Abstract
The polycomb group (PcG) proteins are key epigenetic regulators in stem cell maintenance. PcG proteins have been thought to act through one of two polycomb repressive complexes (PRCs), but more recent biochemical analyses have challenged this model in the identification of noncanonical PRC1 (nc-PRC1) complexes characterized by the presence of Rybp or Yaf2 in place of the canonical Chromobox proteins. However, the biological significance of these nc-PRC1s and the potential mechanisms by which they mediate gene repression are largely unknown. Here, we explore the functional consequences of Yaf2 disruption on stem cell regulation. We show that deletion of Yaf2 results in compromised proliferation and abnormal differentiation of mouse embryonic stem cells (mESCs). Genome-wide profiling indicates Yaf2 functions primarily as a transcriptional repressor, particularly impacting genes associated with ectoderm cell fate in a manner distinct from Rybp. We confirm that Yaf2 assembles into a noncanonical PRC complex, with deletion analysis identifying the region encompassing amino acid residues 102–150 as required for this assembly. Furthermore, we identified serine 166 as a Yaf2 phosphorylation site, and we demonstrate that mutation of this site to alanine (S166A) compromises Ring1B-mediated H2A monoubiquitination and in turn its ability to repress target gene expression. We therefore propose that Yaf2 and its phosphorylation status serve as dual regulators to maintain the pluripotent state in mESCs.
Keywords: embryonic stem cell, polycomb, phosphorylation, epigenetics, differentiation, H2AK119ub1, PRC1, Yaf2
Introduction
PcG3 proteins are epigenetic modifiers that are found to be integral to the maintenance of stem cells (1, 2). PcG proteins mainly reside in two multisubunit complexes termed the Polycomb repressive complex 1 (PRC1) and complex 2 (PRC2) (2, 3). The PRC2 complex consists of the histone methyltransferase EZH1 or EZH2, which together with SUZ12 and EED trimethylates lysine 27 on histone H3 (H3K27me3) (4, 5). The PRC1 complex has each of five subunits (PCGF1–6, PHC1–3, CBX2/4/6/7/8, SCMH1/L2, and RING1A/B) (6) and displays RING1A/B-mediated monoubiquitination of histone H2A at lysine 119 (H2AK119ub1) (7, 8). The classical view on Polycomb-mediated gene repression is a consecutive action where the PRC1 component CBX could bind specifically to the product of PRC2 catalysis, H3K27me3, via its chromodomain, supporting a model in which PRC1 acts downstream of PRC2 (2). In line with this model, many PcG target loci often exhibit co-occupancy of PRC1 and PRC2 in Drosophila and mammalian cells (9, 10). Although the hierarchical model for PRC1 recruitment is well-established, it has been challenged recently by the biochemical characterization of noncanonical PRC1 (nc-PRC1) complexes, characterized by the presence of RYBP or YAF2 instead of a CBX subunit (11–14). To date, the mechanisms of nc-PRC1–dependent gene silencing, independently of H3K27me3, are poorly understood.
How the PcG proteins contribute to gene silencing or activation in diverse physiological states is of interest. The Ring–Pcgf module constitutes a minimal core on which the different PRC1s are assembled and deposits H2AK119ub1, a histone modification that impedes RNA polymerase II elongation (2, 15), through the E3 protein ligase subunits Ring1A/B. Indeed, our recent studies suggest that of the Pcgf family members, only Pcgf3/5 are essential for PRC1-mediated deposition of H2AK119ub1 in mESC (16), supporting a conclusion that some Pcgf proteins confer differing E3 ligase activities on PRC1. Furthermore, the E3 ligase activity of the Pcgf4/Ring1B complex has been reported to be modulated by its auxiliary subunits RYBP, PHC2, or CBX2/8 (6). Interestingly, it has been shown that Mel-18 phosphorylation is required for targeting the Mel-18/Ring1B complex to chromatin (17). In contrast, phosphorylation of Ring1B by CK2 impairs its E3 ubiquitin-ligase activity (18). Therefore, post-translational modifications such as phosphorylation add another layer of regulation to E3 ligase activities on PRC1. Although several PcG proteins of the PRC1 complexes have been found to be phosphorylated (17, 19, 20), the defined contribution of this modification to Polycomb chromatin domain function in vivo has remained largely unknown.
In this study, we provide evidence for a phosphorylation-based mechanism that affects Yaf2-mediated pluripotency programs in mESCs. Yaf2 protein was first identified in a yeast two-hybrid screen for Yin-Yang-1 (YY-1) transcriptional factor (21) and was further shown to modify the transcriptional activity of E4TF1/hGABP and Myc-mediated transactivation and transformation (22, 23). Genetic studies in zebrafish have implicated Yaf2 as a survival factor during early development and organogenesis (24). Although Yaf2 has recently been identified as a component of noncanonical PRC1 complexes (6), its defined contribution and relevance to Polycomb function in vivo still await elucidation. Using the CRISPR/Cas9 technique, we found that deletion of Yaf2 in mESCs resulted in defective proliferation and differentiation, revealing critical roles for Yaf2 in the maintenance of pluripotency in mESCs. Furthermore, Yaf2 is phosphorylated at serine 166, and this modification mediates its transcriptional repression and is essential for fully catalyzing monoubiquitination of histone H2A in vivo. In summary, these findings therefore provide new insights into the functional and mechanistic role of Yaf2 in regulation of pluripotency.
Results
Yaf2 has an indispensable function in mESC self-renewal
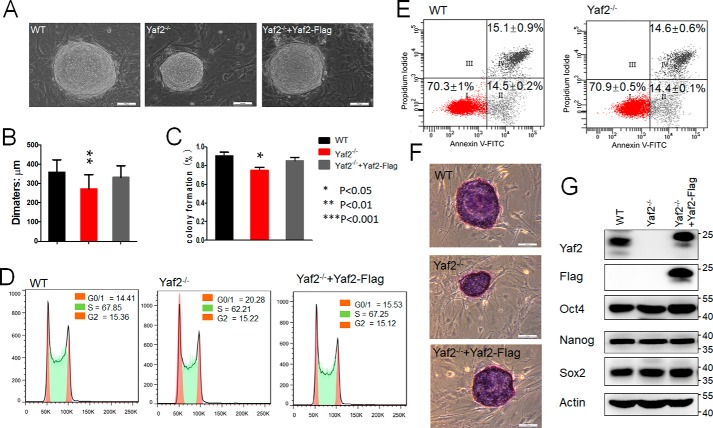
To investigate potential functions of Yaf2, a characteristic subunit of noncanonical PRC1 complexes in mESCs, several Yaf2-deficient mESC clones (referred to as Yaf2−/−) were established by utilizing CRISPR/Cas9 technique (Fig. S1) (25). The Yaf2 knockout mESCs were viable and exhibited typical mESC morphological features when grown on a feeder layer of mouse embryonic fibroblasts (MEFs), indistinguishable from those formed by WT counterparts (Fig. 1A). They did, however, display a stable and mild proliferation defect and moderate reduction in colony-forming ability relative to their controls (Fig. 1, B and C). The knockout mESCs accumulated in the G0/1 phase of the cell cycle, increasing from ∼14% of WT in G0/1 to 20% of the Yaf2 knockout cells (Fig. 1D). Remarkably, lentivirus-based re-introduction of Yaf2 back into Yaf2−/− mESCs rescued the population doubling time and G0/1 phenotypes. The smaller size of Yaf2-deficient mESC colonies was not due to increased apoptosis as we found no significant difference in the percentage of mESCs undergoing apoptosis between the WT and Yaf2−/− when assessed by annexin-V staining (Fig. 1E). Thus, the impaired growth of Yaf2−/− mESCs was likely due to their altered cell cycle profile. Notably, Yaf2 knockout mESCs continued to express high levels of undifferentiated state-specific markers such as Oct4, Nanog, Sox2, and alkaline phosphatase (Fig. 1, F and G). Taken together, these results indicate that Yaf2 is required to retain proper self-renewal capacity of mESCs.
Figure 1.
Yaf2 is essential to maintain mESC self-renewal. A, representative phase images of mESC colonies of the indicated genotypes. mESC colonies corresponding to the indicated genotypes were photographed at day 7 after seeding the cells as a suspension of single cells on feeder cell layers. Scale bar, 100 μm. B, bar graphs show the mean diameter of 30 random mESC colonies. C, percentage of isolated single mESCs of the indicated genotypes giving rise to undifferentiated colonies. D, analysis of cell cycle distribution of mESC of the indicated genotypes by flow cytometry. The percentage of cells in the different stages of the cell cycle is shown at the top right corner. E, Yaf2 deletion results in a degree of apoptosis similar to that in WT mESCs. F, AP staining images of mESC colonies corresponding to the indicated genotypes. The scale bar, 100 μm. G, Oct4, Nanog, and Sox2 protein levels determined by Western blotting in mESCs of the indicated genotypes.
Yaf2 controls differentiation programs in mESCs
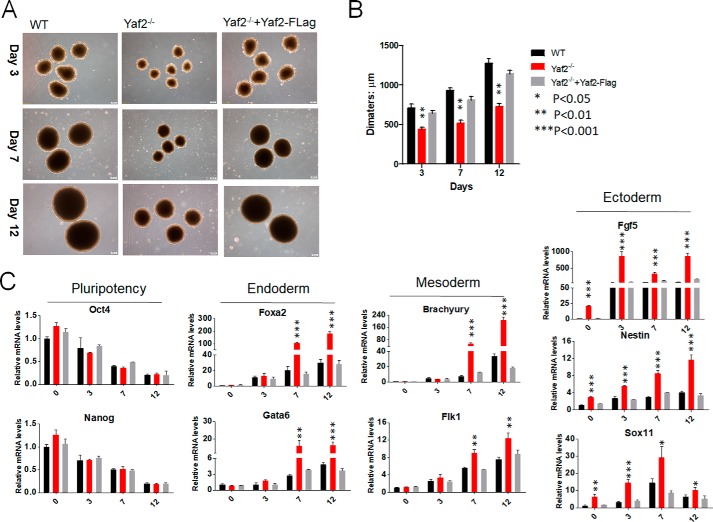
To test the potential of Yaf2−/− mESCs to undergo in vitro differentiation, mutant and control mESCs were tested for their capacity to form embryoid bodies (EBs). As shown in Fig. 2A, Yaf2−/− mESCs did form EBs, but they were reduced in size compared with those of the controls during differentiation (Fig. 2B). RT-PCR analysis of these EBs, which were kept in differentiation medium for 3, 7, and 12 days, revealed up-regulation of genes associated with each of the three germ layers (26), including Gata6 and Foxa2 for endoderm; Fgf5, Nestin, and Sox11 for ectoderm; and Brachyury and Flk1 for mesoderm (Fig. 2C). Further analysis indicated that these changes were accompanied by diminished expression of mESC pluripotency markers, including Oct4 and Nanog. Importantly, throughout Yaf2−/− EB differentiation and even in undifferentiated Yaf2−/− mESCs, the expression of Fgf5, Nestin, and Sox11 was markedly increased with respect to control EBs, whereas higher levels of endoderm and mesoderm markers were only observed at days 7 and 12 of Yaf2−/− EBs, indicating that Yaf2 mainly prevents the differentiation of mESCs into ectoderm (Fig. 2C). Strikingly, lentiviral expression of FLAG-tagged Yaf2 was able to rescue EB size and the pattern of expression of these germ layer markers to levels similar to those in control cells (Fig. 2, A–C). To further investigate the differentiation capacity of Yaf2−/− mESCs, WT and mutant mESCs were subcutaneously injected into the flanks of 6-week-old immune-deficient mice to form teratomas (Fig. S2). Immunohistochemistry showed that various cell types and tissues derived from all three germ layers were present in WT mESC-derived teratomas. However, teratomas from Yaf2−/− mESCs were significantly smaller, and we noted a larger proportion of cells with an ectodermal origin, as evidenced in an immunostaining using antibody specific to glial fibrillary acidic protein (ectoderm marker), when compared with WT teratomas. Notably, Yaf2 deletion has no effect on endoderm and mesoderm specification, as demonstrated by TROMA1 and SMA1, respectively, staining (Fig. S2). These results indicate that Yaf2 is necessary for the differentiation or maintenance of the ectodermal lineage.
Figure 2.
Targeted disruption of the Yaf2 gene results in unscheduled mESC differentiation in vitro. A, representative phase images of EBs derived from mESCs of the indicated genotypes at different time points following differentiation. Images were taken at ×4 magnification at days 3, 7, and 12. B, bar graphs indicate mean diameter of 20 random EBs of the indicated genotypes. C, real-time qPCR was used to assess the expression levels of pluripotency and lineage-specific markers in mRNA samples derived from mESCs and differentiated EBs of the indicated genotypes at days 3, 7, and 12. Bar graphs represent the mean of three independent biological repeats. Error bars indicate ± S.D. Student's t test analysis.
Genome-wide profiling of gene expression in Yaf2−/− mESCs
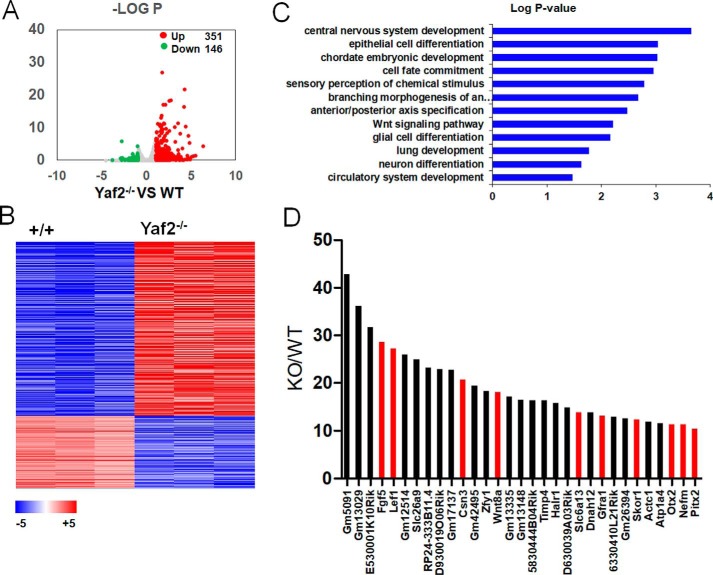
To identify the downstream target genes of Yaf2, we used RNA-seq to compare the transcriptome profiling of WT and Yaf2−/− mESCs. 351 genes were up-regulated, and 146 genes (Fig. 3A and Table S3) were down-regulated based on a fold-change cutoff of 2 and a p value less than 0.05, indicating that Yaf2 functions predominantly as a transcriptional repressor in mESCs (Fig. 3, A and B). Gene ontology (GO) analysis indicated that the up-regulated genes in Yaf2−/− are statistically associated with ectoderm differentiation genes (e.g. central nervous system development and epithelial cell differentiation) (Fig. 3C), implying a particular role for Yaf2 in ectoderm cell fate. Consistently, of the top 31 up-regulated genes with >10-fold increase in expression, 10 have been reported to be essential or are expected to have a function in ectoderm development (Fig. 3D). To determine the potential cross-talk between Yaf2 and its homolog Rybp, Rybp null mESCs were also generated, and their transcriptional status was analyzed by RNA-seq (Fig. S3). There were 904 differentially expressed genes between the WT and Rybp−/− mESCs, with 609 up- and 295 down-regulated genes in Rybp−/− mESCs (Table S4). Of the 48 up-regulated genes with >10-fold increase in expression, 16 were strongly associated with germline development (meiosis and spermatogenesis), consistent with a role of Rybp in PRC1.6 activity (26, 27). Further analysis revealed no significant overlap between the genes regulated by Yaf2 and Rybp (Fig. S3), indicating Yaf2 and Rybp serve distinct functions in mESCs. Together, these results strongly suggest that the repression of developmental genes by Yaf2 may contribute to the maintenance of mESC identity.
Figure 3.
Yaf2 represses the expression of developmental genes in mESCs. A, volcano plots depicting significant differentially expressed genes in Yaf2−/− versus WT mESCs. Up-regulated genes were labeled in red and down-regulated genes were labeled in green if they had a log2 fold change of >1 or less than −1, respectively. The number of differentially expressed genes they represented is indicated. B, RNA-seq heat map of the 497 expressed transcripts with >2-fold expression differences in WT and Yaf2−/− mESCs. Red indicates high expression, and blue indicates low expression. C, Gene Ontology analysis of biological functions of Yaf2 target genes. D, fold changes in the expression of the top 31 genes, each of which displayed more than 10-fold up-regulation in Yaf2−/− compared with WT. Ectoderm-specific genes are highlighted in red.
Yaf2 assembles a noncanonical PRC1 in mESCs
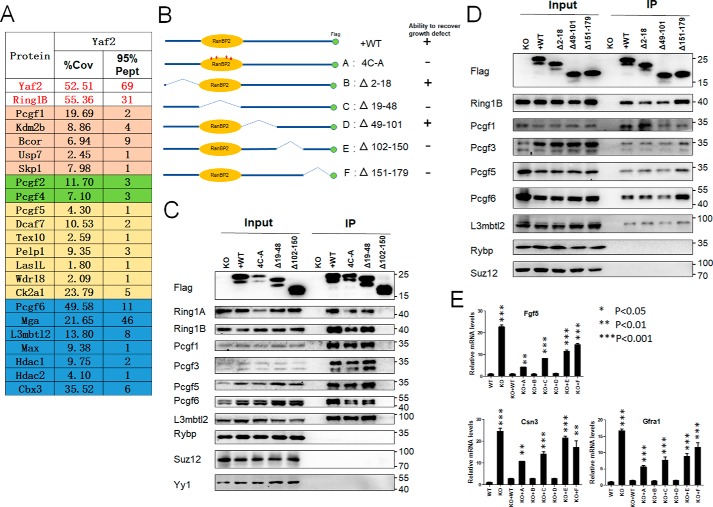
To investigate the molecular background of Yaf2 function, we characterized the yet-unknown physical interactome of Yaf2 in mESCs. Yaf2−/− mESCs were stably transfected with a C-terminal FLAG-tagged form of Yaf2, and nuclear extracts were subjected to immunoprecipitation with anti-FLAG antibody, and then the immunoprecipitates were analyzed by mass spectrometric analysis to determine associated proteins. Fig. 4A shows a summary of Yaf2 interactome, where we focused on known canonical and noncanonical PRC1 complexes. Yaf2 co-purified many proteins that reside in noncanonical PRC1 complexes (PRC1.1, PRC1.3/5, and PRC1.6). In FLAG–Yaf2 immunoprecipitates, Rybp was not detected, in line with previous reports showing that Yaf2 and Rybp are mutually exclusive in PRC1 complexes. Interestingly, we found that Yaf2 specifically co-purified with Pcgf2 and Pcgf4, but not with canonical components, such as polyhomeotic homolog proteins (Phcs), chromobox proteins (Cbxs), and Sex comb on midleg proteins (Scms), which is consistent with the finding that Cbx proteins and Yaf2 are mutually exclusive within the PRC1 complex context, as both compete for the same surface on the C-terminal domain of Ring1B (28). These interactions were confirmed using specific antibodies with deficient mESCs rescued with Yaf2–FLAG (Fig. 4, C and D). Of note, we did not detect Yaf2 interactions with Rybp by Western blotting, consistent with the MS results.
Figure 4.
Yaf2 is physically associated with nc-PRC1 complexes in mESCs. A, list of Yaf2-interacting proteins identified by mass spectrometry (MS) from FLAG purifications of nuclear extracts from Yaf2−/− mESCs re-expressed FLAG–Yaf2. Proteins, percent coverage (% Cov), and 95% unique peptides (95% Pept) are indicated. B, shown is a schematic of Yaf2 deletion and point mutants and summary of their ability to rescue growth defect in Yaf2−/− mESCs. C and D, validation by immunoprecipitation (IP)–Western blotting of selected Yaf2 protein partners identified by MS analysis. Additionally, the ability of Yaf2 mutants to assemble into the PRC1 complexes is also shown. 5% of the cell lysates (Input) used in each assay was loaded as input. Rybp, Suz12, and Yy1 are shown as negative controls. Of note, the C-terminal region of 29 residues (151–179) is required for keeping the doublet of Yaf2 protein on SDS-polyacrylamide gels. E, quantitative RT-PCR detecting Fgf5, Csn3, and Gfra1 expression levels in Yaf2−/− mESCs following re-expression of WT and Yaf2 mutants. Error bars indicate ± S.D. Bar graphs represent the mean of three independent biological repeats. Student's t test analysis.
To define the domains in Yaf2 that mediate its association with other members of the complex, we constructed a series of deletion mutants of Yaf2 and first tested their ability to interact with other subunits in the complex in mESCs by immunoprecipitation followed by immunoblotting. As summarized in Fig. 4B, a deletion mutant of Yaf2 lacking amino acids 102–150 (Δ102–150) failed to interact with other components of the complexes, consistent with its inability to rescue growth defects associated with Yaf2 deficiency in mESCs. The C-terminal deletion mutant (Δ151–179) and the RanBP2-type zinc finger domain deletion (Δ19–48) and point mutants (4C-A, in which Cys-25, Cys-28, Cys-39, and Cys-42 sites were mutated from cysteine to alanine), however, retained their ability to incorporate into the complexes in vivo, although they failed to restore the proliferation defect observed in Yaf2−/− mESCs (Fig. 4, B–D). These studies thus define a novel domain in Yaf2 that mediates its interaction with other components of the complexes. Additionally, these deletion mutants were also assessed for their ability to inhibit the expression of Yaf2 target genes in mESCs. Three Yaf2 deletion mutants, including Δ19–48, Δ102–150, and Δ151–179 (Fig. 4E), failed to restore target gene repression. In contrast, mutants deleted of amino acids 2–18 or 49–101 (Δ2–18 and Δ49–101, respectively) showed a WT level of gene repression.
Critical role for Yaf2 phosphorylation in maintaining mESC identity
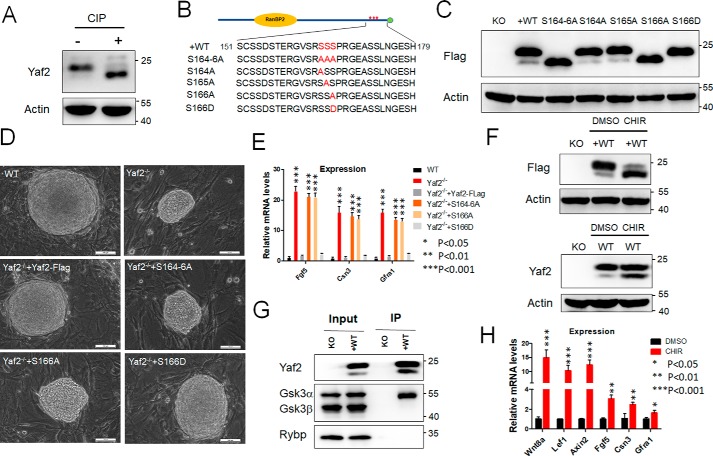
The WT Yaf2 protein migrated as a doublet on SDS-PAGE, and deletion of amino acids 151–179, but not other regions, caused the disappearance of the top band of the doublet (Fig. 4, C and D). Furthermore, calf intestine phosphatase (CIP) treatment resulted in the same change in WT Yaf2 protein (Fig. 5A), indicating that phosphorylation within the region between amino acids 151 and 179 is causal to the motility shift. Strikingly, mutation of three consecutive serines (164–166) within this region to alanines abolished the slowly migrating band (Fig. 5, B and C). To identify the exact amino acid(s) that is phosphorylated, we substituted these three serine residues with alanine individually to prevent phosphorylation. We found that only the S166A mutant migrated as a single band with mobility identical to the lower band of the doublet, whereas the S166D mutant, where the serine was mutated to aspartic acid to mimic phosphoserine, migrated as a single band with mobility identical to the slower migrating band of WT Yaf2 (Fig. 5C).
Figure 5.
Essential role of Yaf2 Ser-166 phosphorylation in the repression of its transcriptional activity. A, treatment with calf alkaline phosphatase abolished the upper band of the Yaf2 doublet and increased the intensity of the lower one. Lysates prepared from the WT mESCs were treated with CIP and analyzed by Western blotting. B and C, up-shift of gel mobility of Yaf2 is due to the phosphorylation at Ser-166. The potential phosphorylated residues in the area of residues 164–166 were mutated to alanine(s) (B) and their band patterns were assessed by Western blotting (C). Notably, only the S166A mutant showed a single (lower) band, and mutation of Ser-166 to aspartate results in the upper band only. D, representative phase images of mESC colonies of the indicated genotypes grown on mitomycin-C inactivated feeders. Scale bar, 100 μm. E, quantitative RT-PCR detecting Fgf5, Csn3, and Gfra1 expression levels in mESCs of the indicated genotypes. F, inhibition of Gsk3 leads to a decrease in the phosphorylation of Yaf2. mESCs of the indicated genotypes were treated with the indicated chemicals in culture medium. Cell lysates were prepared for Western blotting. G, examination of the interaction between Yaf2 and Gsk3 by immunoprecipitation (IP) and Western blotting with antibodies against either Gsk3α/β or FLAG in Yaf2−/− (KO) and Yaf2−/− rescued FLAG–Yaf2 (+WT) mESCs. Whole-cell lysate (Input) was also subjected to Western blotting to determine the expression of Gsk3α/β and Yaf2. H, quantitative RT-PCR analysis to determine mRNA expression of Yaf2 target genes in mESCs treated with the Gsk3 inhibitor CHIR (6 μm) for 24 h. Expression levels of these genes were normalized and compared with those of the untreated control cells. Error bars indicate ± S.D. Bar graphs represent the mean of three independent biological repeats. Student's t test analysis was used.
To evaluate the role of Yaf2 Ser-166 phosphorylation in regulating mESC self-renewal and differentiation, we stably introduced full-length or phosphorylation mutant Yaf2 into Yaf2−/− mESCs. As shown in Fig. 5D, S166D fully restored WT levels of growth in mESCs. However, phosphodefective S166A abolished its ability to rescue the colony-growth defect associated with the deficiency of the Yaf2 gene in mESCs. We examined the ability of the S166A mESCs to undergo differentiation by method of EB and teratoma formation. These S166A mESC-derived EB exhibited higher expression of the ectodermal marker Nestin, Fgf5, and Sox11 throughout EB differentiation (Fig. S4). Consistent with these observations, teratomas derived from S166A mESCs contained more advanced neuron-selected structures (Fig. S2). Furthermore, S166A also failed to suppress Yaf2-mediated target gene repression in undifferentiated mESCs (Fig. 5E). Thus, these results provide strong evidence that the Ser-166 can be phosphorylated and highlight the pivotal role of this residue for maintaining the function of Yaf2 protein in mESCs.
Close examination of the amino acid residues surrounding Ser-166 revealed the presence of a glycogen synthesis kinase 3 (Gsk3) consensus sequence (Ser/Thr-(Xaa-Xaa-Xaa)-Ser/Thr, with Xaa being any amino acid) (29). To gain further support that Gsk3 is responsible for Ser-166 phosphorylation, we used a most selective Gsk3 inhibitor, CHIR99021. To this end, mESCs expressing FLAG-tagged Yaf2 were treated in the presence or absence of CHIR99021. Results shown in Fig. 5F demonstrated that phosphorylation of Yaf2 was greatly inhibited by the presence of CHIR99021 as evidenced by Western blotting. Similar analysis also demonstrated that CHIR99021 treatment resulted in decreased endogenous phospho-Yaf2 levels in WT mESCs (Fig. 5F), indicating that Gsk3 at least partially contributes to Yaf2 phosphorylation. This notion was further supported by the co-immunoprecipitation experiment that detected an interaction between Yaf2 and Gsk3α, but not Gsk3β, in mESCs (Fig. 5G). In addition, treatment of mESCs with CHIR99021 also derepressed the activity of Yaf2-mediated target gene expression (Fig. 5H). Taken together, Gsk3 is likely responsible for the phosphorylation of Yaf2 at the Ser-166 site to control developmental gene expression.
Yaf2 phosphorylation status correlates with levels of H2AK119ub1 at specific promoters
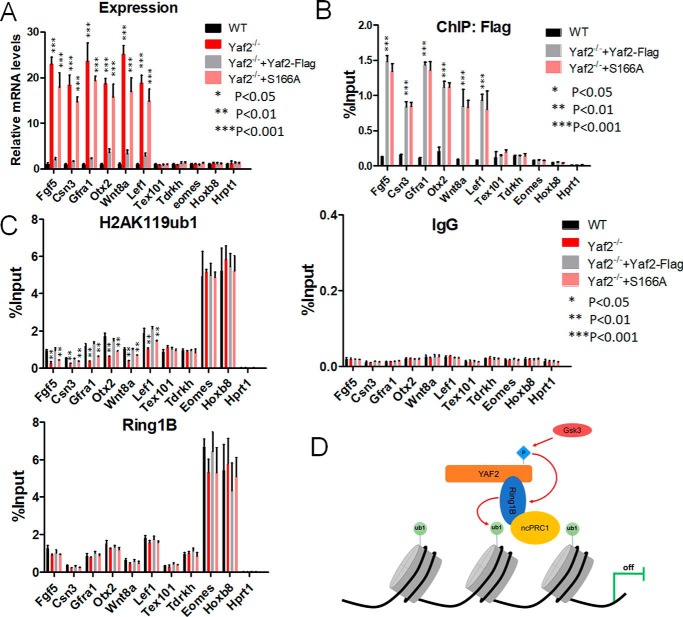
To test whether Yaf2 can directly regulate target genes, we examined Yaf2 occupancy on the promoter of a panel of selected target genes, whose expressions were significantly derepressed upon Yaf2 deletion (Fig. 6A), with the use of the ChIP–qPCR assay. ChIP analysis revealed robust binding by Yaf2 to target promoters in WT mESCs, but the signal was dramatically reduced to background levels in Yaf2−/− mESCs (Fig. 6B). As expected, the PRC1.6 (Tdrkh and Tex101) and canonical PRC1 (Emoes and Foxa2) target genes were not significantly enriched and served as a control in our ChIP experiments. As an additional control for specificity, we showed that an irrelevant antibody (IgG) failed to immunoprecipitate Yaf2 target promoter sequences in ChIP experiments (Fig. 6C). These strongly suggest that Yaf2 targets to its target promoters directly in mESCs. Similar ChIP experiments were also performed with Yaf2−/− and WT mESCs to determine whether the absence of Yaf2 protein would have an impact on the recruitment of Ring1B, Rybp (components of PRC1), or Suz12 (PRC2 complex). Unexpectedly, ChIP data indicate that recruitment of these components was minimally affected by loss of Yaf2 (Fig. 6C and Fig. S5), which is inconsistent with the results obtained in HeLa cells (30).
Figure 6.
Phosphorylation of Yaf2 promotes H2AK119 monoubiquitination and represses target gene expression. A, quantitative RT-PCR analysis to determine mRNA expression of target genes of Yaf2 (Fgf5, Csn3, Gfra1, Otx2, Wnt8a, and Lef1), PRC1.6 (Tex101 and Tdrkh), and canonical PRC1/2 (Eomes and Hoxb8) in mESCs of the indicated genotypes. Hprt1 was used as a control. Expression was normalized to actin. B, anti-FLAG ChIP–qPCR assays were done using Yaf2−/− and Yaf2−/− rescued with FLAG–Yaf2 mESCs. C, ChIP-qPCR was used to analyze the occupancy of H2AK119ub1 and Ring1B on the promoters of Yaf2 target genes in mESCs of the indicated genotypes. Error bars indicate ± S.D. Bar graphs represent the mean of three independent biological repeats. p value: Student's t test analysis. D, model for Yaf2-mediated transcriptional repression. See under “Discussion” for details.
Importantly, despite our observation that promoter occupancy by H3K27me3 was not affected by loss of Yaf2, we observed that H2AK119ub1 enrichment was reduced by 2–3-fold in the Yaf2−/− mESCs at these specific targets of Yaf2 (Fig. 6C and Fig. S5). Furthermore, deletion of Yaf2 also resulted in moderate reduction in global H2AK119ub1 as measured by Western blotting (Fig. S5). To verify that the reduction of H2AK119ub1 levels at genes bound by Yaf2 in knockout cells is only due to the lack of Yaf2, we reexpressed both WT and mutant Yaf2 in the knockout mESCs using lentiviral expression vectors. WT Yaf2 displayed full rescue of H2AK119ub1 levels as well as target gene derepression. However, a Yaf2 phosphorylation-defective mutant S166A virtually failed to restore H2AK119ub1 levels at target promoters or target gene repression in null mESCs, although it displayed a binding affinity for target promoters similar to that of the WT (Fig. 6, A–C). This indicated that phosphorylation of Yaf2 at Ser-166 enhances Ring1B-mediated deposition of H2AK119ub1, thus contributing to target gene repression.
Discussion
The central conclusion from this study is the demonstration that Yaf2 contributes to the maintenance of mESC pluripotency in a manner that depends upon its phosphorylation state. Interestingly, we find that phosphorylation of Yaf2 is essential for Ring1B-mediated E3 ubiquitin ligase activity. Thus, this study suggests a mechanism for how signaling affects PcG-mediated chromatin-based epigenetic processes to ensure proper regulation of cell fate at the molecular level and how the enzymatic activity of PRC1 is regulated.
Recent studies show that the composition of PRC1 complexes is considerably far more diverse than previously recognized. In mammals, six major groups of PRC1 complexes, referred to as PRC1.1–1.6 based on available Pcgf subunits, and their potential roles in H2AK119ub1 have been characterized (6). Although distinct types of PRC1 are believed to catalyze H2AK119ub1 due to the presence of a common subunit Ring1B, they do not appear to have an equal effect on enzymatic activity. Indeed, we have shown that among Pcgf family members only, Pcgf3/5 are required for maintaining global H2AK119ub1 levels in mESCs (16). In vitro ubiquitylation assays demonstrated that Ring1A/B with either Pcgf2 or Pcgf4 can catalyze H2AK119ub1 (17, 31), but incorporation of the auxiliary subunits Rybp, CBX2/8, or PHC2 into these minimal complexes has been reported to affect their E3 ubiquitin ligase activity (6). According to our findings, PRC1-mediated H2AK119ub1 in mESCs is significantly reduced after deletion of Yaf2. This scenario is reminiscent of the reductions in H2AK119ub1 at PRC1 targets that occur after deletion of Rybp in mESCs (32), suggesting that Yaf2 exert a function similar to that of Rybp in mESCs.
Recent literature has revealed that the properties of self-renewal and pluripotency are governed by an intricate set of cell-intrinsic programs in response to the extracellular signals, including developmental cues (33). These intracellular signaling cascades ultimately converge to the chromatin directly (i.e. via histone phosphorylation) or indirectly (via post-translational modifications of chromatin-modifying complexes) to enable gene regulation (33, 34). Recent observations have shown that PcG proteins, as central players in above epigenetic programming events, are targeted for post-translational modifications such as phosphorylation (17, 19, 20, 35). However, the functional relevance of these phosphorylation events has remained largely unknown. Here, we report Yaf2 was found to be phosphorylated at serine 166 by, at least partially, Gsk3. Intriguingly, Yaf2 Ser-166 phosphodefective mutants were still assembled into PRC1 complexes and did not significantly differ in its promoter tethering activity from WT Yaf2. However, this mutant failed to rescue H2AK119ub1 levels at Yaf2-specific target promoters and thus lost its ability to repress target gene expression in null mESCs. Collectively, these findings suggest Yaf2 is a phosphoprotein that may integrate diverse signals and directly converts them into chromatin modifications (i.e. H2AK119ub1) that control developmental gene expression and the regulation of mESC maintenance and lineage commitment (Fig. 6D). Our findings also implicate PcG-mediated epigenetic silencing as a dynamically controlled process.
Our data demonstrate that phosphorylation of Yaf2 on serine 166 is at least partially regulated through the Gsk3 pathway. Previous studies (35) revealed that phosphorylation of EZH2 on Ser-21 by the Akt/Gsk3 pathway inhibits its methyltransferase activity. We found that Gsk3-mediated phosphorylation of Yaf2 on serine 166 supports Ring1B-mediated H2AK119ub1 and is critical for maintenance of pluripotency in mESCs. Gsk3 signaling has not been fully explored in regulation of stem cell pluripotency, although a combination of small molecule inhibitors for the mitogen-activated protein kinase kinase (MEK) and Gsk3 (2i conditions) has been implicated in the maintenance of mESC pluripotency under minimal growth conditions (36, 37). Therefore, our findings that this signaling directly modifies PRC1 activity warrant further investigation of this signaling cue in control of these processes.
It has been shown by transgenic studies that Yy1, the mammalian homologue of Drosophila PHO, can phenotypically correct PHO mutant flies (38) and is required for recruitment of PRC1/2 to certain Polycomb-responsive elements (PRE) in Drosophila (39, 40). Because of its potential interaction with Yaf2, it has been proposed that Yaf2 might act as a bridge between Yy1 and a subset of PRC1 to target these complexes to specific DNA sequences (PREs) (41, 42). Intriguingly, however, a stable association of Yy1 with PRC1 has never been detected in studies from us and other groups (Fig. 4) (6, 26, 43). It is possible that the interaction between Yy1 and PRC1 components may be transient. Thus, further experiments are needed to carefully examine the influence of Yy1 on the recruitment of PRC1 to chromatin, at least in mESCs.
Transcriptome analysis reveals that Yaf2 deficiency in mESCs resulted in selective derepression of ectoderm-specific genes (Fig. 3D). Moreover, Yaf2−/− EBs expressed increased levels of ectodermal markers (Fig. 2C). Consistent with these observations, Yaf2 deletion skewed differentiation toward neural ectoderm lineages in a teratoma assay (Fig. S2). Importantly, ChIP analysis revealed direct binding by Yaf2 to target promoters in mESCs (Fig. 6), suggesting a potential requirement for Yaf2 in repression of polycomb-controlled developmental regulators. Given the evidence that Yaf2 regulates gene expression, particularly by repressing essential developmental genes involved in neural ectoderm differentiation, it is possible that subtle deregulation of these developmental regulators interferes with timely progression of neural ectoderm development. Further studies using mice with a conditional Yaf2 mutant in the nervous system will shed light about the in vivo contributions of Yaf2 during nervous system development.
Experimental procedures
Plasmid construction
The plasmid of Cas9 vector with single guide RNA (sgRNA) was prepared as described previously (44). Briefly, sgRNAs were designed by CRISPR design tool (http://crispr.mit.edu/)4 (45) and cloned into pX330(Addgene plasmid ID 42230) vector, which was modified to express Cas9 and sgRNA.
The full-length cDNA of mouse Yaf2 (Q99LW6) was added to a C-terminal FLAG-tag (DYKDDDDK) sequence by PCR and inserted into pBS KS II vector. Deletions and point mutations of the Yaf2 cDNA were generated by QuikChange II site-directed mutagenesis kit (Agilent). All the verified products were introduced into the lentiviral vector as described previously (26).
Cell culture
mESCs were grown on gelatin-treated cell culture plates or on mitomycin-treated feeder MEFs and cultured in DMEM high glucose (Gibco) with 15% fetal calf serum (Gibco), 1000 units/ml leukemia inhibitory factor, nonessential amino acids (Gibco), l-glutamine (Gibco), β-mercaptoethanol (Sigma), and penicillin/streptomycin (Gibco). 293T cells were cultured in DMEM with 10% fetal calf serum and penicillin/streptomycin. Embryoid body medium consists of Iscove's modified Dulbecco's medium, 15% fetal calf serum, 2 mm l-glutamine, nonessential amino acids, 50 μg/ml ascorbic acid, 200 μg/ml iron-saturated holo-transferrin, sodium pyruvate, 450 μm monothioglycerol, and penicillin/streptomycin.
Generation of mutant mESCs
Knockout mESCs were generated as described previously (44). Briefly, constructs expressing a pair of sgRNAs were co-transfected along with a puromycin resistance plasmid into mESCs by Lipofectamine 2000 (Life Technologies, Inc.). After 24 h of transfection, mESCs were selected by puromycin for 2 days and then seeded on mitomycin-irradiated feeder cells on gelatinized tissue culture plates in mESC medium. Correctly targeted knockout mESC clones were identified by genomic PCR analysis. Subsequently, the null mESCs were confirmed by Western blotting.
Lentiviral supernatant production and infections
Lentiviral supernatants were prepared as described (26). The mESCs were infected by lentiviral supernatants with Polybrene (Sigma, final concentration of 8 μg/ml). After 24 h, positive mESCs were selected by puromycin.
Colony formation, AP staining, and flow cytometry
WT or mutant mESCs (about 1000 cells) were dissociated by trypsin into single-cell suspension and plated onto 10-cm MEF-covered dishes and incubated for 7 days before imaging by microscopy.
AP staining of mESC colonies was done following the manufacturer's instructions (Yeasen). Cell cycle analysis has been described previously (46). Propidium iodide (PI) staining of mESCs was performed according to the manufacturer's instructions (Vazyme). Apoptosis was detected using an annexin V-FITC/PI detection kit (Yeasen) and followed by flow cytometry analysis on a FACS LSRF or tessa (BD Biosciences).
Embryoid body and teratoma formation and analysis
Embryoid body formation was performed as described previously (26). Briefly, trypsinized mESCs were resuspended in Embryoid body medium. A 30-μl sample (300–500 cells) was pipetted onto the 15-cm Petri-dish plate, which was then placed upside down, and the hanging drops were cultured for about 3 days. Then EBs were collected and cultured on a rotating shaker. Total RNA was collected (TRIzol, ThermoFisher Scientific) on days 3, 7, and 12.
Teratoma formation was performed as described previously (26). Briefly, mESCs were trypsinized, washed twice with PBS, and then injected subcutaneously into the flanks of 8-week-old immunodeficient mice. After 4 weeks, mature teratomas were fixed in 4% phosphate-buffered formaldehyde. Paraffin-embedded tissue was sliced and stained with hematoxylin and eosin (H&E) and then immunohistochemistry. The experimental animal facility has been accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC), and the Institutional Animal Care and Use Committee (IACUC) of the Model Animal Research Center of Nanjing University approved all animal protocols used in this study.
Expression analysis
Total RNA was isolated from mESCs or EBs using TRIzol reagent (ThermoFisher Scientific) and was reverse-transcribed according to the manufacturer's protocol. Briefly, RT-PCRs were carried out with the HiScriptTM First Strand cDNA synthesis kit (Vazyme). Quantitative RT-PCR was performed in triplicate from independent biological samples using PowerUpTM SYBR® Green Master Mix (Life Technologies, Inc.) on a StepOneTM software version 2.3 (Applied Biosystems). Expression levels of each gene were normalized to actin. Primers for RT-PCR are listed in Table S1.
Whole-cell lysates, histone extraction, and Western blot analysis
Whole-cell lysates of mESCs were prepared in RIPA buffer as described (16). Proteins were separated via SDS-PAGE and analyzed by Western blotting. Histone extraction was performed as described (47). Briefly, mESCs were resuspended in 1 ml of Hypotonic lysis buffer and incubated for 30 min on a rotator at 4 °C. The intact nuclei were spun by centrifuge at 10,000 × g for 10 min at 4 °C. The nuclear pellet was resuspended in 200 μl of 0.2 n HCl. Histone modifications were analyzed by immunoblotting. List of the antibodies used in this study is shown in Table S2.
RNA-seq analysis
RNA-seq was performed as described previously (16) and supported by Annoroad Gene Tech. Total RNA was isolated using TRIzol reagent (ThermoFisher Scientific). Sequencing libraries were implemented by the Next® Ultra RNA Library Prep Kit for Illumina (New England Biolabs), and then three independent biological samples were sequenced on an Illumina Hiseq4000 platform and generated PEI150 strategy. The expression levels of each gene were calculated by fragments per kilobase of transcript per million fragments mapped. Then differential gene expression analysis between WT and knockout mESCs was measured by DESeq (version 1.16). The list of deregulated genes in knockout is shown in Tables S3 and S4. In addition, RNA-seq data were deposited at the Gene Expression Omnibus under accession number GSE113830.
CHIR99021 treatment
Gsk3 inhibitor CHIR99021 was obtained from Sigma. mESC lines were treated with CHIR99021 (6 μm) for 24 h.
Dephosphorylation assay
mESCs were resuspended in the dephosphorylation buffer (100 mm NaCl, 50 mm Tris-HCl, pH 7.9, 10 mm MgCl2, 1 mm DTT, pH 7.9 at 25 °C) with protease inhibitors. Then, CIP (New England Biolabs) was added to the lysates and incubated up to 60 min at 37 °C. The reaction was stopped by the addition of phosphatase inhibitors (10 mm sodium fluoride, 10 mm sodium vanadate, 10 mm potassium pyrophosphate, and 5 mm sodium phosphate). The dephosphorylated lysates were processed for SDS-PAGE.
Immunoprecipitation and mass spectrometry (MS)
Immunoprecipitation and MS were performed as described previously (16). The nuclear lysates of Yaf2 −/− mESCs rescued with FLAG–Yaf2 were used for immunoprecipitation by M2 beads. For MS identification, the products were run into the SDS-polyacrylamide gel about 1–2 cm, followed by Coomassie Blue staining. The gel slice, including all proteins, was cut and identified by LC-MS analysis (2D-NanoLC/TripleTOF5600).
ChIP
ChIP analysis was performed as described previously (26). The proteins and DNAs of mESCs were cross-linked in 1% fresh formaldehyde solution diluted from 37% formaldehyde (Sigma) followed by 0.125 m glycine to quench. Cells were lysed in ChIP buffer, and then sonication was performed using Bioruptor system (Diagenode) to produce DNA fragments of 200–1000 bp. For immunoprecipitation, 1 volume of chromatin solution was mixed with 4 volumes of ChIP dilution buffer and appropriate antibodies. Immunocomplexes were captured by protein A/G-Sepharose overnight at 4 °C. The beads were washed twice by low-salt buffer and high-salt buffer, respectively. Immunocomplexes were eluted from beads using eluent buffer, and then the DNAs were decross-linked and purified using DNA gel extraction kit (Axygen). Quantitative PCRs were performed using PowerUpTM SYBR® Green Master Mix (ThermoFisher Scientific) on a StepOneTM software version 2.3 (Applied Biosystems). The enrichment was calculated as 2−ΔCt, where ΔCt = Ct (ChIP) − Ct(input). Primers utilized for ChIP are listed in supporting Table 1.
Author contributions
W. Z., M. L., Q. J., and J. Q. conceptualization; W. Z., M. L., H. J., Y. Z., C. W., Y. H., X. M., G. X., Y. X., Q. J., and J. Q. resources; W. Z. and M. L. data curation; W. Z. software; W. Z. formal analysis; W. Z. validation; W. Z. and M. L. investigation; W. Z., M. L., and H. J. methodology; W. Z. and J. Q. writing-original draft; G. X., Y. X., Q. J., and J. Q. supervision; J. Q. funding acquisition; J. Q. project administration; J. Q. writing-review and editing.
Supplementary Material
Acknowledgments
We thank Dr. Xiang Gao for providing lentivirus expression vector. We are indebted to Drs. Yun Shi and Zhaoyu Lin for their helpful suggestions and for kindly providing reagents.
This work was supported by National Natural Science Foundation of China Grants 31671532 and 31471387 and the 2015 Shuangchuang Program of Jiangsu Province (to J. Q.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S5 and Tables S1–S4.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- PcG
- polycomb group
- mESC
- mouse embryonic stem cells
- EB
- embryoid body
- MEF
- mouse embryonic fibroblast
- sgRNA
- single guide RNA
- CIP
- calf intestine phosphatase
- RNA-seq
- RNA-sequencing
- DMEM
- Dulbecco's modified Eagle's medium
- AP
- alkaline phosphatase
- PI
- propidium iodide
- qPCR
- quantitative PCR
- nc-
- noncanonical
- PRE
- Polycomb-responsive element
- CBX
- Chromobox.
References
- 1. Di Croce L., and Helin K. (2013) Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 20, 1147–1155 10.1038/nsmb.2669 [DOI] [PubMed] [Google Scholar]
- 2. Simon J. A., and Kingston R. E. (2009) Mechanisms of polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 10, 697–708 10.1038/nrm2763 [DOI] [PubMed] [Google Scholar]
- 3. Steffen P. A., and Ringrose L. (2014) What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory. Nat. Rev. Mol. Cell Biol. 15, 340–356 10.1038/nrm3789 [DOI] [PubMed] [Google Scholar]
- 4. Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R. S., and Zhang Y. (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298, 1039–1043 10.1126/science.1076997 [DOI] [PubMed] [Google Scholar]
- 5. Czermin B., Melfi R., McCabe D., Seitz V., Imhof A., and Pirrotta V. (2002) Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111, 185–196 10.1016/S0092-8674(02)00975-3 [DOI] [PubMed] [Google Scholar]
- 6. Gao Z., Zhang J., Bonasio R., Strino F., Sawai A., Parisi F., Kluger Y., and Reinberg D. (2012) PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell 45, 344–356 10.1016/j.molcel.2012.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cao R., Tsukada Y., and Zhang Y. (2005) Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol. Cell 20, 845–854 10.1016/j.molcel.2005.12.002 [DOI] [PubMed] [Google Scholar]
- 8. Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R. S., and Zhang Y. (2004) Role of histone H2A ubiquitination in Polycomb silencing. Nature 431, 873–878 10.1038/nature02985 [DOI] [PubMed] [Google Scholar]
- 9. Boyer L. A., Plath K., Zeitlinger J., Brambrink T., Medeiros L. A., Lee T. I., Levine S. S., Wernig M., Tajonar A., Ray M. K., Bell G. W., Otte A. P., Vidal M., Gifford D. K., Young R. A., and Jaenisch R. (2006) Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353 10.1038/nature04733 [DOI] [PubMed] [Google Scholar]
- 10. Bracken A. P., Dietrich N., Pasini D., Hansen K. H., and Helin K. (2006) Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 20, 1123–1136 10.1101/gad.381706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Turner S. A., and Bracken A. P. (2013) A “complex” issue: deciphering the role of variant PRC1 in ESCs. Cell Stem Cell 12, 145–146 10.1016/j.stem.2013.01.014 [DOI] [PubMed] [Google Scholar]
- 12. Tavares L., Dimitrova E., Oxley D., Webster J., Poot R., Demmers J., Bezstarosti K., Taylor S., Ura H., Koide H., Wutz A., Vidal M., Elderkin S., and Brockdorff N. (2012) RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 148, 664–678 10.1016/j.cell.2011.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Morey L., Pascual G., Cozzuto L., Roma G., Wutz A., Benitah S. A., and Di Croce L. (2012) Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell 10, 47–62 10.1016/j.stem.2011.12.006 [DOI] [PubMed] [Google Scholar]
- 14. Schwartz Y. B., and Pirrotta V. (2014) Ruled by ubiquitylation: a new order for polycomb recruitment. Cell Rep. 8, 321–325 10.1016/j.celrep.2014.07.001 [DOI] [PubMed] [Google Scholar]
- 15. Stock J. K., Giadrossi S., Casanova M., Brookes E., Vidal M., Koseki H., Brockdorff N., Fisher A. G., and Pombo A. (2007) Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat. Cell Biol. 9, 1428–1435 10.1038/ncb1663 [DOI] [PubMed] [Google Scholar]
- 16. Zhao W., Huang Y., Zhang J., Liu M., Ji H., Wang C., Cao N., Li C., Xia Y., Jiang Q., and Qin J. (2017) Polycomb group RING finger proteins 3/5 activate transcription via an interaction with the pluripotency factor Tex10 in embryonic stem cells. J. Biol. Chem. 292, 21527–21537 10.1074/jbc.M117.804054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Elderkin S., Maertens G. N., Endoh M., Mallery D. L., Morrice N., Koseki H., Peters G., Brockdorff N., and Hiom K. (2007) A phosphorylated form of Mel-18 targets the Ring1B histone H2A ubiquitin ligase to chromatin. Mol. Cell 28, 107–120 10.1016/j.molcel.2007.08.009 [DOI] [PubMed] [Google Scholar]
- 18. Gao Z., Lee P., Stafford J. M., von Schimmelmann M., Schaefer A., and Reinberg D. (2014) An AUTS2-Polycomb complex activates gene expression in the CNS. Nature 516, 349–354 10.1038/nature13921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Voncken J. W., Niessen H., Neufeld B., Rennefahrt U., Dahlmans V., Kubben N., Holzer B., Ludwig S., and Rapp U. R. (2005) MAPKAP kinase 3pK phosphorylates and regulates chromatin association of the polycomb group protein Bmi1. J. Biol. Chem. 280, 5178–5187 10.1074/jbc.M407155200 [DOI] [PubMed] [Google Scholar]
- 20. Roscic A., Möller A., Calzado M. A., Renner F., Wimmer V. C., Gresko E., Lüdi K. S., and Schmitz M. L. (2006) Phosphorylation-dependent control of Pc2 SUMO E3 ligase activity by its substrate protein HIPK2. Mol. Cell 24, 77–89 10.1016/j.molcel.2006.08.004 [DOI] [PubMed] [Google Scholar]
- 21. Kalenik J. L., Chen D., Bradley M. E., Chen S. J., and Lee T. C. (1997) Yeast two-hybrid cloning of a novel zinc finger protein that interacts with the multifunctional transcription factor YY1. Nucleic Acids Res. 25, 843–849 10.1093/nar/25.4.843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sawa C., Yoshikawa T., Matsuda-Suzuki F., Deléhouzée S., Goto M., Watanabe H., Sawada J., Kataoka K., and Handa H. (2002) YEAF1/RYBP and YAF-2 are functionally distinct members of a cofactor family for the YY1 and E4TF1/hGABP transcription factors. J. Biol. Chem. 277, 22484–22490 10.1074/jbc.M203060200 [DOI] [PubMed] [Google Scholar]
- 23. Bannasch D., Mädge B., and Schwab M. (2001) Functional interaction of Yaf2 with the central region of MycN. Oncogene 20, 5913–5919 10.1038/sj.onc.1204747 [DOI] [PubMed] [Google Scholar]
- 24. Stanton S. E., McReynolds L. J., Evans T., and Schreiber-Agus N. (2006) Yaf2 inhibits caspase 8-mediated apoptosis and regulates cell survival during zebrafish embryogenesis. J. Biol. Chem. 281, 28782–28793 10.1074/jbc.M603348200 [DOI] [PubMed] [Google Scholar]
- 25. Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A., and Zhang F. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Qin J., Whyte W. A., Anderssen E., Apostolou E., Chen H. H., Akbarian S., Bronson R. T., Hochedlinger K., Ramaswamy S., Young R. A., and Hock H. (2012) The polycomb group protein L3mbtl2 assembles an atypical PRC1-family complex that is essential in pluripotent stem cells and early development. Cell Stem Cell 11, 319–332 10.1016/j.stem.2012.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huang Y., Zhao W., Wang C., Zhu Y., Liu M., Tong H., Xia Y., Jiang Q., and Qin J. (2018) Combinatorial control of recruitment of a variant PRC1.6 complex in embryonic stem cells. Cell Rep. 22, 3032–3043 10.1016/j.celrep.2018.02.072 [DOI] [PubMed] [Google Scholar]
- 28. Wang R., Taylor A. B., Leal B. Z., Chadwell L. V., Ilangovan U., Robinson A. K., Schirf V., Hart P. J., Lafer E. M., Demeler B., Hinck A. P., McEwen D. G., and Kim C. A. (2010) Polycomb group targeting through different binding partners of RING1B C-terminal domain. Structure 18, 966–975 10.1016/j.str.2010.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cohen P., and Frame S. (2001) The renaissance of GSK3. Nat. Rev. Mol. Cell Biol. 2, 769–776 10.1038/35096075 [DOI] [PubMed] [Google Scholar]
- 30. Basu A., Wilkinson F. H., Colavita K., Fennelly C., and Atchison M. L. (2014) YY1 DNA binding and interaction with YAF2 is essential for Polycomb recruitment. Nucleic Acids Res. 42, 2208–2223 10.1093/nar/gkt1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Buchwald G., van der Stoop P., Weichenrieder O., Perrakis A., van Lohuizen M., and Sixma T. K. (2006) Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 25, 2465–2474 10.1038/sj.emboj.7601144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rose N. R., King H. W., Blackledge N. P., Fursova N. A., Ember K. J., Fischer R., Kessler B. M., and Klose R. J. (2016) RYBP stimulates PRC1 to shape chromatin-based communication between Polycomb repressive complexes. eLife 5, e18591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hackett J. A., and Surani M. A. (2014) Regulatory principles of pluripotency: from the ground state up. Cell Stem Cell 15, 416–430 10.1016/j.stem.2014.09.015 [DOI] [PubMed] [Google Scholar]
- 34. Atlasi Y., and Stunnenberg H. G. (2017) The interplay of epigenetic marks during stem cell differentiation and development. Nat. Rev. Genet. 18, 643–658 10.1038/nrg.2017.57 [DOI] [PubMed] [Google Scholar]
- 35. Cha T. L., Zhou B. P., Xia W., Wu Y., Yang C. C., Chen C. T., Ping B., Otte A. P., and Hung M. C. (2005) Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science 310, 306–310 10.1126/science.1118947 [DOI] [PubMed] [Google Scholar]
- 36. Ying Q. L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., and Smith A. (2008) The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 10.1038/nature06968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sim Y. J., Kim M. S., Nayfeh A., Yun Y. J., Kim S. J., Park K. T., Kim C. H., and Kim K. S. (2017) 2i maintains a naive ground state in ESCs through two distinct epigenetic mechanisms. Stem Cell Rep. 8, 1312–1328 10.1016/j.stemcr.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Atchison L., Ghias A., Wilkinson F., Bonini N., and Atchison M. L. (2003) Transcription factor YY1 functions as a PcG protein in vivo. EMBO J. 22, 1347–1358 10.1093/emboj/cdg124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Srinivasan L., and Atchison M. L. (2004) YY1 DNA binding and PcG recruitment requires CtBP. Genes Dev. 18, 2596–2601 10.1101/gad.1228204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wilkinson F. H., Park K., and Atchison M. L. (2006) Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proc. Natl. Acad. Sci. U.S.A. 103, 19296–19301 10.1073/pnas.0603564103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wilkinson F., Pratt H., and Atchison M. L. (2010) PcG recruitment by the YY1 REPO domain can be mediated by Yaf2. J. Cell. Biochem. 109, 478–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Woo C. J., Kharchenko P. V., Daheron L., Park P. J., and Kingston R. E. (2010) A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell 140, 99–110 10.1016/j.cell.2009.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Trojer P., Cao A. R., Gao Z., Li Y., Zhang J., Xu X., Li G., Losson R., Erdjument-Bromage H., Tempst P., Farnham P. J., and Reinberg D. (2011) L3MBTL2 protein acts in concert with PcG protein-mediated monoubiquitination of H2A to establish a repressive chromatin structure. Mol. Cell 42, 438–450 10.1016/j.molcel.2011.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhao W., Tong H., Huang Y., Yan Y., Teng H., Xia Y., Jiang Q., and Qin J. (2017) Essential role for polycomb group protein Pcgf6 in embryonic stem cell maintenance and a noncanonical polycomb repressive complex 1 (PRC1) integrity. J. Biol. Chem. 292, 2773–2784 10.1074/jbc.M116.763961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., and Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yan Y., Zhao W., Huang Y., Tong H., Xia Y., Jiang Q., and Qin J. (2017) Loss of polycomb group protein Pcgf1 severely compromises proper differentiation of embryonic stem cells. Sci. Rep. 7, 46276 10.1038/srep46276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shechter D., Dormann H. L., Allis C. D., and Hake S. B. (2007) Extraction, purification and analysis of histones. Nat. Protoc. 2, 1445–1457 10.1038/nprot.2007.202 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.