1) | 79.5 (78.1, 80.9) | 81.2 (79.9, 82.5) | .028 | .206 | .206 |
| ACLT only | 0-100 | 78.3 (77.0, 79.6) | 78.7 (76.8, 80.6) | 0.052 | 0.95 | 1 | |
| RECON only | 0-100 | 77.1 (75.4, 78.7) | 80.6 (79.9, 81.4) | 0.011 | 0.99 | 0.853 | |
| REPAIR only | 0-100 | 83.1 (82.6, 83.6) | 84.4 (83.6, 85.1) | 1 | 1 | 1 | |
| Synovial Transcriptome | Mean (95% CI) | Mean (95% CI) | Mean (95% CI) | P value | P value | P value | |
| Transcripts >1RPKM in thousands | |||||||
| Pooled | 0-∞ | 12.6 (12.5, 12.8) | 12.8 (12.7, 12.9) | 12. 6 (12.4, 12.7) | .460 | .694 | .072 |
| ACLT only | 0-∞ | 12.8 (12.6, 13.0) | 12.4 (12.3, 12.5) | 1 | 1 | 0.372 | |
| RECON only | 0-∞ | 12.7 (12.4, 13.0) | 12.7 (12.5, 13.0) | 1 | 1 | 1 | |
| REPAIR only | 0-∞ | 12.9 (12.7, 13.1) | 12.5 (12.2, 12.8) | 1 | 1 | 1 | |
| Quantitative Cartilage Assessment | Mean (95% CI) | Mean (95% CI) | Mean (95% CI) | P value | P value | P value | |
| Safranin-O saturation | |||||||
| Pooled | 0-255 | 220 (217, 223) | 195 (182, 208) | 190 (175, 205) | 0.072 | 0.040 | 0.587 |
| ACLT only | 0-255 | 209 (191, 226) | 211 (196, 226) | 1 | 1 | 1 | |
| RECON only | 0-255 | 196 (178, 213) | 186 (177, 196) | 0.859 | 0.115 | 1 | |
| REPAIR only | 0-255 | 181 (156, 210) | 172 (134, 210) | 0.311 | 0.180 | 1 | |
Statistical analysis
Group-wise comparisons of macroscopic and microscopic outcomes were carried out in two iterations using the Kruskal-Wallis test, followed by Dunn's post-hoc test with Holm's p-value adjustment to correct for multiple comparisons. First, all possible combinations of factors, ACL status, post-surgery time and treatment, were compared (i.e. INTACT, 1W ACLT, 1W Reconstruction, 1W Repair, 4W ACLT, 4W Reconstruction and 4W Repair (n=6 in each group)). Second, groups defined by ACL status and post-surgery time only were compared with surgical treatments being pooled (INTACT n=6, 1W n=18 and 4W m=18). Adjusted p-values<.05 were considered statistically significant.
Differential expression analysis
Differential expression analyses were performed using the edgeR package25 in R version 3.2.3 (The R Foundation for Statistical Computing) by comparing the counts of reads uniquely aligned to exons between groups. P-values were adjusted for transcriptome-wide testing using the Benjamini and Hochberg method to represent the false discovery rate26. Adjusted p-values<.05 were only considered significant when the expression levels were >1RPKM in at least one of the compared groups. To eliminate the risk of a single outlier producing a significant result, p-values were required to be reproducibly <.05 while each single sample was left out during replications of the tests.
Linear regression analyses were carried out on the data from the surgical animals only (n=36). Transcripts that were significantly differentially expressed in the comparison of 1W and 4W post-surgery animals with INTACT controls were evaluated for associations with the site-matched cartilage GAG loss16. The associations of medial femoral condyle articular cartilage Safranin-O saturation (SAF-O; ranging from 0-absent staining to 255-full red saturation throughout the entire cartilage section) with synovial transcript expression levels after adjustment for post-surgery time were tested. Transcripts with mean abundances <1RPKM in the surgical samples were eliminated prior to performing the test. P-values were used to calculate the false-discovery rate (FDR) according to Benjamini and Hochberg.26 Associations with both p-value<.05 and FDR≤.20 were defined statistically significant.
Pathway enrichment analyses
The differentially expressed genes were used to analyze the enrichment of specific pathways using the MetaCore bioinformatics suite (Thomson Reuters).27 Two ontologies were used: The Process Networks ontology (Thomson Reuters) and the GO Biological Process ontology (Gene Ontology Consortium). Sus scrufa Ensembl identifiers and, if not recognized, gene symbols were used for the upload of the transcript lists. A similar list containing Ensembl identifiers and gene symbols of all 25,322 transcripts that could possibly be detected using our RNA-seq workflow was uploaded in an identical fashion and used as background list. Enrichment was considered significant when FDR<.05.
Marker genes of monocytes, macrophages, lymphocytes, inflammation and wound healing
A specific set of genes including markers of monocytes, macrophages, and lymphocytes and their function was analyzed in detail (Supplementary Table 6). Monocyte surface markers and molecules involved in their trafficking were evaluated as summarized by Shi and Pamer28. Commonly used macrophage surface markers as summarized by Murray and Wynn and their proposed combinatorial marker systems for the phenotyping of activated macrophages were included29. Lymphocyte markers included commonly used T cell, B cell, and natural killer cell surface markers. Furthermore, we assessed additional genes related to inflammation and wound healing (Supplementary Table 7).”
Results
There were no significant differences in baseline age, baseline weight and sex distribution between all groups as previously reported16 (see Supplementary Table 1). No adverse events were observed during surgery or follow-up.
Synovitis outcomes
At both post-surgery time points, there were no significant differences in any macroscopic or microscopic synovitis outcomes between groups with ligament reconstruction, repair surgery, or without additional treatment (see Supplementary Table 1). The results were thus pooled within each time-point for comparison to the age-matched INTACT control knees.
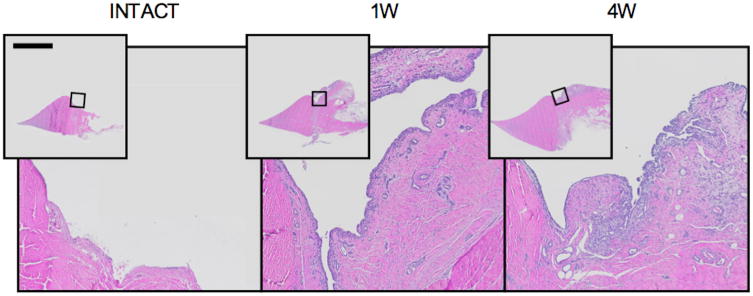
The microscopic synovitis sum score increased significantly from the INTACT controlsto the 1W and 4W samples (Table 1; Figure 1). In addition, increased lining and stromal cellularity scores (i.e. synovial lining thickening and increased number of stromal cells) were observed at 1 and 4W post-surgery (Table 1). There were no significant increases in the infiltration score (i.e. no significant lymphocytic infiltrates) in the macroscopic synovitis score at either post-surgery time. In the quantitative microscopic assessment, the synovial cell count and tissue area both increased significantly from the INTACT group to the 1W and 4W groups (Table 1).
Figure 1.
The histologic response of synovium to ACL surgery. Hematoxylin and eosin stained frontal plane sections of medial meniscus, attached synovium and capsule, corresponding to medians of the microscopic sum score. Bar indicates 5 mm in overview photomicrographs. An increase in synovial tissue area, intimal thickness and stromal cellularity is apparent post-surgery.
RNASeq data for leukocyte markers (Supplementary Table 6) revealed that relevant expression of monocyte (such as CSF1R expressed at 48.5 RPKM) and macrophage surface markers (CD68 expressed at 110.3 RPKM), but not of T-, B- and plasma cell surface markers (CD3, CD19, and CD20<1 RPKM, respectively), was present in the synovium of the INTACT group. Monocyte and macrophage surface markers were downregulated post-surgery, but upregulations of genes indicating macrophage M1 activation (including MARCO, IDO1, AND SOCS3) were observed.
Transcriptional Profile of Healthy Synovium
12,792 out of 25,322 transcripts were expressed with an average ≥1 RPKM in the INTACT control group, the threshold set for relevant expression. RPKM values ranged over 4 orders of magnitude. The majority of transcripts were of low abundance (i.e. 7147 transcripts expressed at 1 to 10 RPKM) or moderate abundance (i.e. 4912 expressed at 10 to 100 RPKM), while fewer transcripts were of high abundance (i.e. 620 transcripts expressed at 100-1000 RPKM and 113 transcripts expressed at >1000 RPKM). Conversely, the 113 and 620 transcripts expressed at high abundance contributed 54.36% and 22.39% of the cumulative RPKMs, while moderate and low abundant transcripts contributed only 18.90% and 4.09% respectively.
The highest expression levels for genes coding for secreted proteins (i.e. proteins annotated to be secreted into the extracellular space in the Uniprot database) were observed for DCN, APOR, MGP and FN. The top abundant non-mitochondrial, non-ribosomal protein-coding transcripts with expression levels >2000 RPKM are displayed in Table 2 (see Supplementary Table 2 for the full transcriptional profiles of all groups).
Table 2.
Transcriptional profile of healthy synovium and synovium at 4 weeks following the surgical induction of osteoarthritis. Non-mitochondrial, non-ribosomal protein coding genes with RPKM>2000 (see Supplementary Table 2 for full list).
| Rank | Gene symbol * | Protein name | Intact RPKM ** | Secreted *** |
|---|---|---|---|---|
| Healthy Synovium | ||||
| 1 | DCN | Decorin | 9165.4 | Yes |
| 2 | APOR | Apolipoprotein R | 8253.7 | Yes |
| 3 | EF1ALPHA | Elongation factor 1-alpha 1 | 8239.9 | - |
| 4 | UBC | Ubiquitin C | 6830.0 | - |
| 5 | MGP | Matrix Glaprotein | 6133.0 | Yes |
| 6 | FN1 | Fibronectin | 5739.8 | Yes |
| 7 | CLU | Clusterin | 4577.6 | Yes |
| 8 | COL3A1 | Collagen alpha-1(III) chain | 3970.5 | Yes |
| 9 | FTL | Ferritin light chain | 3840.3 | - |
| 10 | CST3 | Cystatin-C | 3567.2 | Yes |
| 11 | FTH1 | Ferritin heavy chain | 3207.1 | - |
| 12 | VIM | Vimentin | 2948.8 | Yes |
| 13 | S100A6 | Protein S100-A6 | 2796.7 | Yes |
| 14 | C1QC | Complement C1q subcomponent subunit C | 2775.9 | Yes |
| 15 | ENSSSCG00000004579 | Uncharacterizedprotein | 2729.1 | - |
| 16 | PRG4 | Proteoglycan 4 | 2502.1 | Yes |
| 17 | TMSB10 | Thymosin beta-10 | 2311.3 | - |
| 18 | CTSB | Cathepsin B | 2272.9 | Yes |
| Synovium at 4W post-surgery | ||||
| 1 | COL3A1 | Collagen alpha-1(III) chain | 12194.9 | Yes |
| 2 | COL1A1 | Collagen alpha-1(I) chain | 6962.5 | Yes |
| 3 | EF1ALPHA | Elongation factor 1-alpha 1 | 6205.8 | - |
| 4 | FN1 | Fibronectin | 5488.4 | Yes |
| 5 | UBC | Ubiquitin C | 5352.5 | - |
| 6 | DCN | Decorin | 4084.1 | Yes |
| 7 | SPARC | SPARC (Secreted protein acidic and rich in cysteine) | 3832.4 | Yes |
| 8 | FTH1 | Ferritin heavy chain | 3398.3 | - |
| 9 | PRG4 | Proteoglycan 4 | 3211.0 | Yes |
| 10 | VIM | Vimentin | 3093.6 | Yes |
| 11 | SPARC **** | SPARC (Secreted protein acidic and rich in cysteine) | 2990.2 | Yes |
| 12 | COL1A2 | Collagen alpha-2(I) chain | 2963.0 | Yes |
| 13 | ENSSSCG00000004579 | Uncharacterizedprotein | 2868.5 | - |
| 14 | TMSB10 | Thymosin beta-10 | 2817.0 | - |
| 15 | CLU | Clusterin | 2779.3 | Yes |
| 16 | S100A6 | Protein S100-A6 | 2518.4 | Yes |
| 17 | FTL | Ferritin light chain | 2507.1 | - |
| 18 | APOR | Apolipoprotein R | 2488.2 | Yes |
| 19 | MGP | Matrix Glaprotein | 2363.8 | Yes |
Ensemble identifier is provided when gene symbol is not available,
RPKM = reads per kilobase of transcript per million mapped reads,
Encoding secreted proteins based on annotations in the Uniprot database,
Orthologue.
Transcriptional response of synovium to ACL surgery
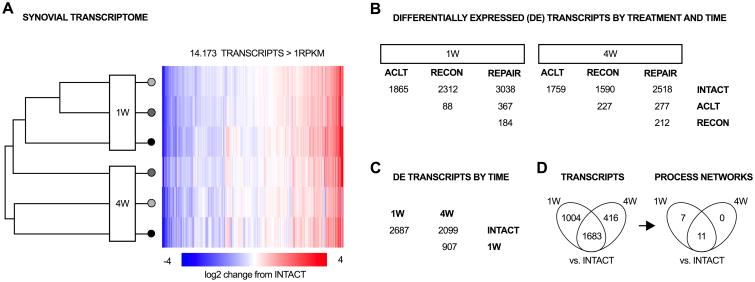
At 1W post-surgery, 1865, 2260 and 2872 transcripts were differentially abundant in the comparison of ACLT, RECON and REPAIR groups with the INTACT control group, while only 82 out of 25,322 (0.3%) transcripts were differentially abundant between RECON and ACLT group, and 133 transcripts (0.5%) were differentially abundant between REPAIR and RECON group (Figure 2B). At 4W post-surgery, 1767, 1525 and 2466 transcripts were differentially abundant in the comparison of ACLT, RECON and REPAIR groups with the INTACT control group, while only 192 (0.7%) transcripts were differentially abundant between RECON and ACLT group, and 185 (0.7%) transcripts were differentially abundant between REPAIR and RECON group (Figure 2B). Since the majority of changes were observed due to ACL transection surgery independent of treatment, the results were thus pooled for each time-point for this report; however, lists of all differentially expressed genes by treatment group are also provided (see Supplementary Table 3).
Figure 2.
The transcriptional response of synovium to surgical induction of osteoarthritis. (A) 14,173 transcripts were expressed >1 RPKM in at least one of the 7 groups defined by ACL status, time and treatment. Log2 fold-changes compared to INTACT controls are depicted as heatmap. (B) Number of differentially expressed genes by time-point and treatment. Most differentially expressed genes were detected in the comparison of post-surgery groups with intact controls, rather than between the surgical groups (each group with n=6). (C) Number of differentially expressed genes by time-point (with surgical groups pooled, 1W and 4W with n=18 each, Intact with n=6). Vast changes in gene expression were observed between 1 or 4 week post-surgery with controls and between 1 and 4 weeks post-surgery. (D) 1683 transcripts were concordantly differentially expressed in the comparison of 1W and 4W with intact controls (One disconcordant between 1 and 4 weeks). Those transcripts enriched 11 pathways (process network ontology), including terms related to cell cycle, cytoskeleton, cell adhesion, inflammation, proteolysis, development, and signal transduction.
With surgical treatment groups pooled, 13,052 and 12,908 out of 25,322 transcripts were expressed with an average ≥1 RPKM in the 1W and 4W post-surgery groups, respectively. 15 of the 18 genes expressed at high levels (over 2000 RPKM) in intact synovium were also expressed at high levels in the synovium at 4 weeks after surgery. While CST3, C1QC and CTSB were in the high expression list for intact, they were replaced by COL1A1, COL1A2, and SPARC at 4 weeks, and in addition, COL3A1 tripled in abundance between intact and 4 weeks after surgery (Table 2). In comparison to intact controls, 2687 transcripts were differentially expressed at 1W post-surgery and 2099 transcripts were differentially expressed at 4W post-surgery. Amongst genes encoding for secreted proteins, the largest upregulations were observed for MMP13 (upregulated by 767- and 381-fold at 1 and 4 weeks post-injury). The 20 most significantly changed transcripts are shown in Table 3 (see Supplementary Table 3 for full list).
Table 3.
Transcriptional response of synovium to the surgical induction of osteoarthritis. Concordantly differentially expressed genes at 1 and 4 weeks (ranked by p-value; excluding mitochondrial, ribosomal and non-protein coding genes; see Supplementary Table 3 for full list).
| Rank | Gene symbol * | Description | Intact RPKM** | 1W/Intact log2FC | 4W/Intact log2FC | 4W/Intact PVALUE | Secreted *** |
|---|---|---|---|---|---|---|---|
| 1 | CALCB **** | Calcitonin gene-relatedpeptide 2 | 82.3 | -5.66 | -6.88 | 1.51E-107 | Yes |
| 2 | CRSP2 | Calcitonin receptor-stimulating peptide 2 | 14.0 | -7.06 | -8.55 | 3.65E-104 | Yes |
| 3 | NPY5R | Neuropeptide Y receptor type 5 | 7.9 | -7.28 | -6.55 | 7.76E-86 | - |
| 4 | LUZP2 | Leucine zipper protein 2 | 3.9 | -6.63 | -5.46 | 1.32E-75 | Yes |
| 5 | MYOT | Myotilin | 0.1 | 4.94 | 8.45 | 7.81E-61 | - |
| 6 | ENSSSCG00000023896 | Uncharacterized protein | 22.0 | -4.54 | -4.72 | 1.44E-59 | - |
| 7 | MMP-13 | Collagenase 3 | 0.1 | 9.58 | 8.57 | 1.04E-58 | Yes |
| 8 | MYH2 **** | Myosin-2 | 0.1 | 5.64 | 7.69 | 5.14E-56 | - |
| 9 | CDHR5 | Cadherin-related family member 5 | 15.3 | -4.46 | -4.27 | 8.43E-56 | - |
| 10 | MYH2 | Myosin-2 | 0.1 | 6.21 | 7.59 | 2.70E-55 | - |
| 11 | MYBPC1 | Myosin-binding protein C, slow-type | 0.4 | 4.06 | 7.21 | 8.48E-54 | - |
| 12 | ACTA1 | Actin, alpha skeletal muscle | 6.3 | 2.18 | 6.89 | 1.95E-52 | - |
| 13 | TNI-S4 | Troponin I, slow skeletal muscle | 0.5 | 2.98 | 7.21 | 9.03E-52 | - |
| 14 | ENSSSCG00000022671 | Uncharacterized protein | 11.9 | -4.29 | -4.56 | 3.76E-50 | - |
| 15 | MYL1 | Myosin light chain 1/3, skeletal muscle isoform | 2.8 | 4.06 | 6.61 | 1.19E-48 | - |
| 16 | XIRP2 | Xinactin-binding repeat-containing protein 2 | 0.0 | 5.55 | 6.72 | 1.87E-47 | - |
| 17 | MYOZ1 | Myozenin-1 | 0.7 | 3.55 | 6.59 | 1.93E-47 | - |
| 18 | VIT | Vitrin | 29.1 | -4.75 | -3.88 | 3.07E-46 | Yes |
| 19 | ENSSSCG00000016397 | Uncharacterized protein | 0.2 | 4.79 | 6.40 | 3.11E-46 | - |
| 20 | SMPX | Small muscular protein | 0.2 | 4.44 | 7.10 | 2.07E-45 | - |
Ensemble identifier is provided when gene symbol is not available,
RPKM = reads per kilobase of transcript per million mapped reads,
Encoding secreted proteins based on annotations in the Uniprot database,
Orthologue.
The set of concordantly differentially expressed genes enriched pathways (Process network ontology) related to six terms including proteolysis, cartilage development and inflammation (see Supplementary Table 4 for details). The proteolysis pathway was enriched by upregulation of genes coding for 6 extracellular matrix molecules (ACAN, COL3A1, COL16A1, FN1, SPARC and TNC; 5203 total RPKM in INTACT to 21,732 RPKM at 4W post-surgery), as well as 5 matrix-metalloproteinases (MMP1, MMP2, MMP9, MMP13 and MMP15; total 590 RPKM in INTACT to 2421 RPKM at 4W post-surgery), and 3 aggrecanases (ADAMTS1, ADAMTS4, ADAMTS9, total 9 RPKM in INTACT to 16 RPKM at 4W post-surgery).
The cartilage development pathway was enriched by upregulation of genes coding for 7 extracellular matrix molecules (ACAN, COL12A1, COL1A1, COL1A2, COL3A1, FBN1, FN1, total 5460 RPKM in INTACT to 27,835 RPKM at 4W post-surgery) and one matrix metalloproteinase (MMP13, 0.1 PRKM in INTACT to 43 RPKM at 4W post-surgery). The genes for the transcription factors RUNX2 and SOX9 were both upregulated in the synovium after ACL surgery (total of 2.2 RPKM in INTACT to 11RPKM at 4W post-surgery).
Amongst pathways pertinent to inflammation, the complement system pathway was the only one found to be significantly enriched. The enrichment was due to downregulation of the genes coding for complement components (C1QC, C2, C3, C8G), positive regulators of complement activation (CFD, CFP), and negative regulators of complement activation (C4BPA) and ITGAM. Although several members of the matrix metalloproteinase family and genes associated with the complement pathway were differentially expressed after surgery, many of the genes associated with classical inflammation were not found to be differentially expressed in the synovium in this model (Supplementary Table 7). These include IL1A, TNFA, NOS2, and PTGS2 (Supplementary Tables 6 and 7). IL1B was downregulated at 4 weeks after surgery.
Correlations between synovitis, synovial gene expression and cartilage glycosaminoglycan loss
The mean articular cartilage Safranin-O saturation decreased from the INTACT controls to the 1W (p=0.072) and 4W samples (p=0.040, see Table 1). In post-surgical animals (n=36) the synovial cell count (p<.001), but not the tissue area (p=.103), was significantly associated with the Safranin-O saturation, with high synovial cell counts being present in individuals with a greater loss of Safranin-O saturation (Table 4). Further, the synovial expression levels of 138 transcripts were associated with the cartilage Safranin-O saturation with unadjusted p-values<.05. Of those, 20 transcripts were associated with a FDR≤.20 (Table 4). The highest R2 value was found for ITGA8, which encodes Integrin alpha-8. Higher post-surgery expression levels in the synovium were associated with a loss of articular cartilage Safranin-O saturation (Table 4).
Table 4.
Relations of microscopic synovitis and synovium gene expression with articular cartilage glycosaminoglycan loss in post-surgery animals (Transcripts with p < 0.05 and FDR ≤ 0.20).
| Gene symbol | Description | Slope * | R̂2 | p | FDR |
|---|---|---|---|---|---|
| Synovial cell count | - | 0.31 | 5.23E-04 | ||
| Synovial tissue area | + | 0.09 | .103 | ||
| ITGA8 | Integrin alpha-8 | - | 0.44 | 1.51E-05 | 0.02 |
| GATM | Glycine amidinotransferase, mitochondrial | - | 0.40 | 5.11E-05 | 0.04 |
| U6 | - | - | 0.37 | 1.27E-04 | 0.06 |
| FGF7 | Fibroblast growth factor 7 | - | 0.32 | 4.87E-04 | 0.12 |
| ARHGAP28 | Rho GTPase-activating protein 28 | - | 0.32 | 4.99E-04 | 0.12 |
| VMP1 | Vacuole membrane protein 1 | - | 0.31 | 5.69E-04 | 0.12 |
| RUNX2 | Runt-related transcription factor 2 | - | 0.30 | 6.85E-04 | 0.12 |
| ENSSSCG00000020975 ** | Novel protein coding | - | 0.30 | 7.26E-04 | 0.12 |
| EYA3 *** | Eyes absent homolog 3 | - | 0.30 | 7.75E-04 | 0.12 |
| PCOLCE | Procollagen C-endopeptidase enhancer 1 | + | 0.28 | 1.12E-03 | 0.16 |
| ATRAID | All-trans retinoic acid-induced differentiation factor | + | 0.27 | 1.44E-03 | 0.17 |
| ssc-mir-21 | - | - | 0.27 | 1.49E-03 | 0.17 |
| TFPI | Tissue factor pathway inhibitor | - | 0.27 | 1.64E-03 | 0.17 |
| PLXDC1 | Plexin domain-containing protein 1 | + | 0.27 | 1.74E-03 | 0.17 |
| PCDH17 | Protocadherin-17 | - | 0.26 | 2.14E-03 | 0.20 |
| R3HDM1 | R3H domain-containing protein 1 | - | 0.25 | 2.30E-03 | 0.20 |
| EFR3A | Protein EFR3 homolog A | - | 0.25 | 2.74E-03 | 0.20 |
| SHOX2 | Short stature homeobox protein 2 | + | 0.25 | 2.82E-03 | 0.20 |
| LEF1 | Lymphoid enhancer-binding factor 1 | - | 0.24 | 2.88E-03 | 0.20 |
| KIAA1217 | Sickle tail protein homolog | - | 0.24 | 2.91E-03 | 0.20 |
A negative slope (-) indicates that higher expression levels are associated with lower Safranin-O saturation values (i.e. GAG loss), a positive slope (+) indicates that higher expression levels are associated with higher Safranin-O saturation values;
gene symbol not available;
orthologue
The 20 transcripts with significant association to articular cartilage glycosaminoglycan loss enriched nine pathways (GO biological process ontology), including embryonic organ development, embryonic skeletal system development, embryonic appendage morphogenesis, embryonic limb morphogenesis, cell-cell adhesion, chondrocyte development and osteoblast differentiation (see Supplementary Table 5). This enrichment was due to ITGA8, RUNX2, LEF1, PCDH17, EFR3A, VMP1, KIAA1217, GATM and SHOX2 of which all but GATM and SHOX2 were upregulated post-surgery.
Discussion
This study established that microscopic synovitis is present during the molecular stage of PTOA in the porcine model, and that the observed increase in synovial cellularity is associated with a broad transcriptional response in the synovium and correlates directly with the loss of glycosaminoglycans in the articular cartilage. The extent of microscopic synovitis and cartilage damage was not affected by the surgical treatment. Between 12,532 and 13,150 transcripts expressed with an average ≥1 RPKM in the various conditions examined constitute the synovial transcriptome (see Supplementary Table 2), corresponding to 49.5% and 51.9% of the 25,322 assessed transcripts. The synovial transcriptional response to surgery involved significant changes in the expression of 1590 to 3038 genes, or 6.3 to 12.0% of all assessed genes, whereas treatment-related differential expression involved only 88 to 367 genes, or 0.3 to 1.4% of all assessed genes. Detailed analysis of all treatment-related effects on gene expression are provided (see Supplementary Table 3); however, the focus here was to report the vast, treatment-independent, transcriptional response to ACL transection surgery and to identify associations between synovial expression levels of these genes with the deterioration of adjacent articular cartilage. With treatment groups pooled, a set of 1683 genes (6.6%) was identified, which was concordantly differentially expressed at 1 and 4 weeks post-surgery. This set predominantly included genes involved in extracellular matrix remodeling and cartilage development. Amongst those differentially expressed genes, several genes were identified for which the synovial post-surgery expression directly correlated with the loss of glycosaminoglycans in the articular cartilage. This set of genes particularly included those involved in skeletal development. As there were no genes expressed by the chondrocytes that were similarly correlated with the Safranin-O staining16, our findings suggest that the response of the synovium to the ACL surgery may have a role in the early glycosaminoglycan loss seen in articular cartilage. Whether the glycosaminoglycan loss in the articular cartilage triggers upregulation of genes related to the developmental pathways in the synovium or the pathway activation results in glycosaminoglycan loss remains unclear.
The synovial transcriptional response to surgery included significant expression changes of genes associated with extracellular matrix remodeling, including several that have been previously reported to be differentially expressed by chondrocytes in late osteoarthritis, including MMP1, MMP1330, and ADAMTS-431. MMP1 expression (encoding MMP-1, which also degrades type II collagen) was also upregulated in the synovium after surgery. MMP13 (which is thought to be the collagenase that plays a significant role in the pathogenesis of OA)32 was upregulated by several hundred fold in the synovium. Previous studies have reported MMP-133 and ADAMTS-431 in the synovial fluid of injured joints, and MMP-1 and MMP-13 in the fluid of osteoarthritic joints34. In addition, ADAMTS-4 has been found to correlate with the presence of aggrecan fragments in the synovial fluid.35 Thus, synovial production of MMP-1, MMP-13 or ADAMTS-4 may represent a future target for reducing the synovial fluid levels of these proteases in the molecular phase of osteoarthritis.
The transcriptomic response also resulted in significant changes in genes associated with cartilage matrix formation and cartilage development, including the ACAN, COL2A1, SOX9 and RUNX2 genes. SOX9 and RUNX2 are genes encoding Transcription factor SOX-9 and Runt-related transcription factor 2 (master regulators of cartilage and bone formation, respectively)36,37 and increased SOX-9 deposition was previously observed in areas of ectopic cartilage metaplasia within the synovium of osteoarthritic knee joints of mice38. This suggests that the synovium may not only respond with a fibrotic healing response after joint injury, but may also be activating pathways important for cartilage development and cartilage matrix production. As no cartilage tissue formation was seen in the synovium, it is possible the upregulation of the cartilage formation pathway resulted in the expression of cartilage-associated extracellular matrix proteins by the synoviocytes which could perhaps influence the articular cartilage via the synovial fluid. This hypothesis is supported by prior reports of synovial fluid from knees with OA having increased concentrations of these proteoglycans.39,40 Further studies would be needed to explore this hypothesis.
The changes observed in the synovium, both on a histologic and transcriptomic basis, were also directly associated with glycosaminoglycan loss in the articular cartilage16. Specifically, a higher cell count in the perimeniscal synovium and expression of genes related to skeletal system development were greatest in the knees with the highest loss of glycosaminoglycans in the articular cartilage of the femoral condyle. A similar relation between arthroscopically graded synovitis and chondropathy was detected in the medial tibiofemoral compartment in patients with primary painful knee osteoarthritis fulfilling the American College of Rheumatology criteria, with more severe chondropathy being present in knees with synovial inflammation1. Further, a faster progression of chondropathy within one year was observed in the group with synovitis at baseline1. However, the underlying molecular mechanisms of these associations remained elusive. Intra-articular inflammation has also been previously reported to be present in patients with acutely ACL injured knee joints38–41. In this study, inflammatory pathways were also found to be enriched in both cartilage (Complement system, Interferon signaling and Macrophage migration inhibitory factor (MIF) signaling) and the synovium (Complement system) after the surgical induction of OA. In the cartilage, the observed pathway enrichment was primarily due to the upregulation of pro-inflammatory genes (i.e. C2, C3, C7, CCL2, CCL8, PTGS2, and TLR4 amongst others), while in the synovium, it was primarily due to the downregulation of genes encoding complement components (i.e. C2 and C3). Of note, synovial gene expression levels of the typical pro-inflammatory mediators were not associated with the articular cartilage status, while synovial expression levels of genes pertinent to skeletal system development were associated (see Table 4). These findings should highlight that synovitis not only includes changes related to cell proliferation, pro-inflammatory cytokine and protease expression, but also changes related to metaplasia, with the latter being particularly important due to their relation to the articular cartilage status observed here.
These findings need to be interpreted in the light of several limitations. First, the porcine model is different from the clinical condition. Pigs are quadrupeds and their post-surgery knee joint biomechanics might differ from those in humans. It is also possible that their biologic pathways differ from those in humans to some degree. Nonetheless, many of the wound healing mechanisms and the patterns of post-traumatic OA development after an ACL injury have been shown to be reflective of what occurs in patients.19,45,46 Second, in order to reduce false positive detection rates, we filtered all transcripts that were expressed <1 RPKM in all groups. This threshold might include relevant transcripts, such as those encoding cytokines.
Lastly, although there were only a relatively small number of differentially expressed genes among the surgical groups (between 88 and 367 genes; Provided in Supplementary Table 3), justifying pooling of these groups for this analysis, the number of differentially expressed genes between the individual surgical groups and intact controls varied widely (see Figure 2B). For example, looking at the ACLT and REPAIR groups at 1 week reveals that the repair group had a 63% higher number of genes that were differentially expressed over the INTACT group than the ACLT only group (1865 vs 3038). These numbers correspond to 1526 genes that were differentially expressed in the comparison of the REPAIR and INTACT groups, but not differentially expressed in the comparison of the ACLT and INTACT groups (and 353 genes differentially expressed in the comparison of ACLT and INTACT groups, but not REPAIR and INTACT groups). However, it is important to note that the expression of these genes is not necessarily different between REPAIR and ACLT groups at the conventional level of certainty (i.e. 5% false discovery rate). In fact, only 367 genes were significantly differentially expressed between REPAIR and ACLT groups at 1 week post-surgery in the direct comparison. These genes predominantly included those encoding skeletal muscle components (including higher expression of MYBPH and TNNC2 in the REPAIR group). At 4 weeks post surgery, 277 genes were significantly differentially expressed, which were predominantly related to cell matrix interactions (including higher expression of COL1A1, COL5A1, COL16A1, MMP9, and MMP13 in the REPAIR group). Further studies evaluating the role of these pathways in the early PTOA response are planned.
In conclusion, a strong transcriptional response of the synovium was observed during the molecular disease stage of post-traumatic OA following surgical induction of this disease. We confirmed that synovitis and the upregulation of genes associated with developmental pathways, such as embryonic skeletal system development, were greatest in the knees with the highest loss of proteoglycans in the articular cartilage of the femoral condyle. Whether the synovial gene expression leads to the increased proteoglycan loss in the cartilage or if the proteoglycan loss leads to the observed gene upregulation in the synoviocytes is not yet known. However, our data suggest that reducing synovitis and targeting specific synovial protease production (particularly for MMP-1 and ADAMTS-4 inhibition) may serve as reasonable treatment targets for the amelioration of early post-traumatic OA.
Supplementary Material
Acknowledgments
The authors thank Ugur Ayturk, PhD for providing R scripts that were modified to perform the RNA-seq data analysis, Scott McAllister, BS, Alison Biercevicz, PhD, Andrew Rohan, MS, Katherine Larson, BS, Veronica Bouvier, Roxanne Burrill, Pam Norberg, James Harper, DVM, Lara Helwig, DVM, Tiffany Borgeson, DVM for their assistance and care in handling the minipigs and their engagement in surgery and tissue collection, and Paul Monfils for supporting the histological analyses. This investigation was supported by the National Institutes of Health under NIAMS 2R01-AR056834 and 1R01-AR065462, Children's Hospital Translational Research Program, and the Lucy Lippitt Endowment. Bioanalyzer analysis was performed in the BCH IDDRC Molecular Genetic Core that is supported by National Institutes of Health award NIH-P30-HD18655. Whole Slide Imaging was performed in the Neurobiology Imaging Facility / HNDC Enhanced NeuroImaging Core (NIFENC) which is supported by NIH-NINDS-P30-NS072030. It should be noted that Dr. Murray is an inventor on patents held by Boston Children's Hospital related to one of the ACL surgical procedures described herein, and that Drs. Murray and Fleming recently founded a company (Miach Orthopaedics Inc) to translate that ACL surgical procedure to clinical use. The company currently has no assets.
Footnotes
Authors have made substantial contributions to the study design, acquisition, analysis or interpretation of the data, have contributed to drafting of the manuscript, and have read and approved the final submitted version.
References
- 1.Ayral X, Pickering EH, Woodworth TG, Mackillop N, Dougados M. Synovitis: a potential predictive factor of structural progression of medial tibiofemoral knee osteoarthritis – results of a 1 year longitudinal arthroscopic study in 422 patients. Osteoarthritis and Cartilage. 2005;13:361–367. doi: 10.1016/j.joca.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Felson DT, et al. Synovitis and the risk of knee osteoarthritis: the MOST Study. Osteoarthritis and Cartilage. 2016;24:458–464. doi: 10.1016/j.joca.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fell HB, Jubb RW. The effect of synovial tissue on the breakdown of articular cartilage in organ culture. Arthritis Rheum. 1977;20:1359–1371. doi: 10.1002/art.1780200710. [DOI] [PubMed] [Google Scholar]
- 4.Sellam J, Berenbaum F. The role of synovitis in pathophysiology and clinical symptoms of osteoarthritis. Nat Rev Rheumatol. 2010;6:625–635. doi: 10.1038/nrrheum.2010.159. [DOI] [PubMed] [Google Scholar]
- 5.Beynnon BD, et al. Rehabilitation After Anterior Cruciate Ligament Reconstruction A Prospective, Randomized, Double-Blind Comparison of Programs Administered Over 2 Different Time Intervals. Am J Sports Med. 2005;33:347–359. doi: 10.1177/0363546504268406. [DOI] [PubMed] [Google Scholar]
- 6.Heard BJ, et al. Changes of early post-traumatic osteoarthritis in an ovine model of simulated ACL reconstruction are associated with transient acute post-injury synovial inflammation and tissue catabolism. Osteoarthritis and Cartilage. 2013;21:1942–1949. doi: 10.1016/j.joca.2013.08.019. [DOI] [PubMed] [Google Scholar]
- 7.Roemer FW, Frobell R, Lohmander LS, Niu J, Guermazi A. Anterior Cruciate Ligament OsteoArthritis Score (ACLOAS): Longitudinal MRI-based whole joint assessment of anterior cruciate ligament injury. Osteoarthritis and Cartilage. 2014;22:668–682. doi: 10.1016/j.joca.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 8.Kumahashi N, et al. Type II collagen C2C epitope in human synovial fluid and serum after knee injury--associations with molecular and structural markers of injury. Osteoarthr Cartil. 2015;23:1506–1512. doi: 10.1016/j.joca.2015.04.022. [DOI] [PubMed] [Google Scholar]
- 9.Heard BJ, et al. Single intra-articular dexamethasone injection immediately post-surgery in a rabbit model mitigates early inflammatory responses and post-traumatic osteoarthritis-like alterations. J Orthop Res. 2015;33:1826–1834. doi: 10.1002/jor.22972. [DOI] [PubMed] [Google Scholar]
- 10.Sieker JT, et al. Immediate Administration of Intraarticular Triamcinolone Acetonide After Joint Injury Modulates Molecular Outcomes Associated With Early Synovitis. Arthritis & Rheumatology. 2016;68:1637–1647. doi: 10.1002/art.39631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lattermann C, et al. A Multicenter Study of Early Anti-inflammatory Treatment in Patients With Acute Anterior Cruciate Ligament Tear. Am J Sports Med. 2016 doi: 10.1177/0363546516666818. 0363546516666818. [DOI] [PubMed] [Google Scholar]
- 12.Poole AR, et al. Type II collagen degradation and its regulation in articular cartilage in osteoarthritis. Ann Rheum Dis. 2002;61 Suppl 2:ii78–81. doi: 10.1136/ard.61.suppl_2.ii78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poole AR, et al. Proteolysis of the collagen fibril in osteoarthritis. Biochem Soc Symp. 2003:115–123. doi: 10.1042/bss0700115. [DOI] [PubMed] [Google Scholar]
- 14.Little CB, et al. Matrix metalloproteinase 13–deficient mice are resistant to osteoarthritic cartilage erosion but not chondrocyte hypertrophy or osteophyte development. Arthritis & Rheumatism. 2009;60:3723–3733. doi: 10.1002/art.25002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glasson SS, et al. Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature. 2005;434:644–648. doi: 10.1038/nature03369. [DOI] [PubMed] [Google Scholar]
- 16.Sieker JT, et al. Transcriptional Profiling of Articular Cartilage in a Porcine Model of Early Post-traumatic Osteoarthritis. J Orthop Res. 2017 doi: 10.1002/jor.23644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krenn V, et al. Synovitis score: discrimination between chronic low-grade and high-grade synovitis. Histopathology. 2006;49:358–364. doi: 10.1111/j.1365-2559.2006.02508.x. [DOI] [PubMed] [Google Scholar]
- 18.Little CB, et al. The OARSI histopathology initiative – recommendations for histological assessments of osteoarthritis in sheep and goats. Osteoarthritis and Cartilage. 2010;18(3):S80–S92. doi: 10.1016/j.joca.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 19.Murray MM, Fleming BC. Use of a Bioactive Scaffold to Stimulate Anterior Cruciate Ligament Healing Also Minimizes Posttraumatic Osteoarthritis After Surgery. Am J Sports Med. 2013;41:1762–1770. doi: 10.1177/0363546513483446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 2012;9:676. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grant GR, et al. Comparative analysis of RNA-Seq alignment algorithms and the RNA-Seq unified mapper (RUM) Bioinformatics. 2011;27:2518–2528. doi: 10.1093/bioinformatics/btr427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ayturk UM, et al. An RNA-seq protocol to identify mRNA expression changes in mouse diaphyseal bone: Applications in mice with bone property altering Lrp5 mutations. Journal of Bone and Mineral Research. 2013;28:2081–2093. doi: 10.1002/jbmr.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgan M, Pages H, Obenchain V, Hayden N. Rsamtools: Binary alignment (BAM), FASTA, variant call (BCF), and tabix file import. R Package version 1.24.0. 2016 URl: http://bioconductor.org/packages/release/bioc/html/Rsamtools.html.
- 24.Lawrence M, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. 1995;57:289–300. [Google Scholar]
- 27.Bessarabova M, Ishkin A, JeBailey L, Nikolskaya T, Nikolsky Y. Knowledge-based analysis of proteomics data. BMC Bioinformatics. 2012;13 Suppl 16:S13. doi: 10.1186/1471-2105-13-S16-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi C, Pamer EG. Monocyte recruitment during infection and inflammation. Nat Rev Immunol. 2011;11:762–774. doi: 10.1038/nri3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lambert C, et al. Gene Expression Pattern of Cells From Inflamed and Normal Areas of Osteoarthritis Synovial Membrane. Arthritis & Rheumatology. 2014;66:960–968. doi: 10.1002/art.38315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts S, et al. ADAMTS-4 activity in synovial fluid as a biomarker of inflammation and effusion. Osteoarthr Cartil. 2015;23:1622–1626. doi: 10.1016/j.joca.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dahlberg L, et al. Selective enhancement of collagenase-mediated cleavage of resident type II collagen in cultured osteoarthritic cartilage and arrest with a synthetic inhibitor that spares collagenase 1 (matrix metalloproteinase 1) Arthritis Rheum. 2000;43:673–682. doi: 10.1002/1529-0131(200003)43:3<673::AID-ANR25>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 33.Tchetverikov I, et al. MMP protein and activity levels in synovial fluid from patients with joint injury, inflammatory arthritis, and osteoarthritis. Ann Rheum Dis. 2005;64:694–698. doi: 10.1136/ard.2004.022434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pozgan U, et al. Expression and activity profiling of selected cysteine cathepsins and matrix metalloproteinases in synovial fluids from patients with rheumatoid arthritis and osteoarthritis. Biol Chem. 2010;391:571–579. doi: 10.1515/BC.2010.035. [DOI] [PubMed] [Google Scholar]
- 35.Zhang E, et al. Aggrecanases in the human synovial fluid at different stages of osteoarthritis. Clin Rheumatol. 2013;32:797–803. doi: 10.1007/s10067-013-2171-0. [DOI] [PubMed] [Google Scholar]
- 36.Akiyama‡ H, et al. Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. PNAS. 2005;102:14665–14670. doi: 10.1073/pnas.0504750102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kempf H, Ionescu A, Udager AM, Lassar AB. Prochondrogenic signals induce a competence for Runx2 to activate hypertrophic chondrocyte gene expression. Dev Dyn. 2007;236:1954–1962. doi: 10.1002/dvdy.21205. [DOI] [PubMed] [Google Scholar]
- 38.Kurth TB, et al. Functional mesenchymal stem cell niches in adult mouse knee joint synovium in vivo. Arthritis & Rheumatism. 2011;63:1289–1300. doi: 10.1002/art.30234. [DOI] [PubMed] [Google Scholar]
- 39.Gobezie R, et al. High abundance synovial fluid proteome: distinct profiles in health and osteoarthritis. Arthritis Research & Therapy. 2007;9:R36. doi: 10.1186/ar2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamphorst JJ, et al. Profiling of endogenous peptides in human synovial fluid by NanoLC-MS: method validation and peptide identification. J Proteome Res. 2007;6:4388–4396. doi: 10.1021/pr0704534. [DOI] [PubMed] [Google Scholar]
- 41.Bigoni M, et al. Acute and late changes in intraarticular cytokine levels following anterior cruciate ligament injury. Journal of Orthopaedic Research. 2013;31:315–321. doi: 10.1002/jor.22208. [DOI] [PubMed] [Google Scholar]
- 42.Irie K, Uchiyama E, Iwaso H. Intraarticular inflammatory cytokines in acute anterior cruciate ligament injured knee. The Knee. 2003;10:93–96. doi: 10.1016/s0968-0160(02)00083-2. [DOI] [PubMed] [Google Scholar]
- 43.Cuellar VG, Cuellar JM, Golish SR, Yeomans DC, Scuderi GJ. Cytokine Profiling in Acute Anterior Cruciate Ligament Injury. Arthroscopy: The Journal of Arthroscopic & Related Surgery. 2010;26:1296–1301. doi: 10.1016/j.arthro.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 44.Marks PH, Donaldson MLC. Inflammatory Cytokine Profiles Associated With Chondral Damage in the Anterior Cruciate Ligament–Deficient Knee. Arthroscopy: The Journal of Arthroscopic & Related Surgery. 2005;21:1342–1347. doi: 10.1016/j.arthro.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 45.Seaton M, Hocking A, Gibran NS. Porcine models of cutaneous wound healing. ILAR J. 2015;56:127–138. doi: 10.1093/ilar/ilv016. [DOI] [PubMed] [Google Scholar]
- 46.Okafor EC, et al. The effects of femoral graft placement on cartilage thickness after anterior cruciate ligament reconstruction. J Biomech. 2014;47:96–101. doi: 10.1016/j.jbiomech.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.