Abstract
We investigate the effect of biofouling in a microfluidic filtration system. The microfluidic platform consists of cylindrical microposts with a pore-spacing of 2 μm, which act as the filtration section of the device. One of our key findings is that there exists a critical pressure difference above which pronounced streamer formation is observed, which eventually leads to rapid clogging of the device with an accompanying exponential decrease in permeate flow. Moreover, when streamers do form, de-clogging of pores also occurs intermittently, which leads to small time scale fluctuations [O(101 s)] superimposed upon the large time scale [O(102 min)] clogging of the system. These de-clogging phenomena lead to a sharp increase in water permeation through the microfluidic filtration device but rates the water quality as biomass debris is transported in the permeate. Streamer-based clogging shares similarities with various fouling mechanisms typically associated with membranes. Finally, we also show that the pH of the feed strongly affects biofouling of the microfluidic filtration system.
INTRODUCTION
Recently, several researchers have shown that bacterial biofilm formation in microfluidic devices, with sustained hydrodynamic flow, is accompanied by the formation of filamentous structures called streamers.1–4 Bacterial streamers, just like biofilms, consist of bacterial cells encased in a matrix of self-secreted extra-cellular polymeric substances (EPS); however, they have distinct filamentous morphology that distinguishes them from surface-hugging biofilms. Streamers are tethered at one or both ends to walls, while the rest of the structure can extend significantly with the flow.1 This structure allows streamers to rapidly proliferate microfluidic devices despite the often characteristically low Reynolds number (Re) flows in these systems.4–7 This behavior makes bacterial streamers to assume significance for applications, such as water filtration,7,8 biomedical devices,6,9,10 and bacterial colonization of porous media.4,5 The generalization of the phenomena of streamer formation beyond the biophysical domain has made it imperative to conduct exhaustive studies to understand the impact of streamers on natural and artificial systems.11
Amongst the different application domains that bacterial streamers can impact is the domain of biofouling of membranes. Biofouling is one of the most common types of fouling for membranes.5,12–14 Biofilm and streamer formation on membranes can lead to a host of issues including reduced membrane performance and longevity, as well as low permeate flux and water quality, and subsequently increases operating costs.14,15 Moreover, bacterial growth itself is also affected by the feed water properties (e.g., pH and ionic strength),1,12 the physico-chemical properties of the surface (e.g., hydrophilicity and charge),13,15 and hydrodynamic variables (e.g., flow rate).1 Inspired by the need to understand the impact of various environmental factors on biofouling of membranes, researchers have investigated bacterial streamer formation in microfluidic filtration systems that mimic membranes. For instance, Marty et al.7 designed a microfluidic device to mimic a membrane system and showed that significant bacteria cell accumulation and streamer formation in the downstream area occur during filtration. They studied the effect of different pore-sizes and filtration modes, (dead-end and pseudo cross flow) on the lengths of streamers and reported that smaller pore-sizes and low flow rates promoted streamer formation in the microfluidic device. The formation of streamers in the downstream section of the filtration unit stands in contrast to some reports of mass accumulation in inorganic fouling.16 In another study, they reported the effects of pore tortuosity and secondary flows on streamer formation as well.17 Biswas et al.2 studied the deformation and failure behavior of bacterial streamer in a microfluidic filtration device. They discovered instability in the streamers, like deformation regimes, as they transitioned slowly through a complex creep process, and ultimately underwent failure. The studies related to the microfluidic filtration devices would have been more interesting if the connection between the dynamics of streamer formation and the transport through microfluidic pores had been explored. Recent investigations, particularly those utilizing the porous microfluidic mimic platforms,2–5 indicate that, during experiments, there are at least two distinct time phases of streamer formation: the initial phase when the streamer forms and second phase that occurs after the initial streamers have formed. The dynamics of streamers post formation are important because, upon completion of the initial formation, the streamers undergo a rapid mass accrual mainly due to their “web” like structures. During this phase, they can completely clog the system,3,4 undergo a catastrophic nonlinear deformation,2 or suffer localized failure leading to an extended water channel throughout the system.5 Despite these studies, both the formation and impact of biofilm streamers in natural habitats and artificial environments remain sparsely investigated.5 Significantly, the time-dependent deformation tendency of bacterial streamers in creeping flows (Reynolds number, Re,≪ 1) can have important implications for filtration systems and yet only few studies till date have focused on this issue.
In this study, we report an in situ observation of post-formation dynamics of bacterial streamers by using a pressure-driven microfluidic filtration platform that is designed to operate in the dead-end filtration mode.18 The device has a 2 μm pore size, which was inspired by microfiltration membranes that are most commonly used for filtering out biological substances.18 Bacterial streamers were formed in the channel by using a culture laden with bacterial flocs—a previously studied and characterized mode of streamer formation.2,3,5 An advantage of floc-mediated streamer formation is that streamers form rapidly [∼O (100 s)],2,3 whereas biofilm-mediated streamers can take hours to form.4,6 Rapid formation allows this biophysical to be dominated by physical aspects. In our work, we focused on the post-formation dynamics of streamer formation, i.e., the time period following the formation of the streamer, and observe rapid clogging of the system because of streamer formation. This clogging results in an exponential decrease in flow-rate through the device. We find that superimposed on this exponential decrease are smaller time-scale fluctuations in flow rate, which coincide with deformation/breaking of streamers. Moreover, de-clogging of pores, deformation and breaking of the streamer occurred, leading to the transport of bacterial biomass downstream. We also found that there exists a lower bound on the applied pressure difference below which streamer based clogging is not dominant. Streamer-based clogging also shows simultaneous similarities with different forms of membrane clogging models, suggesting that this is a novel mode of clogging for filtration systems. We further investigated the effect of pH on the filtration system.
MATERIALS AND METHODS
Microfluidic chip fabrication
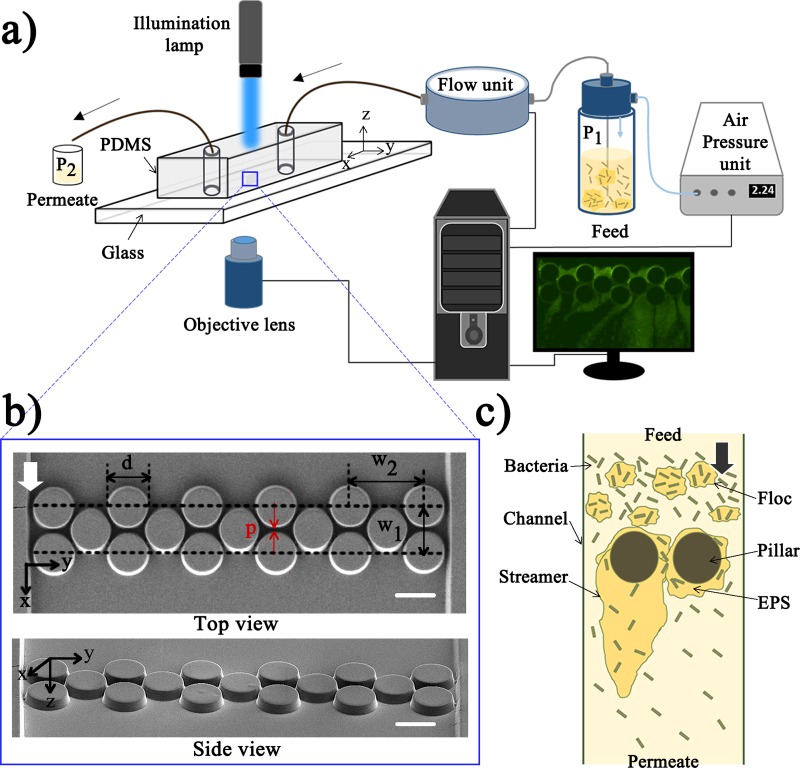
The microfluidic chip is created from a Polydimethylsiloxane (PDMS) stamp sealed with a glass coverslip [Fig. 1(a)]. In the fabrication of the PDMS stamp, first a silicon master mold was prepared by using standard photo-lithography techniques.19 We used positive photoresist “HPR 504” to coat the silicon wafer. A small aspect ratio of 5:1 for the pillar height and pore-size was considered to avoid collapse of PDMS pillars.20–22 The microfluidic design consists of a straight channel of length 11.5 mm with a single inlet and a single outlet. 17 cylindrical micropillars of 50 μm diameter are placed in a staggered pattern at a distance of 6.6 mm from the inlet. The pitch length (p) is 2 μm (Fig. S1) and the height of the pillars (along z-axis) is 10 μm [Fig. 1(b)]. To create this design from the silicon master, PDMS (Sylgard 184, Dow Corning, NY, USA) was poured on the silicon wafer and cured for 2 h. Subsequently, the PDMS stamps were cut and bonded with glass cover slips (thickness 0.13–0.17 mm) (Fisher Scientific, ON, Canada). To enable good sealing between the PDMS and the glass, both were exposed to oxygen plasma for s and the device (PDMS and glass cover slip) was then annealed at for min to seal the channel. We considered PDMS because of its optically transparency, electrically and thermally insulating, mechanically elastomeric (Young's modulus ∼750 kPa), low surface free energy ∼20 erg/cm2, impermeability to liquid water, permeability to gas and nonpolar organic solvents, reactivity to oxygen plasma, nontoxic nature, and easy to make smooth and nonpolar surface.23 PDMS has been widely used as a microfluidic membrane material for modeling the transport phenomena at pore scale.7,16,17,24
FIG. 1.
Experimental set-up. (a) Schematic of the set-up. The pressure unit maintains the required pressure difference and the flow unit directly measures the flow rate inside the device. (b) Field emission scanning electron microscopy (FESEM) images of the micropillars inside the microfluidic channel. The flow is along the x-axis as indicated by the arrow. The geometric dimensions are: and height of the pillars is μm (along z-axis). The scale bar is μm. (c) Schematic of the biofouling process and streamer formation around the micro-pillars.
Bacteria culture preparation
We used Pseudomonas fluorescens CHA0 (wild type)25 and Pseudomonas aeruginosa MPAO1 (wild type) (P. aeruginosa Mutant Library—University of Washington) bacteria for this study. P. fluorescens are gram-negative aerobic bacteria which can be found in soil and water and play an important role in plant health.26 The strain of bacteria here is green fluorescent as they express green fluorescent protein (GFP) constitutively. The typical bacterial cell is cylindrical with length ∼2 μm and diameter ∼1 μm. Both bacterial strains were taken from collections and streaked on 2 different LB (Luria Bertani) agar plates in a zigzag pattern. The plates of P. fluorescens and P. aeruginosa were incubated for h at and , respectively. A single colony was taken from each plate and put into ml of LB broth (pH = 7). P. fluorescens was incubated for h in a shaking incubator (VWR, PA, USA) at rpm and . P. aeruginosa was incubated for h in a shaking incubator at rpm and . The incubation time-period was carefully chosen so that flocculation of the bacteria occurred during this period. Flocs consist of bacteria encased in EPS and are considered an aggregative form of bacterial growth. Flocs are typically found suspended in a fluid, unlike biofilms which are surface-associated communities.1–3,5 The incubation time-period adopted here results in large mesoscopic flocs with an average diameter of ∼26 μm (also see Fig. S2). 5 ml of this bacterial culture (pH = 7) was mixed with ml LB broth (pH = 3–12) so that the pH of the feed, prior to injection in the microfluidic platform, lay between 5 and 10 (pH was measured by using pH paper strips). The optical density at 600 nm (OD600) was measured by a spectrophotometer (Novaspec II, MA, USA). OD600 is a widely used parameter for measuring the density of bacteria in a sample.27–30 Permeate was sampled at intervals ranging between 10 and 20 min to evaluate OD600 and the permeate volume (). Permeate volume was further used to measure the permeate flux.
Microscopy
For imaging of bacterial flocs in quiescent conditions, the bacterial culture was poured into an imaging chamber (Lab-Tek® chambers, Fisher Scientific, ON, Canada), which was then placed on an inverted optical microscope (Nikon Eclipse Ti). Fluorescence imaging was performed with a GFP long-pass green filter cube. The equivalent diameter () of flocs was measured through image processing by using the NIS-Element AR software interface (Nikon). The equivalent diameter is defined as where A is the area of a floc. For the microfluidic experiments, the same microscope was used [Fig. 1(a)]. Pressure-driven flow was created in the microfluidic channel with a pump (Fluigent, MA, USA) as shown in Fig. 1(a). and denote the air pressures in the feed that was controlled by the pump and normal atmospheric pressure in chambers, respectively [Fig. 1(a)]. For each experiment, the pressure difference ( was maintained at a constant value. The volumetric flow rate (Q) was measured directly using the flow unit [Fig. 1(a)] and the corresponding values are obtained from the flow-rate-control-module software (Fluigent, MA, USA). All experiments were performed at room temperature.
RESULTS
Our microfluidic filtration system was designed to operate in the dead-end mode [Fig. 1(a)]. In this filtration mode, a pressure difference across the pillar-array causes the feed (input fluid) to flow through the filtration zone and the foulant materials thus accumulate around the pore-walls. As discussed earlier, the pitch length (p) or the pore-length scale of the filtration zone was 2 μm [Fig. 1(b); also see Fig. S1]. This pore size was inspired by the range of a typical microfiltration membrane, where the pore length scale lies between μm and μm. Microfiltration membranes are widely used to remove biological matter from contaminated water.18,31,32 In the work discussed here, all experiments were performed under constant applied pressure across the microfluidic filtration system. To conduct a parametric study on the influence of the applied pressure difference , the pressure difference was varied from kPa to kPa. For each experiment, a diluted culture of bacterial flocs was used as feed [Fig. 1(c)]. The clean fluid that passes through the pillar-array because of the pressure gradient across the pillars is referred to as the permeate. The streamers form on the downstream side of the device, which also represents the permeate side. Previous reports have established that bacterial flocs lead to rapid-streamer formation in the microfluidic systems, thus providing a suitable experimental method to evaluate the impact of streamers on various systems.2,3,5 The flocs in the feed attached to the pillar walls inside the device causing biofouling and the fluidic loading caused them to stretch out in the form of the thin filaments called streamers.
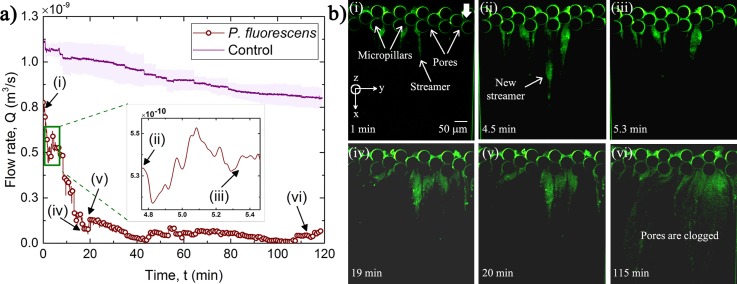
Figure 2 shows the impact of biofouling and streamer formation in the filtration zone of the microfluidic device. Initially, a control experiment was performed, where Phosphate-buffered saline (PBS) without any bacterial cells was used as a feed liquid and a constant pressure of 138 kPa was applied across the microfluidic channel. The resulting temporal variation of the flow rate (Q) is shown in Fig. 2(a). Although, no bacterial cell was present in the PBS fluid, some debris would inevitably accumulate at the pore-space during the experiment (see Fig. S3) leading to a decrease in the flow rate. When the bacterial culture is used as feed, bacterial biomass causes clogging of the pore-spaces in the filtration zone. In the case of 138 kPa pressure difference, the resulting flow causes streamer formation in the device and in such situations, Q decreases sharply with time; a representative graph is shown in Fig. 2(a). The initial flow rate of m3/s decreases to almost m3/s after approximately 100 min of the experiment. Equivalently, we also calculated the additional resistance to fluid flow (resistance of cake layer and pore clogging) due to the fouling by using Darcy's law. The results show that the resistance of membrane increases with the decrease in the permeate volume (see, Fig. S4). Fluorescent microscopy images corresponding to certain temporal points (marked in roman numerals) are shown in Fig. 2(b). It must be noted that visible biomass corresponds only to fluorescing bacteria and that EPS is not visible under fluorescent imaging. As the flow is initiated, biofouling of the filtration zone and streamer formation occur almost immediately (∼1 min). After several minutes, streamers become thicker, i.e., their y-axis span increases, and they extend for ∼200 μm from the micro-pillar posts. Streamers are formed from strongly viscoelastic biomass and can undergo phenomena such as creep and failure. Failures and breakage of streamers were observed to cause short time scale (∼101 s) fluctuations in the flow rate. The inset of Fig. 2(a) captures such an instance when Q increases sharply because of partial de-clogging of the pore-space. Such fluctuations coincide with de-clogging of pore-space, when some biomass detaches fully or partially from the filtration zone (see supplementary material, Video 1). Partially detached biomass is stretched by shear forces leading to streamer formation, while complete detachments lead to biomass entering the post-filtration zone into the permeate. Biswas et al.2 have shown that bacterial biomass shows strong material non-linearity and a complex creep response; these responses likely contribute to this phenomenon. The short time scale (∼101 s) enhancements in Q do not affect the long-time (∼101–102 min) decay behavior because of the continuous accumulation of biomass at the filtration zone.
FIG. 2.
(a) The variation of flow rate with time at a constant pressure difference, kPa. The control experiment with PBS is also plotted and shaded portion shows the error during experiment. Q decreases due to rapid pore-blockage by streamers. (Inset) Short time scale [∼ s)] fluctuations can also be seen, which occur due to de-clogging of the pores. The pH of the feed is 7. (b) Fluorescent microscopy images of entire width on the channel along with the pore-scale phenomena corresponding to the time-points delineated by roman numerals in “(a).”
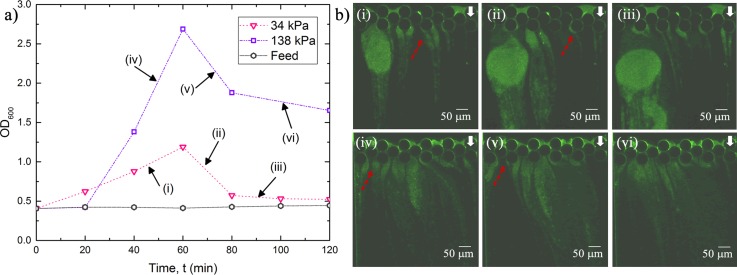
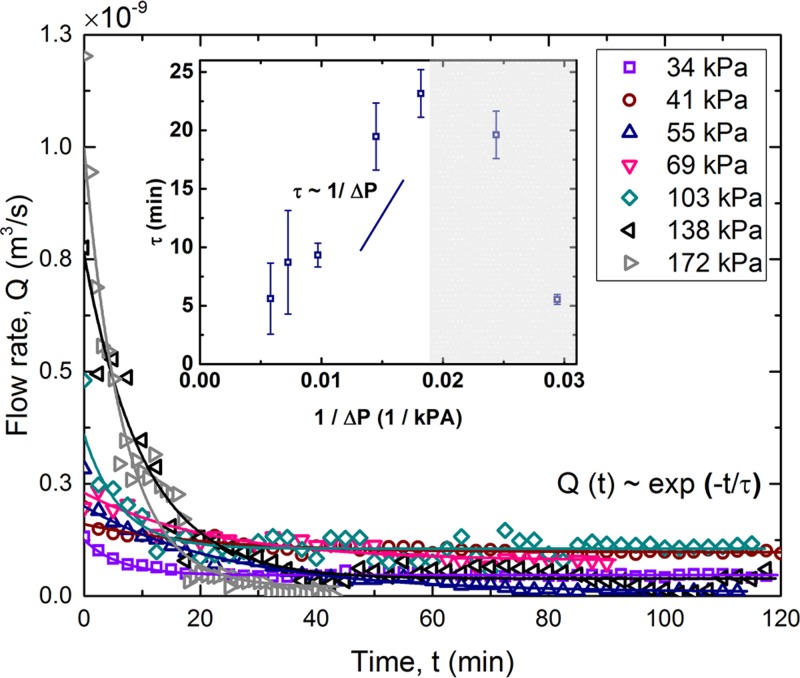
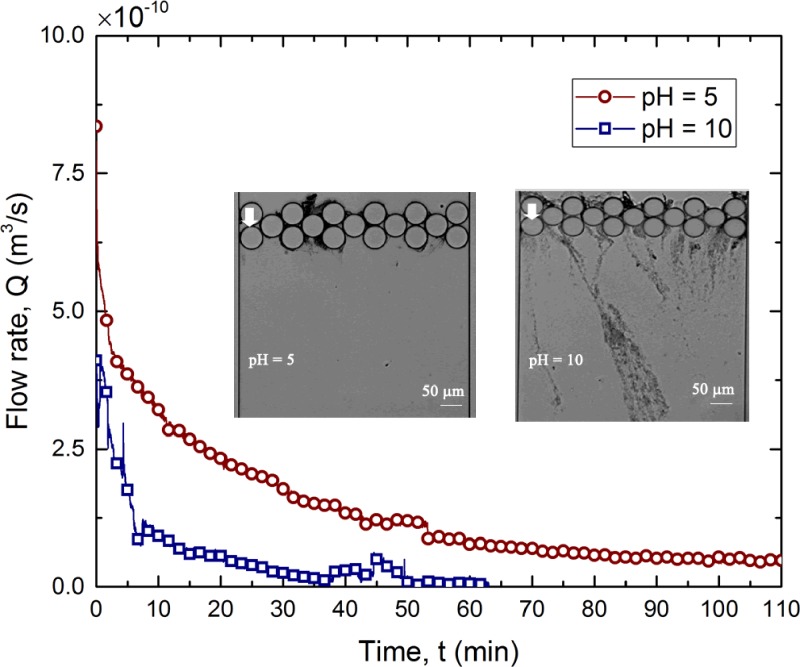
In order to assess contamination of the permeate due to streamer breakage and filtration efficiency, we quantified the optical density at 600 nm (OD600) of the permeate. Figure 3(a) shows the change of OD600 of the permeate as a function of time for two different applied pressures. As can be seen, the optical density of permeate increases for about 60 min, after which it shows a decreasing trend. This initial increase is likely due to the streamer breakage events, which generate biomass debris and subsequently contaminates the permeate. It is now known that streamers can break due to different kinds of instabilities. Biswas et al.2,33 have reported on two different kinds of instabilities leading to streamer failure events, which occur due to the creep response of the bacterial soft matter. After the initial increase, the flow rate decreases substantially, which should result in a corresponding decrease in streamer breakage events (as shear forces decrease). This should result in a decrease in OD, which is also observed in the latter half of the experiment [Fig. 3(a)]. Such a decrease in OD can also be attributed to the common cake filtration in the membrane process where the pore blockage and formation of a cake layer of foulant contribute itself to the separation of contaminants.31,34 Moreover, it is clear that the permeate is substantially more contaminated at a higher operating pressure difference. The corresponding fluorescent microscopy images are shown in Fig. 3(b). Both cake layer formation and pore blocking can be observed clearly from the images. From the image [Fig. 3(b)], instances of streamer breaking can be seen which result in a corresponding increase in OD600 [Fig. 3(a)]. Figure 3(a) also shows the change of OD600 of the feeding sample for 2 h. However, the change of OD600 was insignificant compared to the permeate OD600 as can be seen in Fig. 3(a). Therefore, the effect of room temperature in growing these bacteria was neglected. To further explore the role of the operating pressure difference (), we conducted filtration experiments for a range of applied pressure differences. Applied transmembrane pressure for a typical microfiltration process is in the range of 10 kPa–200 kPa.18,35 Hence, in the present work, the performance of the microfluidic filtration system is investigated from 34 kPa to 172 kPa pressure. The resulting temporal variation of flow rates for these values is plotted in Fig. 4. The long time scale behavior of the flow rates obeys an exponential decaying trend, i.e., , where is a constant and is the decay time-scale, for different (also see Table S1). Exponential decaying law is commonly used to estimate the bacterial streamer dynamics in creeping flow2,6,36 (see Fig. S5 for the corresponding OD600 values). This exponential decay behavior was extremely repeatable (see Fig. S6). The inset of Fig. 4 depicts the change in the decay time scale () as a function of the applied pressure difference . Interestingly, the decay time scale has a linear relation with the inverse of applied pressure difference, i.e., , when pressure is higher than a certain lower bound. For applied pressure values, where the scaling applies agrees well with the theoretical models for streamer-based clogging proposed by Drescher et al.6 and Das and Kumar.36 Based on this figure, can be treated as a critical pressure zone, below which streamer-based clogging is not dominant. This deviation from the linear scaling is most significant for and hence we denote this pressure as the critical pressure difference below which clogging is not dominated by streamer-based clogging. For pressure differences greater than 55 kPa, streamer based clogging dominates the system.
FIG. 3.
(a) Optical Density (OD600) of feed and permeate plotted at two different pressure differences of kPa and kPa. (b) Fluorescent microscopy images of the filtration section corresponding to the time-points delineated by roman numerals in “(a).” The red arrows show the formation and breaking of streamers.
FIG. 4.
Change of flow rate with at different . The markers represent experimental data, whereas the lines represent exponential curve fits. (Inset) The time scale () obtained from the regression fit is plotted against inverse of . For pressures kPa, shows a linear scaling with . In the grey zone, the linear scaling does not apply.
Here, we draw a parallel between our microfluidic filtration system and microfiltration membranes. In membrane studies, the critical flux is defined as the permeate flux above which an irreversible fouling occurs. In general, operation at the sub-critical region (below the critical flux) is desired as no fouling occurs and the membrane functions in an entirely clean regime.37,38 The critical flux in our microfluidic filtration system is calculated by using (), where Q is flow rate ∼ m3/s, , width of the channel, is , and h, height of the pillars, is . The approximate critical flux for this system is ∼ (or ). This observation provides a valuable insight into the proper operating pressure for microfiltration membranes with ∼2 μm pores size when applied to removal of bacteria and bacterial biomass. Higher initial flux does not necessarily result in optimum production as it leads to more frequent cleaning. Hence, it is important to not exceed ∼ as this is counter-productive and increases overall cost in long term.
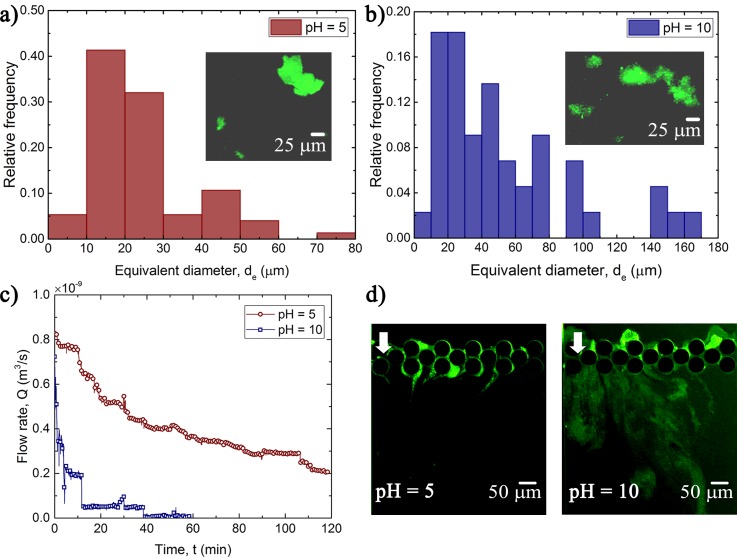
In filtration systems, pH of the feed is an important experimental variable, as it can strongly affect deposition on membranes, fouling potential, and performance of membranes.13,18,39 In order to assess the impact of pH, we carried out further experiments by changing the pH of the feed media. Prior to the microfluidic experiments, the effect of pH on floc size was assessed. Figures 5(a) and 5(b) depict the P. fluorescens floc diameter histograms for the acidic (pH = 5) and alkaline (pH = 10) environments, respectively. For pH = 5, the average floc diameter was found to be approximately 21 μm, whereas for the alkaline environment (pH = 10), it was found to be 42 μm. Hence, we can see that pH clearly affects bacterial behavior, and we can expect pH to affect biofouling of the microfluidic filtration system. Figure 5(c) shows the temporal change in Q at two different pH values under a constant applied pressure difference of 138 kPa (see, Fig. S5). For the highly alkaline feed (pH = 10), the pores become almost completely clogged and the flow rate decreases to practically zero after approximately 60 min of operation. However, for the acidic feed (pH = 5), the system continues to operate well beyond 120 min from the initiation of the experiment. This observation is also corroborated by visualization of the filtration section [Fig. 5(d)], which compares the filtration zone for two different pH conditions. Compared to the highly alkaline feed, the acidic feed causes significantly less biofouling. This significant variation in biofouling under different pH conditions likely occurs due to higher EPS secretion under alkaline conditions.40,41 Similar behavior was observed when we used P. aeruginosa bacteria in the feed (Fig. 6). This indicates that the effect of pH is not only restricted to one specific bacterial strain but also likely a more generic response. Based on these findings, we suggest that in membrane applications, the pH of the feed must be adjusted to the acidic condition before treatment. This pretreatment of feed water significantly reduces the risk of abrupt membrane clogging and thus replacement of membranes. The exact mechanism by which pH affects the bacterial floc size is not known. However, it is well known that the EPS matrix is composed of biopolymers such as polymeric sugars, nuclear acids, proteins, and lipid.42 Literature suggests that EPS of gram-negative bacteria often contain charged carboxylate groups.43 It is likely that at low pH these carboxylate groups are neutralized through protonation, which can cause the contraction of floc/biofilm and reduction of the thickness.43 It has also been observed that the increase in pH can swell up the EPS44 and lead to a higher biofilm/floc formation.45 We leave it to future investigations to determine the full extent of this generality.
FIG. 5.
Histograms for the equivalent diameter () of flocs at (a) pH and (b) pH . The total number of flocs considered are 60 and 46 and the medians are μm and μm at pH and pH , respectively. Inset images show sample flocs imaged using optical microscopy. (c) Comparison of the flow rate with time at different pH conditions under constant kPa. (d) Fouling at the filtration zone for the two different pH conditions at 50 min. The white arrow shows that the flow direction is from top to bottom.
FIG. 6.
Comparison of the flow rate with time at different pH conditions under constant kPa using P. aeruginosa bacteria. Inset images show fouling at the filtration zone for the two different pH conditions after 50 min of initiation of the experiment. The white arrows show the flow direction (top to bottom).
DISCUSSION
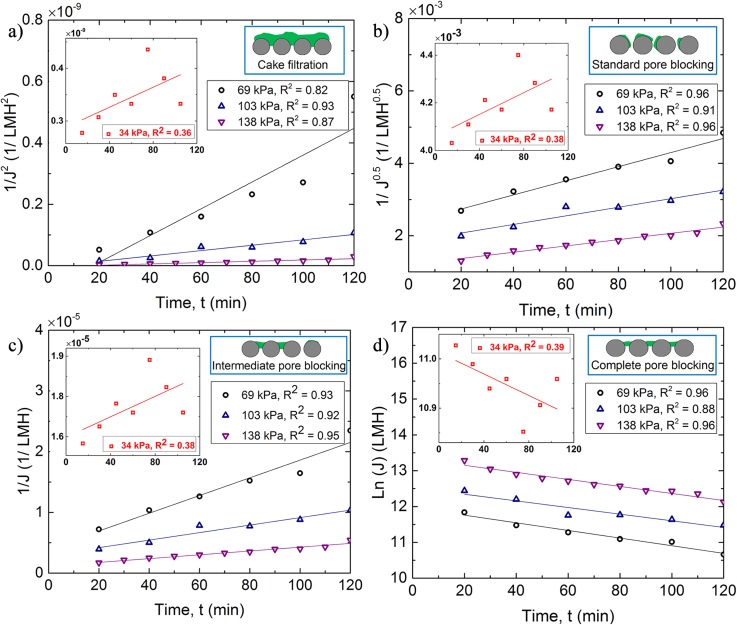
Hermia's model is typically used to describe the fouling behavior of porous membranes at constant-pressure dead-end filtration modes.46 This model explains four general membrane fouling categories including pore blocking, complete pore blocking intermediate pore blocking, and cake filtration.46,47 To analyze the governing fouling mechanisms in our microfluidic filtration system by using Hermia's model, we calculated the permeate flux (J) as, = (,48–52 where is the permeate volume, is the cross-sectional area of membrane, and is the time to collect the permeate volume. The permeate volume was measured every 20 min of each experiment. We used this model for 4 different . Figures 7(a)–7(d) show the permeate flux values with time that describe the general membrane fouling categories.46,47 The schematics shown in the plot insets show the corresponding fouling mechanisms at the pore-scale. It must be noted that in our microfluidic system streamer formation and fouling happen at the same time. The formation of streamer is a new phenomenon, which is not captured by Hermia's model. The results, therefore, need to be interpreted with caution. Figure 7 suggests that all 4 types of fouling occur for all pressure differences except for . This result matches well with our previous observation (Fig. 2). This is an extremely interesting and counter-intuitive result, suggesting the presence of an entirely new mode of fouling in our microfluidic filtration device. Recently, it has been suggested that bacterial streamers belong to a more generic category of “colloidal streamer”33 formation as morphologically similar fouling modes have been observed in particle laden polymeric flows.11 It has also been observed that the dynamics of these colloidal streamers is extremely complex and non-linear.2,5,33 Figure 7 suggests that that biofouling due to streamer formation is likely a separate category of porous membranes fouling that calls for further investigation.
FIG. 7.
Permeate flux trends plotted against time for different pressure differences at pH = 7 by using Hermia's model. Flux appears to follow trends for all different models except at (a) for cake filtration, (b) for standard pore blocking, (c) intermediate pore blocking, and (d) complete pore blocking. R2 value in the legend is the regression coefficient. The inset schematics show the corresponding expected pore-scale fouling.
CONCLUSION
In conclusion, our work shows that the post-formation dynamics of bacterial streamers can affect filtration efficiency in a microfluidic filtration system. We investigated the effect of de-clogging of the pore-space and the streamer failure on the quality of the permeate. Partially detached biomasses result in new streamer formation in the device, while fully de-clogged biomass is transported into the permeate. Streamer formation and clogging dynamics were strongly dependent on the applied pressure difference. We found that when streamer-based clogging dominates, the permeate flux exponentially decreases with time, but this decrease is accompanied by smaller time scale fluctuations caused by the failure of streamer biomass. Our results suggest that clogging due to streamer formation possibly belongs to a new class of filtration system fouling. A critical pressure difference was also found below which streamer-based clogging did not dominate. We also found that the pH of the feed can affect bacterial biomass and can affect biofouling of the membrane. We observed that acidic feed can increase the permeate flux compared to the highly alkaline feed; thus, the biofouling under acidic condition is lesser. Thus, the post-formation dynamics of bacterial streamer being a very complex process can be affected by many factors and still remains to be fully understood.
SUPPLEMENTARY MATERIAL
See supplementary material for supplementary figures and video caption.
ACKNOWLEDGMENTS
I.B. and M.S. gratefully acknowledge the financial support provided by the Natural Sciences and Engineering Research Council of Canada (NSERC) and Natural Resources Canada (NRCan). A.K. acknowledges partial support from the Saroj Poddar Young Investigator Grant.
References
- 1. Karimi A., Karig D., Kumar A., and Ardekani A. M., Lab Chip 15, 23–42 (2015). 10.1039/C4LC01095G [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Biswas I., Ghosh R., Sadrzadeh M., and Kumar A., Sci. Rep. 6, 32204 (2016). 10.1038/srep32204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hassanpourfard M., Nikakhtari Z., Ghosh R., Das S., Thundat T., Liu Y., and Kumar A., Sci. Rep. 5, 13070 (2015). 10.1038/srep13070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Valiei A., Kumar A., Mukherjee P. P., Liu Y., and Thundat T., Lab Chip 12, 5133–5137 (2012). 10.1039/c2lc40815e [DOI] [PubMed] [Google Scholar]
- 5. Hassanpourfard M., Ghosh R., Thundat T., and Kumar A., Lab Chip 16, 4091–4096 (2016). 10.1039/C6LC01055E [DOI] [PubMed] [Google Scholar]
- 6. Drescher K., Shen Y., Bassler B. L., and Stone H. A., Proc. Natl. Acad. Sci. U. S. A. 110, 4345–4350 (2013). 10.1073/pnas.1300321110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marty A., Roques C., Causserand C., and Bacchin P., Biofouling 28, 551–562 (2012). 10.1080/08927014.2012.695351 [DOI] [PubMed] [Google Scholar]
- 8. Houari A., Picard J., Habarou H., Galas L., Vaudry H., Heim V., and Di Martino P., Biofouling 24, 235–240 (2008). 10.1080/08927010802023764 [DOI] [PubMed] [Google Scholar]
- 9. Kim M. K., Drescher K., Pak O. S., Bassler B. L., and Stone H. A., New J. Phys. 16, 065024 (2014). 10.1088/1367-2630/16/6/065024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weaver W. M., Milisavljevic V., Miller J. F., and Di Carlo D., Appl. Environ. Microbiol. 78, 5890–5896 (2012). 10.1128/AEM.01139-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Debnath N., Hassanpourfard M., Ghosh R., Trivedi J., Thundat T., Sadrzadeh M., and Kumar A., Soft Matter 13, 8698–8705 (2017). 10.1039/C7SM01771E [DOI] [PubMed] [Google Scholar]
- 12. Paquet-Mercier F., Parvinzadeh Gashti M., Bellavance J., Taghavi S. M., and Greener J., Lab Chip 16, 4710–4717 (2016). 10.1039/C6LC01101B [DOI] [PubMed] [Google Scholar]
- 13. Guo W., Ngo H. H., and Li J., Bioresour. Technol. 122, 27–34 (2012). 10.1016/j.biortech.2012.04.089 [DOI] [PubMed] [Google Scholar]
- 14. Miura Y. W. Y. and Okabe S., Environ. Sci. Technol. 41, 632–638 (2007). 10.1021/es0615371 [DOI] [PubMed] [Google Scholar]
- 15. Matin A., Khan Z., Zaidi S. M. J., and Boyce M. C., Desalination 281, 1–16 (2011). 10.1016/j.desal.2011.06.063 [DOI] [Google Scholar]
- 16. Bacchin P., Marty A., Duru P., Meireles M., and Aimar P., Adv. Colloid Interface Sci. 164, 2–11 (2011). 10.1016/j.cis.2010.10.005 [DOI] [PubMed] [Google Scholar]
- 17. Marty A., Causserand C., Roques C., and Bacchin P., Biomicrofluidics 8, 014105 (2014). 10.1063/1.4863724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mulder M., Basic Principles of Membrane Technology ( Springer, 1998), Vol. 72, p. 564. [Google Scholar]
- 19. Hassanpourfard M., Sun X., Valiei A., Mukherjee P., Thundat T., Liu Y., and Kumar A., J. Visualized Exp. 90, e51732 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shao G., Wu J., Cai Z., and Wang W., Sens. Actuators A 178, 230–236 (2012). 10.1016/j.sna.2012.01.034 [DOI] [Google Scholar]
- 21. Perl A., Reinhoudt D. N., and Huskens J., Adv. Mater. 21, 2257–2268 (2009). 10.1002/adma.200801864 [DOI] [Google Scholar]
- 22. Kung Y. C., Huang K. W., Fan Y. J., and Chiou P. Y., Lab Chip 15, 1861–1868 (2015). 10.1039/C4LC01211A [DOI] [PubMed] [Google Scholar]
- 23. Cooper McDonald J. and Whitesides G. M., Acc. Chem. Res. 35, 491–499 (2002). 10.1021/ar010110q [DOI] [PubMed] [Google Scholar]
- 24. Liu M., Sun J., Sun Y., Bock C., and Chen Q., J. Micromech. Microeng. 19, 035028 (2009). 10.1088/0960-1317/19/3/035028 [DOI] [Google Scholar]
- 25. Workentine M. L., Wang S., Ceri H., and Turner R. J., BMC Microbiol. 13(1–7), 175 (2013). 10.1186/1471-2180-13-175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zuber F. C. S., Keel C., Mattart A., Blumer C., Pessi G., Gigot-Bonnefoy C., Schnider-Keel U., Heeb S., Reimmann C., and Haas D., Mol. Plant-Microbe Interact. 16, 634–644 (2003). 10.1094/MPMI.2003.16.7.634 [DOI] [PubMed] [Google Scholar]
- 27. Lin H. L., Lin C. C., Lin Y. J., Lin H. C., Shih C. M., Chen C. R., Huang R. N., and Kuo T. C., Appl. Environ. Microbiol. 76, 1683–1685 (2010). 10.1128/AEM.00824-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maia M. R., Marques S., Cabrita A. R., Wallace R. J., Thompson G., Fonseca A. J., and Oliveira H. M., Front. Microbiol. 7, 1381 (2016). 10.3389/fmicb.2016.01381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stevenson K., McVey A. F., Clark I. B., Swain P. S., and Pilizota T., Sci. Rep. 6, 38828 (2016). 10.1038/srep38828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van Heerden J. H., Kempe H., Doerr A., Maarleveld T., Nordholt N., and Bruggeman F. J., Sci. Rep. 7, 16094 (2017). 10.1038/s41598-017-15895-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Babel S. and Takizawa S., Desalination 261, 46–51 (2010). 10.1016/j.desal.2010.05.038 [DOI] [Google Scholar]
- 32. Ho C. C. and Zydney A. L., J. Colloid Interface Sci. 232, 389–399 (2000). 10.1006/jcis.2000.7231 [DOI] [PubMed] [Google Scholar]
- 33. Biswas I., Ghosh R., Sadrzadeh M., and Kumar A., J. Colloid Interface Sci. 522, 249–255 (2018). 10.1016/j.jcis.2018.03.074 [DOI] [PubMed] [Google Scholar]
- 34. Hayatbakhsh M., Sadrzadeh M., Pernitsky D., Bhattacharjee S., and Hajinasiri J., Desalin. Water Treat. 57, 14869–14887 (2015). 10.1080/19443994.2015.1069216 [DOI] [Google Scholar]
- 35. Van der Bruggen B., Vandecasteele C., Van Gestel T., Doyen W., and Leysen R., Environ. Prog. 22, 46–56 (2003). 10.1002/ep.670220116 [DOI] [Google Scholar]
- 36. Das S. and Kumar A., Sci. Rep. 4, 7126 (2014). 10.1038/srep07126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang Z., Wu Z., Yin X., and Tian L., J. Membr. Sci. 325, 238–244 (2008). 10.1016/j.memsci.2008.07.035 [DOI] [Google Scholar]
- 38. He Z., Miller D. J., Kasemset S., Paul D. R., and Freeman B. D., J. Membr. Sci. 525, 25–34 (2017). 10.1016/j.memsci.2016.10.002 [DOI] [Google Scholar]
- 39. Khorshidi B., Hajinasiri J., Ma G., Bhattacharjee S., and Sadrzadeh M., J. Membr. Sci. 500, 151–160 (2016). 10.1016/j.memsci.2015.11.015 [DOI] [Google Scholar]
- 40. Lijie Zhou, Xia S., Zhang Z., Ye B., Xu X., Gu Z., and Wang X., J. Water Sustainability 4, 90–100 (2014). 10.11912/jws.4.2.91-100 [DOI] [Google Scholar]
- 41. Xia S., Zhou L., Zhang Z., and Li J., J. Environ. Sci. 24, 2035–2040 (2012). 10.1016/S1001-0742(11)61060-7 [DOI] [PubMed] [Google Scholar]
- 42. Poustia M. and Greener J., Surface Science. 676, 56–60 (2018). 10.1016/j.susc.2018.01.004 [DOI] [Google Scholar]
- 43. Stoodley P., Debeer D., and Lappin-Scott H. M., Antimicrob. Agents Chemother. 41, 1876–1879 (1997) PMCID: PMC164028; PMID: 9303377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dogsa I., Kriechbaum M., Stopar D., and Laggner P., Biophys. J. 89, 2711–2720 (2005). 10.1529/biophysj.105.061648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hostacka A., Čiznar I., and Štefkovicova M., Folia Microbiol. 55, 75–78 (2010). 10.1007/s12223-010-0012-y [DOI] [PubMed] [Google Scholar]
- 46. Maiti A., Sadrezadeh M., Guha Thakurta S., Pernitsky D. J., and Bhattacharjee S., Energy Fuels 26, 5604–5612 (2012). 10.1021/ef300865e [DOI] [Google Scholar]
- 47. Hermia J., Trans. IChemE 60, 183–187 (1982). [Google Scholar]
- 48. Khorshidi B., Biswas I., Ghosh T., Thundat T., and Sadrzadeh M., Sci. Rep. 8, 784 (2018). 10.1038/s41598-017-18724-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Khorshidi B., Thundat T., Pernitsky D., and Sadrzadeh M., J. Membr. Sci. 535, 248–257 (2017). 10.1016/j.memsci.2017.04.052 [DOI] [Google Scholar]
- 50. Khorshidi B., Soltannia B., Thundat T., and Sadrzadeh M., J. Membr. Sci. 523, 336–345 (2017). 10.1016/j.memsci.2016.09.062 [DOI] [Google Scholar]
- 51. Miller D. J., Kasemset S., Wang L., Paul D. R., and Freeman B. D., J. Membr. Sci. 452, 171–183 (2014). 10.1016/j.memsci.2013.10.037 [DOI] [Google Scholar]
- 52. Li F., Meng J., Ye J., Yang B., Tian Q., and Deng C., Desalination 344, 422–430 (2014). 10.1016/j.desal.2014.04.011 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See supplementary material for supplementary figures and video caption.