Abstract
Neutrophils are white blood cells that are mobilized to damaged tissues and to sites of pathogen invasion, providing the first line of host defense. Chemokines displayed on the surface of blood vessels promote migration of neutrophils to these sites, and tissue- and pathogen-derived chemoattractant signals, including N-formylmethionylleucylphenylalanine (fMLP), elicit further migration to sites of infection. Although nearly all chemoattractant receptors use heterotrimeric G proteins to transmit signals, many of the mechanisms lying downstream of chemoattractant receptors that either promote or limit neutrophil motility are incompletely defined. Here, we show that regulator of G protein signaling 5 (RGS5), a protein that modulates G protein activity, is expressed in both human and murine neutrophils. We detected significantly more neutrophils in the airways of Rgs5−/− mice than WT counterparts following acute respiratory virus infection and in the peritoneum in response to injection of thioglycollate, a biochemical proinflammatory stimulus. RGS5-deficient neutrophils responded with increased chemotaxis elicited by the chemokines CXC motif chemokine ligand 1 (CXCL1), CXCL2, and CXCL12 but not fMLP. Moreover, adhesion of these cells was increased in the presence of both CXCL2 and fMLP. In summary, our results indicate that RGS5 deficiency increases chemotaxis and adhesion, leading to more efficient neutrophil mobilization to inflamed tissues in mice. These findings suggest that RGS5 expression and activity in neutrophils determine their migrational patterns in the complex microenvironments characteristic of inflamed tissues.
Keywords: regulator of G protein signaling, RGS, chemotaxis, neutrophil, inflammation, trafficking, GPCR, G protein–coupled receptor, leukocyte, innate immunity
Introduction
Leukocyte accumulation at sites of infection and tissue damage is a highly coordinated process. Formyl peptides, lipids, and other pathogen-derived chemoattractants act in concert with host-derived mediators, principally chemokines, leading to recruitment of both innate and adaptive immune cells. Working together, these cells ultimately serve to facilitate pathogen destruction, inflammatory resolution, and tissue repair (1, 2).
In inflamed tissues, cytokine-activated microvascular endothelial cells (ECs)3 generate chemokine gradients that induce tethering, rolling, and, ultimately, firm adhesion and EC transmigration (3). Neutrophils must then navigate away from chemokine gradients toward a second tier of chemoattractants localized in tissues, many of which are generated from pathogen by-products (e.g. fMLP) or the host response itself (e.g. C5a and leukotriene B4). Chemoattractant receptors of both groups typically signal through heterotrimeric G proteins of the Gi subfamily, whose actions are highly sensitive to inhibition by pertussis toxin (4). Ligand binding to chemokine G protein–coupled receptors (GPCRs) promotes exchange of guanosine diphosphate (GDP), which is bound constitutively to the inactive α subunit of the G protein (Gαi), for guanosine triphosphate (GTP). GTP–Gαi is activated and dissociates from the remaining G protein subunits (Gβγ). Both Gαi–GTP and Gβγ activate distinct downstream effectors that modify cell polarity and induce motility, such as phospholipase Cβ, which leads to activation of effectors crucial to chemotaxis, including Ca2+, extracellular signal-regulated kinase (ERK), and phosphoinositide 3-kinase (PI3K) (5). Desensitization of most GPCRs occurs through binding to β-arrestins, which leads to receptor phosphorylation and internalization. In neutrophils, remodeling of the actin cytoskeleton may also desensitize signals by physically separating receptor and G protein (6). However, the mechanism(s) by which neutrophils regulate responsiveness to chemoattractant signals has not been fully delineated.
Another mechanism of GPCR desensitization involves the regulator of G protein signaling (RGS) proteins, a large family of complex, heterogeneous intracellular proteins with pleiotropic functions in both innate and adaptive immune responses (7). Among their critical functions, canonical RGS proteins inactivate Gαi and Gαq subunits through their GTPase-accelerating (GAP) activity. By augmenting the intrinsic capacity of Gα to hydrolyze GTP, RGS proteins decrease the lifetime of activated (GTP-bound) Gα, thereby inhibiting the signaling pathways leading to leukocyte migration (7, 8).
Several physiological functions of RGS proteins in the immune system have been identified. RGS1 and -13 regulate localization of B cells in lymphoid follicles and humoral immune responses (9, 10). Genetic manipulation of RGS13 levels in mast cells has strong effects on chemotaxis and chemokine-induced signaling (11). However, expression and functions of RGS proteins in neutrophils have not been evaluated directly. Interestingly, Cho et al. (12) found that mice carrying mutant alleles encoding a point mutation in Giα2 (G184S), which renders the G protein globally resistant to RGS binding, exhibit diminished neutrophil chemotaxis in vitro and in vivo. These findings suggest an important role for RGS proteins in the modulation of neutrophil trafficking.
RGS5 is a member of the R4/B subfamily of RGS proteins. RGS5 is among the simplest members of the superfamily as it contains only an RGS box, the motif common to all RGS proteins mediating binding to Gα (13), but no other well defined domains. RGS5 is expressed constitutively in relatively few tissues in mice, including vascular and bronchial smooth muscle (14, 15) and cardiomyocytes (16). Rgs5−/− mice at baseline were underweight and hypotensive compared with controls, suggesting critical roles for RGS5 in metabolism and cardiovascular homeostasis (17). Routine immunophenotyping uncovered reduced neutrophil counts in peripheral blood of Rgs5−/− mice compared with controls. Furthermore, RNA-Seq and microarray analysis of FACS-purified leukocytes from C57BL/6 mice revealed expression of RGS5 in neutrophils from spleen and bone marrow (Immunological Genome (ImmGen) Project Consortium). These results prompted us to examine the expression of RGS5 in mouse neutrophils and explore its role in chemotaxis and trafficking in vivo.
Results
Immunoreactive RGS5 is detected in neutrophils
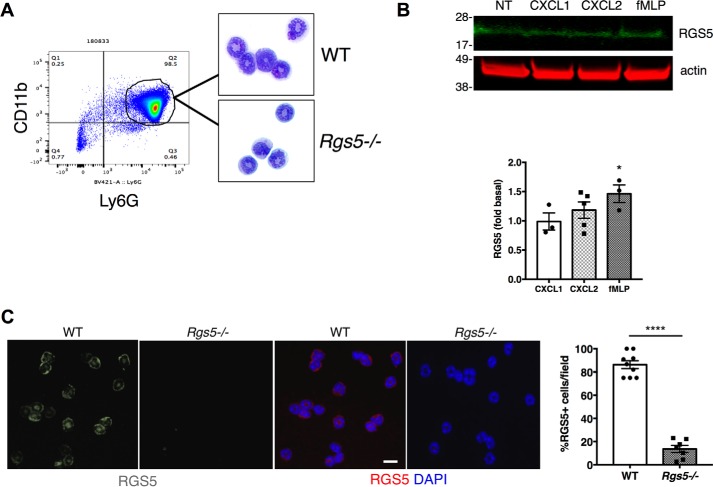
We first characterized RGS5 expression in neutrophils purified from bone marrow (BM) of wildtype (WT) mice using Ly6G microbeads. On average, cells isolated by this method were of 93.2 ± 3% purity based on high forward and side scatter and high CD11b surface expression as indicated by flow cytometry (n = 3) (Fig. 1A). Cells from WT and Rgs5−/− mice had comparable polymorphonuclear morphology by light microscopy (Fig. 1A). We detected relatively low amounts of RGS5 in BM-derived neutrophils by immunoblotting, which might be due to rapid degradation by the proteasome (18). To increase steady-state levels of RGS5 and to enhance detection, we treated cells with a proteasome inhibitor (MG132), which permitted detection of RGS5 in unstimulated neutrophils from bone marrow (Fig. 1B). RGS proteins are highly regulated at the transcriptional level, and expression of some RGS proteins changes rapidly in response to cellular stimulation (19). Although RGS5 expression was not significantly affected by treatment of neutrophils with chemokines, including CXCL1 and CXCL2, fMLP treatment was associated with modest up-regulation of the RGS5 polypeptide. Immunoreactive RGS5 was detected in both the nucleus and cytoplasm of purified neutrophils of WT but not Rgs5−/− mice (Fig. 1C). Cytoplasmic localization of RGS5 has been reported previously for RGS5 detected in airway smooth muscle cells (15), and nuclear localization has been observed for RGS5 and other RGS proteins (14, 20, 21).
Figure 1.
Immunoreactive RGS5 is detected in mouse neutrophils. A, neutrophils were isolated from bone marrow of naïve mice using Ly6G microbeads. Flow cytometry was used to assess purity based on CD11b expression. Neutrophils were dispersed by cytospin and identified by modified Giemsa staining. The plot is from a single experiment representative of three experiments (purity of 93.2 ± 3%). B, BM-derived neutrophils were left untreated (NT) or stimulated with CXCL1 or CXCL2 (100 ng/ml) or fMLP (1 μm) for 2 h followed by cell lysis and immunoblotting. The bar graph shows the relative RGS5 expression (normalized by β-actin signal; error bars indicate mean ± S.E.) of three to five independent experiments using one to two mice/experiment. *, p = 0.04, one-way ANOVA, Dunnett's post hoc test versus control (unstimulated condition). C, CXCL2-treated neutrophils from WT or Rgs5−/− mice were stained with anti-RGS5 (grayscale in first two images on the left and red in the color images) and counterstained with DAPI to identify nuclei (blue). The bar graph shows the percentage of cells/field containing immunoreactive RGS5 (error bars indicate mean ± S.E. of >3 fields/experiment, each containing >10 cells/field) analyzed in three independent experiments using one mouse of each genotype/experiment. ****, p < 0.0001, unpaired t test. Scale bar, 6 μm.
Rgs5 gene deletion has no effect on neutrophil development in vivo
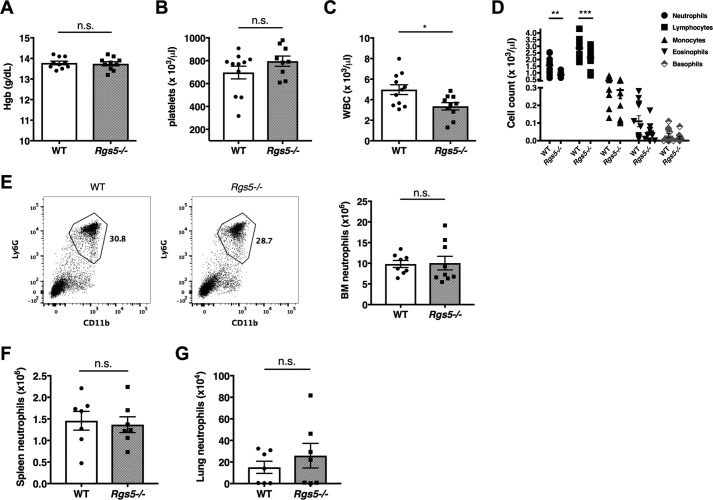
To evaluate neutrophil hematopoiesis and homeostatic neutrophil localization in the absence of inflammatory challenge, we compared numbers of mature neutrophils in bone marrow, peripheral blood, and various tissues of WT and Rgs5−/− mice by flow cytometry. Examination of peripheral blood revealed comparable hemoglobin concentrations, hematocrit (not shown), and platelet counts in WT and Rgs5−/− mice, results suggesting that these mice did not have a global hematopoietic defect (Fig. 2, A and B). By contrast, total peripheral white blood cell counts were significantly lower in Rgs5−/− mice than in WT controls (Fig. 2C). Although there were equivalent numbers of monocytes, basophils, and eosinophils in the blood of WT and mice, Rgs5−/− mice had both peripheral blood neutropenia and lymphopenia relative to controls (Fig. 2D). At the same time, we detected comparable numbers of Ly6GhiCD11b+ mature neutrophils in BM; these results suggest that neutrophil maturation was not affected by the loss of RGS5 (Fig. 2E). Given the abundant evidence that RGS proteins regulate leukocyte trafficking in vivo (23), we hypothesized that the reduction in circulating neutrophils in Rgs5−/− mice represented increased migration into tissues and/or clearance by the reticuloendothelial system. However, under baseline conditions, there were similar numbers of neutrophils in spleen (site of clearance by reticuloendothelial system) or in lungs of naïve WT and Rgs5−/− mice (Fig. 2, F and G).
Figure 2.
Hematopoiesis and neutrophil localization at homeostasis in WT and Rgs5−/− mice. A–D, serum hemoglobin (Hgb; A), platelet (B), total leukocyte (WBC; C), and differential leukocyte (D) counts were determined from peripheral blood. Error bars indicate mean ± S.E. A–C, *, p = 0.01, t test; n.s., not significant. D, **, p = 0.001; ***, p = 0.0008, two-way ANOVA, Sidak multiple comparisons. E–G, neutrophil numbers were determined in BM (tibia and femur of both lower extremities) (E), spleen (F), or lungs (G) by flow cytometry–based assessment of percentages of Ly6GhiCD11b+ cells, respectively, as shown in the representative plot from bone marrow. All results are mean ± S.E.; each symbol represents an individual mouse.
Trafficking of neutrophils from WT and Rgs5−/− mice to lung in response to respiratory virus infection
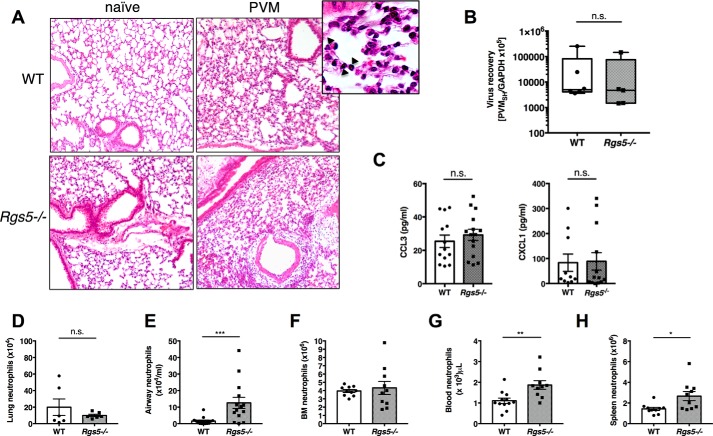
To assess neutrophil migration in response to an inflammatory stimulus, we infected mice with pneumonia virus of mice (PVM), a natural mouse pathogen that replicates in mouse lung tissue and elicits production of neutrophil chemoattractants, notably CCL3 and CXCL1 (24, 25). The severity of PVM infection was similar in WT and Rgs5−/− mice as assessed by peribronchial and alveolar accumulation of neutrophils in lungs (Fig. 3A), equivalent virus recovery (Fig. 3B), and similar levels of bronchoalveolar lavage fluid (BALF) cytokines (CXCL1 and CCL3) (Fig. 3C). However, although PVM infection elicited robust neutrophil recruitment to lung tissue in both strains, there were nearly 8 times more neutrophils in the airways (BALF) of Rgs5−/− mice than in WT controls at the same time point (Fig. 3, D and E). Likewise, although the numbers of neutrophils detected in BM in WT and Rgs5−/− mice were indistinguishable from one another (Fig. 3F), there were significantly more neutrophils in peripheral blood and spleens of Rgs5−/− mice (Fig. 3, G and H). These findings raise the possibility that alveolar reverse transmigration (26, 27) and/or demargination, rather than increased exit from BM, could account for increased numbers of circulating neutrophils following PVM infection.
Figure 3.
RGS5 deficiency results increased in airway neutrophils in acute respiratory virus infection. A, hematoxylin and eosin–stained lung sections from WT or Rgs5−/− mice at baseline (naïve) and 5 days after inoculation with PVM. Images are from a single mouse representative of three to four mice/group. Inset, diffuse neutrophilic alveolitis characteristic of acute PVM infection (40× magnification); arrows denote neutrophils. B, virus recovery was assessed by qPCR detection of virus-specific SH gene. C, chemokines CCL3 and CXCL1 in BALF of PVM-infected WT and Rgs5−/− mice. D–G, neutrophils in lung (D), airways (E), bone marrow (F), peripheral blood (G), or spleens (H) at day 5 of PVM infection. *, p = 0.02; **, p = 0.002; ***, p = 0.0007, Mann–Whitney; n.s., not significant. All error bars indicate mean ± S.E.; each symbol represents one mouse.
Neutrophil-intrinsic defects lead to anomalous trafficking in Rgs5−/− mice
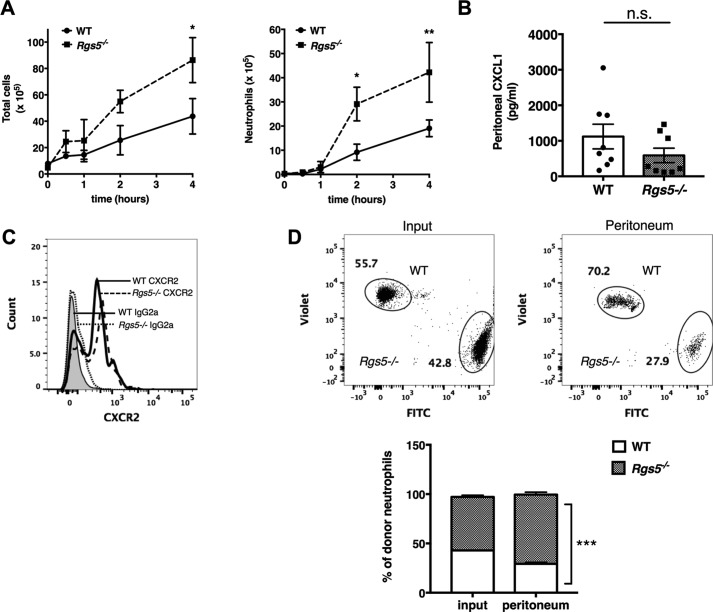
The increased accumulation of neutrophils in the airways of PVM-infected Rgs5−/− mice suggests enhanced extravasation into the airway lumen due to increased chemotaxis, adhesion, demargination, transendothelial migration, or a combination of these mechanisms. The lung microvasculature has specific, unique properties; the diameter of some alveolar capillary segments (6–7 μm) is frequently smaller than that of neutrophils (6–7.5 μm), which can prolong transendothelial transit times (28, 29). To determine whether neutrophils from Rgs5−/− mice exhibited a similar behavior in tissues with larger blood vessels, we used a model of sterile peritonitis induced by thioglycollate (TG). Intraperitoneal injection of TG induces rapid, transient infiltration of both macrophages and neutrophils into the peritoneal space through local production of chemoattractants, such as CXCL1 and CXCL2 (30). Injection of TG induced a dramatic increase in the total number of peritoneal cells within 2–4 h; the numbers of total cells and total neutrophils were nearly double in Rgs5−/− mice compared with WT (Fig. 4A). There were no significant differences in levels of CXCL1 in the peritoneum post-TG injection (Fig. 4B) nor any difference in surface CXCR2 expression on TG-elicited peritoneal neutrophils of WT or Rgs5−/− mice (Fig. 4C). Thus, we hypothesized that enhanced responsiveness to TG-evoked chemokines accounts for the increased recruitment of neutrophils observed in the RGS5-gene deleted mice. To assess the effects of RGS5 deficiency on TG-induced neutrophil migration directly, we isolated BM neutrophils from WT and Rgs5−/− mice and labeled cells from the two strains with different fluorophores. We then injected a 50:50 mixture of cells into WT recipients and assessed accumulation in the peritoneum by flow cytometry following intraperitoneal injection of TG. We detected more than double the number of RGS5-deficient neutrophils compared with WT in the peritoneum after TG injection (70.2 ± 2.5 versus 29.3 ± 1.5%) (Fig. 4D). This result is consistent with the interpretation that deficiency of RGS5 in neutrophils, rather than the microenvironment of Rgs5−/− mice, accounts for their increased recruitment to sites of inflammation.
Figure 4.
Neutrophils from RGS5-deficient mice are intrinsically capable of enhanced migration. A, mice were injected i.p. with 4% sodium thioglycollate. At the indicated time points, total cells (left) and neutrophils (right) were determined by flow cytometry as in Figs. 1 and 2. Error bars indicate mean ± S.E. of six to eight mice/genotype evaluated in three to four experiments. *, p = 0.02; **, p = 0.004, two-way ANOVA, Sidak multiple comparisons; n.s., not significant. B, chemokine CXCL1 levels were evaluated in peritoneal fluid following TG injection. Results are mean ± S.E.; each symbol represents one mouse. C, surface CXCR2 in TG-elicited peritoneal neutrophils determined by flow cytometry using the antibodies as indicated (IgG2a, isotype control). Histograms are from a single mouse representative of four mice/group assessed in two to three independent experiments. D, BM-derived neutrophils were stained with violet (Rgs5−/−) or green (WT) fluorophores. Cells were mixed at a 1:1 ratio and injected i.v. into mice previously administered TG. Numbers of cells of each genotype were determined by flow cytometry. Error bars indicate mean ± S.E. of cells from four mice/group evaluated in two independent experiments. ***, p = 0.0005, two-way ANOVA, Sidak multiple comparisons.
Loss of RGS5 enhances neutrophil chemotaxis and chemokine signaling
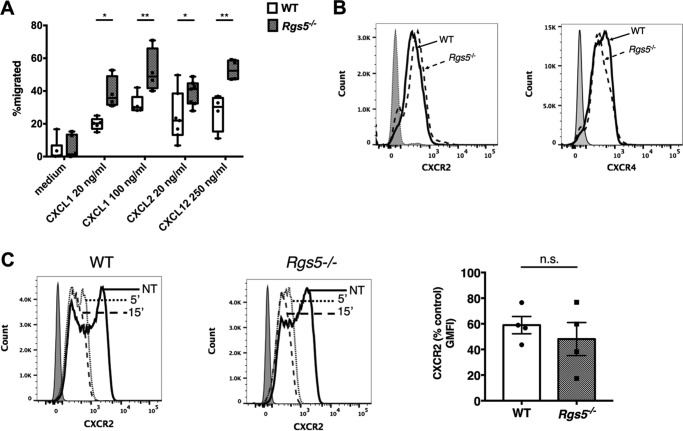
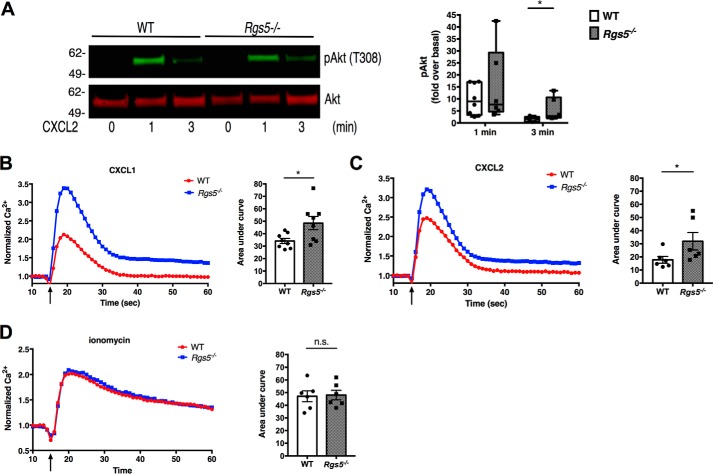
Our cumulative results using two distinct models of inflammation suggested that neutrophils from Rgs5−/− mice migrate much more effectively into inflamed tissue than do their WT counterparts. We hypothesized that RGS5-deficient neutrophils have increased chemotactic and/or adhesive properties that could account for these findings. To assess the chemotaxis of RGS5-deficient neutrophils, we first measured migration of BM-derived neutrophils in Transwell assays. Neutrophils from WT mice underwent chemotaxis to CXCL1 or CXCL2 (CXCR2 ligands) and CXCL12 (CXCR4 ligand) (Fig. 5A). Neutrophils from Rgs5−/− mice demonstrated significantly increased chemotaxis to all three chemokines compared with WT neutrophils, consistent with their responses in vivo. Expression of both CXCR2 and CXCR4 was similar in naïve bone marrow–derived neutrophils from WT and Rgs5−/− mice at baseline (Fig. 5B). In addition, both the magnitude and kinetics of down-regulation of CXCR2 in neutrophils from WT and Rgs5−/− mice following chemokine stimulation were comparable with one another (Fig. 5C). To assess chemokine-induced signaling downstream of receptors, we measured Akt phosphorylation induced by CXCL2 by immunoblotting. Ligand-bound chemokine receptor activates Gαi, the target of RGS5, which dissociates from Gβγ. Free Gβγ then activates PI3Kγ, which catalyzes production of phosphatidylinositol 3,4,5-trisphosphate at the cell membrane, a key step in cytoskeletal rearrangements leading to polarization of the neutrophil in the direction of a chemoattractant gradient (31). Akt phosphorylation induced by phosphatidylinositol 3,4,5-trisphosphate binding is a surrogate for PI3K activity and is typically increased in cells with reduced RGS protein expression compared with WT (11). Although CXCL2 induced comparable peak Akt phosphorylation in neutrophils from WT and Rgs5−/− mice at 1 min, Akt phosphorylation was significantly prolonged in RGS5-deficient cells compared with WT (Fig. 6A). To confirm that chemokine-induced signaling was increased in RGS5-deficient neutrophils, we analyzed intracellular Ca2+ flux. Gβγ released from activated Gα stimulates phospholipase Cβ, which in turn leads to the generation of inositol 3,4,5-trisphosphate and release of Ca2+ from intracellular stores (32). Both CXCL1 and CXCL2 induced a rapid rise in intracellular Ca2+ in neutrophils from WT mice, but the magnitude and duration of the response were significantly greater in RGS5-deficient neutrophils (Fig. 6, B and C). By contrast, receptor-independent Ca2+ flux evoked by ionomycin stimulation was equivalent in neutrophils from WT and Rgs5−/− mice (Fig. 6D). These studies support the hypothesis that that increased chemotaxis of RGS5-deficient neutrophils was due to augmented chemokine-evoked signaling.
Figure 5.
Increased chemotaxis of RGS5-deficent neutrophils. A, chemotaxis of BM-derived neutrophils was assessed in Transwell assays as described under “Experimental procedures.” Error bars indicate mean ± S.E. of four to six experiments using cells from one to two mice of each genotype/experiment. *, p < 0.02; **, p < 0.006, two-way ANOVA, Sidak multiple comparisons. B, surface CXCR2 or CXCR4 in untreated BM-derived neutrophils was assessed by flow cytometry. Histograms represent a single mouse/genotype representative of two to three mice evaluated in separate experiments; the shaded peak represents cells stained with isotype control antibody. C, surface CXCR2 expression in BM-derived neutrophils pre- and poststimulation with CXCL2 (100 ng/ml) for the indicated times. The bar graph is CXCR2 geometric mean fluorescence intensity (GFMI) after treatment with CXCL2 for 15 min as a percentage of that measured in untreated cells (error bars indicate mean ± S.E. of four independent experiments using cells from one mouse of each genotype/experiment). n.s., not significant.
Figure 6.
RGS5 negatively regulates chemokine-induced Ca2+ signaling and Akt phosphorylation in neutrophils. A, Akt phosphorylation (pAkt) in BM-derived neutrophils was evaluated by immunoblotting. The blot is from a single experiment representative of seven similar experiments. Error bars indicate mean ± S.E. *, p = 0.03, Mann–Whitney. B–D, Ca2+ flux in response to CXCL1 (B), CXCL2 (C), or ionomycin (D). Bar graphs represent area under the curve (error bars are mean ± S.E.) of five to eight independent measurements in four separate experiments each using cells from one mouse of each genotype. *, p = 0.04, Mann–Whitney. The arrow represents time of agonist addition. n.s., not significant.
Neutrophil adhesion in the absence of RGS5
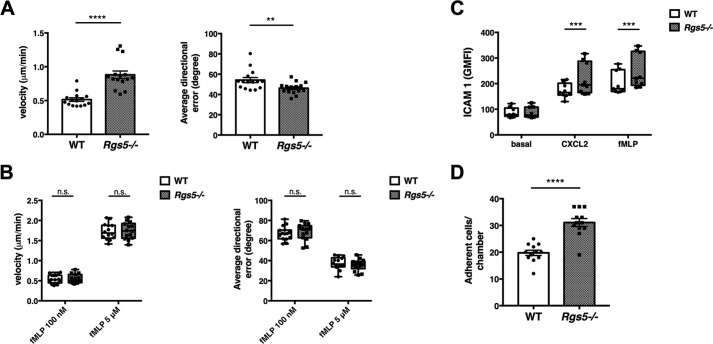
To characterize the chemotactic response of neutrophils in the absence of RGS5 more precisely, we visualized cell migration in the presence of a chemokine gradient by microscopy using Dunn chambers, which permitted us to quantify directional velocity. In this assay, the velocity determined for neutrophils from Rgs5−/− mice responding to CXCL2, but not fMLP, was significantly greater than that of WT mice under the same conditions, even at submaximal concentrations of fMLP (Fig. 7, A and B). Directional error, a measure of the ability of the cells to faithfully track a linear gradient of chemoattractant, was reduced in neutrophils from Rgs5−/− mice exposed to CXCL2 gradients compared with control, whereas directional error in response to fMLP was equivalent. Next, we characterized adhesion of RGS5-deficient and WT neutrophils by measuring binding to immobilized ICAM-1. This interaction is mediated by β2 integrins, such as LFA1, expressed on the neutrophil surface (33). We observed significantly increased binding of neutrophils lacking RGS5 to ICAM-1 in the presence of either CXCL2 or fMLP compared with neutrophils from WT mice (Fig. 7C). Firm adhesion to the endothelium requires strong interaction of neutrophil integrins with ICAM-1 and other ligands expressed on the surface of endothelial cells (34). To confirm the ICAM-1 binding assay results, we studied the adhesion of neutrophils to the endothelium using flow chambers. This assay introduces fluid shear stress that models fluid dynamics within a blood vessel, allowing quantification of cellular adhesive properties toward the endothelium in a more physiological setting. Endothelial cells were preactivated with tumor necrosis factor α to up-regulate surface expression of neutrophil ligands. With this model, neutrophils from Rgs5−/− mice adhered to endothelial cells significantly more than those from WT mice (Fig. 7D).
Figure 7.
RGS5-deficient neutrophils demonstrate increased chemokine-induced directional velocity and adhesion to endothelial cells. A and B, migrational velocity and directional error of BM-derived neutrophils were assessed in Dunn chambers in the presence of CXCL2 (10 μm; A) or fMLP (100 nm or 5 μm; B). Error bars indicate mean ± S.E. of three independent experiments using cells from one mouse/genotype and three to five separate chambers/condition in which 50–110 cells/chamber were individually tracked. ****, p < 0.00001, unpaired t test; **, p = 0.009, Mann–Whitney. C, neutrophil adhesion was assessed by binding to immobilized ICAM-1 as described under “Experimental procedures” in the presence of CXCL2 (10 μm) or fMLP (5 μm) in three independent experiments using cells from one mouse of each genotype. ***, p < 0.0005, one-way ANOVA, Sidak multiple comparisons. Error bars indicate mean ± S.E. D, EC adhesion was assessed in flow chambers as described under “Experimental procedures.” Error bars indicate mean ± S.E. of four independent experiments using cells from one mouse of each genotype. ****, p < 0.00001, unpaired t test. IL-8, interleukin-8.
RGS5 overexpression inhibits chemotaxis of human neutrophils
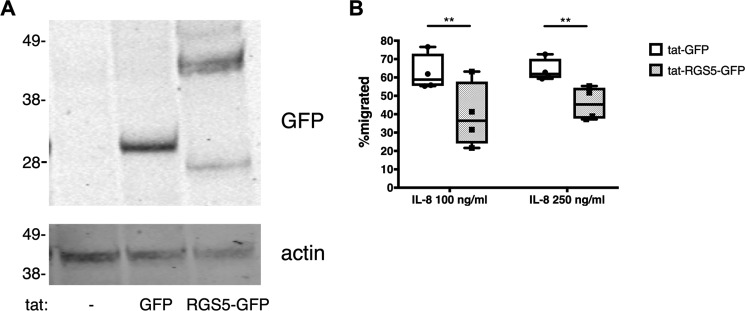
Our findings thus far indicate that loss of RGS5 function in mouse neutrophils leads to increased motility and adhesion in vitro, which in turn increase their effective migration toward an inflammatory stimulus in vivo. To extend these findings to humans, we assessed RGS5 expression in human neutrophils isolated from peripheral blood. Quantification of RGS5 mRNA expression in neutrophils isolated from five separate healthy donors by quantitative PCR revealed very low (but detectable) RGS5 quantities in cells from all donors (Table 1). Thus, we hypothesized that RGS5 gain of function would inhibit chemotaxis of human neutrophils. To test this, we introduced HIV-1 tat fusion proteins encoding an irrelevant protein (GFP) or RGS5-GFP into these cells as we have done successfully in other cell types (15). Using this strategy, we detected intracellular expression of tat-GFP and tat-RGS5-GFP in cell lysates by immunoblotting (Fig. 8A). In Transwell assays, cells transduced with tat-GFP responded with robust chemotaxis to interleukin-8, whereas the migration of tat-RGS5-GFP–transduced cells was significantly weaker to equivalent concentrations of interleukin-8 (Fig. 8B). These results indicate that increased RGS5 expression leads to impaired chemotaxis of human neutrophils.
Table 1.
RGS5 expression in human neutrophils
| Healthy donor | Expression (Ct value, mean ± S.D.) |
RQa | |
|---|---|---|---|
| RGS5 | ACTB | ||
| D1 | 35.78 ± 0.91 | 14.91 ± 0.19 | 0.03 |
| D2 | 33.10 ± 0.45 | 14.97 ± 0.29 | 0.22 |
| D3 | 34.58 ± 0.54 | 19.10 ± 0.35 | 1.36 |
| D4 | 35.84 ± 0.85 | 19.92 ± 0.19 | 1.00 |
| D5 | 33.01 ± 0.18 | 17.41 ± 0.05 | 1.24 |
a Relative expression.
Figure 8.
RGS5 overexpression inhibits chemotaxis of human neutrophils. A, human peripheral blood neutrophils were isolated and transduced with tat fusion proteins as described under “Experimental procedures.” Cell lysates were prepared, and intracellular expression of tat GFP fusion proteins was assessed by immunoblotting. B, chemotaxis of human neutrophils transduced with tat fusion proteins was assessed in Transwell assays. Error bars indicate mean ± S.E. of three independent experiments using neutrophils from one to two donors/experiment. **, p < 0.008, two-way ANOVA, Sidak multiple comparisons.
Discussion
Few studies have explored the role of RGS proteins in innate immunity, and this is the first work to address the functions of an RGS protein in neutrophils. We found that RGS5 is expressed at relatively low levels in mouse neutrophils under homeostatic conditions. Neutrophil hematopoiesis was normal in RGS5-deficient mice; however, fewer mature neutrophils were detected in peripheral blood of these mice than in their WT counterparts. The tissue reservoir for these cells in naïve mice remains to be identified.
Using distinct models of acute inflammation in two different tissues, we demonstrated that neutrophils from RGS5-deficient mice were mobilized much more efficiently to inflamed tissues than those from WT mice, largely due to increased chemotaxis and adhesion. These results are consistent with recent studies demonstrating that RGS proteins may limit transit of other leukocytes to and from specific inflammatory sites. A notable example of this is provided by Patel et al. (35), who found that global RGS1 deficiency enhanced recruitment of macrophages to atherosclerotic plaques in arteries of mice but also reduced retention of these cells in the lesions over time due to prolonged chemokine responsiveness.
In contrast to the phenotype of neutrophils lacking RGS5, cells from mice expressing a Giα2 mutant globally resistant to binding of all RGS proteins had peripheral neutropenia but exhibited a myelokathexis-like phenotype with poor margination of neutrophils from bone marrow upon inflammatory (TG) challenge (12). Neutrophils from these mice had increased basal motility but poor responses to chemokines (CXCL2 and CXCL12) in vitro and exhibited poor adhesion to blood vessels and transmigration into laser-damaged tissues as assessed directly by in vivo imaging. Underlying these findings, these neutrophils had decreased surface expression of CXCR2 following TG challenge due to increased G protein–coupled receptor kinase 2 expression and phosphorylation-mediated internalization. By contrast, we found that, compared with neutrophils from WT mice, RGS5-deficient neutrophils had 1) no evidence of increased spontaneous motility, 2) increased chemotaxis to ELR-4 motif chemokines CXCL1 and CXCL2 but not fMLP, and 3) equivalent chemokine receptor expression at baseline and following CXCL2 or TG challenge. Because neutrophil trafficking depends strongly on Giα2 (36), overall these studies suggest that loss of Giα2 interactions with RGS proteins other than (or in addition to) RGS5 is required to trigger chemokine receptor down-regulation. Alternatively, global RGS5 deficiency could result in changes in expression of a distinct set of chemotaxis-related genes than that induced by the absence of all RGS–Giα2 interactions. Cho et al. (12) also reported expression of several RGS proteins in murine neutrophils, including RGS2, -14, -18, and -19, suggesting the possibility that other RGS family members contribute to neutrophil trafficking and adhesion.
Neutrophils develop from myeloid progenitors in the bone marrow where the actions of two opposing chemokine receptors, CXCR4 (promoting retention) and CXCR2 (stimulating egress), determine quantities of circulating neutrophils circulating at any given time (37) with a dominant role for CXCR4 (38). Although chemotactic responses to both CXCL1/2 and CXCL12 were increased in neutrophils from Rgs5−/− mice relative to WT, we detected neither increased retention of neutrophils in bone marrow nor impaired neutrophil mobilization to tissues following an inflammatory stimulus. Of interest, this phenotype somewhat resembles warts, hypogammaglobulinemia, infections, myelokathexis (WHIM) syndrome in humans, a disorder that is due to a gain-of-function mutation in CXCR4 (39). WHIM patients have peripheral blood neutropenia due to increased bone marrow retention but are nonetheless able to mobilize neutrophils to sites of inflammation.
The most significant aspect of our study is the finding that RGS5 does not control neutrophil responses to chemoattractants equivalently; it inhibits chemokine-evoked chemotaxis and adhesion robustly in vitro, whereas only adhesion, but not chemotaxis, in response to fMLP was increased in the absence of RGS5. The reasons for this discrepancy are not immediately clear, although discordant behavior of neutrophils in solution- versus solid phase–based migration assays has been described previously, possibly because the latter involves cycles of adhesion–deadhesion from a substrate to propel cells forward (40). Published studies of RGS protein–deficient leukocytes have frequently, but not universally, demonstrated increased chemotaxis (35, 41–43). RGS5 may also have differential efficacy toward certain G protein–receptor combinations. Although most of the RGS proteins in the R4 subfamily bind Gαi and Gαq family members promiscuously, there is substantial evidence for direct interactions with GPCRs (44, 45), which may specify selectivity for certain receptor–G protein combinations. A previous study found that chemotaxis of neutrophils elicited by fMLP, but not chemokines, is partially dependent on Gαq (46). RGS proteins have the capacity to regulate Gq signaling both through GAP activity and inhibition of G protein–effector interactions (e.g. Gq–phospholipase Cβ) independently of GAP activity (47). A direct comparative study of the relative potency of various RGS proteins to act as GAPs versus effector antagonists found that RGS5 was a less potent antagonist than other RGS proteins, such as RGS2 and -3, in overexpression studies (48).
Neutrophils arrested on ECs clearly display hierarchical responses to chemoattractant receptors, gradually becoming nonresponsive to stimuli in the blood vessel lumen (e.g. chemokines) and to ligands of different receptors in the same class (6); this provides them with the capacity to migrate toward a second wave of tissue-based chemoattractants, including those associated with pathogens (e.g. fMLP) (49). Further in vivo studies of chemokine- versus fMLP-mediated responses will be required to determine the significance of RGS5's differential capacity to mediate each of these responses in vitro.
We hypothesize that the increased number of neutrophils in the airways and peripheral blood of PVM-infected mice was due to increased transmigration through alveolar capillaries possibly combined with reverse transendothelial migration back into circulation. Reverse transendothelial transmigration of neutrophils has been documented in vivo imaging in several types of inflammation, such as that associated with wound repair, which may aid in resolution (26, 27). Adherent neutrophils can traverse the activated endothelium through both transcellular and paracellular routes, both of which require active participation of blood vessel cells through Gαi signaling (50, 51). RGS5 is also expressed in pericytes, contractile smooth muscle-like cells that encircle microvascular ECs (52). Neutrophil crawling and transmigration both depend on pericyte function (53).
The expression of RGS5 in other cell types aside, our adoptive transfer studies suggest that the enhanced migration of neutrophils from Rgs5−/− mice to sites of inflammation relative to controls was due to intrinsically increased neutrophil chemotaxis and adhesion. In separate assays, we detected increased adhesion of RGS5-deficient neutrophils in the presence of CXCL2 and fMLP. Although these events depend on chemokine-induced activation of Gαi, which is the primary target of RGS5, our findings do not preclude other mechanisms of regulation. For example, other RGS5-interacting proteins, such as PI3Kδ, may also have functions in neutrophil adhesion and integrin activation (54). We showed previously that RGS5 inhibited Toll-like receptor–induced PI3Kδ activation in airway smooth muscle cells, which may be independent of RGS5 GAP activity (15). Future studies of cells and/or animals expressing RGS5 containing a point mutation that selectively abolishes GAP activity (55) should clarify how RGS5 regulates its full interactome and alters neutrophil migration in inflamed tissues.
Experimental procedures
Mice
C57BL/6 Rgs5−/− mice were generated as described previously (17) and backcrossed onto a BALB/c background for 10 generations. Rgs5−/− mice and their WT littermates were used for all experiments. All studies were performed in accordance with institutional guidelines provided by the National Institute of Allergy and Infectious Diseases Animal Use and Care Committee under approved studies (Animal Study Protocols LAD3e and LAD8e).
Reagents, antibodies, and flow cytometry
Antibodies used in this study were purchased from the following sources: rabbit anti-phospho-Akt (Thr-308) and mouse anti-Akt (Cell Signaling Technology), mouse anti-β-actin (Sigma), and biotinylated mouse anti-GFP and mouse monoclonal anti-RGS5 (H-1) (Santa Cruz Biotechnology). CXCL1, CXCL2, and CXCL12 were from BD Biosciences. fMLP, sodium thioglycollate, and Histopaque 1019/1177 were purchased from Sigma. Antibodies used for flow cytometry were purchased from the following sources (dilutions indicated): Ly6G-BV421 (BioLegend, clone 1A8; 1:40); CD11b-FITC (BioLegend, clone M1/70; 1:25), CXCR4-PE (BD Biosciences; 1:100), and CXCR2-APC (BioLegend, clone SA044G4; 1:40). For receptor internalization studies, cells were stimulated with CXCL2 (100 ng/ml) for the indicated time periods at 37 °C followed by fixation with PBS with 4% paraformaldehyde and analysis of surface CXCR2 expression by flow cytometry.
Tat fusion proteins
Recombinant proteins fused to GPF containing HIV-1 tat peptide and His6 sequences were expressed in Escherichia coli and purified using Ni2+ affinity chromatography as described previously (15). We added tat proteins to cells in culture medium for 2 h (final concentration, 200 nm) prior to functional assays.
Ca2+ measurements
Bone marrow–derived neutrophils were resuspended in Hanks' balanced salt solution containing Ca2+ and Mg2+ plus 20 mm HEPES, pH 7.5, and plated in 96-well flat-bottom plates (Corning) (4 × 105 cells/well). An equivalent volume of FLIPR6 Ca2+ assay buffer (Molecular Devices) was added to the wells followed by 2-h incubation at 37 °C. Intracellular Ca2+ before and after agonist addition was evaluated in a FlexStation III automated fluorimeter (Molecular Devices). Relative fluorescence units were divided by initial baseline relative fluorescence unit readings to give normalized Ca2+ values over time.
Cytokine measurements
CXCL1 and CCL3 in BALF and/or peritoneal lavage supernatants were measured by ELISA (R&D Biosystems).
Complete blood counts
Leukocyte counts and hematological parameters were analyzed using an automated Hemavet 950 Multispecies Analyzer (Drew Scientific/Erba Diagnostics) according to the manufacturer's instructions.
PVM infection
Mice under brief isoflurane anesthesia were inoculated with PVM (0.2 tissue culture ID50 units) (56) in a 50-μl volume of Iscove's modified Dulbecco's medium. At day 5 postinoculation, blood was withdrawn by retro-orbital puncture followed by euthanasia. BALF was extracted by lavage of lungs with PBS with 0.2% BSA (1-ml total volume). Spleen, lungs, and bone marrow (right tibia and femur) were extracted followed by generation of single cell suspensions as described (57). Total numbers of viable (trypan blue–negative) cells were determined using a Countess II hemocytometer (Thermo Fisher Scientific). Neutrophils were enumerated by flow cytometry as above. Portions of lung were preserved in formalin for histological studies.
PVM titers
Virus was evaluated in whole lung tissue using a quantitative real-time PCR (qPCR) assay that targets the PVM SH protein as described previously (58).
RNA isolation and quantitative RT-PCR
RNA was isolated using a Direct-zolTM RNA kit (Zymo Research) according to the manufacturer's instructions. Total RNA (500 ng) was reverse transcribed into cDNA using SuperScriptTM IV VILOTM Master Mix (Thermo Fisher) according to the manufacturer's protocol. qPCR was performed using gene-specific TaqMan probes (Applied Biosystems) according to the manufacturer's guidelines. The TaqMan probes were: RGS5, Hs01555175_m1; and ACTB, Hs99999903_m1.
Chemotaxis assays
Neutrophils were isolated from total bone marrow cells extracted from femora using Ly6G microbeads or mouse neutrophil negative isolation microbeads (Miltenyi Biotech) according to the manufacturer's instructions or by Ficoll-Hypaque density gradient isolation as described (59). Ly6G-purified neutrophils were cultured overnight in RPMI 1640 medium plus 10% fetal calf serum plus granulocyte/macrophage colony-stimulating factor (25 ng/ml). Cells (3–5 × 105) were then loaded into the top chamber of Transwell inserts (Corning, 3-μm pore size, 6.5-mm wells). The inserts were placed into 24-well plates containing medium alone or the indicated concentration of chemokine for 45 min at 37 °C. EDTA (final concentration, 5 mm) was added to the lower chamber for 10 min at 4 °C to detach cells from the Transwell filters. Viable cells in the lower chamber were counted using a Countess II automated cell counter by trypan blue exclusion. The percentage of cells migrated was determined by dividing by the number cells in a well lacking insert (input).
Thioglycollate-induced peritonitis and adoptive neutrophil transfer
Mice were injected intraperitoneally (i.p.) with 3–4% sodium TG in PBS for the indicated times. Mice were sacrificed at arbitrary time points followed by injection of PBS plus 2 mm EDTA i.p. The peritoneal cavity was agitated prior to extraction of PBS and counting of total cells as above. Neutrophil percentages were determined by staining with Ly6G/CD11b antibodies and flow cytometry. For adoptive transfer, neutrophils were purified as above using Ly6G microbeads. Cells were labeled separately with either CellTracker Green 5-chloromethylfluorescein diacetate (CMDFA; WT) or violet 2,3,6,7-tetrahydro-9-bromomethyl-1H,5H-quinolizino(9,1-gh)coumarin (BMQC; Rgs5−/−) dyes (Thermo Fisher Scientific) at a final concentration of 1.25 μm for 10 min at 37 °C. Cells were washed once with PBS and mixed together at a 1:1 ratio. An aliquot of cells was saved to quantify input. 100 μl (2–3 × 106 cells) of the cell admixture was injected intravenously by tail vein into WT mice that had been injected i.p. 1 h earlier with 3% TG. Two hours later, peritoneal lavage was performed as above followed by enumeration of neutrophil percentages by flow cytometry.
Immunoblotting and immunofluorescence
Purified BM-derived neutrophils (1 × 106/time point) were resuspended in Hank's balanced salt solution containing 2 mm EDTA and incubated on ice for 1 h. Cells were stimulated with CXCL1 or CXCL2 (100 ng/ml) or with fMLP (1 μm) for the indicated times followed by protein extraction using 0.6 n perchloric acid. Proteins were pelleted, washed once in water, and resuspended in radioimmunoprecipitation lysis buffer containing protease and phosphatase inhibitors (Complete protease inhibitor mixture and PhosSTOP tablets, Roche Applied Science) and sodium orthovanadate (1 mm). Lysates were sonicated briefly before addition of NuPAGE SDS sample buffer (Thermo Fischer Scientific), boiling at 95 °C for 5 min, and brief centrifugation. Clarified lysates were electrophoresed on 12% NuPAGE Tris-glycine gels and immunoblotted with the indicated antibodies. For immunofluorescence staining, purified neutrophils were incubated with CXCL2 for 2 h at 37 °C in medium containing MG132 (Sigma). Cells were then dispersed on glass microscope slides by cytospin. Cells were then fixed with 4% paraformaldehyde in PBS for 10 min at room temperature. Cells were permeabilized with PBS containing 0.2% Triton X-100 for 10 min and then blocked in 5% BSA in PBS plus 2% goat serum for 1 h at room temperature. Cells were incubated with mouse anti-RGS5 (1:250) overnight at 4 °C followed by staining with Alexa Fluor 594 goat anti-mouse immunoglobulin G (IgG) (1:200; Thermo Fisher) for 1 h at room temperature. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; Sigma; 1 μg/ml). A minimum of three fields (∼50–100 cells at 40× magnification) per condition were evaluated with a Leica DMI4000B fluorescence microscope.
Adhesion assays
The ICAM-1–Fc–F(ab′)2 complexes were generated by incubating Cy5-conjugated AffiniPure goat anti-human Fc fragment–specific IgG F(ab′)2 fragments (Jackson ImmunoResearch Laboratories) and ICAM-1–Fc (100 mg/ml; R&D Systems) at 4 °C for 30 min in PBS. Neutrophils were resuspended at a concentration of 0.5 × 106 cells/ml in PBS containing 0.5% BSA, 0.5 mm Mg2+, and 0.9 mm Ca2+ and incubated with ICAM-1–Fc–F(ab′)2 in the presence of chemokine for the indicated time periods. Cells were fixed with 4% paraformaldehyde and analyzed by flow cytometry.
In vitro chemotaxis assay in a Dunn chamber
Migrational velocity and directional error were examined in a Dunn chamber as described previously (60). We analyzed WT and mutant neutrophils simultaneously by labeling the cells with different tracing dyes. Images were acquired at 30-s intervals for 30 min and analyzed using MetaMorph image analysis software.
Flow chamber assays
To examine neutrophil adherence to endothelial cells under shear stress, we established confluent monolayers of immortalized mouse embryonic ECs (22) on coverslips coated with fibronectin (10 mg/ml). ECs were stimulated with tumor necrosis factor α (50 ng/ml) for 4 h prior to being placed in a flow chamber apparatus (GlycoTech). Neutrophils from WT or Rgs5−/− mice labeled with distinct fluorophores were mixed at a 1:1 ratio and flowed into the chamber at a shear flow rate of 1 dyne/cm2. Adherent cells were quantified by fluorescence microscopy.
Statistical analysis
Values are reported as mean ± S.E. unless otherwise specified. Normally distributed data were analyzed by t test (two groups) or one- or two-way ANOVA (multiple groups). For non-normally distributed data, nonparametric Mann–Whitney U test or Kruskal–Wallis tests were used. p values <0.05 were considered to be significant.
Author contributions
E. C. C., C. R., Z. X., J. A. J., T. B., R. A. P., D. W., H. F. R., and K. M. D. conceptualization; E. C. C., C. R., Z. X., J. A. J., T. B., C. A. K.-W., M. M., R. A. P., H. F. R., and K. M. D. data curation; E. C. C., C. R., Z. X., J. A. J., T. B., R. A. P., D. W., and K. M. D. formal analysis; E. C. C., R. A. P., D. W., H. F. R., and K. M. D. supervision; E. C. C., C. R., Z. X., R. A. P., D. W., H. F. R., and K. M. D. validation; E. C. C., C. R., Z. X., J. A. J., T. B., C. A. K.-W., M. M., R. A. P., D. W., H. F. R., and K. M. D. investigation;tk;1 E. C. C., C. R., Z. X., J. A. J., T. B., C. A. K.-W., R. A. P., H. F. R., and K. M. D. methodology; E. C. C. and K. M. D. writing-original draft; E. C. C., C. R., Z. X., J. A. J., T. B., C. A. K.-W., R. A. P., D. W., H. F. R., and K. M. D. writing-review and editing; R. A. P., D. W., H. F. R., and K. M. D. resources; R. A. P., D. W., H. F. R., and K. M. D. project administration.
Acknowledgment
We thank Rebecca Drummond (NIAID, National Institutes of Health) for technical assistance.
This work was supported by the Intramural Research Program, NIAID, National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- EC
- endothelial cell
- fMLP
- N-formylmethionylleucylphenylalanine
- RGS
- regulator of G protein signaling
- CXCL
- CXC motif chemokine ligand
- GPCR
- G protein–coupled receptor
- PI3K
- phosphoinositide 3-kinase
- GAP
- GTPase-activating protein
- BM
- bone marrow
- PVM
- pneumonia virus of mice
- BALF
- bronchoalveolar lavage fluid
- TG
- thioglycollate
- CXCR
- CXC motif chemokine receptor
- ICAM-1
- intracellular adhesion molecule-1
- WHIM
- warts, hypogammaglobulinemia, infections, myelokathexis
- PE
- phycoerythrin
- APC
- allophycocyanin
- qPCR
- quantitative real-time PCR
- SH
- small hydrophobic
- ANOVA
- analysis of variance.
References
- 1. Naumenko V., Turk M., Jenne C. N., and Kim S. J. (2018) Neutrophils in viral infection. Cell Tissue Res. 371, 505–516 10.1007/s00441-017-2763-0 [DOI] [PubMed] [Google Scholar]
- 2. Laurent P., Jolivel V., Manicki P., Chiu L., Contin-Bordes C., Truchetet M. E., and Pradeu T. (2017) Immune-mediated repair: a matter of plasticity. Front. Immunol. 8, 454 10.3389/fimmu.2017.00454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Massena S., Christoffersson G., Hjertström E., Zcharia E., Vlodavsky I., Ausmees N., Rolny C., Li J. P., and Phillipson M. (2010) A chemotactic gradient sequestered on endothelial heparan sulfate induces directional intraluminal crawling of neutrophils. Blood 116, 1924–1931 10.1182/blood-2010-01-266072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cyster J. G., and Goodnow C. C. (1995) Pertussis toxin inhibits migration of B and T lymphocytes into splenic white pulp cords. J. Exp. Med. 182, 581–586 10.1084/jem.182.2.581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. de Oliveira S., Rosowski E. E., and Huttenlocher A. (2016) Neutrophil migration in infection and wound repair: going forward in reverse. Nat. Rev. Immunol 16, 378–391 10.1038/nri.2016.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Holdfeldt A., Dahlstrand Rudin A., Gabl M., Rajabkhani Z., König G. M., Kostenis E., Dahlgren C., and Forsman H. (2017) Reactivation of Gαi-coupled formyl peptide receptors is inhibited by Gαq-selective inhibitors when induced by signals generated by the platelet-activating factor receptor. J. Leukoc. Biol. 102, 871–880 10.1189/jlb.2A0317-086RR [DOI] [PubMed] [Google Scholar]
- 7. Druey K. M. (2017) Emerging roles of regulators of G protein signaling (RGS) proteins in the immune system. Adv. Immunol. 136, 315–351 10.1016/bs.ai.2017.05.001 [DOI] [PubMed] [Google Scholar]
- 8. Sprang S. R. (2016) Invited review: activation of G proteins by GTP and the mechanism of Gα-catalyzed GTP hydrolysis. Biopolymers 105, 449–462 10.1002/bip.22836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hwang I. Y., Park C., Harrision K. A., Huang N. N., and Kehrl J. H. (2010) Variations in Gnai2 and Rgs1 expression affect chemokine receptor signaling and the organization of secondary lymphoid organs. Genes Immun. 11, 384–396 10.1038/gene.2010.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hwang I. Y., Hwang K. S., Park C., Harrison K. A., and Kehrl J. H. (2013) Rgs13 constrains early B cell responses and limits germinal center sizes. PLoS One 8, e60139 10.1371/journal.pone.0060139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bansal G., DiVietro J. A., Kuehn H. S., Rao S., Nocka K. H., Gilfillan A. M., and Druey K. M. (2008) RGS13 controls G protein-coupled receptor-evoked responses of human mast cells. J. Immunol. 181, 7882–7890 10.4049/jimmunol.181.11.7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cho H., Kamenyeva O., Yung S., Gao J. L., Hwang I. Y., Park C., Murphy P. M., Neubig R. R., and Kehrl J. H. (2012) The loss of RGS protein-Gαi2 interactions results in markedly impaired mouse neutrophil trafficking to inflammatory sites. Mol. Cell. Biol. 32, 4561–4571 10.1128/MCB.00651-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xie Z., Chan E. C., and Druey K. M. (2016) R4 regulator of G protein signaling (RGS) proteins in inflammation and immunity. AAPS J. 18, 294–304 10.1208/s12248-015-9847-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Arnold C., Demirel E., Feldner A., Genové G., Zhang H., Sticht C., Wieland T., Hecker M., Heximer S., and Korff T. (2018) Hypertension-evoked RhoA activity in vascular smooth muscle cells requires RGS5. FASEB J. 32, 2021–2035 10.1096/fj.201700384RR [DOI] [PubMed] [Google Scholar]
- 15. Balenga N. A., Jester W., Jiang M., Panettieri R. A. Jr, and Druey K. M. (2014) Loss of regulator of G protein signaling 5 promotes airway hyperresponsiveness in the absence of allergic inflammation. J. Allergy Clin. Immunol 134, 451–459 10.1016/j.jaci.2014.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang Z., Huang H., He W., Kong B., Hu H., Fan Y., Liao J., Wang L., Mei Y., Liu W., Xiong X., Peng J., Xiao Y., Huang D., Quan D., et al. (2016) Regulator of G-protein signaling 5 protects cardiomyocytes against apoptosis during in vitro cardiac ischemia-reperfusion in mice by inhibiting both JNK1/2 and P38 signaling pathways. Biochem. Biophys. Res. Commun. 473, 551–557 10.1016/j.bbrc.2016.03.114 [DOI] [PubMed] [Google Scholar]
- 17. Cho H., Park C., Hwang I. Y., Han S. B., Schimel D., Despres D., and Kehrl J. H. (2008) Rgs5 targeting leads to chronic low blood pressure and a lean body habitus. Mol. Cell. Biol. 28, 2590–2597 10.1128/MCB.01889-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bodenstein J., Sunahara R. K., and Neubig R. R. (2007) N-terminal residues control proteasomal degradation of RGS2, RGS4, and RGS5 in human embryonic kidney 293 cells. Mol. Pharmacol. 71, 1040–1050 10.1124/mol.106.029397 [DOI] [PubMed] [Google Scholar]
- 19. Alqinyah M., and Hooks S. B. (2018) Regulating the regulators: epigenetic, transcriptional, and post-translational regulation of RGS proteins. Cell. Signal. 42, 77–87 10.1016/j.cellsig.2017.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Waugh J. L., Lou A. C., Eisch A. J., Monteggia L. M., Muly E. C., and Gold S. J. (2005) Regional, cellular, and subcellular localization of RGS10 in rodent brain. J. Comp. Neurol. 481, 299–313 10.1002/cne.20372 [DOI] [PubMed] [Google Scholar]
- 21. Xie Z., Geiger T. R., Johnson E. N., Nyborg J. K., and Druey K. M. (2008) RGS13 acts as a nuclear repressor of CREB. Mol. Cell 31, 660–670 10.1016/j.molcel.2008.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang Z., Liu B., Wang P., Dong X., Fernandez-Hernando C., Li Z., Hla T., Li Z., Claffey K., Smith J. D., and Wu D. (2008) Phospholipase Cβ3 deficiency leads to macrophage hypersensitivity to apoptotic induction and reduction of atherosclerosis in mice. J. Clin. Investig. 118, 195–204 10.1172/JCI33139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kehrl J. H. (2016) The impact of RGS and other G-protein regulatory proteins on Gαi-mediated signaling in immunity. Biochem. Pharmacol. 114, 40–52 10.1016/j.bcp.2016.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Domachowske J. B., Bonville C. A., Gao J. L., Murphy P. M., Easton A. J., and Rosenberg H. F. (2000) The chemokine macrophage-inflammatory protein-1 alpha and its receptor CCR1 control pulmonary inflammation and antiviral host defense in paramyxovirus infection. J. Immunol. 165, 2677–2682 10.4049/jimmunol.165.5.2677 [DOI] [PubMed] [Google Scholar]
- 25. Percopo C. M., Rice T. A., Brenner T. A., Dyer K. D., Luo J. L., Kanakabandi K., Sturdevant D. E., Porcella S. F., Domachowske J. B., Keicher J. D., and Rosenberg H. F. (2015) Immunobiotic Lactobacillus administered post-exposure averts the lethal sequelae of respiratory virus infection. Antiviral Res. 121, 109–119 10.1016/j.antiviral.2015.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hirano Y., Aziz M., and Wang P. (2016) Role of reverse transendothelial migration of neutrophils in inflammation. Biol. Chem. 397, 497–506 10.1515/hsz-2015-0309 [DOI] [PubMed] [Google Scholar]
- 27. Burn T., and Alvarez J. I. (2017) Reverse transendothelial cell migration in inflammation: to help or to hinder? Cell. Mol. Life Sci. 74, 1871–1881 10.1007/s00018-016-2444-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gane J., and Stockley R. (2012) Mechanisms of neutrophil transmigration across the vascular endothelium in COPD. Thorax 67, 553–561 10.1136/thoraxjnl-2011-200088 [DOI] [PubMed] [Google Scholar]
- 29. Doerschuk C. M., Beyers N., Coxson H. O., Wiggs B., and Hogg J. C. (1993) Comparison of neutrophil and capillary diameters and their relation to neutrophil sequestration in the lung. J. Appl. Physiol. 74, 3040–3045 10.1152/jappl.1993.74.6.3040 [DOI] [PubMed] [Google Scholar]
- 30. Call D. R., Nemzek J. A., Ebong S. J., Bolgos G. L., Newcomb D. E., and Remick D. G. (2001) Ratio of local to systemic chemokine concentrations regulates neutrophil recruitment. Am. J. Pathol. 158, 715–721 10.1016/S0002-9440(10)64014-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang F. (2009) The signaling mechanisms underlying cell polarity and chemotaxis. Cold Spring Harb. Perspect. Biol. 1, a002980 10.1101/cshperspect.a002980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Khan S. M., Sleno R., Gora S., Zylbergold P., Laverdure J. P., Labbé J. C., Miller G. J., and Hébert T. E. (2013) The expanding roles of Gβγ subunits in G protein-coupled receptor signaling and drug action. Pharmacol. Rev. 65, 545–577 10.1124/pr.111.005603 [DOI] [PubMed] [Google Scholar]
- 33. Lomakina E. B., and Waugh R. E. (2010) Signaling and dynamics of activation of LFA-1 and Mac-1 by immobilized IL-8. Cell. Mol. Bioeng. 3, 106–116 10.1007/s12195-009-0099-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hyun Y. M., and Hong C. W. (2017) Deep insight into neutrophil trafficking in various organs. J. Leukoc. Biol. 102, 617–629 10.1189/jlb.1RU1216-521R [DOI] [PubMed] [Google Scholar]
- 35. Patel J., McNeill E., Douglas G., Hale A. B., de Bono J., Lee R., Iqbal A. J., Regan-Komito D., Stylianou E., Greaves D. R., and Channon K. M. (2015) RGS1 regulates myeloid cell accumulation in atherosclerosis and aortic aneurysm rupture through altered chemokine signalling. Nat. Commun. 6, 6614 10.1038/ncomms7614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zarbock A., Deem T. L., Burcin T. L., and Ley K. (2007) Gαi2 is required for chemokine-induced neutrophil arrest. Blood 110, 3773–3779 10.1182/blood-2007-06-094565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Borregaard N. (2010) Neutrophils, from marrow to microbes. Immunity 33, 657–670 10.1016/j.immuni.2010.11.011 [DOI] [PubMed] [Google Scholar]
- 38. Eash K. J., Greenbaum A. M., Gopalan P. K., and Link D. C. (2010) CXCR2 and CXCR4 antagonistically regulate neutrophil trafficking from murine bone marrow. J. Clin. Investig. 120, 2423–2431 10.1172/JCI41649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Heusinkveld L. E., Yim E., Yang A., Azani A. B., Liu Q., Gao J. L., McDermott D. H., and Murphy P. M. (2017) Pathogenesis, diagnosis and therapeutic strategies in WHIM syndrome immunodeficiency. Expert Opin. Orphan Drugs 5, 813–825 10.1080/21678707.2017.1375403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Surve C. R., To J. Y., Malik S., Kim M., and Smrcka A. V. (2016) Dynamic regulation of neutrophil polarity and migration by the heterotrimeric G protein subunits Gαi-GTP and Gβγ. Sci. Signal. 9, ra22 10.1126/scisignal.aad8163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Han J. I., Huang N. N., Kim D. U., and Kehrl J. H. (2006) RGS1 and RGS13 mRNA silencing in a human B lymphoma line enhances responsiveness to chemoattractants and impairs desensitization. J. Leukoc. Biol. 79, 1357–1368 10.1189/jlb.1105693 [DOI] [PubMed] [Google Scholar]
- 42. Shankar S. P., Wilson M. S., DiVietro J. A., Mentink-Kane M. M., Xie Z., Wynn T. A., and Druey K. M. (2012) RGS16 attenuates pulmonary Th2/Th17 inflammatory responses. J. Immunol. 188, 6347–6356 10.4049/jimmunol.1103781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee J. K., Kannarkat G. T., Chung J., Joon Lee H., Graham K. L., and Tansey M. G. (2016) RGS10 deficiency ameliorates the severity of disease in experimental autoimmune encephalomyelitis. J. Neuroinflammation 13, 24 10.1186/s12974-016-0491-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Miyamoto-Matsubara M., Chung S., and Saito Y. (2010) Functional interaction of regulator of G protein signaling-2 with melanin-concentrating hormone receptor 1. Ann. N. Y. Acad. Sci. 1200, 112–119 10.1111/j.1749-6632.2010.05507.x [DOI] [PubMed] [Google Scholar]
- 45. Croft W., Hill C., McCann E., Bond M., Esparza-Franco M., Bennett J., Rand D., Davey J., and Ladds G. (2013) A physiologically required G protein-coupled receptor (GPCR)-regulator of G protein signaling (RGS) interaction that compartmentalizes RGS activity. J. Biol. Chem. 288, 27327–27342 10.1074/jbc.M113.497826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shi G., Partida-Sánchez S., Misra R. S., Tighe M., Borchers M. T., Lee J. J., Simon M. I., and Lund F. E. (2007) Identification of an alternative Gαq-dependent chemokine receptor signal transduction pathway in dendritic cells and granulocytes. J. Exp. Med. 204, 2705–2718 10.1084/jem.20071267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hepler J. R., Berman D. M., Gilman A. G., and Kozasa T. (1997) RGS4 and GAIP are GTPase-activating proteins for Gqα and block activation of phospholipase Cβ by γ-thio-GTP-Gqα. Proc. Natl. Acad. Sci. U.S.A. 94, 428–432 10.1073/pnas.94.2.428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Anger T., Zhang W., and Mende U. (2004) Differential contribution of GTPase activation and effector antagonism to the inhibitory effect of RGS proteins on Gq-mediated signaling in vivo. J. Biol. Chem. 279, 3906–3915 10.1074/jbc.M309496200 [DOI] [PubMed] [Google Scholar]
- 49. Kolaczkowska E., and Kubes P. (2013) Neutrophil recruitment and function in health and inflammation. Nat. Rev. Immunol. 13, 159–175 10.1038/nri3399 [DOI] [PubMed] [Google Scholar]
- 50. Pero R. S., Borchers M. T., Spicher K., Ochkur S. I., Sikora L., Rao S. P., Abdala-Valencia H., O'Neill K. R., Shen H., McGarry M. P., Lee N. A., Cook-Mills J. M., Sriramarao P., Simon M. I., Birnbaumer L., et al. (2007) Gαi2-mediated signaling events in the endothelium are involved in controlling leukocyte extravasation. Proc. Natl. Acad. Sci. U.S.A. 104, 4371–4376 10.1073/pnas.0700185104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Muller W. A. (2016) Transendothelial migration: unifying principles from the endothelial perspective. Immunol. Rev. 273, 61–75 10.1111/imr.12443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shen J., Shrestha S., Yen Y. H., Scott M. A., Soo C., Ting K., Peault B., Dry S. M., and James A. W. (2016) The pericyte antigen RGS5 in perivascular soft tissue tumors. Hum. Pathol. 47, 121–131 10.1016/j.humpath.2015.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ayres-Sander C. E., Lauridsen H., Maier C. L., Sava P., Pober J. S., and Gonzalez A. L. (2013) Transendothelial migration enables subsequent transmigration of neutrophils through underlying pericytes. PLoS One 8, e60025 10.1371/journal.pone.0060025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sadhu C., Masinovsky B., Dick K., Sowell C. G., and Staunton D. E. (2003) Essential role of phosphoinositide 3-kinase δ in neutrophil directional movement. J. Immunol. 170, 2647–2654 10.4049/jimmunol.170.5.2647 [DOI] [PubMed] [Google Scholar]
- 55. Tamirisa P., Blumer K. J., and Muslin A. J. (1999) RGS4 inhibits G-protein signaling in cardiomyocytes. Circulation 99, 441–447 10.1161/01.CIR.99.3.441 [DOI] [PubMed] [Google Scholar]
- 56. Percopo C. M., Dubovi E. J., Renshaw R. W., Dyer K. D., Domachowske J. B., and Rosenberg H. F. (2011) Canine pneumovirus replicates in mouse lung tissue and elicits inflammatory pathology. Virology 416, 26–31 10.1016/j.virol.2011.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Percopo C. M., Brenner T. A., Ma M., Kraemer L. S., Hakeem R. M., Lee J. J., and Rosenberg H. F. (2017) SiglecF+Gr1hi eosinophils are a distinct subpopulation within the lungs of allergen-challenged mice. J. Leukoc. Biol. 101, 321–328 10.1189/jlb.3A0416-166R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Percopo C. M., Dyer K. D., Karpe K. A., Domachowske J. B., and Rosenberg H. F. (2014) Eosinophils and respiratory virus infection: a dual-standard curve qRT-PCR-based method for determining virus recovery from mouse lung tissue. Methods Mol. Biol. 1178, 257–266 10.1007/978-1-4939-1016-8_22 [DOI] [PubMed] [Google Scholar]
- 59. Swamydas M., Luo Y., Dorf M. E., and Lionakis M. S. (2015) Isolation of mouse neutrophils. Curr. Protoc. Immunol. 110, 3.20.1–3.20.15 10.1002/0471142735.im0320s110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yuan Q., Ren C., Xu W., Petri B., Zhang J., Zhang Y., Kubes P., Wu D., and Tang W. (2017) PKN1 directs polarized RAB21 vesicle trafficking via RPH3A and is important for neutrophil adhesion and ischemia-reperfusion injury. Cell Rep. 19, 2586–2597 10.1016/j.celrep.2017.05.080 [DOI] [PMC free article] [PubMed] [Google Scholar]