Abstract
Ferroptosis is an iron-dependent form of programmed cell death characterized by the accumulation of lipid-targeting reactive oxygen species that kill cells by damaging their plasma membrane. The lipid repair enzyme GSH peroxidase 4 (GPX4) protects against this oxidative damage and enables cells to resist ferroptosis. Recent work has revealed that matrix-detached carcinoma cells can be susceptible to ferroptosis and that they can evade this fate through the signaling properties of the α6β4 integrin, which sustains GPX4 expression. Although these findings on ferroptosis are provocative, they differ from those in previous studies indicating that matrix-detached cells are prone to apoptosis via a process referred to as anoikis. In an effort to reconcile these discrepant findings, here we observed that matrix-detached epithelial and carcinoma cells cluster spontaneously via a mechanism that involves the cell adhesion protein PVRL4 (also known as Nectin-4). We found that this clustering process allows these cells to survive by stimulating a PVRL4/α6β4/Src signaling axis that sustains GPX4 expression and buffers against lipid peroxidation. In the absence of α6β4, PVRL4-mediated clustering induced an increase in lipid peroxidation that was sufficient for triggering ferroptosis. When the clustering was inhibited, single cells did not exhibit a significant increase in lipid peroxidation in the absence of α6β4, and they were more susceptible to apoptosis than to ferroptosis. These results indicate that ferroptosis induction depends on cell clustering in matrix-detached cells that lack α6β4 and imply that the fate of matrix-detached cells can be determined by the state of their cell–cell interactions.
Keywords: breast cancer, cell death, cell–cell interaction, extracellular matrix, integrin, ferroptosis, glutathione peroxidase 4, lipid peroxidation
Introduction
Ferroptosis is defined as an iron-dependent form of programmed cell death that is characterized by the accumulation of intracellular soluble and lipid reactive oxygen species (ROS)3 that damage the plasma membrane by peroxidation of polyunsaturated fatty acids (1). At a mechanistic level, ferroptosis is triggered by the loss of activity of the lipid repair enzyme GSH peroxidase 4 (GPX4), which catalyzes the reduction of lipids and other peroxides and is a target of several ferroptosis inducers (2). The antiporter system XC−, which imports cystine into the cell in exchange for glutamate, also has a critical role in protecting against ferroptosis because cysteine, the monomeric form of cystine, is a building block for the antioxidant GSH, which is a co-factor for GPX4 (3, 4). Molecules that inhibit system XC−, such as erastin, trigger ferroptosis and have proven to be useful for studying this process in detail (4, 5).
Rapid progress is being made in unraveling the biochemical nature of ferroptosis and determining its role in specific biological and pathological processes. A central theme that is emerging from these studies is the essential role of GPX4 in enabling cells to resist ferroptosis, a role that is particularly important for more aggressive tumor cells (6). Less is known, however, about the cell biological context of the mechanisms used to either promote or evade ferroptosis. Recently, we discovered that ferroptosis is a component of cell death induced by the detachment of epithelial and carcinoma cells from the matrix (7). We also observed that the α6β4 integrin has the ability to confer ferroptosis resistance under these conditions. In other terms, matrix-detached cells undergo ferroptosis but only in the absence of α6β4. The mechanism underlying this phenomenon involves the ability of α6β4 to sustain GPX4 expression and activity in matrix-detached cells and buffer the increase in lipid peroxidation that occurs in these cells, which can trigger ferroptosis (7).
Our findings on ferroptosis are provocative, but they differ from prior studies concluding that matrix-detached cells are prone to apoptosis, a process referred to as anoikis, unless survival mechanisms are enabled (8, 9). In an effort to reconcile these discrepant findings, we observed that matrix-detached epithelial and carcinoma cells cluster spontaneously by a mechanism that involves PVRL4 (also known as Nectin-4). If this clustering is inhibited, then single cells are more susceptible to apoptosis in the absence of α6β4. Based on these observations, we explored the possibility of a functional connection between cell clustering and ferroptosis. Indeed, the data we report reveal that ferroptosis depends on cell clustering in matrix-detached cells that lack α6β4.
Results
Matrix-detached cells cluster spontaneously by a mechanism that is independent of α6β4
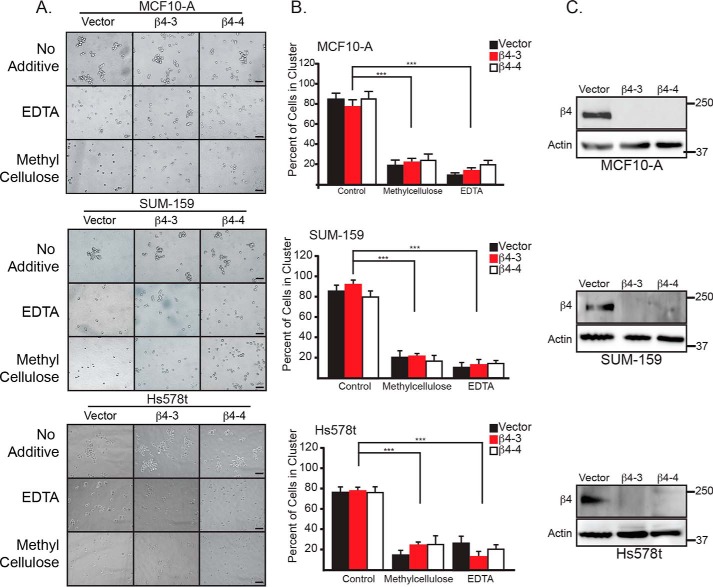
Our interest in understanding the distinction between ferroptosis and apoptosis in matrix-detached carcinoma cells was piqued by the observation that immortalized mammary epithelial cells (MCF10-A) and breast carcinoma cells (SUM-159 and Hs578t) cluster spontaneously upon detachment (Fig. 1, A and B). These clusters could be dissociated into single cells with either 0.5% methylcellulose or 2 mm EDTA (Fig. 1, A and B). Interestingly, we did not observe significant differences in cluster number upon depletion of α6β4 using CRISPR/Cas9 to knock down the β4 subunit using two guide RNAs that target different regions of the first exon of ITGB4 (referred to as β4-3 and β4-4) (Fig. 1, B and C), indicating that this integrin does not contribute to clustering.
Figure 1.
Breast epithelial and carcinoma cells preferentially cluster upon matrix detachment. A, MCF10-A, SUM-159, and Hs578t cells (vector and β4-depleted) were detached and allowed to cluster spontaneously or treated with either 2 mm EDTA or 0. 5% methylcellulose to generate single-cell suspensions. Photomicrographs were taken 2 h post-detachment. Scale bars = 10 microns. B, quantification of MCF10-A, SUM-159, and Hs578t clusters was performed by counting the total number of single and clustered cells, total number of clusters (defined as ≥ 3 cells). Six independent photos of each cell line and each condition were quantified. ***, p < 0.005. C, the β4 integrin subunit was depleted in Hs578t cells by CRISPR/Cas9 using two independent guide RNAs (β4-3 and β4-4). Depletion of β4 expression was verified by immunoblotting.
Clustering of matrix-detached cells influences the relative levels of ferroptosis and apoptosis in the absence of α6β4
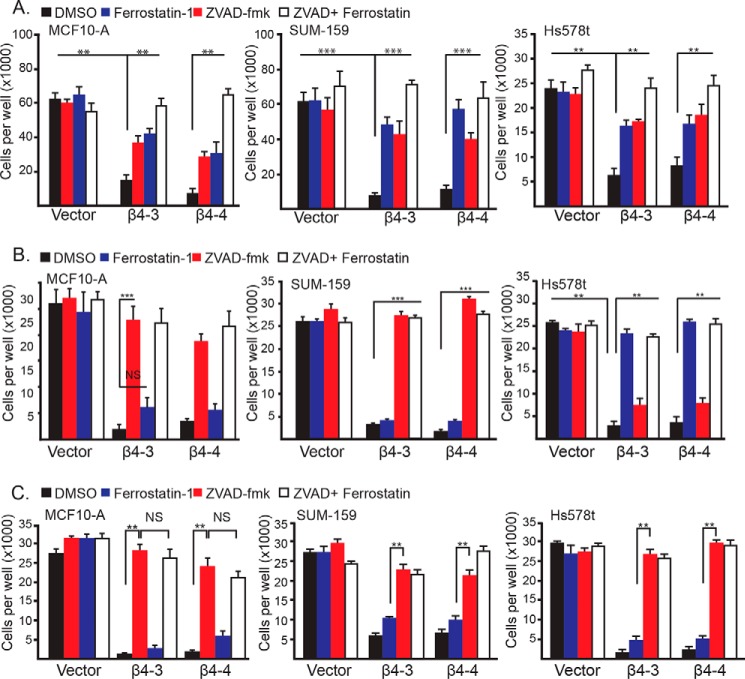
The observations described above prompted us to assess the role of cell clustering in ferroptosis that is triggered by the loss of α6β4. For this purpose, we assayed cell viability 24 h after matrix detachment as a function of α6β4 expression in clustered cells or single cells (MCF10-A, SUM-159, and Hs578t) that had been generated using either methylcellulose or EDTA. These cells, which express α6β4, remained viable 24 h after matrix detachment regardless of their clustering status (Fig. 2, A–C). In response to loss of α6β4 by CRISPR/Cas9-mediated knockdown of the β4 subunit, however, a dramatic decrease in viability was seen under all conditions (Fig. 2, A–C), confirming the importance of the α6β4 integrin in maintaining survival in response to stress. It is important to note that CRISPR-mediated depletion of the β4 subunit in these cells does affect expression of the α6 integrin subunit, and they express the α6β1 integrin (7). Specificity is also evidenced by our previous finding that expression of the β4 construct that cannot be targeted by these CRISPRs in β4-depleted cells rescued their viability under matrix-detached conditions (7). Moreover, the α3β1 integrin does not promote ferroptosis resistance in these cells (7).
Figure 2.
Clustering of matrix-detached cells influences the relative levels of ferroptosis and apoptosis in the absence of α6β4. A, control and β4-depleted cells were detached for 24 h in the presence of either DMSO, ferrostatin-1 (2 μm), Z-VAD-fmk (25 μm), or ferrostatin-1 and Z-VAD-fmk, and the number of viable cells was quantified. B, control and β4-depleted cells were detached for 24 h with 2 mm EDTA in the presence of either DMSO, ferrostatin-1 (2 μm), Z-VAD-fmk (25 μm), or ferrostatin-1 and Z-VAD-fmk, and the number of viable cells was quantified. C, control and β4-depleted cells were detached for 24 h with 0.5% methylcellulose in the presence of either DMSO, ferrostatin-1 (2 μm), Z-VAD-fmk (25 μm), or ferrostatin-1 and Z-VAD-fmk, and the number of viable cells was quantified. NS, not significant. **, p < 0.01, ***, p < 0.005.
To determine whether the nature of cell death differed between clustered and single detached cells in the absence of α6β4, we compared the ability of ferrostatin-1, Z-VAD-fmk, or both inhibitors to rescue the viability of detached, α6β4-depleted cells in the presence or absence of either methylcellulose or EDTA. In the absence of methylcellulose or EDTA, either inhibitor alone yielded a partial rescue of viability, and the use of both inhibitors resulted in a complete rescue (Fig. 2A), indicating that ferroptosis and apoptosis contribute to the death of these cells. Interestingly, though, ferrostatin-1 was unable to rescue viability in the presence of methylcellulose or EDTA (single cells), but Z-VAD-fmk did rescue viability almost completely (Fig. 2, B and C). These results indicate that cell clustering predisposes cells to ferroptosis in the absence of α6β4 and that disruption of these clusters results in apoptosis. Moreover, the finding that α6β4-depleted cells are prone to both ferroptosis and apoptosis in the absence of EDTA and methylcellulose probably reflects the fact that this population is comprised of a mixture of clustered and single cells (Fig. 1A).
Lipid peroxidation is significantly higher in clustered, detached cells compared with single cells in the absence of α6β4
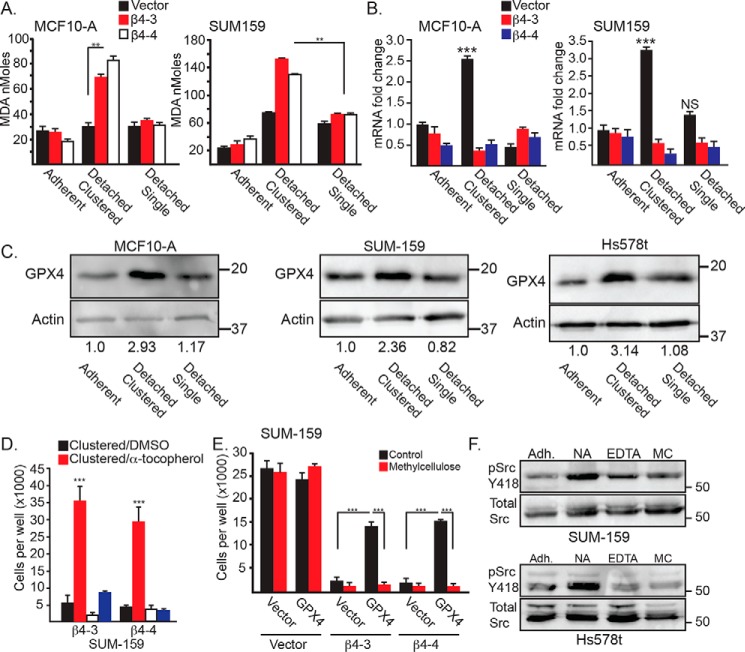
To understand why clustered but not single cells are prone to ferroptosis in the absence of α6β4, we quantified lipid peroxidation, which is the root cause of ferroptosis, using the malondialdehyde (MDA) assay. Indeed, depletion of α6β4 resulted in a marked increase in lipid peroxidation in clustered but not single cells (Fig. 3A). This finding suggests that the clustering of detached cells has the potential to increase lipid peroxidation to levels that are sufficient to trigger ferroptosis and that α6β4 mitigates these effects on clustered cells. The observation that detached single cells do not exhibit this increase in lipid peroxidation is consistent with our finding that these cells are not susceptible to ferroptosis.
Figure 3.
Clustering of matrix-detached cells influences lipid peroxidation, GPX4 expression, and Src activity. A, lipid peroxidation was quantified using the MDA assay in control and β4-depleted MCF10-A and SUM-159 cells under either adherent, detached clustered (no additive), or detached single (0.5% methylcellulose) conditions for 4 h. Similar results were obtained using 2 mm EDTA. B, GPX4 mRNA expression was quantified by qPCR in control and β4-depleted MCF10-A and SUM-159 cells under either adherent, detached clustered (no additive) or detached single (0.5% methylcellulose) conditions for 2 h. C, GPX4 protein expression was assessed by immunoblotting in control MCF10-A, SUM-159, and Hs578t cells under detached clustered (no additive) or detached single (0.5% methylcellulose) conditions for 4 h. Immunoblots were quantified by densitometry, and the ratio of the intensity of the GPX4/actin bands relative to adherent cells is shown under the blots. D, SUM-159 β4-depleted cells were detached for 24 h with either no additive (clustered) or 2 mm EDTA (single) in the presence of either DMSO or 500 mm α-tocopherol, and the number of viable cells was quantified. E, control and β4-depleted cells SUM-159 cells that had been transfected with either a vector control or a GPX4 expression vector were detached for 24 h with no additive (Control) or 0.5% methylcellulose, and the number of viable cells was quantified. F, SUM-159 and Hs578t control cells were assessed for phosphorylated (Tyr-418) Src by immunoblotting under adherent conditions (Adh) as well as following 2 h of matrix detachment with no additive (NA), 2 mm EDTA (EDTA), or 0.5% methylcellulose (MC). NS, not significant. **, p < 0.01, ***, p < 0.005.
The above results suggest that α6β4 function differs between detached clustered and single cells and that this integrin facilitates a mechanism to evade ferroptosis in detached, clustered cells. Based on our previous work (7), we focused on the ability of α6β4 to induce GPX4 expression and activity in these distinct populations. We observed that GPX4 mRNA (Fig. 3B) and protein (Fig. 3C) expression increased significantly upon matrix detachment of clustered cells (2 h), but this increase was less evident in single cells. The increase in GPX4 expression depends on α6β4, consistent with our previous results (7). Moreover, the viability of the clustered, α6β4-depleted cells was rescued by either the lipophilic antioxidant α-tocopherol (Fig. 3D) or exogenous expression of GPX4 (Fig. 3E). In contrast, neither α-tocopherol nor GPX4 re-expression in α6β4-depleted, single cells rescued viability. These data infer that α6β4 signaling differs between clustered and single, matrix-detached cells. Given that the induction of GPX4 expression upon matrix detachment depends on α6β4/Src signaling (7), we assessed Src activation in detached clustered and single cells as a function of α6β4 expression. Matrix detachment caused an increase in activated Src in clustered cells, as assayed by Tyr-418 immunoblotting, that was dependent on α6β4 expression and was not detected in either EDTA- or methylcellulose-treated cells (Fig. 3F).
PVRL4 mediates clustering of matrix-detached cells and ferroptosis resistance
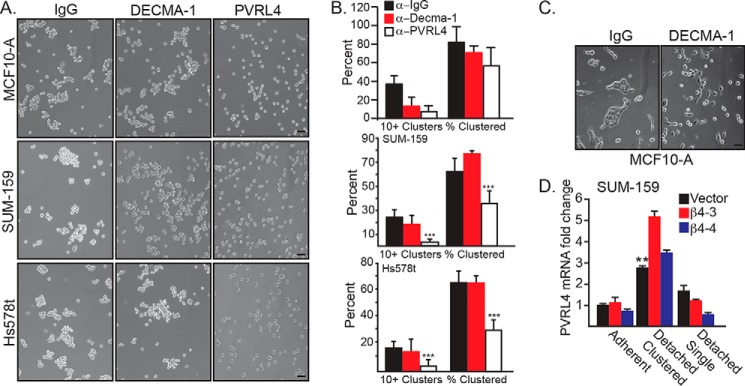
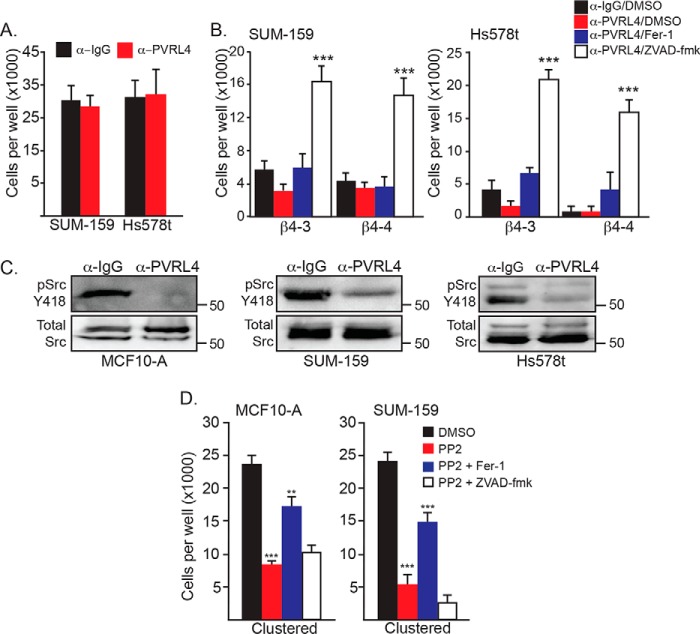
An important issue that arose from our data concerns the mechanism by which matrix-detached cells cluster and the contribution of this mechanism to ferroptosis resistance. The possibility that this clustering is mediated by E-cadherin is negated by the fact that two of the cell lines we studied (SUM-159 and Hs578t) have a mesenchymal phenotype and express little, if any, E-cadherin. We confirmed this assumption by treating matrix-detached cells with a function-blocking E-cadherin Ab (DECMA-1) and observed no effect on clustering (Fig. 4A). Interestingly, however, DECMA-1 had no effect on the percentage of MCF10-A cells in clusters, although it did cause a significant decrease in the number of large (10+ cells) clusters (Fig. 4B). In contrast, treatment of adherent MCF10-A cells with DECMA-1 disrupted cell–cell adhesion (Fig. 4C). For this reason, we focused on the cell surface receptor PVRL4 (also referred to as nectin-4) because it has been shown to mediate the clustering of matrix-detached carcinoma cells by interacting in trans with PVRL1 (10). Importantly, PVRL4 also maintains the survival of these cells by interacting with α6β4 in cis and enabling Src activation (10). These seminal studies, however, did not consider a potential role for PVRL4 in ferroptosis resistance. For this reason, we initially examined the ability of a function-blocking PVRL4 Ab to disrupt the clustering of matrix-detached cells and found that this Ab was as effective as either EDTA or methylcellulose in disrupting clusters into single cells (Fig. 4A). Depletion of β4 did not significantly affect PVRL4 expression in β4-depleted cells (Fig. 4D).
Figure 4.
PVRL4 mediates clustering upon matrix detachment. A, MCF10-A, SUM-159, and Hs578t vector control cells were detached and allowed to aggregate in the presence of 4 μg/ml of either IgG, DECMA-1, or a PVRL4-blocking Ab. Photomicrographs were taken 2 h post-detachment. Scale bars = 10 μm. B, quantification of MCF10-A, SUM-159, and Hs578t clusters was performed by counting the total number of single and clustered cells, the total number of clusters (defined as ≥ 3 cells), and the number of large clusters (defined as ≥ 10 cells). **, p < 0.01, ***, p < 0.005. C, MCF10-A cells were detached and allowed to readhere in the presence of either IgG or DECMA-1. Photomicrographs were taken at 24 h. Scale bar = 10 μm. D, SUM-159 control and β4-depleted cells under adherent, detached clustered, or detached single cell conditions were assessed for expression of PVRL4 mRNA by qPCR.
The above finding prompted us to assess the contribution of PVRL4 to the cell death that occurs upon matrix detachment. Matrix-detached cells that express α6β4 exhibited no significant loss of viability in response to treatment with the PVRL4 function-blocking antibody compared with the IgG control (Fig. 5A), supporting our conclusion that this integrin promotes the survival of clustered and single, matrix-detached cells. As we reported previously (7), matrix-detached cells that lack α6β4 exhibit a dramatic loss of viability that is rescued by ferrostatin-1. In marked contrast, α6β4-depleted cells treated with the PVRL4 function-blocking antibody exhibited a significant loss of viability that was rescued by Z-VAD-fmk and not ferrostatin-1 (Fig. 5B). This result indicates that disruption of PVRL4-mediated clustering of α6β4-depleted cells renders them sensitive to apoptosis as opposed to ferroptosis and that PVRL4 is necessary to execute ferroptosis in this population of cells.
Figure 5.
PVRL4-mediated cell clustering is necessary for α6β4-dependent Src activation. A, SUM-159 and Hs578t control cells were detached for 24 h in the presence of either IgG or a PVRL4 Ab, and the number of viable cells was quantified. B, SUM-159 and Hs578t β4-depleted cells were detached for 24 h in the presence of either IgG and DMSO, PVRL4 Ab, PVRL4 Ab and ferrostatin-1 (2 μm), or PVRL4 and Z-VAD-fmk (25 μm), and the number of viable cells was quantified. C, MCF10-A, SUM-159 and Hs578t control cells were detached for 2 h in the presence of either IgG or a PVRL4-blocking Ab, and phosphorylation of Src (Tyr-418) was assessed by immunoblotting. D, cells (MCF10-A and SUM-159) were assessed for viability after 24 h of detachment (clustered conditions) in the presence of either DMSO, PP2 (10 μm), or PP2 in combination with either ferrostatin-1 (2 μm) or Z-VAD-fmk (25 μm). **, p < 0.01, ***, p < 0.005.
Given that α6β4-mediated Src signaling maintains the survival of matrix-detached cells (7), we examined the impact of the PVRL4 function-blocking antibody on Src activation in matrix-detached cells. Indeed, disruption of cell clustering by this antibody resulted in a significant decrease in Src activation compared with control cells, as assessed by phospho-Src (Tyr-418) immunoblotting (Fig. 5C). A causal role for Src activation in ferroptosis resistance is indicated by the observation that Src inhibition using PP2 decreased the viability of clustered, matrix-detached cells significantly and that this loss of viability was rescued by ferrostatin-1 but not Z-VAD-fmk. (Fig. 5D). Together, these results indicate that PVRL4-mediated clustering is necessary for α6β4-mediated Src activation.
Discussion
The results of this study reveal an intimate relationship between the clustering of matrix-detached carcinoma cells and ferroptosis. We conclude from our data that matrix-detached carcinoma cells cluster spontaneously and that this clustering triggers an increase in lipid peroxidation that is sufficient to induce ferroptosis. Ferroptosis can be evaded under these conditions when these cells express the α6β4 integrin, which functions to buffer lipid peroxidation by sustaining GPX4 expression. In marked contrast, dissociation of clustered, detached cells into single cells results in apoptosis in the absence of α6β4. An important and novel inference from these data is that the fate of matrix-detached cells can be determined by the state of their cell–cell interactions.
Our findings build upon the seminal study by Elledge and co-workers (10) that identified PVRL4 in a gain-of-function screen for genes that promote the survival of matrix-detached carcinoma cells. Not only did they implicate PVRL4 in the clustering of matrix-detached carcinoma cells and anoikis resistance, they also demonstrated that PVRL4 enables α6β4-mediated Src activation and the importance of this pathway in the survival of these detached cells. Our work substantiates these findings and extends them to ferroptosis resistance. Indeed, we demonstrate that the PVRL4/α6β4/Src axis allows matrix-detached cells to resist ferroptosis by sustaining GPX4 expression and minimizing lipid peroxidation, establishing a novel role for PVRL4 in ferroptosis resistance mechanisms. Interestingly, however, we also show that PVRL4 can promote ferroptosis in the absence of α6β4 by mediating cell clustering and contributing, either directly or indirectly, to increased lipid peroxidation. In other terms, this observation implies that the process of cell clustering mediated by PVRL4 stimulates an increase in lipid peroxidation by a mechanism to be determined and that this scenario does not occur in single cells, which do not engage PVRL4-mediated adhesion.
Previous studies have noted the role of cell “aggregation” in promoting apoptosis resistance in matrix-detached squamous carcinoma and ErbB2-expressing MCF10-A mammary epithelial cells (11, 12). These studies focused on cells that expressed E-cadherin, and they implicated E-cadherin as the mediator of cell aggregation and apoptosis resistance. For this reason, our finding that PVRL4 mediates the clustering of matrix-detached MCF10-A cells is intriguing and raises the need to investigate the relationship between E-cadherin- and PVRL4-mediated cell–cell adhesion. Our primary interest, however, is survival mechanisms used by aggressive carcinoma cells, which often exhibit a mesenchymal phenotype and express little, if any, E-cadherin (13). In fact, the carcinoma cell lines used in our study are mesenchymal when grown as adherent cells but retain the ability to engage in cell–cell adhesion upon matrix detachment, a scenario that mimics the transition from aggressive primary carcinomas to circulating tumor cells, which tend to cluster (14).
Although studies of the nature of cell death triggered by matrix detachment have focused largely on apoptosis, it is becoming apparent that other modes of cell death can occur that involve elevated ROS levels (15, 16). Our findings establish ferroptosis as one such ROS-mediated form of cell death. They also provide insight into the cell biological parameters that influence ferroptosis by revealing how the clustering of matrix-detached carcinoma cells can promote ferroptosis as opposed to apoptosis. On a broader level, the role of ferroptosis in eliminating matrix-detached carcinoma cells is timely and significant based on the recent realization that aggressive carcinoma cells with a mesenchymal phenotype are highly sensitive to ferroptosis unless they enable mechanisms that sustain GPX4 expression (6). Based on our data, PVRL4/α6β4/Src signaling in the context of cell clustering could be one such mechanism.
Experimental procedures
Cell lines and reagents
MCF10-A cells were obtained from the Barbara Ann Karmanos Cancer Institute, and SUM-159 cells were provided by Dr. Stephen Ethier (Medical College of South Carolina). Hs578T cells were provided by Dr. Dohoon Kim (UMass Medical School). MCF10-A cells were maintained in Dulbecco's modified Eagle's medium/F-12 (Gibco) with 5% horse serum, 1% pen/strep (Gibco), 20 ng/ml epidermal growth factor (Peprotech), 0.5 mg/ml hydrocortisone (Gibco), 100 ng/ml cholera toxin (Sigma), and 10 μg/ml insulin (Sigma). SUM-159 cells were maintained in F-12 (Gibco) medium with 5% fetal bovine serum (Hyclone), 1% pen/strep, 0.5 mg/ml hydrocortisone, and 10 μg/ml insulin. Hs578t cells were maintained in Dulbecco's modified Eagle's medium high-glucose (Gibco) with 10% fetal bovine serum, 1% pen/strep, and 10 μg/ml insulin. All cells were checked quarterly for mycoplasma and authenticated using the University of Arizona Genetic Core. The following antibodies were used: GPX4 (Abcam, ab125066), actin (Sigma-Aldrich), integrin β4 (505; Ref. 18), phospho-Src Y418 (R&D Systems, MAB2685), total Src (Santa Cruz Biotechnology, sc-8056), DECMA-1 (Abcam, ab11512), PVRL4 (R&D Systems, 17402; Cell Signaling Technology, 17402). Other reagents used were as follows: Z-VAD-fmk (SelleckChem), ferrostatin-1 (Sigma-Aldrich), α-tocopherol (Sigma-Aldrich), and PP2 (SelleckChem).
Matrix detachment assays
Twenty-four-well plates were coated with 800 μl of polyHEMA (30 mg/ml, Sigma-Aldrich) and dried overnight at room temperature. Cells were trypsinized, and complete medium was used to quench the trypsin. Cells were counted, and the remaining suspension was diluted to 2.5 × 104 cells/100 μl and plated (100 μl/well) in cell-line specific serum-free medium with the reagents indicated in the figure legends for the times noted. Four wells were plated per condition as technical replicates. Cells were counted with a hemocytometer using trypan blue exclusion. Total cell number was calculated by multiplying average cells per square by dilution factor and chamber volume.
Biochemical experiments
For all experiments, three wells were plated per condition for each independent experiment as technical replicates, and at least three independent experiments were performed. For immunoblotting, cells collected in to prechilled tubes and spun down before washing twice in ice-cold PBS. Protein was extracted using radioimmune precipitation assay buffer containing protease and phosphatase inhibitors (Boston Bioproducts, Worcester, MA) by lysing cells for 15 min on ice, followed by centrifugation at maximum speed for 15 min at 4 °C. Supernatant was collected, and protein concentration was quantified using Bradford reagent (Bio-Rad). Samples (25 μg of protein) were separated by SDS-PAGE and immunoblotted using the Abs specified in the figure legends. For qPCR, RNA was isolated from cells using the NucleoSpin Gel and PCR Clean-Up kit (Macherey-Nagel). Briefly, cells were collected and lysed immediately following centrifugation, and RNA was isolated following the manufacturer's specifications. RNA concentrations were determined by Nanodrop, and complementary DNA (1 μg/sample) was produced using All-In-One cDNA Synthesis SuperMix (BioScript). qPCR was performed using Cyber Green Master Mix (BioTool). The sequences for primers used are as follows: 18S, 5′-AACCCGTTGAACCCCATT-3′ (forward) and 5′-CCATCCAATCG GTAGTAGCG-3′ (reverse), derived from NT_167214.1; GPX4, 5′-GAGGCAAGACCGAAGTAAACTAC-3′ (forward) and 5′-CCGAACTGGTTACACGGGAA-3′ (reverse), derived from NC_000019.10.
To assay lipid peroxidation, the MDA lipid peroxidation microplate assay (Sigma-Aldrich) was used according to the manufacturer's specifications. Briefly, cells (1 × 106) were cultured in cell-line specific, serum-free medium on polyhydroxyethylmethacrylate (polyHEMA)–coated plates for 4 h. Three wells were plated per condition for each independent experiment as technical replicates. Cells were collected, lysed, and reacted with thiobarbituric acid for 1 h at 95 °C. An aliquot of lysate (5 μl) was saved for protein concentration determination using the Bradford assay prior to reaction with thiobarbituric acid. Fluorescence was read at 532 (excitation)/590 (emission). Lipid peroxidation levels were normalized to protein concentration.
Molecular biology experiments
For CRISPR-mediated deletion of the β4 integrin subunit, guide RNAs (sgRNA) targeting exon 1 of the β4 sequence were selected using CRISPR Design (http://crispr.mit.edu)4 and CRISPRdirect (https://crispr.dbcls.jp; 17).4 Four guide RNAs were tested, and the two most efficient knockouts were selected. The guide RNA sequences used were as follows: B4-3, 5′-CACCGTTGTCCAGATCATCGGACA-3′ and 5′-AAACTGTCCGATGATCTGG ACAAC-3′; B4-4, 5′-CACCGAAATCCAATAGTGTAGTCGC-3′ and 5′-AAACGCGACTACACTAT TGGATTTC-3′. Cells were subcloned by FACS and screened for loss of protein expression by immunoblotting. New CRISPR-depleted cells were generated every 3 months to control for potential secondary effects caused by long-term gene depletion. To express GPX4, a plasmid construct for GPX4 was purchased from Origene (RC208065). Cells were transfected with 10 μg of DNA using Lipofectamine 2000 (Thermo Fisher) and incubated for 48 h prior to use.
Statistical analysis
The error bars in graphs represent mean ± S.D. All experiments were repeated at least three times. The p values were calculated using ANOVA, and a p value of less than 0.05 was considered significant.
Author contributions
C. W. B. and A. M. M. conceptualization; C. W. B. and J. J. A. data curation; C. W. B. formal analysis; C. W. B. and A. M. M. funding acquisition; C. W. B. investigation; C. W. B. and J. J. A. methodology; C. W. B. and A. M. M. writing-original draft; C. W. B. and A. M. M. writing-review and editing; J. J. A. software; A. M. M. supervision; A. M. M. project administration.
This work was supported by Department of Defense Grant W81XWH-17-1-0009. The authors declare that they have no conflicts of interest with the contents of this article.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party–hosted site.
- ROS
- reactive oxygen species
- Z-VAD-fmk
- benzyloxycarbonyl-VAD-fluoromethyl ketone
- MDA
- malondialdehyde
- Ab
- antibody
- pen/strep
- penicillin/streptomycin
- qPCR
- quantitative PCR.
References
- 1. Stockwell B. R., Friedmann Angeli J. P., Bayir H., Bush A. I., Conrad M., Dixon S. J., Fulda S., Gascón S., Hatzios S. K., Kagan V. E., Noel K., Jiang X., Linkermann A., Murphy M. E., Overholtzer M., et al. (2017) Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell 171, 273–285 10.1016/j.cell.2017.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yang W. S., SriRamaratnam R., Welsch M. E., Shimada K., Skouta R., Viswanathan V. S., Cheah J. H., Clemons P. A., Shamji A. F., Clish C. B., Brown L. M., Girotti A. W., Cornish V. W., Schreiber S. L., and Stockwell B. R. (2014) Regulation of ferroptotic cancer cell death by GPX4. Cell 156, 317–331 10.1016/j.cell.2013.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dixon S. J., Patel D. N., Welsch M., Skouta R., Lee E. D., Hayano M., Thomas A. G., Gleason C. E., Tatonetti N. P., Slusher B. S., and Stockwell B. R. (2014) Pharmacological inhibition of cystine–glutamate exchange induces endoplasmic reticulum stress and ferroptosis. eLife 3, e02523 10.7554/eLife.02523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang W. S., and Stockwell B. R. (2016) Ferroptosis: death by lipid peroxidation. Trends Cell Biol. 26, 165–176 10.1016/j.tcb.2015.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dixon S. J., Lemberg K. M., Lamprecht M. R., Skouta R., Zaitsev E. M., Gleason C. E., Patel D. N., Bauer A. J., Cantley A. M., Yang W. S., Morrison B. 3rd, and Stockwell B. R. (2012) Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072 10.1016/j.cell.2012.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Viswanathan V. S., Ryan M. J., Dhruv H. D., Gill S., Eichhoff O. M., Seashore-Ludlow B., Kaffenberger S. D., Eaton J. K., Shimada K., Aguirre A. J., Viswanathan S. R., Chattopadhyay S., Tamayo P., Yang W. S., Rees M. G., et al. (2017) Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature 547, 453–457 10.1038/nature23007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brown C. W., Amante J. J., Goel H. L., and Mercurio A. M. (2017) The α6β4 integrin promotes resistance to ferroptosis. J. Cell Biol. 216, 4287–4297 10.1083/jcb.201701136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Meredith J. E. Jr., Fazeli B., and Schwartz M. A. (1993) The extracellular matrix as a cell survival factor. Mol. Biol. Cell 4, 953–961 10.1091/mbc.4.9.953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Frisch S. M., and Francis H. (1994) Disruption of epithelial cell-matrix interactions induces apoptosis. J. Cell Biol. 124, 619–626 10.1083/jcb.124.4.619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pavlova N. N., Pallasch C., Elia A. E., Braun C. J., Westbrook T. F., Hemann M., and Elledge S. J. (2013) A role for PVRL4-driven cell-cell interactions in tumorigenesis. eLife 2, e00358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shen X., and Kramer R. H. (2004) Adhesion-mediated squamous cell carcinoma survival through ligand-independent activation of epidermal growth factor receptor. Am. J. Pathol. 165, 1315–1329 10.1016/S0002-9440(10)63390-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rayavarapu R. R., Heiden B., Pagani N., Shaw M. M., Shuff S., Zhang S., and Schafer Z. T. (2015) The role of multicellular aggregation in the survival of ErbB2-positive breast cancer cells during extracellular matrix detachment. J. Biol. Chem. 290, 8722–8733 10.1074/jbc.M114.612754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Neve R. M., Chin K., Fridlyand J., Yeh J., Baehner F. L., Fevr T., Clark L., Bayani N., Coppe J. P., Tong F., Speed T., Spellman P. T., DeVries S., Lapuk A., Wang N. J., et al. (2006) A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10, 515–527 10.1016/j.ccr.2006.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Aceto N., Bardia A., Miyamoto D. T., Donaldson M. C., Wittner B. S., Spencer J. A., Yu M., Pely A., Engstrom A., Zhu H., Brannigan B. W., Kapur R., Stott S. L., Shioda T., Ramaswamy S., et al. (2014) Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 158, 1110–1122 10.1016/j.cell.2014.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schafer Z. T., Grassian A. R., Song L., Jiang Z., Gerhart-Hines Z., Irie H. Y., Gao S., Puigserver P., and Brugge J. S. (2009) Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature 461, 109–113 10.1038/nature08268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hawk M. A., Gorsuch C. L., Fagan P., Lee C., Kim S. E., Hamann J. C., Mason J. A., Weigel K. J., Tsegaye M. A., Shen L., Shuff S., Zuo J., Hu S., Jiang L., Chapman S., et al. (2018) RIPK1-mediated induction of mitophagy compromises the viability of extracellular-matrix-detached cells. Nat. Cell Biol. 20, 272–284 10.1038/s41556-018-0034-2 [DOI] [PubMed] [Google Scholar]
- 17. Naito Y., Hino K., Bono H., and Ui-Tei K. (2015) CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31, 1120–1123 10.1093/bioinformatics/btu743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rabinovitz I., Toker A., and Mercurio A. M. (1999) Protein kinase C-dependent mobilization of the α6β4 integrin from hemidesmosomes and its association with actin-rich cell protrusions drive the chemotactic migration of carcinoma cells. J. Cell Biol. 146, 1147–1160 10.1083/jcb.146.5.1147 [DOI] [PMC free article] [PubMed] [Google Scholar]